Abstract
The long non-coding RNA Xist is only expressed from the paternal X chromosome in mouse pre-implantation female embryos and leads to its transcriptional silencing. In females, absence of Xist leads to post-implantation lethality. Here we report that the initiation of imprinted XCI absolutely requires Xist using single-cell RNA-sequencing of early pre-implantation mouse embryos. Lack of paternal Xist leads to genome-wide transcriptional misregulation in the early blastocyst, with failure to activate the extra-embryonic pathway that is essential for post-implantation development. We also demonstrate that the expression dynamics of X-linked genes depends both on strain and parent-of-origin, as well as on location along the X chromosome, particularly at Xist’s first “entry” sites. This study demonstrates that dosage compensation failure has an impact as early as the blastocyst stage and reveals genetic and epigenetic contributions in orchestrating the transcriptional silencing of the X chromosome during early embryogenesis.
Introduction
In mammals, differences in sex-chromosome constitution between males (XY) and females (XX) have led to the evolution of dosage compensation strategies, including transcriptional silencing of one X chromosome in females1. In mice, X-chromosome inactivation (XCI) first initiates in the pre-implantation embryo. The non-coding Xist RNA is expressed only from the paternal allele leading to paternal X (Xp) inactivation2. The Xp remains inactive in extra-embryonic tissues, but is reactivated in the inner cell mass followed by random XCI in the embryo proper3,4. In early mouse embryos, XCI has been shown to be very dynamic and its requirements, both in cis at the level of the X-inactivation center (Xic) and in trans, have been debated5. Imprinted XCI has been proposed to initiate de novo2,9 following the onset of zygotic genome activation (ZGA) and Xist expression. One study proposed that Xp inactivation is initially Xist-independent and that Xist may only be required for early maintenance of silencing6, while another reported a lack of Xp gene silencing in the absence of Xist7. These studies were all based on the analysis of just a few genes, however. Two recent single cell transcriptomic studies exploited inter-specific crosses to investigate XCI in female pre-implantation embryos8 and differentiating ESCs9. These revealed that imprinted XCI indeed initiates between the 4-8-cell stage8 and that progression of random XCI is correlated with differentiation9. However, the extent to which initiation of Xp-linked gene silencing is dependent on Xist RNA, or is influenced by strain- or parent-of-origin (eg imprinted X-linked genes) were not explored.
In this study, we set out to explore the precise kinetics of paternal and maternal X-linked gene expression during pre-implantation embryogenesis, using inter-specific crosses and single cell RNA sequencing (scRNAseq). This allowed us to investigate differences in the dynamics of imprinted XCI that were due to genetic background and/or to parental origin. By investigating X-linked gene expression in female embryos derived from Xist KO males, we also demonstrate the absolute Xist dependence of early, imprinted XCI and report the genome-wide transcriptional consequences induced by a lack of dosage compensation. Overall, this study provides important insights into the transcriptional and allelic dynamics of XCI, as well as the nature of the requirement for dosage compensation during the first stages of mammalian development.
Results
Allele-specific scRNAseq during pre-implantation development
To investigate the extent and requirements of gene silencing during imprinted XCI in early embryogenesis, we profiled the expression kinetics of genes on the Xp and Xm chromosomes, using scRNAseq10. F1 embryos were derived from inter-specific crosses, of either wild-type (wt) or Xist paternally deleted mutant (Xistpat∆) origin, between the 2-cell and blastocyst (approximately 60-64-cell) stages. Reciprocal crosses between highly polymorphic Mus musculus castaneus (Cast/EiJ) and Mus musculus domesticus (C57BL6/J) strains, herein referred to as Cast and B6 respectively, were used (Figure 1a) and a minimum of 5 embryos, and 6 single cells per stage for BC and CB wt embryos (Supplementary Data Set 1). Of 24,499 referenced mouse genes, 15,581 were found expressed in at least one developmental stage, including 580 X-linked genes.
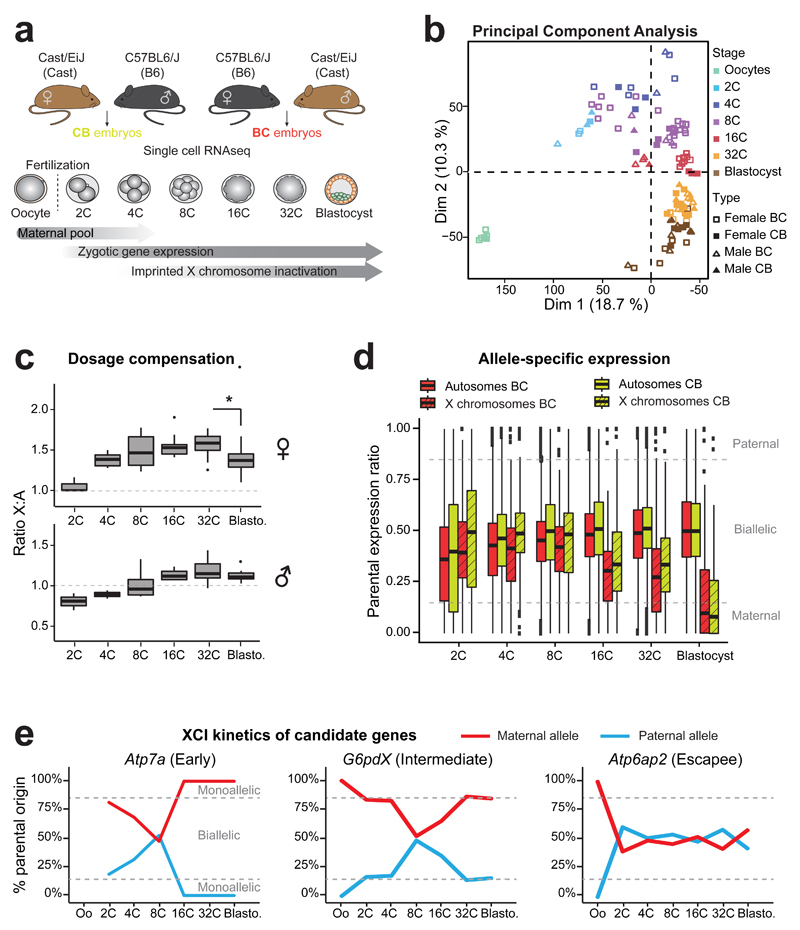
Figure 1. Single cell RNA sequencing of early hybrid embryos and dosage compensation mechanisms.
(a) Schematic illustration of the single cell experiment and the harvested stages during pre-implantation mouse development. Time windows showing the persistence of maternal mRNA pool, activation of zygotic gene expression and Xp inactivation are indicated.
(b) Principal component analysis (PCA) of single oocytes and pre-implantation blastomeres (2C to blastocysts) based on scRNA data. Different stages are designed by different colors. n= 6 to 30 cells per stage (details of each single cell are in Supplementary Data Set 1).
(c) Differences in ratio of X-chromosome expression levels by autosomal expression levels, between 2-cell stage to blastocyst, using Dunn¹s test (Kruskal-Wallis), p<0.001 to **. Boxplots represent median with lower and upper quartiles.
(d) Allele-specific expression ratios for genes on autosomes (plain red, BC or yellow CB) and on X chromosomes (dashed red, BC or yellow, CB) in female single blastomeres (2-cell to blastocyst) from BC and CB crosses. Allele-specific proportion represents the number of reads mapped to the paternal genome divided by the total number of paternal and maternal reads mapped for each gene. Boxplots represent medians with lower and upper quartiles.
e) Examples of scRNA expression dynamics of three X-linked genes with their classification as “early inactivated”, “intermediate inactivated” or “escapee” (as used in Patrat et al, 2009 14) (see also Supplementary Figure 2). Mean percentage of parental origin transcripts is represented between oocytes and blastocyst.
We first assessed the extent to which transcriptomes of single cells were associated by stage, sex or cross, by performing principal component analyses (PCA) and hierarchical clustering (Figure 1b and Supplementary Figure 1). The primary source of variability between all cells was developmental stage, as expected based on previous studies8, thus validating the quality of our data. Single cell transcriptomes clustered to a lesser extent by cross (BC and CB), and then by sex (XX and XY) (Supplementary Figure 1), with the differences between the sexes reaching a minimum by the 32-cell and blastocyst stages, presumably due to dosage compensation.
Timing of dosage compensation and imprinted XCI
To assess the precise timing of dosage compensation in male and female embryos, we examined autosomal and X-linked transcripts at each stage in both sexes. According to Ohno’s law11 average X-linked gene expression should be equivalent to the expression of autosomal genes. Furthermore, equal expression of X-linked genes between females and males is expected through XCI. We compared X:Autosomes (X:A) expression ratios in single blastomeres of each sex (Figure 1c). Expected X:A ratios would be 1 in females and 0.5 in males in the absence of any dosage compensation (ie no X overexpression compared to autosomes, and no XCI). We found that the X:A ratios were significantly above the expected ratios as early as the 4-cell stage (p<9*10-4 for males and females after 4-cell stage, t-test) and continued to rise until the 32-cell stage, suggesting that there is a progressive increase in expression of the X compared to autosomes at the same time as, or soon after ZGA. In females, the X:A ratio rose to 1.58, by the 32 cell stage and then significantly dropped to 1.37 by the early blastocyst stage (p=1.96*10-2 between 32-cell and blastocyst, Kruskal-Wallis (KW) test), presumably due to XCI by this stage (see below). This suggests that X:A ratios in female blastocysts progressively reach 1, although even at the early blastocyst stage, they were still slightly higher compared to males (p=2.03*10-3, KW), in agreement with previously published data12;13.
We next investigated allele-specific X-linked gene expression and the timing of XCI in BC and CB female embryos. At the 2-cell stage, ZGA and massive degradation of the maternal pool of mRNAs occur. Here, transcripts are maternally biased genome-wide as expected given the residual maternal pool (Figure 1d). At subsequent stages, while autosomal transcripts reached parity for both parental genomes, with a parental ratio in blastocysts of 0.5 in both crosses, X-linked transcripts displayed maternal skewing even at the 16-cell stage. By the blastocyst stage global transcription of the Xp was significantly reduced in both crosses (p<2.2*10-16, KW) indicating that XCI was fairly complete, as previously reported7,8,14. We compared the kinetics of Xp silencing for 13 X-linked genes previously analyzed by nascent RNA-FISH14 and found that most (12/13) genes showed very similar patterns (Figure 1e and Supplementary Figure 2), giving us confidence that our scRNAseq data, bioinformatics pipeline and expression thresholds were valid. The one gene (out of the 13) for which slightly earlier Xp silencing was found by scRNAseq compared to previous reports was Atrx. We confirmed that Atrx is inactivated on the Xp in most cells by the morula stage using RNA FISH with a gene-specific probe (Supplementary Figure 3a). We also confirmed its previously reported Xp reactivation in the blastocyst14 (Supplementary Figure 2).
Strain-specific XCI and escape
We established an in vivo chromosome-wide map of X-linked gene activity between the 4-cell and blastocyst stages. Of the 580 X-linked genes expressed in our scRNAseq, we focused on the 164 (BC cross) and 134 (CB cross) most highly expressed and informative genes (RPRT>4 and expressed in at least 25% of the cells of each stage and cross with a minimum of 2 cells), for which we could establish allelic expression profiles with confidence (Supplementary Figure 3b and Figure 2 for the 125 common genes between BC and CB crosses, see Online Methods for allelic expression threshold details). A striking switch from biallelic (grey, pale pink or pale blue) expression at the 4-cell stage, to monoallelic, maternal (red) expression at the blastocyst stage can be observed for most X-linked genes. Several genes underwent only partial or no XCI (escapees) and will be discussed later. As expected, Xist expression was exclusively of paternal origin throughout (Figure 2 and Supplementary Figures 3b and 4). Another gene showing only paternal expression was Fthl17f, part of the ferritin, heavy polypeptide-like 17 family (also known as Gm5635), which has previously been reported to be exclusively paternally expressed and imprinted15. By the blastocyst stage Fthl17f expression was no longer detectable, presumably due to XCI (Figure 2 and Supplementary Figures 3b and 4).
Figure 2. Kinetics of silencing of X-linked genes over the entire X chromosome during imprinted XCI in different strains.
The mean allele-specific expression ratios per embryonic stage for each informative and expressed X-linked gene in 4-cell to blastocyst stage female embryos are represented as heatmaps, with strictly maternal expression (ratio ≤0.15) in red and strictly paternal expression (ratio ≥0.85) in blue. Color gradients are used in between these two values as shown in the key. Genes are ordered by genomic position (centromere top, telomere bottom). Data from CB (left) and BC (right) female embryos are shown (for thresholds see Online Method) and arrows highlight examples of early silenced or escapee genes. n= 125 informative X-linked genes in common for CB and BC crosses.
We categorized genes into different groups with respect to their timing of XCI for each cross (early ≤ 16-cell; intermediate ≤32-cell; late = blastocyst; Figure 3a, SI Table 2 and Online Methods). Even by the 8-cell stage, XCI is complete for a few genes (eg Rnf12, Pnma5, Figure 2 and Supplementary Figure 3b). By the blastocyst stage, Xp reached a very similar state of inactivation in both BC and CB crosses (respectively 83.5% and 84.3% of the 164 and 134 X-linked informative and expressed genes are either silenced or maternally biased at the blastocyst stage, Figures 2, 3a and 3b). However, when comparing gene expression in embryos derived from BC and CB crosses (125 common genes), marked differences were seen between crosses, with just 71.2% (89 of 125 genes) of X-linked genes falling into the same or a similar category between BC and CB crosses (eg early and mid or late and biased). The degree of non-consistency in silencing kinetics between crosses was evaluated if more than one developmental stage separated the same gene between BC and CB crosses (Supplementary Table 1 and Online Methods for classification details). Several genes also show strain-specific escape (Figure 3b). Some of these have previously been described16 or reported to escape in a tissue-specific fashion at later stages of development or in somatic tissues (eg Ddx3x, Idh3g)16–18. On the other hand, several genes remain biallelically expressed even at the blastocyst stage and tend to show escape independent of strain (Figure 3b and Supplementary Table 2). Many of these also show escape in somatic tissues19 (eg Eif2s3x, Kdm5c, Utp14a). Finally, some genes with biallelic ratios (represented as black dots in Figure 3b), correspond to genes that previously underwent Xp silencing prior to the blastocyst stage but then became re-expressed, as previously described for Atrx.
Figure 3. Different genes show different kinetics of silencing associated with their chromosomal position and Xist “entry” site localization.
(a) X-linked genes are clustered based on their silencing kinetics as “early” (silenced at 16-cell or earlier), “intermediate” (silenced at 32-cell), “late” (silenced at blastocyst), “biased” (maternally biased) and “escapee” (Esc, not silenced). The allelic ratio of each gene represents the number of reads mapped on the paternal genome divided by the total number of reads mapped and is represented at 4-cell, 16-cell, 32-cell and blastocyst stages from single female blastomeres. Further information is provided in Supplementary Table 1 and Online Methods. n= 137 X-linked genes (89 with consistent silencing kinetics between BC and CB crosses and 48 BC or CB-specific).
(b) Parental expression ratios of X-linked genes in female blastocysts in BC and CB strains. Each dot represents a single gene. The upper and lower sections represent data respectively from BC and CB embryos. Xist is represented by a red dot. Green and orange dots represent genes that escape from early XCI respectively in both strains or in strain-specific manner. Further information on escapees is found in Supplementary Table 2. n= 125 common X-linked genes.
(c) Box plot representing the distribution of the genomic distances to Xist locus (in Mb) for the different clusters of genes. “Transcription Start Site (TSS) of each gene has been used to measure the distance to Xist. p<0.05 corresponds to * by Dunn’s test.”
(d) Allelic expression of X-linked genes classified by their relative position to Xist “entry” sites (as identified during XCI induction in ESCs20): “inside” (TSS located in a Xist “entry” site), “next to” (TSS located less than 100 kb to an “entry” site) and “outside” (over 100 kb from an “entry” site). By Dunn’s test; p<0.05 corresponds to *. Consistent or strain-specific genes have been used.
Boxplot represent median with lower and upper quartiles.
Xist “entry” sites and early silenced genes
We next assessed whether gene-silencing kinetics was correlated with genomic position along the X chromosome. We first focused on the 71.2% (n=89 genes) of genes with correlated kinetics between crosses and the strain-specific genes (n=48). Although early and intermediate silenced genes do tend to lie closer to the Xic compared to late silenced genes (Figure 3c), gene silencing does not appear to occur as a simple linear gradient from the Xic according to our allele-specific expression heatmap, with the presence of some escapees close to the Xic (Figure 2). Rather, we noted that several regions across the X chromosome contain early-silenced genes (eg Pnma5, Kif4, Magt1), from which silencing appears to “spread” locally (Figure 2). A recent study in ES cells showed that Xist RNA initially binds to specific genomic regions (Xist “entry” sites) along the X chromosome, dependent on 3D proximity to the Xist locus20. This binding has been hypothesized to silence genes locally and to then spread along the rest of the X chromosome by Engreitz et al20. We found that X-linked genes lying within the predicted Xist entry regions (8 and 11 genes respectively in 32-cell and blastocysts), or close to (20 and 23 respectively in 32-cell and blastocysts) these regions showed the earliest silencing and strongest maternal imbalance (p=0.02 and p=0.03, KW, respectively in 32-cell and blastocysts, Figure 3d). Thus, we show that Xist RNA “entry” sites as defined in ESCs20 could potentially correspond to XCI initiation sites in vivo during imprinted XCI.
Fully Xist-dependent imprinted XCI
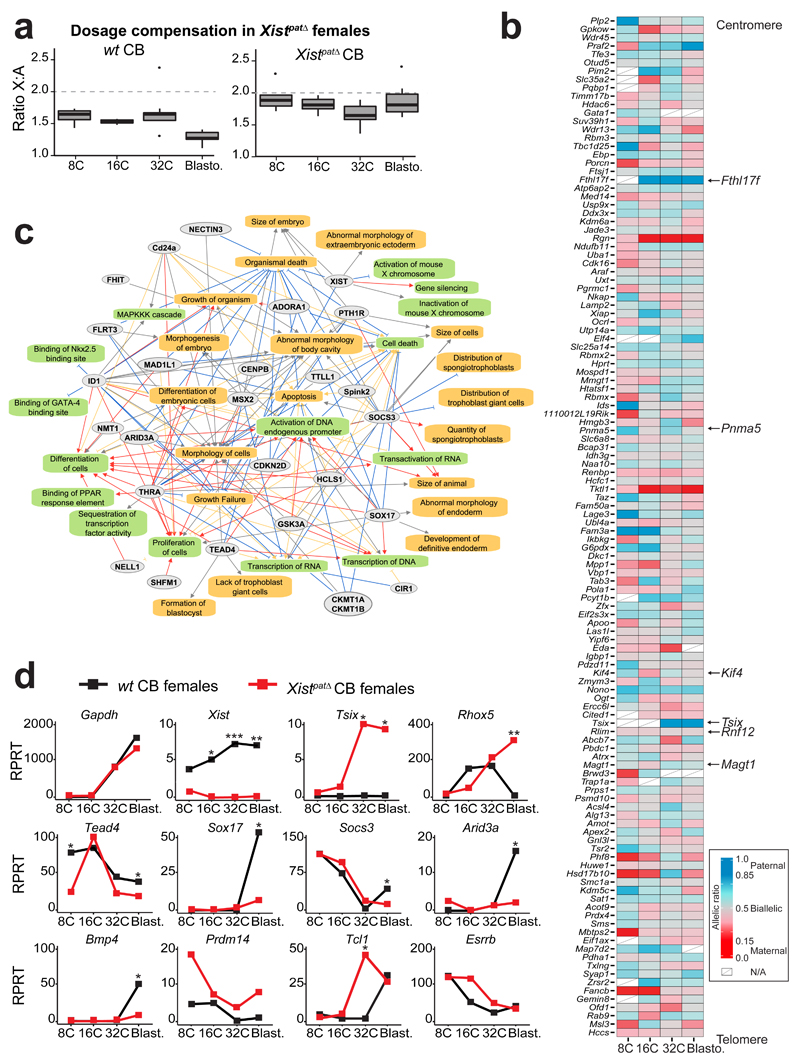
The above findings suggested that Xist RNA plays an early role in triggering gene silencing during imprinted XCI. This contrasts with a previous report suggesting that initiation of imprinted XCI is Xist-independent6. Indeed, although Xistpat∆ females die around E10.5, with major growth delay21, mutant and wt females appear morphologically indistinguishable during pre-implantation development (data not shown). To evaluate whether XCI can be established, even in the absence of Xist expression as previously reported6, we examined X-linked gene expression profiles in single cells of pre-implantation female embryos carrying a paternal Xist deletion (Xistpat∆)21,22. Xist is normally expressed exclusively from the Xp in pre-implantation embryos2 (Figure 2). Transcriptomes of single blastomeres from hybrid F1 embryos (Cast females crossed with Xistmat∆ B6 males) were compared to CB wt embryos from the 8-cell stage (when XCI normally initiates for some genes) to the blastocyst stage. We found similar X:A expression ratios between mutant and control female embryos up to the 32-cell stage (Figure 4a). However at the blastocyst stage, X:A ratios remained much higher in mutants compared to wt embryos where this ratio normally decreases due to XCI (p=1.77*10-4, KW). This indicated that Xp silencing is not initiated in Xistpat∆ female blastocysts. Bioinformatics analysis on the Xist-mutant single cell transcriptomes was used to produce an allele-specific expression heatmap (see Online Methods) and, as expected given the absence of apparent dosage compensation in the mutants, we found that X-linked genes remained significantly biallelically expressed in Xistpat∆ embryos (Figure 4b). Only 2 genes (Rgn and Tktl1) out of 122 assessed (ie 1.6%), showed maternal monoallelic expression in Xistpat∆ mutant blastocysts, compared to 84.3% in CB wt controls. One of these, Tktl1, has been hypothesized to be imprinted23. Moreover, Fthl17f, a well-known imprinted gene was aberrantly expressed in Xistpat∆ blastocysts, suggesting a lack of Xp silencing.
Figure 4. Paternal knockout of Xist impaired XCI, dosage compensation and differentiation pathways.
(a) Differences in ratio of X-chromosome expression levels by autosomal expression levels, between 8-cell stage to blastocyst in CB females (left panel) and Xistpat∆ CB females (with a paternally inherited knock-out allele) (right panel). Boxplots represent median with lower and upper quartiles.
(b) Heatmap representing allele-specific mean expression from 8-cell to blastocyst stage of X-linked genes (as in Figure 2) in Xistpat∆ mutant single cells. Strictly maternally expressed genes (allelic ratio ≤0.15) are represented in red and strictly paternally expressed genes (allelic ratio ≥0.85) in blue. Color gradients are used in between and genes have been ordered by genomic position. Tsix was included in the heatmap if it was expressed in at least 2 single cells per stage, even though it did not reach the expression threshold used (RPRT>4 and expressed in at least 25% of the cells of each stage and cross with a minimum of 2 cells). n = 122 genes.
(c) Major down-regulated genes and pathways detected between CB wt and CB Xistpat∆ females extracted from Supplementary Data Set 2, using QIAGEN’s Ingenuity Pathway Analysis (IPA) software (Supplementary Data Set 3). Color code for arrows, red: leads to inhibition; blue: leads to activation; orange: findings consistent with state of downstream molecule; grey: effect not predicted.
(d) Expression data of candidate genes from wt CB (black) and Xistpat∆ CB (red) females, extracted from scRNAseq. Mean of expression is represented in Reads Per Retro-Transcribed length per million mapped reads (RPRT) during early development (8-cell to blastocyst stages). Gapdh gene is a control housekeeping gene. n= 4 to 30 cells per stage and genotype. By Kruskal-Wallis test; p<0.05 corresponds to *.
We thus found no evidence for Xist-independent XCI (Supplementary Figure 5a), even for X-linked genes previously proposed to be silenced independently of Xist6 (11 out of 14 assayed by Kalantry et al, of which they found only Rnf12, Abcb7 and Atrx to be Xist-dependent). Three of the genes assayed by Kalantry et al (Abcb7, Fmr1 and Pgk1) showed a slight maternal bias at the 16-cell or 32-cell stages in the Xistpat∆ cells in our study (left column, Supplementary Figure 5a). However this is probably due to variability in their parental-origin expression, also observed in CB controls (Abcb7 and Fmr1, Supplementary Figures 3, 4 and 5a) rather than to Xp silencing. Instead, our data is in agreement with the Namekawa et al study7 where Xist-dependent Xp silencing was proposed to occur based on nascent RNA-FISH on 2-cell to blastocyst stage embryos, although their study was only based on 8 genes, 4 of which were in common with ref 5. The discrepancies between these previous studies were likely due to technical differences. The scRNAseq analysis we provide here represents chromosome-wide evidence for Xist-dependent gene silencing during pre-implantation embryogenesis and corroborates recent findings about Xist-dependent X-linked gene dosage13.
Improper gene expression in Xist mutant embryos
The transcriptome of Xistpat∆ embryos provided us with a unique opportunity to explore the molecular defects that occur in the absence of paternal XCI. A genome-wide differentially expressed (DE) gene analysis was performed in wt and Xistpat∆ embryos (Supplementary Data Set 2). Expression profiles of single blastomeres of controls and mutants were still found to cluster according to developmental stage by PCA (data not shown). However, at the 8-cell and 32-cell stages, a surprisingly elevated number of DE autosomal genes (FDR<0.05) was found in Xistpat∆ embryos compared to wt (Supplementary Figure 5b). By the blastocyst stage, when paternal XCI is normally complete in wt females, 30% of the total up-regulated genes in Xistpat∆ embryos were found to be X-linked, corroborating an XCI defect in the absence of Xist. DE genes included Tsix (the antisense transcript to Xist) which is normally not expressed from the Xp at the 32-64 cell stage24 (Figures 4b and d and Supplementary Figure 5b). The absence of Xist on the paternal X thus releases paternal Tsix repression in cis (without affecting the maternally imprinted Xist/Tsix alleles).
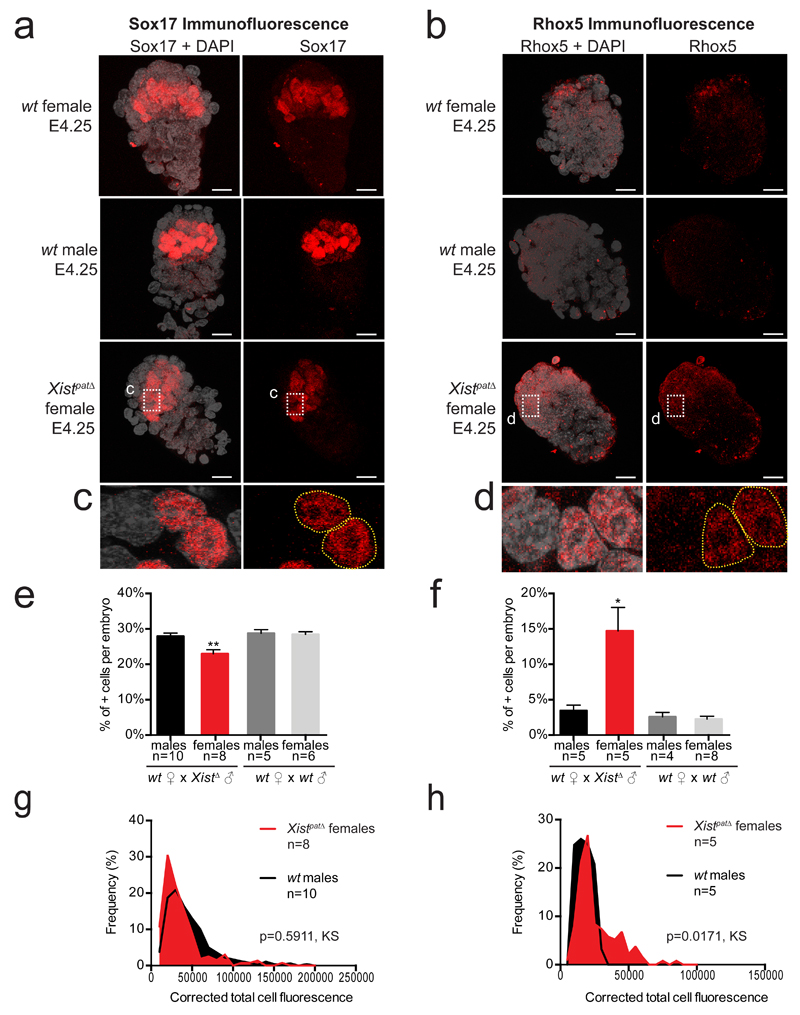
We explored the degree to which transcriptomes were perturbed in the Xist mutant embryos using Ingenuity Pathway Analysis software. We found that many aberrantly down-regulated genes in Xistpat∆ female blastocysts were associated with extra-embryonic tissue pathways, embryonic growth and cell viability (Figure 4c and Supplementary Data Set 3). Key extra-embryonic development genes that were aberrantly down-regulated included Tead4 (trophectoderm)25, Sox1726 (primitive endoderm PrE and ExE), Bmp427 (trophectoderm TE and PrE), Arid3a28 (TE specification) and Socs329 (placental development) (Figure 4d). To confirm the aberrant decrease of Sox17-positive cells in the PrE in Xistpat∆ females, we performed immunofluorescence on late blastocysts (Figure 5a, c, e and g). In Xistpat∆ females, fewer cells express Sox17 compared to their male littermates and the intensity of fluorescence of Sox17 is slightly decreased (Figure 5g), which corroborates the decrease in mRNA expression that we observe by scRNAseq.
Figure 5. Abnormal Sox17 and Rhox5 patterns in Xistpat∆ female blastocysts.
Maximum intensity projection of wt and Xistpat∆ E4.25 blastocysts analyzed by immunofluorescence against Sox17 (a, c) or Rhox 5 (b, d). Staining for Sox17 or Rhox5 is in red, DAPI is in grey. Scale bar represents 20µm. Percentage of positive cells have been assessed and summarized as the median + s.e.m. for Sox17 (e) and Rhox5 (f). Numbers of embryos are indicated under each genotype. By Kruskal-Wallis test; p<0.05 and p<0.001 correspond respectively to * and **. Average distribution of positive single cell fluorescence was represented by measuring the corrected total cell fluorescence using ImageJ software (Fiji, NIH) for Sox17 (g) and Rhox5 (h) and tested by Kolmogorov-Smirnov test. All cells under 10,000 and 5,000 for total cell fluorescence, respectively for Sox17 and Rhox5, have been considered as negative.
Importantly, in addition to aberrant down regulation or repression of extra-embryonic genes, we also found abnormal overexpression of several pluripotency genes including Prdm14, Esrrb and Tcl1 in Xistpat∆ embryos. This suggested an inappropriate activation or lack of repression of such factors in the absence of XCI (Figure 4d). This is relevant to our recent findings showing that the presence of two active X chromosomes delays exit from pluripotency in ESCs, by preventing down-regulation of key genes, such as Prdm14 or Esrrb22. Moreover, aberrant over-expression of Prdm14, Esrrb and Tcl1 was observed in Xist-/- female ESCs induced to differentiate22. Intriguingly, the most significantly up-regulated gene (10 log fold change) in Xistpat∆ female blastocysts was the imprinted Rhox5 gene, also known as Pem-1. Rhox5 is a member of the reproductive X-linked Hox (Rhox) cluster, and is expressed exclusively in the male germ line and in female (but not male) pre-implantation embryos (Xp only)30. Following implantation, its expression switches to the maternal allele and becomes restricted to extra-embryonic tissues30. The human RHOXF1 gene that is hypothesized to be related to the murine Rhox531 shows similar sex-specific and lineage-specific expression in human pre-implantation embryos32. Importantly, previous in vitro studies demonstrated that over-expression of Rhox5 can block differentiation of ESCs by preventing exit from pluripotency33,34. We validated Rhox5 up-regulation at the protein level using immunofluorescence and found that Xistpat∆ female blastocysts show significantly higher Rhox5 staining, particularly in the polar trophectoderm and inner cell mass region of the embryo, compared to wt blastocysts (Figure 5b, d, f and g). Quantification of Rhox5 immunofluorescence showed a significant increase in Rhox5 protein levels (p=0.0171, Kolmogorov-Smirnov KS test, Figure 5h) and in the number of cells stained by Rhox5 antibody (Figure 5f). This correlates well with our scRNAseq data.
We conclude that even by the early blastocyst stage, the lack of initiation of Xp inactivation in Xistpat∆ embryos leads to inappropriate down-regulation of several key genes involved in extra-embryonic development, overexpression of several pluripotency genes and massive overexpression of Rhox5, all of which may interfere appropriate subsequent differentiation.
Discussion
In conclusion, we have demonstrated the key role that Xist RNA plays in initiating imprinted XCI. Although its role in triggering random XCI had previously been established, our study provides evidence that Xist is clearly also essential for initiating early paternal XCI. Furthermore, our scRNAseq enabled us to identify the molecular defects in developmental pathways that emerge from this absence of dosage compensation and result in lethality a few days later. Absence of Xist leads to inappropriate down-regulation of extra-embryonic development, genes, lack of down-regulation of some pluripotency genes and massive overexpression of Rhox5. Together some or all of these defects must ultimately result in compromised extra-embryonic development and redirection towards what appears to be a more pluripotent state, or at least a state from which further differentiation is perturbed. Previous studies22 and a recent scRNA analysis of differentiating ESC9 found that XCI progression is negatively correlated with pluripotency and positively correlated with differentiation. The gene expression perturbations we observe in Xist mutant embryos and their subsequent lethality are consistent with this and point to some of the factors that are potentially implicated.
It is also noteworthy that the previously reported33 aberrant induction of maternal Xist and Xm inactivation in extra-embryonic tissues of blastocysts carrying a maternal Tsix deletion demonstrates that the presence of two active X chromosomes at the blastocyst stage can still be rescued in some females, and suggests that the major defect associated with a lack of paternal XCI is initially in the extra-embryonic lineage.
In this study we also define the influence of chromosomal location, as well as genetic background and parent-of-origin, on XCI kinetics. Our finding that Xist’s predicted initial binding sites on the X chromosome correspond to the earliest regions silenced, between the 8-16 cell stage, with evidence for local spreading in cis at the 32-blastocyst stage should enable exploration of the local features that underlie the spread of silencing along the X chromosome in an in vivo context. Finally, our study demonstrates the critical requirement for accurate X-chromosome gene dosage during early embryo development and uncovers some of the key pathways and factors that are affected in the absence of XCI. Future dissection of these pathways and their relationship to X-linked gene dosage should provide a better understanding of the important role that even small changes in RNA and protein levels can play, not only in development but also in disease.
Online Methods
Mouse crosses and collection of embryos
All experimental designs and procedures were in agreement with the guidelines from French legislation and institutional policies.
All BC and CB embryos were respectively derived from natural meetings between C57BL/6J (B6) females crossed with CAST/EiJ (Cast) males or by the reciprocal cross. The XistpatΔ mutant embryos (Xist+/-) were obtained by mating between Cast females and Xist-/Y males (mixed background: B6D2F1: C57BL/6J and DBA/2J, 129S1/SvlmJ and BALB/cJ). Embryos were harvested at 2-cell, 4-cell, 8-cell, 16-cell, 32-cell and blastocyst (approximately 60 to 64-cell) stages, respectively at E1.5, E2.0, E2.25, E2.75, E3.25 and E3.5. B6 and Cast pure oocytes were collected at E0.5 after matings of females with vasectomized males (Figure 1a). The collected embryos were only included in the analysis if they showed a normal morphology and the right number of blastomeres in relation with their developmental stage.
RNA Fluorescent In Situ Hybridization
RNA FISH on preimplantation embryos was performed as previously described3 using the intron-spanning Fosmid probe WI1-2039P10 (BacPac Consortium at Children's Hospital Oakland Research Institute) for Atrx and the intron-spanning plasmid probe p510 for Xist. Images were acquired using a wide-field Deltavision core microscope (Applied Precision – GE Healthcase) with a 60× objective (1,42 oil PL APO N) and 0.2 μm Z-sections. Images were analyzed using ImageJ software (Fiji, NIH).
Immunofluorescence staining
Immunofluorescence was carried out essentially as described previously35 with an additional step of blocking in 3% FCS before the primary antibody incubation. Immunofluoroscence of embryos either from mutant or control male progeny were always performed in parallel and in suspension. The following antibodies were used: goat anti-mouse Pem-1 (Rhox5)/Santacruz sc-21650/1:50 and goat anti-human Sox17/R&D Systems AF1924/1:100. Images were acquired using an Inverted laser scanning confocal microscope with spectral detection (LSM700 - Zeiss) equipped with a 260nm laser (RappOpto), with a 60X objective and 2 μm Z-sections. Maximum projections and total corrected fluorescence measurements (=integrated density – (area of selected cell x mean fluorescence of background readings) were performed in Figure 5g and 5h with Image J software (Fiji, NIH) using previously described methodology36. The total corrected cellular fluorescence (TCCF) = integrated density – (area of selected cell × mean fluorescence of background readings), was calculated.
Single cell dissociation from pre-implantation mouse embryos
Oocytes and embryos were collected by flushing oviducts (E0.5 to E2.75) or uterus (E3.25 and E3.5) with M2 medium (Sigma). The zona pellucida was removed using acid Tyrode’s solution (Sigma), and embryos were washed twice with M2 medium (Sigma). To isolate individual cells, we then incubated embryos in Ca2+, Mg2+ free M2 medium for 5 to 20 minutes, depending on the embryonic stage. For the blastocyst stage, Ca2+, Mg2+ free M2 free medium was replaced by a 5-minute incubation in TrypLE (Invitrogen). After incubation, each blastomere was mechanically dissociated by mouth pipetting with a thin glass capillary. Single cells were then washed 3 times in PBS/acetylated BSA (Sigma) before being manually picked into PCR tubes with a minimum amount of liquid. We either directly prepared the cDNA amplification or kept the single cells at -80°C for future preparation.
Single cell RNA amplification
PolyA+ mRNA extracted from each single cell was reverse transcribed from the 3’UTR and amplified following the Tang et al protocol10. Care was taken only to process embryos and single blastomeres of the highest quality based on morphology, number of cells and on amplification yield. A total of 72 BC and 110 CB (including 113 wt and 69 XistpatΔ mutant blastomeres) have been processed and passed quality controls.
Quality and sex determination
After cDNA amplification and before size selection and library preparation, the quality of cDNAs from each of the samples was validated by studying expression level of three housekeeping genes: Gapdh, Beta-Actin and Hprt. Primers used for real-time PCR were as follows: Gapdh_F: ccccaacactgagcatctcc; Gapdh_R: attatgggggtctgggatgg; ActB_F: aagtgacgttgacatccg; ActB_R: gatccacatctgctggaagg; Hprt_F: cctgtggccatctgcctagt; Hprt_R: gggacgcagcaactgacatt. Care was taken to process only single cells with consistent amplification rate of the three housekeeping genes in the same developmental stage.
The sex of each embryo was assessed by expression level analysis of Xist (female-specific transcript) and Eif2s3y (male-specific transcript) by real-time PCR. Primers used were: Eif2s3y_F: aattgccaggttattttcattttc Eif2s3y_R: agttcagtggtgcacagcaa; Xist_F: ggttctctctccagaagctaggaa and Xist_R: tggtagatggcattgtgtattatatgg.
Single cell libraries and deep-sequencing
Single-cell libraries were prepared from the 182 samples that passed QC according to the manufacturer’s protocol (Illumina). Sequencing to produce single-end 50bp reads was then performed on an Illumina HiSeq 2500 instrument (Supplementary Data Set 1).
Quality control and filtering of raw data
Quality control was applied on raw data as previously described in (Ancelin et al, 2016)35. Sequencing reads characterized by at least one of the following criteria were discarded from the analysis:
More than 50% of low quality bases (Phred score <5).
More than 5% of N bases.
More than 80% of AT rate.
At least 30% (15 bases) of continuous A and/or T.
SNP calling and allele-specific origin of the transcripts
SNPs collection and strain-specific genome construction
The VCF file (mgp.v5.merged.snps_all.dbSNP142.vcf) reporting all SNP sites from 36 mouse strains based on mm10 was downloaded from the Sanger database. Using SNPsplit tool (v0.3.0)37, these SNPs were filtered based on their quality values (FI value) and used to reconstruct the Cast genome from mm10 genome assembly.
Allele-specific alignments of RNAseq reads
To study the allele-specific gene expression, reads were processed using a pipeline adapted from Gendrel et al, 201438. Single-end reads were first aligned to the mouse mm10 and CAST genomes using the TopHat2 software (v2.1.0)39. Only random best alignments with less than 2 mismatches were reported for downstream analyses. The resulting mapping files for both parental genomes were then merged for each sample, using these following rules:
If a read mapped at the same genomic position on the two genomes with the same number of mismatches, this read will be considered as a common read.
If a read is aligned with less mismatches on one genome, the best alignment will be retained and this read will be considered as a specific read for the corresponding strain.
If a read is aligned with the same number of mismatches on both genomes but at different genomic positions, this read will be discarded.
Allelic imbalance in gene expression and gene classification
SNPs between C57BL/6J (B6) and CAST/EiJ (Cast) were extracted from the VCF file used to reconstruct the Cast genome. After removing common exonic SNPs between Xist and Tsix genes, 20,220,776 SNPs were retained.
The SAMtools mpileup utility (v1.1)40 was then used to extract base-pair information at each genomic position from the merged alignment file. At each SNP position, the number of paternal and maternal allele was counted. Threshold used to call a gene informative was 5 reads mapped per single SNP with a minimum of 8 reads mapped on SNPs per gene to minimize disparity with low polymorphic gene. The allele-specific origin of the transcripts (or allelic ratio) has been measured by the total number of reads mapped on the paternal genome divided by the total number of paternal and maternal reads for each gene: allelic ratio = paternal reads / (paternal+maternal) reads.
Genes are thus classified into two categories:
Monoallelically expressed genes: allelic ratio value ≤ 0.15 or ≥ 0.85.
Biallelically expressed genes: allelic ratio value > 0.15 or < 0.85.
Estimation of gene expression levels
Given that our RNA reverse transcription only allowed sequencing up to on average 3 kilobases from the 3’UTR, half of the expressed genes are only partially covered (less than 50% of the gene size in average). To estimate transcript abundance, read counts are thus normalized based on the amplification size of each transcript (RPRT for Reads Per Retro-Transcribed length per million mapped reads) rather than the size of each gene (RPKM).
Filtering of biased SNPs
As we observed a bias for some polymorphisms in oocytes (maternal reads only) and male cells (maternal X chromosome only), oocytes (autosomes and X-chromosomes) and males (X-chromosome) were used to address the issue. Therefore, SNPs covered by at least 5 reads and having an allelic ratio greater than 0.3 (biallelic or paternally expressed) in at least 2 of these samples were discarded. In total, 275 SNPs were filtered out, including 40 sites located on the X-chromosome.
Generation of XistpatΔ mutant embryos involved the use of a XistΔ/Y stud of mixed background (B6D2F1: C57BL/6J and DBA/2J, 129S1/SvlmJ and BALB/cJ). We therefore had to apply another SNP filtration to the KO samples to remove all B6 polymorphisms that could have been lost on the X chromosome due to the mixed background of the XistΔ/Y stud. To this end, all existing SNPs between B6 and DBA/2J, 129S1/SvlmJ and BALB/cJ on the X chromosome, were removed from our SNP database (34,397 SNPs, which represent 5.5% of X chromosome SNPs between B6 and Cast).
Principal component analysis, hierarchical clustering and differentially expressed genes
Count tables of gene expression were generated using the refSeq annotation and the HTSeq software41 (v0.6.1). Only genes with a RPRT (Reads Per Retro-Transcribed length per million mapped reads) value >1 in at least 25% of the single cells of at least one developmental stage (with a minimum of 2 cells) were kept for the downstream analysis. The TMM method from the edgeR R-package (v3.14.0)42 was first used to normalize the raw counts data. Principal component analysis (PCA) and hierarchical clustering were then used to determine how single cells were clustered to the others though their gene expression profiles, depending of their stage, sex and cross. PCA on normalized data was performed using FactoMineR R-package (v1.33). Hierarchical clustering analysis was based on Spearman correlation distance and the Ward method, using the hclust function implemented in the gplots R-package (v3.0.1). Limma R-package (v3.28.4)43 was applied to identify the differentially expressed genes between 8-cell stage and blastocyst in control and XistpatΔ mutant females. Using the Benjamini-Hochberg correction, genes with an adjusted p-value lower than α=0.05 were called as differentially expressed.
Functional enrichment analysis
Down-regulated genes in XistpatΔ mutant female blastocysts compared to CB female blastocysts were analyzed using QIAGEN's Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity). The Functions and Diseases module has been used to extract the most significantly deregulated pathways and their associated genes.
Heatmap generation for X-chromosome allelic gene expression
For BC and CB heatmaps, data from informative genes were analyzed at each developmental stage only if the gene was expressed (RPRT>4) in at least 25% of single blastomeres (with a minimum of 2 cells) at this particular stage and cross (Figures 2 and 4b and Supplementary Figure 3). To follow the kinetics of expression, we decided to focus only on genes expressed in at least 3 different stages between the 4-cell to blastocyst stages. Mean of the allelic ratio of each gene is represented for the different stages. The same gene candidate list was used to produce the XistpatΔ heatmaps (Figure 4b). A value was given only if the gene reached the threshold of RPRT >4 in at least 25 % of single cells (with a minimum of 2 cells) per stage and cross.
Definition of X-linked gene silencing/escape classes
We have automatically assigned X-linked genes that become strictly maternal (allelic ratio ≤0.15) at the 16-cell stage or before to the “early silenced” gene class; those that become maternal at the 32-cell stage to the “intermediate silenced” class (allelic ratio equals NA or >0.15 at 16C and ≤0.15 at 32C) and those that are silenced only by the blastocyst stage, to the “late silenced” gene class (allelic ratio equals NA or >0.15 at 16C and 32C and ≤0.15 at blastocyst stage). At the blastocyst stage, X-linked genes showing a maternal bias of expression (0.15<allelic ratio≤0.3) are categorized as maternally biased. A final group concerns genes that escape imprinted Xp inactivation (allelic ratio >0.3 at blastocyst stage) (Figure 3a). Genes escaping XCI were separated into two classes: “constitutive escapees” if they were classified as escapees in both CB and BC stages and “strain-specific escapees” if they were escapees in only one cross (Figure 3b and Supplementary Table 2).
Existence of consistency in silencing kinetics between crosses was evaluated if no more than one developmental stage separated the same gene between BC and CB crosses. If the consistent genes belonged to two different classes, a class for all (BC+CB) has been attributed thanks to their parental ratio mean of (BC mean + CB mean) in Figure 3a and 3d.
Dosage compensation, X:A expression ratio
We measured the global X:A expression ratio in females (XX :AA ratio) and males (X :AA ratio) as the level of expression of X-linked genes divided by the global level of expression of the autosomal genes. Only genes with an expression value RPRT >4 were used for subsequent analysis (Figures 1d and 4a). Adjustment of the number of expressed genes between X and autosomes has been published to be critical for X:A expression ratio measurement44. We then added a bootstrapping step and randomly selected, for each sample, an autosomal gene set with the same number of expressed genes compared to the X to estimate the global X:A ratio. This step was repeated 1000 times and the X:A expression ratio was estimated as the median of the 1000 values.
Statistics section
The statistical significance has been evaluated through Dunn's Multiple Comparison Test with Benjamini-Hochberg correction and Kruskal-Wallis analysis of variance. p-values are provided in the figure legends and/or main text. Kruskal-Wallis and Post-hoc test were used to analyze non-parametric and unrelated samples.
Data availability
The Gene Expression Omnibus (GEO) accession number for the data sets reported in this paper is GSE80810.
Source data for Figure 1 (1b, 1c, 1d and 1e), Figure 3 (3a, 3b, 3c and 3d) and Figure 4 (4a and 4d) are available with the paper online.
Alll other data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank S. Bao and N. Grabole for experimental help in single blastomere RNA sequencing and M. Guttman for sharing the Xist “entry” site coordinates. We are grateful to P. Gestraud and V. Sibut respectively for the help in statistical and IPA pathway analysis. We thank the pathogen-free barrier animal facility of the Institut Curie and J. Iranzo for help with the animals and the Cell and Tissue Imaging Platform - PICT-IBiSA (member of France–Bioimaging) of the Genetics and Developmental Biology Department (UMR3215/U934) of Institut Curie for help with light microscopy. We acknowledge E. Schulz, E. Nora, I. Okamoto and the members of E.H. laboratory for help, feedback and critical input. This work was funded by a fellowship of Région Ile-de-France (DIM STEMPOLE) to M.B., the Paris Alliance of Cancer Research Institutes (PACRI-ANR) to LS and ERC Advanced Investigator award (ERC-2010-AdG – No. 250367), EU FP7 grants SYBOSS (EU 7th Framework G.A. no. 242129) and MODHEP (EU 7th Framework G.A. no. 259743), La Ligue, Fondation de France, Labex DEEP (ANR-11-LBX-0044) part of the IDEX Idex PSL (ANR-10-IDEX-0001-02 PSL) and ABS4NGS (ANR-11-BINF-0001) to E.H and France Genomique National infrastructure (ANR-10-INBS-09) to EH, NS, EB.
Footnotes
Author Contributions
M.B., A.S. and E.H. conceived the study. M.B. performed most of the experiments. K.A. and P.D., C.P., M.B. and R.G. performed respectively the IF and the RNA-FISH experiments. T.L., J.B.L. and C.C.C. performed the single cell transcriptome library preparation and sequencing. L.S., M.B., C.C.C., I.V. N.S. and E.B. defined the data processing and bioinformatics analysis. L.S. built the computational pipeline for scRNAseq and analyzed the data with M.B. M.B. and E.H. wrote the paper.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto I, et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 4.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–9. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 5.Galupa R, Heard E. X-chromosome inactivation: New insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30:3187–205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-Cell RNA-Seq Reveals Dynamic, Random Monoallelic Gene Expression in Mammalian Cells. Science (80-. ) 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, et al. Single-cell analyses of X Chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 2016;26:1342–1354. doi: 10.1101/gr.201954.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–35. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockdorff N, Turner BM. Dosage compensation in Mammals. Cold Spring Harb Perspect Biol. 2015;7:a019406. doi: 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, et al. Regulation of X-linked gene expression during early mouse development by Rlim. Elife. 2016;5:e19127. doi: 10.7554/eLife.19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci U S A. 2009;106:5198–203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi S, et al. The X-linked imprinted gene family Fthl17 shows predominantly female expression following the two-cell stage in mouse embryos. Nucleic Acids Res. 2010;38:3672–3681. doi: 10.1093/nar/gkq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calabrese JM, et al. Site-Specific Silencing of Regulatory Elements as a Mechanism of X Inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berletch JB, et al. Escape from X Inactivation Varies in Mouse Tissues. PLOS Genet. 2015;11:e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks H, et al. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16:149. doi: 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balaton BP, Brown CJ. Escape Artists of the X Chromosome. Trends Genet. 2016;32:348–359. doi: 10.1016/j.tig.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 22.Schulz EG, et al. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Nesbitt AM. Genomic imprinting of the X-linked gene transketolase-like 1 in mouse and human. ProQuest Diss Theses A&I. 2010 884624661. [Google Scholar]
- 24.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 25.Nishioka N, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Niakan KK, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham SJL, et al. BMP signalling regulates the pre-implantation development of extra-embryonic cell lineages in the mouse embryo. Nat Commun. 2014;5:5667. doi: 10.1038/ncomms6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee C, et al. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes Dev. 2014;28:2219–2232. doi: 10.1101/gad.247163.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi Y, et al. SOCS3: An essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. EMBO J. 2003;22:372–384. doi: 10.1093/emboj/cdg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi S, et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr Biol. 2006;16:166–172. doi: 10.1016/j.cub.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, O’Malley ME, Bartlett DL, Guo ZS. Homeobox gene Rhox5 is regulated by epigenetic mechanisms in cancer and stem cells and promotes cancer growth. Mol Cancer. 2011;10:63. doi: 10.1186/1476-4598-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petropoulos S, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y, Melhem MF, Chaillet JR. Forced expression of the homeobox-containing gene Pem blocks differentiation of embryonic stem cells. Dev Biol. 1999;210:481–96. doi: 10.1006/dbio.1999.9279. [DOI] [PubMed] [Google Scholar]
- 34.Cinelli P, et al. Expression profiling in transgenic FVB/N embryonic stem cells overexpressing STAT3. BMC Dev Biol. 2008;8:57. doi: 10.1186/1471-213X-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ancelin K, et al. Maternal LSD1 / KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. Elife. 2016 doi: 10.7554/eLife.08851. pii: e0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCloy RA, et al. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozowsky J, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gendrel AV, et al. Developmental dynamics and disease potential of random monoallelic gene expression. Dev Cell. 2014;28:366–380. doi: 10.1016/j.devcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krueger F, Andrews SR, Krueger F, Andrews SR. SNPsplit: Allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Research. 2016;5:1479. doi: 10.12688/f1000research.9037.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kharchenko PV, Xi R, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 2011;43:1167–1169. doi: 10.1038/ng.991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Gene Expression Omnibus (GEO) accession number for the data sets reported in this paper is GSE80810.
Source data for Figure 1 (1b, 1c, 1d and 1e), Figure 3 (3a, 3b, 3c and 3d) and Figure 4 (4a and 4d) are available with the paper online.
Alll other data are available from the corresponding author upon reasonable request.