Abstract
The purpose of this study was to evaluate the contribution of SHISA3 promoter methylation to laryngeal squamous cell carcinoma (LSCC). SHISA3 promoter methylation status and expression were determined using methylation-specific polymerase chain reaction (MSP) and quantitative real-time PCR (qRT-PCR) in 93 paired LSCC and adjacent normal tissues, respectively. Furthermore, the regulatory function of the SHISA3 promoter fragment was analyzed using a luciferase reporter assay. The results reveal that there is a significant increase in SHISA3 methylation in LSCC tissues compared with corresponding nontumor tissues (P = 4.58E − 12). The qRT-PCR results show a significant association between SHISA3 methylation and expression in LSCC (P = 1.67E − 03). In addition, the area under the receiver operating characteristic curve was 0.91. Consequently, a log-rank test and multivariate Cox analysis suggest that SHISA3 promoter hypermethylation is a predictor of poor overall survival for LSCC (log-rank P = 0.024; HR = 2.71; 95% CI = 1.024–7.177; P = 0.047). The results indicate that SHISA3 promoter hypermethylation might increase the risk of LSCC through regulation of gene expression and is a potential diagnostic and prognostic biomarker for LSCC.
1. Introduction
Laryngeal cancer is one of the most common malignant tumors of the head and neck, and the major pathological type is squamous cell carcinoma [1]. Despite great progress in the prevention, diagnosis, and treatment of laryngeal squamous cell carcinoma (LSCC) in recent decades, the global incidence of LSCC remains high and the LSCC survival rate, a key measure of the effectiveness of therapeutic interventions, is still unsatisfactory, especially in Southeast Asia and Eastern Europe [2–4]. Currently, the main diagnostic methods for LSCC are histopathological examination through laryngoscopy and assisted diagnostic systems, including computed tomography and magnetic resonance imaging. However, because of nonspecific symptoms in early stage LSCC, a low rate of early diagnosis makes treatment challenging. It has been reported that LSCC patients' survival rates decrease dramatically when the tumor is diagnosed at an advanced stage [5]. Thus, identification of effective early biomarkers for LSCC is critical to improving patient outcomes.
The pathological mechanism of LSCC is complicated and involves genetic, epigenetic, and environmental factors [6]. DNA methylation, an important epigenetic modification [7], is considered as a hallmark of cancer and is significantly related with various malignancies, including LSCC [8–10]. DNA methylation is precisely regulated by DNA methyltransferases [11]. 5-Methylcytosine mainly occurs among the cytosine-phosphate-guanine (CpG) dinucleotides in regions of densely clustered CpGs, known as CpG islands [11]. The majority of CpG islands are observed in the neighborhood of gene promoters and are often devoid of methylation [12]. Aberrant hypermethylated cytosines among CpG dinucleotides have been proven to be associated with the transcriptional inactivation of genes [12]. Furthermore, aberrant DNA methylation has been shown to play a role in the diagnosis and prognosis of a wide range of cancers, including LSCC [13, 14]. Therefore, the identification of DNA methylation biomarkers specific for LSCC could greatly enhance the power of diagnosis and prognosis of LSCC.
The human shisa family member 3 (SHISA3) is located on chromosome 4p13 and was recently discovered to be a tumor suppressor gene, which suppresses the tumorigenesis, invasion, and metastasis of lung cancer through the degradation of β-catenin of the Wnt signaling pathway [15]. Recent reports have demonstrated that hypermethylation of the SHISA3 promoter region is a common event in colorectal cancer tissues and cell lines, and can serve as an independent predictor of poor overall survival for colorectal cancer patients [16]. However, the association between SHISA3 promoter methylation and LSCC has not yet been fully investigated.
In the present study, the aim was to explore the contribution of SHISA3 promoter methylation to LSCC pathogenesis and its potential diagnostic and prognostic value for LSCC.
2. Materials and Methods
2.1. Patient Characteristics and Tissue Specimen Collection
A total of 93 LSCC patients, who underwent surgery at the Department of Otolaryngology, Head and Neck Surgery, in Ningbo Lihuili Hospital between June 2010 and April 2015, were recruited to the current study. None of the patients underwent chemotherapy or radiotherapy before surgery. All patients have signed the informed consent for the surgical procedure and tissue collection. All of the specimens were collected at the time of surgery and immediately stored in liquid nitrogen at −80°C after excision. In addition, tumor tissues and their paired adjacent tissues were subjected to histological diagnosis by two pathologists according to the World Health Organization classification. There were 45 well-differentiated cases, 35 moderately differentiated cases, and 13 poorly differentiated cases. The clinical stages were confirmed according to the TNM staging system of the AJCC 7th edition (2010). There were 29 Stage I cases, 19 Stage II cases, 12 Stage III cases, and 33 Stage IV cases. Patients were followed up for 60 months. Median follow-up was 41 months, with an interquartile range of 32–52 months. During follow-up, five patients were lost and 34 patients died. All the experiments were approved by the Ethical Committee of Ningbo Lihuili Hospital.
2.2. DNA Extraction and Bisulfite Modification
Genomic DNA was isolated from 93 paired samples using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Then, purified DNA was subjected to bisulfite modification using the ZYMO EZ DNA Methylation-Gold Kit, following the manufacturer's instruction (Zymo Research, Orange, CA, USA).
2.3. Methylation-Specific Polymerase Chain Reaction (MSP) Assay
The methylation status of the SHISA3 promoter region was tested using the MSP assay. The primer sequences are shown in Table 1. Each MSP amplification was performed using 2 μL of bisulfite-modified DNA templates in a 20 μL reaction volume containing 1 μM each of forward and reverse primers, 0.2 mM dNTPs, 10x PCR buffer, and 2.5 U of Hot Start DNA Polymerase (Qiagen, Hilden, Germany). The polymerase chain reaction (PCR) conditions were as follows: denaturation at 95°C for 10 min, followed by 35 cycles of 93°C for 30 s, 56°C for 40 s and 72°C for 50 s, and a final extension at 72°C for 10 min. A water blank was used as a negative control. The generated PCR products were subjected to electrophoresis at 100 V for 30 min, using 2% agarose gel stained with ethidium bromide and visualized under UV illumination. Samples were considered as methylated when there was a clearly visible band (204 bp) on the gel, when using the methylated primers. PCR products were also sequenced to verify the MSP results.
Table 1.
List of all primers used for MSP and qRT-PCR assay.
| Primer | Sequence (5′ to 3′) | Amplification size (bp) |
|---|---|---|
| Methylation | 204 | |
| Forward | AGAGGTGATCGGTAATTTTTTAGTC | |
| Reverse | CCTATTACACAAACTCAAACTCGTT | |
| Unmethylation | 203 | |
| Forward | GAGGTGATTGGTAATTTTTTAGTTG | |
| Reverse | CCTATTACACAAACTCAAACTCATT | |
| SHISA3 qRT-PCR | 85 | |
| Forward | GTCTACGTCCCCTTTCTCATCG | |
| Reverse | AGGTGCAACAATAAATAGCCACT | |
| GAPDH qRT-PCR | ||
| Forward | CCATGGAGAAGGCTGGGG | 194 |
| Reverse | CAAAGTTGTCATGGATGACC |
2.4. Total RNA Extraction and Quantitative Real-Time PCR Assay
Total RNA was extracted from 35 paired LSCC and normal tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. The detailed procedure for the quantitative real-time PCR (qRT-PCR) experiment has been reported previously [17]. The PCR conditions were as follows: 95°C for 10 min and then 45 amplification cycles of 94°C for 20 s, 55°C for 30 s, and 70°C for 30 s. The cycle threshold (Ct) values were recorded for both SHISA3 and GAPDH, which was used as an endogenous control. The expression level of SHISA3 was calculated using the ΔCt method. All the samples were assayed in triplicate. The primer sequences for the qRT-PCR are listed in Table 1.
2.5. Cell Culture
Human HEK293T cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, Utah) supplemented with 10% fetal bovine serum (FBS) (ExCell Biology, Shanghai, China), 100 U/mL penicillin, and 100 µg/mL streptomycin. The incubators were maintained at 37°C in a 5% CO2 atmosphere.
2.6. Construction of Recombinant Plasmids
A fragment of the SHISA3 promoter (−1479 to −276) was amplified, which included the sequence analyzed by MSP. The primer sequences were 5′-CTTACGCGTGCTAGCCCTGTCCTAAGAAATATGTAACTCTAAGAGAG-3′ for the forward primer and 5′-CGCAGATCTCGAGCCCGCTCATAGCGCTCCCCGC-3′ for the reverse primer. The recombinant plasmid (pGL3-SHISA3) concatenated the target fragment of SHISA3 and the pGL3 Basic vector (Promega, Madison city, WI, USA) via a DNA Ligation Kit (TaKaRa, Japan). The pRL-SV40 vector (Promega, Madison city, WI, USA) with the Renilla luciferase gene was used as an internal control in this study.
2.7. Transfection and Reporter Gene Activity Assay
The 293T cells in the exponential growth phase were plated on 24-well plates in DMEM with 10% FBS. After 12 h, cells of 70% attachment were transfected with pGL3-SHISA3 and pRL-SV40 according to the manufacturer's protocol (TransLipid HL Transfection Reagent, TransGen Biotech, Beijing, China). After 4–6 h, the medium was exchanged for fresh DMEM with 10% FBS. After 18–72 h, Renilla and firefly luciferase activity were measured following the manufacturer's protocol (Dual-Luciferase® Reporter Assay Systems, Promega, Madison city, WI, USA).
2.8. Statistical Analyses
All the statistical analysis was performed using SPSS v18.0 (SPSS Inc., Chicago, IL, USA). A chi-squared test or Fisher's exact test was performed to evaluate the difference in SHISA3 promoter methylation frequency between different groups. The comparison of the expression level was analyzed using paired Student's t-tests. The diagnostic value of SHISA3 was assessed using the receiver operating characteristic (ROC) curve and the area under the curve (AUC) [18]. The overall survival (OS) curves were calculated using Kaplan–Meier analysis. Log-rank tests and multivariate Cox proportional hazard models were used to test the prognostic value of SHISA3 methylation for LSCC patients. A two-tailed P value of less than 0.05 was considered statistically significant. All the figures were drawn using the GraphPad Prism 6 software (GraphPad, San Diego, CA).
3. Results
3.1. Methylation Status of SHISA3 in LSCC and Paired Normal Tissues
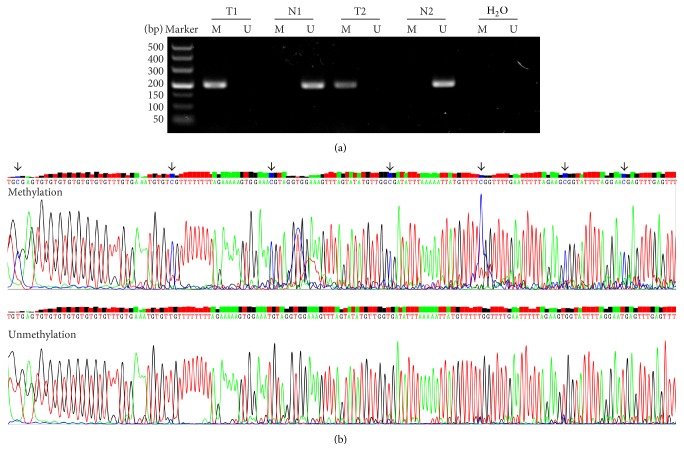
SHISA3 methylation status in 93 LSCC tissue samples and adjacent normal tissue samples was tested using the MSP assay. The representative agarose gel electrophoresis images and the sequencing results are shown in Figure 1. This study reveals that the methylation frequency of the SHISA3 promoter is significantly greater in LSCC tissues than in corresponding normal tissues (P = 4.58E − 12). The SHISA3 promoter was methylated in 66 of 93 (71%) LSCC tissue samples and only in 19 of 93 (20%) adjacent normal tissue samples. Of those patients with methylated SHISA3 in adjacent normal tissues, methylated SHISA3 was also observed in their paired LSCC tissues.
Figure 1.
Agarose gel image and sequencing results of SHISA3. (a) Representative methylation-specific polymerase chain reaction (MSP) results regarding SHISA3 in two paired LSCC and normal tissues. Water was used as a negative control. The methylated CpG sites were highlighted by arrow. T: tumor; N: adjacent normal tissue; M: methylation; U: unmethylation. (b) The sequencing results of methylation and unmethylation PCR products.
3.2. Association between SHISA3 Promoter Methylation and Expression
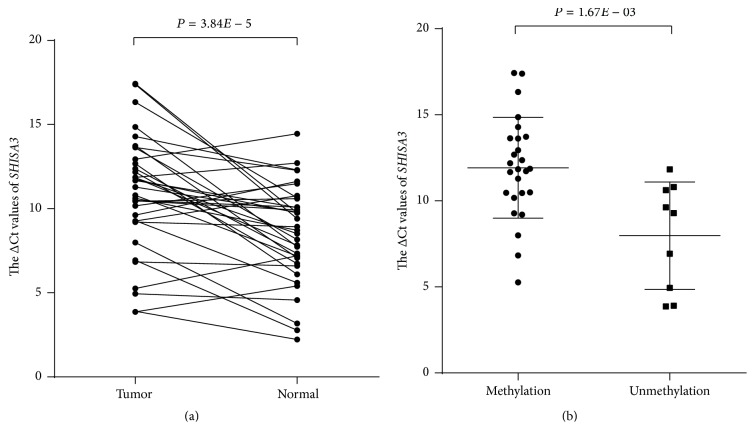
Using qRT-PCR in 35 paired LSCC samples, it was found that the expression of SHISA3 in LSCC tissues was significantly downregulated compared with adjacent normal tissues (P = 3.84E − 05, Figure 2(a)). SHISA3 promoter methylation was observed in 74% (26/35) of these patients. In addition, the analysis showed that the expression of SHISA3 was significantly lower in the methylated group compared with the unmethylated group (P = 1.67E − 03, Figure 2(b)).
Figure 2.
The expression of SHISA3 assessed using quantitative RT-PCR assay. (a) Expression levels of SHISA3 in LSCC and adjacent normal samples (n = 35). The SHISA3 expression level was significantly lower in tumor tissues than in corresponding normal tissues (P = 3.84E − 05). (b) Expression levels of SHISA3 in the methylation group (n = 26) and unmethylation group (n = 9). The SHISA3 expression levels were significantly lower in the methylation group compared with the unmethylation group (P = 1.67E − 03).
3.3. Reporter Gene Activity Assay
In order to verify the transcriptional activity of the tested SHISA3 promoter region, an in vitro luciferase reporter assay was performed. The results show that the pGL3-SHISA3 plasmid, which contains a fragment of the SHISA3 promoter region, had a significantly higher luciferase activity (P < 0.05, Figure 3) and imply that the hypermethylation of the target fragment of SHISA3 may be responsible for its downregulation.
Figure 3.
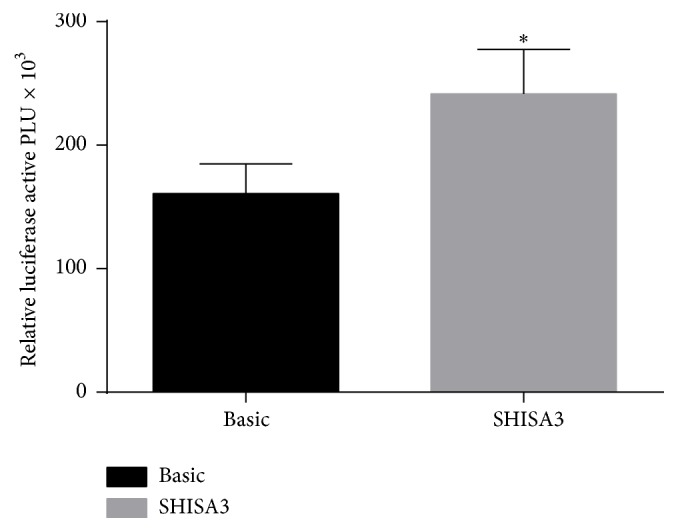
Activity analysis of fragment of promoter region of SHISA3 gene. Asterisk symbol indicates P < 0.05 (∗).
3.4. Association between SHISA3 Promoter Methylation Status and Clinicopathological Parameters
The association between SHISA3 promoter methylation status and the clinicopathological parameters of the LSCC patients, including age, gender, smoking behavior, histological grade, clinical stage, tumor stage, and lymph node metastasis, was assessed (Table 2). The results show that SHISA3 promoter methylation is significantly associated with histological grade (P = 0.02), clinical stage (P = 0.02), tumor stage (P = 0.05), and lymph node metastasis (P = 0.03). However, no significant correlation between SHISA3 promoter methylation and any other parameters was observed.
Table 2.
Association between the SHISA3 methylation in LSCC and the clinicopathological features. N: number; M: methylation; U: unmethylation; #P value calculated by Fisher's exact test.
| Characteristics | SHISA3 methylation | ||||
|---|---|---|---|---|---|
| N | M | U | χ 2 | P | |
| Gender | |||||
| Female | 4 | 3 | 1 | — | 1# |
| Male | 89 | 63 | 26 | ||
| Age | |||||
| <60 | 49 | 36 | 13 | 0.31 | 0.57 |
| ≥60 | 44 | 30 | 14 | ||
| Smoking behavior | |||||
| Yes | 75 | 53 | 21 | 0.08 | 0.78 |
| No | 18 | 13 | 6 | ||
| Histological grade | |||||
| Well/moderately | 80 | 53 | 27 | — | 0.02# |
| Poorly | 13 | 13 | 0 | ||
| Clinical stage | |||||
| I + II | 48 | 29 | 19 | 5.36 | 0.02 |
| III + IV | 45 | 37 | 8 | ||
| Tumor invasion | |||||
| 1 + 2 | 58 | 37 | 21 | 3.85 | 0.05 |
| 3 + 4 | 35 | 29 | 6 | ||
| Lymph node metastasis | |||||
| No | 64 | 41 | 23 | 4.75 | 0.03 |
| Yes | 29 | 25 | 4 | ||
3.5. The Diagnostic and Prognostic Value of SHISA3 Promoter Methylation for LSCC
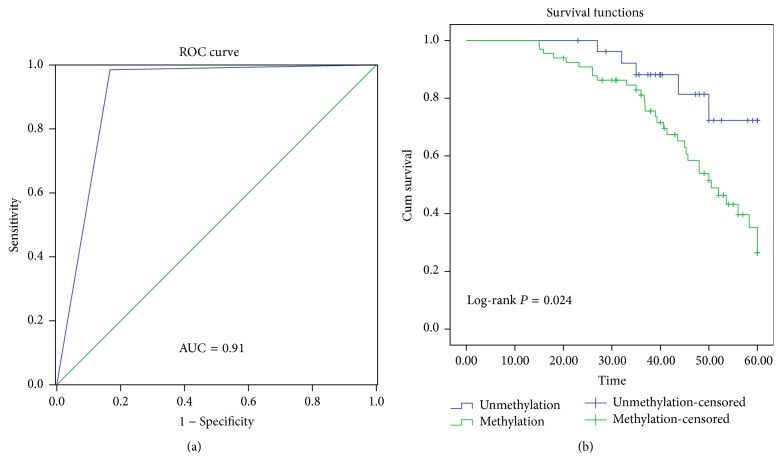
A ROC curve was plotted to evaluate the diagnostic value of SHISA3 promoter methylation (Figure 4(a)). The area under the ROC curve is 0.91 (P < 0.01), while the sensitivity and specificity with the maximum Youden index are 0.99 and 0.83, respectively. In the current study, the prognostic value of SHISA3 methylation status in LSCC was also investigated. As shown in Figure 4(b), the survival curves demonstrate that hypermethylated SHISA3 is significantly associated with poor outcome in LSCC (log-rank P = 0.024). A multivariate Cox proportional hazard analysis was then performed, by adjusting for age, smoking behavior, histological differentiation, clinical stage, and lymphatic metastasis (Table 3), and the results confirm a significantly poor prognosis in LSCC patients with SHISA3 methylation (hazard ratio (HR), 2.71; 95% confidence interval, 1.024–7.177; P = 0.047).
Figure 4.
The receiver operating characteristic (ROC) curve and survival curve. (a) The ROC analyses of the curve. The area under the curve was 0.91. (b) The survival curve of patient groups according to SHISA3 methylation status. The SHISA3 methylation group showed significantly worse survival rates than the SHISA3 unmethylation group (log-rank P = 0.024).
Table 3.
Multivariate Cox proportional hazards analysis in 93 LSCC patients. N: number; Ref: reference category; HR: hazard ratio; CI: confidence interval.
| Characteristics | N | P value | HR | 95% CI |
|---|---|---|---|---|
| Age | 93 | 0.979 | 0.999 | 0.953–1.048 |
| Smoking behavior | ||||
| No (Ref) | 18 | — | 1 | — |
| Yes | 75 | 0.288 | 0.636 | 0.276–1.465 |
| Differentiation | ||||
| Well (Ref) | 45 | — | 1 | — |
| Moderated | 35 | 0.256 | 0.656 | 0.318–1.356 |
| Poorly | 13 | 0.24 | 0.512 | 0.167–1.566 |
| Clinical stage | ||||
| Stage I (Ref) | 29 | — | 1 | — |
| Stage II | 19 | 0.783 | 1.196 | 0.336–4.257 |
| Stage III | 12 | 0.053 | 3.735 | 0.982–14.208 |
| Stage IV | 33 | 0.019 | 4.238 | 1.272–14.117 |
| Lymphatic metastasis | ||||
| No (Ref) | 64 | — | 1 | — |
| Yes | 29 | 0.723 | 1.172 | 0.489–2.809 |
| Methylation | ||||
| No (Ref) | 27 | — | 1 | — |
| Yes | 66 | 0.045 | 2.71 | 1.024–7.177 |
4. Discussion
In cancer research, identification of robust biomarkers, which enable early detection and reliable prognosis of malignancy, is a top priority. DNA methylation is one of the most widely studied epigenetic changes [19]. Previous studies have shown that the inactivation of tumor suppressor genes (TSGs) in numerous cancers, including LSCC, may be attributed to the hypermethylation of CpG islands in the promoter region [20, 21]. DNA methylation can provide an alternative target in the search for biomarkers that could provide a sensitive, specific, and early marker for cancer diagnosis and prognosis [22, 23]. SHISA3 is a newly found TSG in non-small-cell lung cancer, which conducts its tumor suppression activity through the Wnt signaling pathway [15].
In the current study, the methylation status of the SHISA3 promoter in 93 paired LSCC tissue samples was measured using MSP, revealing that the SHISA3 promoter is highly methylated in LSCC. Interestingly, the results indicate that SHISA3 promoter methylation is more frequently observed in poorly differentiated, lymph node metastasis, and advanced clinical stages, as well as advanced tumor invasion LSCC patients. It can be concluded that SHISA3 methylation is associated with LSCC clinicopathological characteristics. Additionally, the qRT-PCR results show that the SHISA3 expression level is significantly lower in LSCC tissues than in corresponding normal tissue. Methylated SHISA3 in LSCC was also associated with a significant reduction in SHISA3 transcriptional activity, when compared with unmethylated SHISA3. The reporter gene activity assay revealed that the SHISA3 promoter fragment analyzed by MSP was able to regulate gene expression, which implied that the dysregulation of SHISA3 may attribute to the hypermethylation of promoter region. All these phenomena may be explained by the fact that the SHISA3 protein can protect from malignant transformation and invasion during early stage disease [15], and the dysregulation of SHISA3 in LSCC, at least partly, results from methylation [16].
The diagnostic power of methylation biomarkers has been well illustrated in many human cancers [24, 25]. In the present study, the summary ROC curve and AUC were applied to determine the diagnostic value of SHISA3 methylation for LSCC [26]. When the AUC is close to 1.0, this signifies a good risk predictor [27]. In this study, the sensitivity, specificity and AUC for SHISA3 methylation in LSCC is 0.99, 0.83 and 0.91, respectively, suggesting that detecting SHISA3 methylation has a good diagnostic accuracy and represents a potential diagnostic biomarker for LSCC. Additionally, accumulating evidences have shown that the combination of several epigenetic biomarkers can improve the sensitivity and specificity of diagnostic testing for cancers [28, 29]. Therefore, it would be logical to combine SHISA3 methylation testing with other methylation biomarkers. This approach requires further studies to determine its potential diagnostic power for LSCC. It is important to point out that according to these findings, SHISA3 methylation is strongly associated with inferior survival outcomes, since the log-rank test analysis showed that the OS in LSCC patients with hypermethylated SHISA3 was statistically lower than in those without methylated SHISA3. Considering the contribution of a variety of factors (such as age, smoking behavior, histological differentiation, clinical stage and lymphatic metastasis, and SHISA3 methylation) to OS, a multivariate Cox proportional hazard analysis was performed to adjust for these factors, and the results confirmed that methylation of SHISA3 could be an independent unfavorable prognostic factor for LSCC. These results are also supported by previous reports in lung cancer and colorectal cancer [15, 16].
In conclusion, this study has revealed that SHISA3 promoter hypermethylation is a common event in LSCC, contributing to its transcriptional inactivation, and may be involved in the invasion, progression, and metastasis of LSCC. These findings strongly indicate that methylated SHISA3 is a new potential epigenetic biomarker for the early diagnosis and prognosis of LSCC. However, further research is needed to elucidate the underlying mechanisms of the SHISA3 gene in the pathogenesis of LSCC.
Acknowledgments
This work was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (no. LY14H160003), the Scientific Innovation Team Project of Ningbo (no. 2012B82019), the Ningbo Social Developmental Key Research Project (no. 2012C5015), the Ningbo Natural Science Foundation (no. 2012A610208 and no. 2012A610217), the Medical and Health Research Project of Zhejiang Province (no. 2012ZDA042), and the Medical and Health Training Project of Zhejiang Province (no. 2014PYA017).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this article.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2016. CA Cancer Journal for Clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.De Angelis R., Sant M., Coleman M. P., et al. Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE-5—a population-based study. The Lancet Oncology. 2014;15(1):23–34. doi: 10.1016/s1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 3.Du L., Li H., Zhu C., Zheng R., Zhang S., Chen W. Incidence and mortality of laryngeal cancer in China, 2011. Chinese Journal of Cancer Research. 2015;27(1):52–58. doi: 10.3978/j.issn.1000-9604.2015.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European Journal of Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Teppo H., Koivunen P., Hyrynkangas K., Alho O.-P. Diagnostic delays in laryngeal carcinoma: professional diagnostic delay is a strong independent predictor of survival. Head and Neck. 2003;25(5):389–394. doi: 10.1002/hed.10208. [DOI] [PubMed] [Google Scholar]
- 6.Maurya S. S., Katiyar T., Dhawan A., et al. Gene-environment interactions in determining differences in genetic susceptibility to cancer in subsites of the head and neck. Environmental and Molecular Mutagenesis. 2015;56(3):313–321. doi: 10.1002/em.21920. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A. P., Tycko B. The history of cancer epigenetics. Nature Reviews Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 8.Brower V. Epigenetics: unravelling the cancer code. Nature. 2011;471(7339):S12–S13. doi: 10.1038/471s12a. [DOI] [PubMed] [Google Scholar]
- 9.Kis A., Tatár T. Z., Gáll T., et al. Frequency of genetic and epigenetic alterations of p14ARF and p16INK4A in head and neck cancer in a Hungarian population. Pathology Oncology Research. 2014;20(4):923–929. doi: 10.1007/s12253-014-9775-9. [DOI] [PubMed] [Google Scholar]
- 10.Zauri M., Berridge G., Thézénas M.-L., et al. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature. 2015;524(7563):114–118. doi: 10.1038/nature14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones P. A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 12.Deaton A. M., Bird A. CpG islands and the regulation of transcription. Genes and Development. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Z., Chen X., Li Q., et al. SSTR2 promoter hypermethylation is associated with the risk and progression of laryngeal squamous cell carcinoma in males. Diagnostic Pathology. 2016;11(1, article 10) doi: 10.1186/s13000-016-0461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephen J. K., Chen K. M., Shah V., et al. DNA hypermethylation markers of poor outcome in laryngeal cancer. Clinical Epigenetics. 2010;1(1-2):61–69. doi: 10.1007/s13148-010-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.-C., Chen H.-Y., Su K.-Y., et al. Shisa3 is associated with prolonged survival through promoting β-catenin degradation in lung cancer. American Journal of Respiratory and Critical Care Medicine. 2014;190(4):433–444. doi: 10.1164/rccm.201312-2256OC. [DOI] [PubMed] [Google Scholar]
- 16.Tsai M.-H., Chen W.-C., Yu S.-L., et al. DNA hypermethylation of SHISA3 in colorectal cancer: an independent predictor of poor prognosis. Annals of Surgical Oncology. 2015;22(supplement 3):1481–1489. doi: 10.1245/s10434-015-4593-1. [DOI] [PubMed] [Google Scholar]
- 17.Shen Z., Li Q., Deng H., Lu D., Song H., Guo J. Long non-coding RNA profiling in laryngeal squamous cell carcinoma and its clinical significance: potential biomarkers for LSCC. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0108237.e108237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pencina M. J., D'Agostino S., D'Agostino J., Vasan R. S. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statistics in Medicine. 2008;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 19.Ouadid-Ahidouch H., Rodat-Despoix L., Matifat F., Morin G., Ahidouch A. DNA methylation of channel-related genes in cancers. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2015;1848(10):2621–2628. doi: 10.1016/j.bbamem.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Niyazi M., Sui S., Zhu K., Wang L., Jiao Z., Lu P. Correlation between Methylation of Human Papillomavirus-16 L1 Gene and Cervical Carcinoma in Uyghur Women. Gynecologic and Obstetric Investigation. 2017;82(1):8. doi: 10.1159/000444585. [DOI] [PubMed] [Google Scholar]
- 21.Teng Y., Fan Y. C., Mu N. N., Zhao J., Sun F. K., Wang K. Serum SOX11 promoter methylation is a novel biomarker for the diagnosis of Hepatitis B virus-related hepatocellular carcinoma. Neoplasma. 2016;63(3):419–426. doi: 10.4149/311_151029N552. [DOI] [PubMed] [Google Scholar]
- 22.Haldrup C., Lynnerup A.-S., Storebjerg T. M., et al. Large-scale evaluation of SLC18A2 in prostate cancer reveals diagnostic and prognostic biomarker potential at three molecular levels. Molecular Oncology. 2016;10(6):825–837. doi: 10.1016/j.molonc.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brock M. V., Hooker C. M., Ota-Machida E., et al. DNA methylation markers and early recurrence in stage I lung cancer. The New England Journal of Medicine. 2008;358(11):1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 24.Kim E. J., Chung W. C., Kim D. B., et al. Long interspersed nuclear element (LINE)-1 methylation level as a molecular marker of early gastric cancer. Digestive and Liver Disease. 2016;48(9):1093–1097. doi: 10.1016/j.dld.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Liu K., Zhang Y., Zhang C., et al. Methylation of S100A8 is a promising diagnosis and prognostic marker in hepatocellular carcinoma. Oncotarget. 2016;7(35):56798–56810. doi: 10.18632/oncotarget.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zweig M. H., Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clinical Chemistry. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 27.Jones C. M., Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Annals of Thoracic Surgery. 2005;79(1):16–20. doi: 10.1016/j.athoracsur.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Shan M., Yin H., Li J., et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7(14):18485–18494. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K., Tian Y., Xu H. Improved noninvasive bladder cancer diagnosis using urine sediments and novel DNA methylation biomarker panels. Clinical Laboratory. 2016;62(3):327–336. doi: 10.7754/clin.lab.2015.150602. [DOI] [PubMed] [Google Scholar]