Abstract
Background
HIV-1 infection remains incurable on antiretroviral therapy (ART) due to virus latency. To date, enhanced co-culture assays, including viral outgrowth assays (VOA), are commonly used to measure HIV-1 latent reservoirs and evaluate latency-reversing agents (LRAs). However, VOA can only reactivate a small fraction of intact proviruses.
Methods
To explore the utility of NOD scid gamma (NSG) mice as an in vivo model to reactivate HIV-1 proviruses from VOA-negative CD4+ T cells, resting CD4+ T cells from an HIV-1 latently infected individual were isolated and the human CD4+ T cells corresponding to VOA-positive and VOA-negative CD4+ T cells were engrafted into NSG mice. Plasma viral load (pVL) and human CD4+ T cells were quantified every other week using qRT-PCR and flow cytometry.
Results
We found that NSG mice reactivated latently infected HIV-1 from VOA-positive CD4+ T cells as well as VOA-negative CD4+ T cells. Engrafted CD4+ T cells proliferated considerably in vivo, peaked prior to provirus reactivation, and lasted for up to 14 weeks. Sequence analyses revealed that reactivated proviruses in VOA-positive and VOA-negative CD4+ T cells are different.
Conclusion
Taken together, NSG mice can support long-term engraftment of human CD4+ T cells and reactivate VOA-positive and VOA-negative proviruses. Therefore, this in vivo model has the potential to be used to study the underlying mechanisms of HIV-1 latency and reactivation.
Keywords: HIV-1 latency, viral outgrowth assay, immune-compromised mice, in vivo provirus reactivation, resting CD4+ T cells, clonal expansion
Introduction
Remarkable progress has been made in reducing the mortality and morbidity of HIV-1 infection through combined antiretroviral therapy (ART); however, HIV-1 infection remains incurable due to virus latency [1–3]. To better understand the mechanisms of HIV-1 latency and develop strategies to purge latent infection, quantitative in vitro assays and in vivo animal models to study HIV-1 latency and evaluate latency-reversing agents (LRAs) are critically needed. As such, major quantitative assays, namely inverse PCR [4,5], improved Alu PCR [6–8] and linker-primer PCR [9], have been previously developed. Although these methods can quantify the frequency of proviruses, they cannot distinguish between defective and functional/intact proviruses, thus often overestimating the size of the latent reservoirs. Enhanced co-culture assays, including quantitative viral outgrowth assay (VOA), are frequently used in quantifying functional proviruses [1,5]. It has been demonstrated that less than 1% of proviruses can be reactivated to produce infectious viruses after stimulation using VOA. It was also found that the frequency of intact proviruses that cannot be reactivated by VOA (VOA-negative intact proviruses) is at least 60-fold higher than the frequency of induced proviruses by VOA (VOA-positive intact proviruses) [10], indicating there is a population of intact proviruses that is unmeasurable by VOA. A study compared the reactivation properties of five in vitro primary T cell latency models and four J-Lat cell latency models in response to 13 stimuli and found that the in vitro model is not able to capture the ex vivo reactivation properties of human infected cells [11]. Recently a more sensitive Tat/rev induced limiting dilution assay (TILDA) was developed to quantify the frequency of cells harbouring proviruses by detecting tat/rev multiply-spliced HIV RNA upon in vitro stimulation [12]. However, a new study showed that HIV-1 ‘defective’ proviruses are capable of transcribing and translating novel open reading frames (ORFs), including the rev ORF [13], which further complicated the measurement of functional proviruses using TILDA.
The new generation of immune-compromised mice, such as NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ Jackson Laboratory, Bar Harbor, Maine, Stock number 005557), are excellent recipients for engrafting human stem cells, cancer cells, lymphocytes, or even peripheral blood mononuclear cells for biomedical research [14–17]. Recently, a report showed that engrafting a large quantity of peripheral blood mononuclear cells (PBMCs) (around 40 million per mouse) from HIV-1 infected patients into NSG mice can reactivate proviruses [18]. In this work, the viraemia after engrafting can be detected up to 6 weeks, or shown to be transiently positive at 1 week post cell injection (wpi). Nevertheless, it remains unknown whether xenograft can reactivate HIV-1 provirus from VOA-negative CD4+ T cells and whether the in vivo model in this study can support HIV-1 viraemia long term after solely engrafting human CD4+T cells.
To further explore the utility of NSG mice as an in vivo model to quantify HIV-1 functional provirus and test whether this in vivo method can be used to reactivate HIV-1 proviruses from VOA-negative CD4+ T cells, we engrafted NSG mice with a limited quantity (2–3.6 million per mouse) of human resting CD4+ T cells. We also included VOA-positive CD4+ T cells and resting CD4+ T cells without VOA (uncultured CD4+ T cells) from an HIV-1 aviraemic patient under ART as a positive control.
We found NSG mice can reactivate not only latently infected HIV-1 from VOA-positive and uncultured CD4+ T cells, but also from VOA-negative CD4+ T cells. Engrafted CD4+ T cells proliferated considerably in vivo starting from 2 wpi and peaked prior to provirus reactivation. The proliferation of engrafted CD4+ T cells in NSG mice can support stable viraemia up to 14 weeks post engraftment. Sequence analyses revealed that reactivated proviruses from VOA-positive and VOA-negative CD4+ T cells are distinct from each other, implying a difference in latency regulation. This study, for the first time, demonstrates that NSG mouse engraftment can reactivate intact proviruses from VOA-negative CD4+ T cells. This in vivo model may be valuable to quantify the size of the HIV-1 latent reservoir through serial dilution of a patient's resting CD4+T cells. Furthermore, this method can be helpful to study the underlying mechanisms of HIV-1 latency through amplification and long-term maintenance of provirus reservoirs.
Methods
Study subject
PBMCs were obtained from an HIV-1 infected individual, who had 14.3 years of diagnosed infection and continuous suppression of plasma HIV-1 with ART (emtricitabine-tenofovir, ritonavir and atazanavir) at undetectable levels (<50 copies/mL) for 6.3 years prior to sampling time. The CD4 cell count was 627 cells/mm3 at the time of this study. The infected donor exhibited 0.518 infectious units per million (IUPM). This study was approved by the Johns Hopkins Institutional Review Board, written informed consent was obtained from all participants as per a previous report [10].
Human resting CD4+ T cell isolation and virus outgrowth assay
Resting CD4+ T cells were isolated from the PBMCs of the HIV-1 aviraemic individual by negative selection using the CD4+ T Isolation Kit II with CD25, CD69 and HLA-DR microbeads (Miltenyi Biotec, San Diego, CA, USA) [1,10]. Some of the resting CD4+ T cells were cryopreserved immediately, using liquid nitrogen, as an uncultured control; the remaining cells were clonally expanded using a previously published protocol [10] and divided into equal portions for VOA and mouse engraftment, respectively. Cells engrafted into mice were grouped based on the VOA results into VOA-positive and VOA-negative groups.
NSG mice
Six-week old female NSG mice were maintained in micro-isolator cages within BSL-2 animal rooms following the protocol approved by the Institutional Animal Care and Use Committee at the University of Nebraska-Lincoln. Cryopreserved cells corresponding to VOA-negative CD4+ T cells (P46N) with VOA-positive (P46P) and uncultured (P46U) controls were thawed at 37°C and washed once using culture medium containing 100 U/mL IL-2. Cell viability was checked using a Vi-Cell XR cell viability analyzer (Beckman Coulter, Indianapolis, IN, USA); cells were resuspended into 200 μL of culture medium and injected into a mouse via a tail vein. Plasma viral load (pVL) and peripheral blood CD4+ T cells were quantified using qRT-PCR and flow cytometry, respectively, every 2 weeks.
Plasma viral load quantification
Viral RNA was extracted from plasma using QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA, USA). pVL was determined by qRT-PCR, wherein cDNA was synthesised using Superscript III Reverse Transcriptase, RNaseOUT and RNase H (Life Technologies, Grand Island, NY, USA) with the gene specific primer: 5’-TTGCTACTTGTGATTGCTCCATGT-3’. The qPCR was run on C1000 Thermal Cycler and CFX96 Real-Time system (Bio-Rad) using TaqMan Fast Virus 1-Step Master Mix (Life Technologies, Grand Island, NY) with primers: forward 5’-GCCTCAATAAAGCTTGCCTTGA-3’, reverse 5’-GGGCGCCACTGCTAGAGA-3’ and probe: 5’-/56-FAM/CCAGAGTCA/ZEN/CACAACAGACGGGC ACA/3IABkFQ/-3’. The detection limit of plasma HIV-1 RNA was 200 copies/mL and was determined through repeating endpoint detection from serial dilution of the AcroMetrix HIV-1 Panel (Life Technologies) [19].
Flow cytometry
Human CD4+ T cells isolated from mouse peripheral blood were measured using FACS Aria II flow cytometer (BD Biosciences, San Jose, CA, USA) after staining with antibodies against human CD45 (FITC conjugated; clone# HI30, BioLegend, San Diego, CA, USA), human CD3 (PE conjugated, clone# HIT3a, BioLegend), and human CD4 (Alexa Fluor 700 conjugated, clone# OKT4, BioLegend). Cells were blocked using Human TruStain FcX and Mouse TruStain fcX Antibodies (BioLegend) prior to staining. Raw data were analysed with FlowJo version 10.0 (FlowJo LLC, Ashland, OR, USA).
Sequencing of reactivated viruses
Viruses from the first pVL-positive samples were amplified using bulk RT-PCR and sequenced. The cDNAs were amplified using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs) with PCR primers: forward 5’–TAGAGCCCTGGAAGCATCCAGGAAG-3’ and reverse 5’–TTGCTACTTGTGATTGCTCCATGT-3’. The amplicons were confirmed by 1.0% agarose gel stained with ethidium bromide and bands were cut and purified by GeneJET gel extraction kit (Thermo Scientific). The amplicons of the 3kb spanning env region were sequenced using Sanger's method at Sequetech (Mountain View, CA, USA) with eight overlapping primers based on primer walking: F1 5’–TTAGGCATCTCCTATGGCAGGAAGAAG-3’, R1 5’-GTCTCGAGATACTGCTCCCACCC-3’, F2 5’-ATGATAC AGAGGTACATAATGTTTGG-3’, F3 5’-AAGTTGTAAC ACCTCAGTCATTACAC-3’, F4 5’- GGAGATATGAGGG ACAATT-3’, F5 5’-GATTTGGGGTTGCTCTGGAAAACT CA-3’, F6 5’-AAGGAATAGAAGAAGAAGGTGGAGAG-3’, and R2 5’-GTTCTACCATGTCATTTTTCCACATG-3’. The sequencing results were manually examined peak by peak and the ends of sequences containing ambiguous nucleotides were trimmed. The sequences were confirmed by overlapping identical regions. Then the sequences were assembled individually to obtain the 3-kb partial genome using Sequencher 5.0 (Gene Codes Corporation Ann Arbor, MI, USA). A phylogenetic tree containing the assembled virus sequences and a HXB-2 homolog region reference sequence was generated using MUSCLE 3.8 [20] and PHYML 3.0 [21] through the maximum likelihood method and displayed with FigTree 1.4.2 ( http://tree.bio.ed.ac.uk/software/figtree/).
HIV-1 sequences obtained from this study have been deposited into GenBank under accession number KT223503, KT223504 and KT223505.
Results
Xenograft of human CD4+T cells into NSG mice reactivated the proviruses from VOA-positive and VOA-negative samples
First, we wanted to test whether NSG mice engrafted with VOA-negative CD4+ T cells could reactivate proviruses. To compare in vivo NSG mouse reactivation of provirus with VOA, CD4+ T cells were first expanded in vitro and split into two equal portions before VOA. One portion was immediately cryopreserved for NSG mouse engraftment and the other portion was used for VOA serving as a surrogate for reactivation of the corresponding cryopreserved cells (Figure 1). After 3 weeks of VOA culture, HIV-1 p24 in the supernatant of each well was quantified using ELISA. The cryopreserved cells corresponding to the VOA-positive and the VOA-negative samples and uncultured positive control were injected into NSG mice via the tail vein. Plasma vRNA was quantified using qRT-PCR every other week.
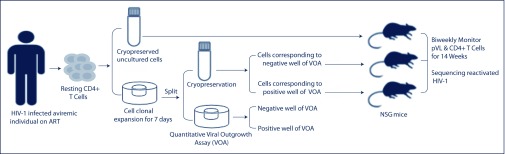
Figure 1.
Experimental design. Resting CD4+ T cells were isolated from an HIV-1 aviraemic individual on ART. After clonal expansion, CD4+ T cells were split into equal parts for cryopreservation and VOA. Cryopreserved cells corresponding to VOA-positive wells (P46P), VOA-negative wells (P46N), and uncultured resting CD4+ T cells (P46U), were thawed and intravenously injected into NSG mice respectively. Plasma viral load (pVL) and peripheral blood CD4+ T cells were measured every other week using qRT-PCR and flow cytometry. HIV-1 env sequences from the first pVL positive samples were sequenced using Sanger's method.
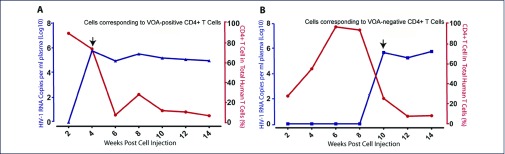
Plasma viral RNA load was detectable at 4 wpi in both the VOA-positive (P46P, Figure 2A) and the uncultured sample (P46U, data not shown) as positive control. The pVL was undetectable at 2 wpi, became positive at 4 wpi (6.06 × 105 copies/mL), and then remained at a plateau until the animal was euthanised at 14 wpi. In contrast, the pVL in the P46N engrafted mouse was undetectable before and at 8 wpi, became positive at 10 wpi (4.78 × 105 copies/mL) (Figure 2B), and remained at a plateau until the animal was euthanised at 14 wpi.
Figure 2.
Kinetics of provirus reactivation and human CD4+ T cell quantification in NSG mouse engrafted with VOA-negative resting CD4+ T cells with positive control. (A) Cryopreserved cells corresponding to VOA-positive well (P46P) were intravenously injected into NSG mouse and (B) VOA-negative well (P46N). Arrows indicate the first time that pVL became positive and the samples for sequencing.
Human CD4+T cells engrafted in NSG mice can proliferate and support stable viraemia for a long time
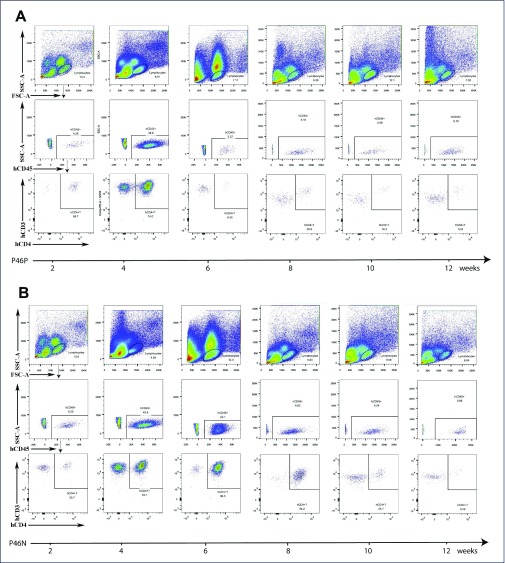
We then checked whether immune-compromised NSG mice can maintain engrafted human CD4+ T cells for a long term using flow cytometry. As shown in Figure 2, we found that engrafted CD4+ T cells proliferated considerably in vivo starting from 2 wpi and peaked before the provirus was reactivated (4 wpi for P46P and P46U, and 10 wpi for P46N). Upon provirus reactivation, CD4+ T cells significantly declined (Figures 2 and 3). Of note, active proliferation of CD4+T cells (4 and 6 wpi) in the VOA-negative sample was several weeks ahead of pVL detection (Figure 3B). This temporal difference between CD4+ T cell proliferation and provirus reactivation indicates cell proliferation alone is not sufficient to reactivate provirus, and cell proliferation and provirus reactivation are independent events.
Figure 3.
Kinetics of human peripheral blood CD4+ T cells in engrafted NSG mice. (A) The NSG mouse was engrafted with cryopreserved VOA-positive (P46P) as a control. (B) The NSG mouse was engrafted with VOA-negative sample (P46N).
Reactivated proviruses from VOA-positive and VOA-negative CD4+ T cells were heterogeneous
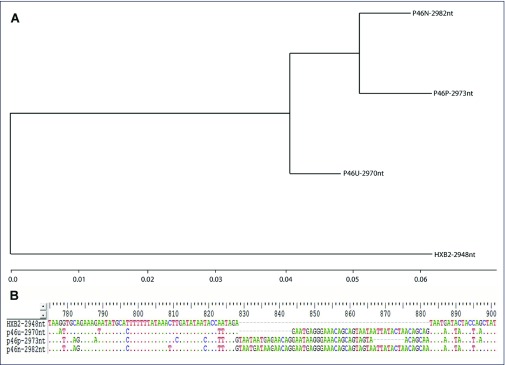
To determine whether the reactivated proviruses from VOA-positive and VOA-negative CD4+ T cells were identical, a 3-kb HIV-1 partial genome spanning the full-length env from the first viraemic plasma samples was amplified using qRT-PCR and sequenced using Sanger's method. The sequences of each sample are homogeneous with a pure single peak for every position. However, for different samples, the reactivated HIV-1 proviruses were heterogeneous (Figure 4A) and contained different in-frame insertions compared to the reference HXB2 sequence (Figure 4B).
Figure 4.
Sequencing results for reactivated HIV-1 from VOA-positive, VOA-negative and uncultured resting CD4+ T cells engrafted into NSG mice. (A) Phylogenetic relationship of reactivated HIV-1 from VOA-positive provirus, VOA-negative provirus, and uncultured resting CD4+ T cells using HXB2 as the reference sequence. (B) A representative region showing the differences between the reactivated viruses from VOA-positive, VOA-negative and uncultured sample contains different in-frame insertions as compared to the reference HXB2 sequence.
Discussion
Curing HIV-1 infection by eradicating virus latency is one of the highest public health priorities. Enhanced co-culture assays, including VOA, have greatly deepened our understanding of HIV-1 latency; however, VOA can only reactivate a small fraction of intact proviruses. Thus, the underlying mechanisms of HIV-1 latency are not completely understood.
This study for the first time shows that engraftment of VOA-negative CD4+ T cells into NSG mice can reactivate proviruses. The results herein also demonstrate that HIV-1 latently infected CD4+ T cells engrafted into NSG mice can proliferate and survive for up to 14 weeks, which provides a prolonged window of opportunity to study HIV-1 latency and its reactivation. While VOA-positive CD4+ T cells and uncultured cells engrafted into NSG mice reactivated latent HIV-1 at 4 wpi, VOA-negative CD4+ T cells reactivated latent HIV-1 at 10 wpi. This delayed reactivation may mirror the delayed HIV-1 rebound after cessation of ART in HIV-1 functional controllers mediated through early initiation of ART in the ‘Mississippi child’ [22] and the ANRS VISCONTI study [23], as well as in the ‘Boston patients’ mediated through allogenic bone marrow stem cell transplant [24]. The delayed reactivation observed in the VOA-negative CD4+ T cells in this study supports the notion of heterogeneity of latently infected HIV-1 in resting CD4+ T cells [25,26]. The distinct sequences of reactivated proviruses from VOA-positive and VOA-negative CD4+ T cells indicate that there are different intact proviruses in resting CD4+ T cells with different reactivation efficiency.
Our study has demonstrated that provirus from engrafted human VOA-negative CD4+ T cells can be reactivated. A plausible explanation is that engrafted human CD4+ T cells underwent proliferation and expansion as shown in Figure 3, which might increase the quantity of provirus-bearing cells; meanwhile xeno-immune activation might also contribute to the reactivation of proviruses. However, the exact mechanism of provirus reactivation in this system remains to be studied. The temporal difference between the engrafted CD4+ T cell expansion and provirus reactivation indicates that cell proliferation does not directly reactivate provirus. Furthermore, the sequencing results revealed that the reactivated proviruses from VOA-negative and VOA-positive CD4+ T cells were different, indicating that latency reactivation regulation maybe different.
We would like to point out the limitation of this case study. We operationally called cryopreserved cells corresponding to VOA-positive/negative cells; however, it is likely the split portions contained clonally expanded cells. Future studies that include more cases with determined clonality of engrafted cells by integration site sequencing analyses will be needed.
Collectively, this study clearly demonstrates that NSG mice can support long-term engraftment of human CD4+ T cells and reactivate proviruses from VOA-negative CD4+ T cells, which not only provide a large window for studying the HIV-1 latency at molecular and cellular detail, but can also be used to quantify the functional latent reservoir.
Acknowledgements
QL and ZY designed the experiments. ZY, GK and WL performed NSG mice experiments. ZY performed plasma viral load, flow cytometry and viral partial genome sequencing and data analysis. ZY and QL wrote the manuscript. The authors would like to thank Dr Robert Siliciano and Dr Ya-Chi Ho for providing patient samples and conducting viral outgrowth assay, and Lance Daharsh and Dr Fangrui Ma for their discussion and critical reading of this manuscript. This work was supported in part by NIH grant R01 AI111862 (to Li Q, Guo J), P30GM103509 and BEAT-HIV: Delaney Collaboratory to Cure HIV-1 Infection by Combination Immunotherapy 1UM1AI126620-01 (L Montaner, J Riley).
Competing interests
The authors declare that there is no conflict of interest in this study.
References
- 1. Finzi D, Hermankova M, Pierson T et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278: 1295– 1300. [DOI] [PubMed] [Google Scholar]
- 2. Wong JK, Hezareh M, Günthard HF et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278: 1291– 1295. [DOI] [PubMed] [Google Scholar]
- 3. Chun T-W, Stuyver L, Mizell SB et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94: 13193– 13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chun T-W, Carruth L, Finzi D et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387: 183– 188. [DOI] [PubMed] [Google Scholar]
- 5. Chun T-W, Finzi D, Margolick J et al. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1: 1284– 1290. [DOI] [PubMed] [Google Scholar]
- 6. O’Doherty U, Swiggard WJ, Jeyakumar D et al. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 2002; 76: 10942– 10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butler SL, Hansen MST, Bushman FD.. A quantitative assay for HIV DNA integration in vivo. Nat Med 2001; 7: 631– 634. [DOI] [PubMed] [Google Scholar]
- 8. Brussel A, Sonigo P.. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol 2003; 77: 10119– 10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vandegraaff N, Kumar R, Burrell CJ, Li P.. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J Virol 2001; 75: 11253– 11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ho Y-C, Shan L, Hosmane Nina N et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155: 540– 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spina CA, Anderson J, Archin NM et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 2013; 9: e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Procopio FA, Fromentin R, Kulpa DA et al. A Novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2015; 2: 874– 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imamichi H, Dewar RL, Adelsberger JW et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A 2016; 113: 8783– 8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL.. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 2012; 12: 786– 798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito R, Takahashi T, Katano I, Ito M.. Current advances in humanized mouse models. Cell Mol Immunol 2012; 9: 208– 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito M, Hiramatsu H, Kobayashi K et al. NOD/SCID/γ mouse: an excellent recipient mouse model for engraftment of human cells. Blood 2002; 100: 3175– 3182. [DOI] [PubMed] [Google Scholar]
- 17. Brehm MA, Wiles MV, Greiner DL, Shultz LD.. Generation of improved humanized mouse models for human infectious diseases. J Immunol Methods 2014; 410: 3– 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metcalf Pate KA, Pohlmeyer CW, Walker-Sperling VE et al. A murine viral outgrowth assay to detect residual HIV type 1 in patients with undetectable viral loads. J Infect Dis 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan Z, Kang G, Ma F et al. Recapitulating cross-species transmission of simian immunodeficiency virus SIVcpz to humans by using humanized BLT mice. J Virol 2016; 90: 7728– 7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32: 1792– 1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S, Dufayard J-F, Lefort V et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. System Biol 2010; 59: 307– 321. [DOI] [PubMed] [Google Scholar]
- 22. Luzuriaga K, Gay H, Ziemniak C et al. Viremic relapse after HIV-1 remission in a perinatally infected child. N Engl J Med 2015; 372: 786– 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sáez-Cirión A, Bacchus C, Hocqueloux L et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 2013; 9: e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henrich TJ, Hu Z, Li JZ et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis 2013; 207: 1694– 1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dahabieh MS, Battivelli E, Verdin E.. Understanding HIV latency: the road to an HIV cure. Ann Rev Med 2015; 66: 407– 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS 2011; 6: 4– 11. [DOI] [PMC free article] [PubMed] [Google Scholar]