Abstract
Ramoplanin is a glycolipodepsipeptide antibiotic obtained from fermentation of Actinoplanes sp. ATCC 33076 that exhibits activity against clinically important multi-drug-resistant, Gram-positive pathogens including vancomycin-resistant Enterococcus (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-intermediate resistant Clostridium difficile. It disrupts bacterial cell wall through a unique mechanism of action by sequestering the peptidoglycan intermediate Lipid II and therefore does not show cross-resistance with other antibiotics. However, while demonstrating excellent antimicrobial activity in systemic use in animal models of infection, ramoplanin presents low local tolerability when injected intravenously. As a consequence of this limitation, new derivatives are desirable to overcome this issue. During a natural product screening program developed to discover compounds that disrupt bacterial cell wall synthesis by inhibiting peptidoglycan transglycosylation through binding to the intermediate Lipid II, 49 actinomycete strains were identified by HR-LCMS as producers of ramoplanin-related compounds. The producing strains were isolated from environmental samples collected worldwide comprising both tropical and temperate areas. To assess the diversity of this microbial population, the 49 isolates were initially identified to the genus level on the basis of their micromorphology, and 16S sequencing confirmed the initial identification of the strains. These analyses resulted in the identification of members of genus Streptomyces, as well as representatives of the families Micromonosporaceae, Nocardiaceae, Thermomonosporaceae, and Pseudonocardiaceae, suggesting that the production of ramoplanins is relatively widespread among Actinomycetes. In addition, all of these isolates were tested against a panel of Gram-positive and Gram-negative bacteria, filamentous fungi, and yeast in order to further characterize their antimicrobial properties. This work describes the diversity of actinomycete strains that produced ramoplanin-related compounds, and the analysis of the antimicrobial activity exhibited by these isolates. Our results strongly suggest the presence of new ramoplanin-analogs among these actinomycete producers.
Keywords: high-throughput screening, microbial natural products, antibacterial activity, actinomycetes, ramoplanin, peptidoglycan intermediate Lipid II
Introduction
New antibiotics with new mechanisms of action are urgently needed because clinically significant bacterial pathogens have not only acquired resistance to nearly all existing antibiotics, but also increasingly exhibit multi-drug resistance (Gerits et al., 2016). Considering that the present antibiotic therapies have become increasingly inefficient, new technologies are required to identify and develop novel classes of antibacterial agents.
While searching for novel antibacterial leads, an attractive alternative to the classical target-based approach is the use of promoter-inducible reporter assays that are amenable to high-throughput screening. These assay systems are based on cells that carry reporters such as β-galactosidase or luciferase genes fused to promoters that specifically respond to certain types of antibiotic stress (Shapiro and Baneyx, 2002; Sun et al., 2002; Fischer et al., 2004). Those genetically modified strains are particularly advantageous for high-throughput assay systems considering the low compound concentrations that they require. The selective induction of the reporter fusion indicates that a compound is at least perturbing the pathway of interest as part of its biochemical impact (Fischer et al., 2004).
DNA microarray analysis of the response of Bacillus subtilis treated with sublethal concentrations of the cell wall antibiotic bacitracin revealed the presence of overlapping regulons controlled by alternative sigma factors and two component systems. One operon strongly induced by bacitracin, which was identified by John Helmann's laboratory at Cornell University, contains the genes liaIHGFSR (liaRS stands for lipid II cycle interfering antibiotic response Regulator and Sensor). The β-galactosidase gene lacZ was placed downstream of the promoter for this operon (PliaI) such that induction of the operon by bacitracin or other compounds that have the same effect on the liaRS two component regulatory system will induce the production of β-galactosidase. This can then be monitored as an indication of antibiotic action that is impacting this liaRS system (Cao et al., 2002; Mascher et al., 2003).
Lipid II is a membrane-bound cell wall precursor which performs the cycle of peptidoglycan “building block” translocation. The efficiency of targeting Lipid II as an antibacterial strategy is highlighted by the fact that it is the target for at least four different classes of antibiotics (lantibiotics, mannopeptimycins, ramoplanin, and vancomycin). Since this pathway is limited to prokaryotes and low toxicity is expected, it is highly desirable to discover novel structural classes that interfere with it. Moreover, the emergent problem of bacterial resistance to drugs such as vancomycin has led to increasing interest in the therapeutic potential of other compounds that target Lipid II (Donadio et al., 2002).
A natural product screening program was developed to detect inhibitors that interfere with stage II peptidoglycan biosynthesis. This screening approach called “LiaRS assay” allowed the identification of positive controls such as ramoplanin.
Ramoplanin is a lipoglycodepsipeptide antibiotic derived from Actinoplanes spp. ATCC 33076 and was first isolated as a complex of three closely related components A1, A2, and A3 (A1, 6–12%; A2 72–86%; A3 8–14%) in which A2 was the most abundant (Cavalleri et al., 1984). The structural difference is in the length of the N-terminal acyl chain. Ramoplanin A2 exhibits activity against clinically important Gram-positive bacteria including vancomycin-resistant Enterococcus sp. (VRE), methicillin-resistant S. aureus (MRSA) and vancomycin-intermediate resistant Clostridium difficile (Finegold et al., 2004; Peláez et al., 2005). Preclinical studies have also demonstrated that ramoplanin exerts a rapid bactericidal effect on S. aureus biofilms (Schmidt et al., 2010) and that a clinical vancomycin-resistant S. aureus strain containing the vanA gene was susceptible to ramoplanin (Bozdogan et al., 2003). Recently, it has been reported (by Nanotherapeutics) to have additional activity against C. difficile spores, both in vitro and in an animal model (Jabes et al., 2014). Ramoplanins have a unique mechanism of action that disrupts bacterial cell wall by interfering with late-stage transglycosylation cross-linking reactions in peptidoglycan biosynthesis. Ramoplanin A2 acts by sequestering the Lipid Intermediate II, which keeps this substrate from accurate use in downstream reactions catalyzed by transglycosylases that produce mature peptidoglycan polymer. This antibiotic works at a site complementary to vancomycin and shows no cross-resistance with other glycopeptides. At present, ramoplanin is being developed for the targeted prophylaxis of recently treated patients with C. difficile infection (CDI) at high risk for infection relapse. Twelve Phase I studies, two Phase II studies (one in CDI and one in VRE) as well as one Phase III study (in VRE) have been conducted (http://www.nanotherapeutics.com/ramoplanin/). Although Phase III study was not completed.
Results presented in this paper illustrate that this screening approach allowed us to detect actinomycete strains that produced ramoplanin-related components. The diversity of those actinomycete strains is described, along with the analysis of the antimicrobial activity exhibited by these isolates. Likewise, HR-LCMS analyses strongly suggest the presence of new ramoplanin-analogs among these actinomycete producers and current large scale fermentations and purifications are being performed for the identification of these novel antimicrobials. To our knowledge, this is the first report that the production of ramoplanin-type natural products is relatively widespread within Actinomycetes.
Materials and methods
Morphological identification of actinomycete strains
Actinomycetes were tentatively identified to the genus or family level after direct observation of the microscopic morphology (400 × and 1000 × magnification with long distance range objectives) of the vegetative and aerial mycelium and characteristic sporulating structures developed upon growth on water agar for 21 days at 28°C (Goodfellow et al., 1984, 2012).
DNA extraction and molecular characterization
Total genomic DNA from the actinomycetes used in this study was purified as previously described (Innis et al., 1990) from strains grown in ATCC-2 liquid medium [0.5% yeast extract (Difco, Franklin Lakes, NJ, USA), 0.3% beef extract (Difco), 0.5% peptone (Difco), 0.1% dextrose (Difco), 0.2% starch from potato (Panreac, Barcelona, Spain), 0.1% CaCO3 (E. Merck, Darmstadt, Germany), and 0.5% NZ amine E (Sigma, St Louis, MO, USA)]. PCR primers fD1 and rP2 were used for amplifying the nearly full-length 16S ribosomal RNA genes of the strains (Weisburg et al., 1991). PCR products were sent to Secugen (http://www.secugen.es/) for sequencing, and were used as a template in sequencing reactions using the primers fD1 and rP2, and 1100R and 926F (Lane, 1991). Partial sequences were assembled and edited using the Assembler contig editor component of Bionumerics (ver 5.10) analysis software (Applied Maths NV, Sint-Martens-Latem, Belgium).
The identification of the closest match sequences was performed against the database of type strains with validly published prokaryotic names (Chun and Int, 2007) which was implemented at the EzTaxon server (http://ezbiocloud.net/eztaxon; Kim et al., 2012).
Characterization of actinomycete strains
The soil samples used for the isolation of the 49 actinomycete strains were collected worldwide comprising both tropical and temperate areas: Costa Rica, French Guyana, Mexico, New Caledonia, South Africa, Spain, and Sri Lanka, including different ecological habitats like agricultural soils, riverbeds, lakes, ponds, swamps, dunes, tropical and sempervirent forests, savanna soil, and rhizospheres.
The strains were tentatively identified on the basis of their macro and micro-morphology and the genus assignment of 39 strains was confirmed by 16S rDNA gene sequencing. The most abundant taxonomic group was the family Micromonosporaceae (71.4%) with 34 strains identified as members of the genus Micromonospora and 1 strain of Actinoplanes. The remaining strains were, identified as Nocardia (3 strains), Streptomyces (3 strains), Actinomadura (1 strain), Amycolatopsis (1 strain), and Lechevalieria (1 strain). Five strains could not be assigned to any genus and were morphologically identified as filamenting Actinobacteria (Table 2).
Natural products extract library generation
Bacteria and fungi were grown in complex liquid media containing different amounts of nutrient sources: carbon sources (e.g., monosaccharides, disaccharides), complex carbon sources (e.g., wheat flour, soluble starch), nitrogen sources, and mineral salts sources (R. Tormo et al., 2003). The secondary metabolites in the broths were extracted with acetone (1:1) and shaking in an orbital shaker for 1 h. The extracts were then centrifuged at 1,500 × g for 15 min. 1.5 mL of DMSO were added to aliquots of 15 mL and the resulting mixture was evaporated in a Turbovap equipment until half of the volume remained, ~7.5 mL, to a final crude concentration of 1xWBE (whole broth equivalents) and 20% DMSO. Five-hundred microliters of these crude extracts were stored at −20°C in 96-well ABgene plates until needed.
LiaRS screening assay
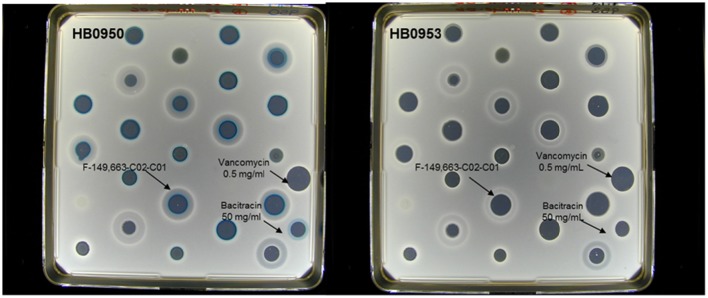
The screening assay was performed against two Bacillus subtilis strains. The lacZ gene was fused to the liaI promoter (P liaI) in both strains such that induction of this promoter results in the production of β-galactosidase. One strain contains the lacZ fusion and additionally has a kanamycin insertion into the liaR gene that inactivates liar. The P liaI-lacZ fusions were licensed from Cornell University by Merck & Co., Inc. (Mascher et al., 2003, 2004). Strains belong to the Merck culture collection with the following codes: HB0950 (MB5826, CU1065 SPb P-liaI74-cat-lacZ) and HB0953 (MB5827, CU1065 SPb P-liaI74-cat-lacZ liaR::kan). Afterwards, Merck & Co., Inc. donated both of them to Foundation MEDINA. Strain HB0953 contains the lacZ fusion but additionally has a kanamycin insertion into the liaR gene which inactivates liaR. B. subtilis strain HB09050 or HB09053 was incubated to stationary phase in LB (1% NaCl) at 37°C and 220 rpm. The culture was adjusted to an optical density of 0.25 at a wavelength of 600 nm. Then, 100-mL of LB agar (1% NaCl) that contained 250 μg/ml of X-gal was inoculated with 1 mL of bacteria. The inoculated agar was poured into a NUNC bioassay plate (245 × 245 mm). After the agar solidified, the plates were dried in a laminar-flow hood for 15 min. After that, 0.02 mL drops of natural product extract were placed on the surface of the agar and the plates were allowed to dry in a laminar-flow hood for 15 min. The plates were incubated for 18 h at 37°C. They were scored for zones of inhibition and for hydrolysis of X-gal (blue precipitate). Bacitracin (50 mg/mL) was used as positive control of the assay while vancomycin (0.5 mg/mL) was used as negative control.
In vitro antimicrobial activity
MIC determination assays
MICs values of samples against a panel of strains from MEDINA collection which are: Staphylococcus aureus (CL 860), Haemophillus influenzae (CLB 21526), Pseudomonas aeruginosa (PAO-1), Escherichia coli (ATCC 25922), Enterococcus faecalis (ATCC 29212), and Candida albicans (MY 1055) strains were determined by the microdilution technique in accordance with the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS, 2007) using inocula of 1 × 105–5 × 105 CFU/mL. Serial dilutions of extracts were performed to assess growth inhibition. Microtiter plates were incubated at 35°C for 20–24 h. Cell growth was monitored by comparing the optical density between the time of treatment and after incubation time. The lowest concentration causing 90% inhibition microbial growth was defined as the MIC.
Aspergillus fumigatus agar-based assay
The A. fumigatus (MF5668: ATCC13073) stock conidial suspension was adjusted by quantitative colony counts at 3.5 × 109 CFU/mL. The conidial suspension was diluted into Yeast Nitrogen Base broth (YNB, 6.75 g L−1 yeast nitrogen base) to 65% transmittance at 660 nm. Then, 10 mL of this inoculum broth was added to 1 L of Yeast Nitrogen Base-Dextrose and 20 mL of the seeded agar media were poured into Omnitray plates. Once the agar solidified, the plates were dried in a laminar flow hood for 15 min. After this time 10 μL of each extract were distributed on the surface of agar plates and incubated at 29°C for 18 h. Finally, the halos of inhibition were measured. Amphotericin B (0.25 mg/mL) was used as a positive control for this assay.
Macromolecular labeling assay
The labeling reaction contained 0.05 mL of a log phase culture of Bacillus subtilis HB0950 in nutrient broth that contained 1% NaCl, 0.025 mL of two-fold concentrated nutrient broth (2% NaCl) that contained 7.5 mCi/L of L-[2, 3-3H]-aspartic acid, 5 mCi/L of [2-14C]-thymidine, 100 mg/L of chloramphenicol or 0.8 mg/L of rifampicin and 0.025 mL of inhibitor (1xWBE). The samples were incubated 37°C on a microtiter plate shaker. After 30 min, 0.025 mL of 25% TCA was added to stop radioactive incorporation. The TCA-insoluble material was collected on a glass microfiber filtermat with a Skatron Cell Harvester. The filtermat was washed with distilled H2O and dried under a stream of hot air. Radioactivity was measured in a Betaplate Scintillation Counter.
Liquid chromatography-high resolution mass spectrometry and data analysis
Two micro liters of the extracts were analyzed by LC-HRMS. Analysis was performed on an Agilent (Santa Clara, CA) 1100 single Quadrupole LC-MS, using a Zorbax SB-C8 column (2.1 × 30 mm), maintained at 40°C and with a flow rate of 300 ul/min. Solvent A consisted of 10% acetronitrile and 90% water with 0.01% trifluoroacetic acid and 1.3 mM ammonium formate, while solvent B was 90% acetronitrile and 10% water with 0.01% trifluoroacetic acid and 1.3 mM ammonium formate. The gradient started at 10% B and went to 100% B in 6 min, kept at 100% B for 2 min, and returned to 10% B for 2 min to initialize the system. Full diode array UV scans from 100 to 900 nm were collected in 4 nm steps at 0.25 s/scan.
HRMS data was acquired in a Thermo Finnigan LTQ-FT with the standard Ion Max API source (without the sweep cone) and ESI probe. Three scan events were used. The ion trap was scanned from 150 to 2,000 first in negative ion mode and then in positive ion mode. The FT was scanned from 200 to 2,000 in the positive ion mode only. In all cases the SID was set to 18 volts to try to reduce multiple ion clusters.
Data were analyzed with Apex software. The standard conditions were; peak width/resolution is set to 100,000 resolution at mass 400, MS search tolerance set between 0.008 and 0.003 Da. Apex software is a product from Sierra Analytics (Modesto, CA).
Results
Validation of LiaRS assay
The LiaRS (Lipid II cycle interfering antibiotic response Regulator and Sensor) two-component system (Figure 1) is one of several antibiotic-sensing systems that coordinate the genetic response to cell wall active antibiotics. Upon addition of inhibitors of stage II peptidoglycan synthesis, LiaRS autoregulates the liaIHGFSR operon. The lia promoter responds strongly and specifically to antibiotics that interfere with the lipid II cycle. The induction of β-galactosidase by these types of antibiotics is detected with X-gal, which turns blue when hydrolyzed by β-galactosidase. It is possible to differentiate specific interference of the lipid II cycle because specific inhibitors will induce the production of β-galactosidase in strain HB0950 but not in HB0953, in which the liaR gene has been disrupted. The LiaRS system and the two Bacillus strains have been previously described by Mascher et al. (2004).
Figure 1.
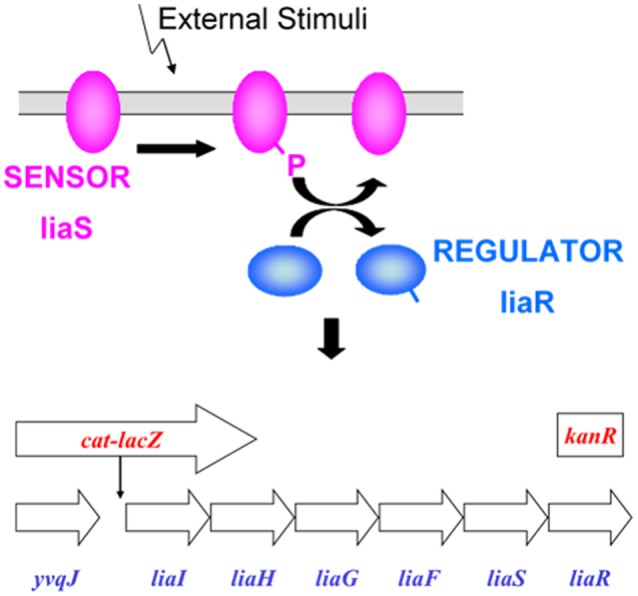
LiaRS—Lipid II cycle interfering antibiotic response Regulator and Sensor: Two-component antibiotic sensing system in Bacillus subtilis that co-ordinates genetic response to inhibitors of peptidoglycan synthesis that interfere with the lipid II cycle. Background strain: Bacillus subtilis CU1065; HB0950: lacZ fusion; HB0953: lacZ fusion, kanR insertion (liaR gene disrupted). LiaRS autoregulates the liaIHGFSR operon. Induction of the liaIH operon mediated by the two-component liaRS sensor/regulator system. Specificity associated with liaRS. lacZ inserted downstream from PliaI. Induction results in production of β-galactosidase. Induction of β-galactosidase in B. subtilis HB0953 indicates that induction was not due to an inhibitor of peptidoglycan synthesis.
B. subtilis HB0950 (MB5826, CU1065 SPb P-liaI74-cat-lacZ) produces β-galactosidase in response to the induction of liaI by certain cell-wall active antibiotics. B. subtilis HB0953 (MB5827, CU1065 SPb P-liaI74-cat-lacZ liaR::kan) is a liaR knock-out mutant. It is also capable of synthesizing β-galactosidase, but not in response to the liaIHGFSR operon.
In B. subtilis HB0950, the lacZ gene has been fused to the liaI promoter (PliaI) such that induction of this promoter results in the production of β-galactosidase. This production can be detected when this strain is grown on Luria agar (LA) medium containing either 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal) or 3,4-cyclohexenoesculetin-β-D-galactopyranoside (S-gal) in the presence of an inducer. Detection of β-galactosidase activity in HB0950 has been shown to be 150–200-fold over background in the presence of bacitracin, ramoplanin, and nisin, and ~35-fold over background in the presence of vancomycin (Mascher et al., 2004). Other antibiotics affecting protein synthesis (chloramphenicol, kanamycin, spectinomycin, streptomycin, tetracycline), RNA synthesis (rifampicin), and peptidoglycan synthesis (β-lactams, moenomycin, D-cycloserine), did not induce β-galactosidase production; neither did detergents (SDS, Triton) nor uncouplers of oxidative phosphorylation (DNP, CCCP). The production of β-galactosidase by HB0950 appears to be a specific response to compounds that bind to or otherwise interfere with recycling of lipid I in stage II peptidoglycan biosynthesis (Table 1).
Table 1.
Non-inducers of liaI Expression and Inducers of liaI Expression.
| Compound | liaI Expression | Disk diffusion assay response |
|---|---|---|
| Kanamycin | Non-inducer | Negative |
| Rifampicin | Non-inducer | Negative |
| Tetracycline | Non-inducer | Negative |
| Chloramphenicol | Non-inducer | Negative |
| Spectinomycin | Non-inducer | Negative |
| Streptomycin | Non-inducer | Negative |
| SDS | Non-inducer | Negative |
| Triton | Non-inducer | Negative |
| DNP | Non-inducer | Negative |
| Lysozyme | Non-inducer | Negative |
| Ampicillin | Inducer | Negative |
| Bacitracin | Inducer | Positive |
| Cepaholsporin | Inducer | Negative |
| D-cycloserine | Inducer | Negative |
| Moenomycin | Inducer | Negative |
| Nisin | Inducer | Positive |
| Penicillin G | Inducer | negative |
| Polymyxin B | Inducer | negative |
| Ramoplanin | Inducer | Positive |
| Tunicamycin | Inducer | Positive |
| Vancomycin | Inducer | Positive |
Activity was assessed by the appearance of a blue ring around the edge of the zone of inhibition on LB agar plates supplemented with X-Gal. (Compounds were tested in a range concentration of 50–0.5 mg/mL).
B. subtilis strain, HB0953, contains the lacZ fusion but additionally has a kanamycin insertion into the liaR gene which inactivates liaR. Since an intact two-component liaRS system is required for induction of the liaIH operon, inactivation of this system by kan insertion into liaR results in the inability of true inducers of P liaI to turn on β-galactosidase production. On X-gal medium, the LiaRS response appears to be very specific for true inducers and no false positive responses on HB0950 have been observed. However, with S-gal, fluoroquinolones, and cephalosporins produce a response on HB0950 that might be interpreted as a weak positive. These can be differentiated from true positives through the use of HB0953. On HB0953, true positives such as bacitracin and ramoplanin give an unambiguously negative response, while fluoroquinolones and cephalosporins give the same weak positive response as they do on HB0950.
Natural products screening for lipid II inhibitors
A set of 37,000 natural products (NP) extracts (50% bacteria and 50% fungi) were screened against the LiaRS assay described above at a single concentration (1xWBE), giving a hit rate of 0.13%. As controls, extracts with bacitracin and vancomycin were applied in each plate onto the agar surface (Figure 2). Extracts that caused induction of β-galactosidase in B. subtilis strain HB0950 were tested for specificity by re-testing them against both strains HB0950 and HB0953. The blue response was rated from 1 to 3 according to their intensity. A total of 49 actinomycetes strains were authentic inducers of LiaRS activity causing a blue response against HB0950 but not against HB0953. The larger and bluer inhibition zones were produced by a series of extracts within the group of actinomycetes.
Figure 2.
Example of the response of some active (blue ring response) extracts in the agar- based LiaRS assay (Nunc-plate).
Characterization of actinomycete strains
The soil samples used for the isolation of the 49 actinomycete strains were collected worldwide comprising both tropical and temperate areas: Costa Rica, French Guyana, Mexico, New Caledonia, South Africa, Spain, and Sri Lanka, including different ecological habitats like agricultural soils, riverbeds, lakes, ponds, swamps, dunes, tropical and sempervirent forests, savanna soil, and rhizospheres.
The most abundant taxonomic group identified by morphological analysis was the family Micromonosporaceae (69.3%, 34 strains most in the genera Micromonospora). Also, 3 strains were assigned to Nocardia, 3 strains to Streptomyces, 1 strain to Actinomadura, 2 strains to Amycolatopsis, and 1 strain to Actinoplanes. A set of 5 strains could not be identified (Table 2).
Table 2.
Taxonomy and number of strains isolated from both tropical and temperate countries.
| Strain | Origin | Ecological habitats | Family | Closest neighbor (EzTaxon result) | Similarity | Accession number |
|---|---|---|---|---|---|---|
| F-178,252 | Mexico | Soil | Micromonosporaceae | Actinoplanes ferrugineus | 97.98 | KY454560 |
| F-151,217 | French Guyana | Rainforest | Micromonospora chaiyaphumensis | 99.35 | KY454536 | |
| F-151,215 | Micromonospora sp. (*) | |||||
| F-175,917 | Mexico | Pond | Micromonospora chaiyaphumensis | 99.06 | KY454552 | |
| F-179,413 | Micromonospora chaiyaphumensis | 99.85 | KY454568 | |||
| F-143,677 | New Caledonia | Savannah | Micromonospora chaiyaphumensis | 99.78 | KY454532 | |
| F-182,202 | South Africa | Rhizosphere | Micromonospora chaiyaphumensis | 99.64 | KY454570 | |
| F-182,239 | Micromonospora chaiyaphumensis | 99.64 | KY454571 | |||
| F-182,241 | Micromonospora chaiyaphumensis | 99.64 | KY454572 | |||
| F-188,416 | Micromonospora chaiyaphumensis | 99.64 | KY454573 | |||
| F-188,417 | Micromonospora chaiyaphumensis | 99.64 | KY454574 | |||
| F-188,442 | Micromonospora chaiyaphumensis | 99.63 | KY454575 | |||
| F-162,175 | French Guyana | Rainforest | Micromonospora chersina | 99.64 | KY454541 | |
| F-140,177 | New Caledonia | Sempervirent forest | Micromonospora sp. (*) | |||
| F-144,426 | Micromonospora chersina | 99.64 | KY454533 | |||
| F-149,663 | Savannah | Micromonospora chersina | 99.72 | KY454535 | ||
| F-143,676 | Micromonospora sp. (*) | |||||
| F-169,627 | Sri Lanka | Mangrove | Micromonospora chersina | 99.71 | KY454545 | |
| F-175,357 | Micromonospora chersina | 99.64 | KY454551 | |||
| F-177,720 | Micromonospora chersina | 99.78 | KY454556 | |||
| F-177,770 | Micromonospora chersina | 99.57 | KY454557 | |||
| F-177,777 | Micromonospora chersina | 99.71 | KY454558 | |||
| F-178,645 | Micromonospora chersina | 99.71 | KY454563 | |||
| F-178,647 | Micromonospora chersina | 99.71 | KY454564 | |||
| F-179,454 | Micromonospora chersina | 99.64 | KY454569 | |||
| F-151,222 | French Guyana | Rainforest | Micromonospora endolithica | 99.57 | KY454537 | |
| F-170,297 | South Africa | Rhizosphere | Micromonospora equina | 99.41 | KY454546 | |
| F-178,247 | Mexico | Soil | Micromonospora fulviviridis | 99.42 | KY454559 | |
| F-179,400 | Pond | Micromonospora halotolerans | 99.64 | KY454567 | ||
| F-161,233 | South Africa | Dune | Micromonospora halotolerans | 99.85 | KY454540 | |
| F-168,651 | Rhizosphere | Micromonospora halotolerans | 99.64 | KY454542 | ||
| F-169,254 | Micromonospora inositola | 99.43 | KY454543 | |||
| F-169,257 | Micromonospora inositola | 99.48 | KY454544 | |||
| F-170,386 | Micromonospora inositola | 99.5 | KY454547 | |||
| F-171,509 | Micromonospora inositola | 99.49 | KY454548 | |||
| F-146,703 | New Caledonia | Sempervirent forest | Nocardiaceae | Nocardia niigatensis | 99.86 | KY454534 |
| F-161,197 | South Africa | Dune | Nocardia testacea | 99.64 | KY454539 | |
| F-186,787 | Rhizosphere | Nocardia sp. (*) | ||||
| F-117,410 | Costa Rica | Agricultural land | Pseudonocardiaceae | Amycolatopsis umgeniensis | 99.71 | KY454530 |
| F-178,253 | Mexico | Soil | Lechevalieria atacamensis | 98.99 | KY454561 | |
| F-177,328 | Lake | Streptomycetaceae | Streptomyces atrovirens | 99.62 | KY454553 | |
| F-177,430 | Soil | Streptomyces atrovirens | 99.57 | KY454554 | ||
| F-152,578 | Spain | River bed | Streptomyces mirabilis | 99.36 | KY454538 | |
| F-151,073 | French Guyana | Rainforest | Thermomonosporaceae | Actinomadura sp. (*) | ||
| F-175,914 | Mexico | Pond | Unidentified | Unidentified (*) | ||
| F-161,198 | South Africa | Dune | Unidentified (*) | |||
| F-161,203 | Unidentified (*) | |||||
| F-161,272 | Unidentified (*) | |||||
| F-171,173 | Rhizosphere | Unidentified (*) |
16S rDNA gene sequence not available.
Evaluation of antimicrobial activity
Most of the extracts exhibited activity against Gram-positive bacteria (S. aureus, Enterococcus faecalis). Their IC90-values oscillated in a range of 0.06–0.015 WBE/mL, together with some extracts which showed a IC90 value below 0.002 WBE/mL. Contrary, none of the extracts displayed activity against Gram-negative bacteria (Haemophilus influenzae, Pseudomonas aeruginosa, Escherichia coli) or against Aspergillus fumigatus, nor against Candida albicans. They showed MIC-values above the first point of dilution (>0.06 WBE/mL) except for extract F-144,426-C03-C01 that showed weak antifungal activity against C. albicans giving a value of 0.06 WBE/mL (Table 3).
Table 3.
Extracts tested in the LiaRS assay, in the susceptibility testing assays, and for High Resolution MS analysis.
| Strain | Extract | IC90 WBE/mL | A. fumigatus | LiaRS_Assay | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aMedium (days) | S. aureus | S. aureus | H. influenzae | E. faecalis | C. albicans | MF_5668 | HB950 | HB953 | ||||
| CL_8260 | EPI_167 | CLB21526 | ATCC_29212 | MY_1055 | ZOI (mm)|Q | ZOI (mm)|Q(#) | ZOI (mm)|Q | LC-MS | ||||
| F-117,410 | F-117,410-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | 0 | 7|C(2) | 7|C | Related to ramoplanin | |
| F-140,177 | F-140,177-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | 0 | 8|C(2) | 8|C | Related to ramoplanin | |
| F-143,676 | F-143,676-C02-C01 | FR23(13) | 0.06 | 0.06 | (−) | 0.06 | (−) | 0 | 9|B(2) | 8|B | Related to ramoplanin | |
| F-143,677 | F-143,677-C02-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | 0 | 8|C(2) | 8|C | Related to ramoplanin | |
| F-144,426 | F-144,426-C03-C01 | GOT(13) | 0.06 | 0.06 | (−) | 0.06 | 0.06 | 11|A | 14A(2) | 14A | Related to ramoplanin | |
| F-146,703 | F-146,703-C02-C01 | GOT(7) | (−) | (−) | (−) | (−) | (−) | 0 | 9|A(3) | 11|A | Related to ramoplanin | |
| F-146,703-C03-C01 | MPG(7) | (−) | (−) | (−) | 0.06 | (−) | 0 | 13|A(3) | 15|A | Related to ramoplanin | ||
| F-149,663 | F-149,663-C01-C01 | CLA(13) | 0.06 | 0.06 | (−) | 0.06 | (−) | 0 | 16|A(3) | 17|A | Related to ramoplanin | |
| F-149,663-C02-C01 | GOT(13) | (−) | 0.06 | (−) | 0.06 | (−) | 0 | 17A(3) | 19A | Related to ramoplanin | ||
| F-149,663-C03-C01 | MPG(13) | (−) | (−) | (−) | (−) | (−) | 0 | 11|A(3) | 11|A | Related to ramoplanin | ||
| F-151,073 | F-151,073-C01-C01 | FR23(13) | 0.06 | 0.06 | (−) | (−) | (−) | 0 | 8|B(2) | 8|B | Related to ramoplanin | |
| F-151,215 | F-151,215-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | 0 | 7|B(2) | 8|B | Related to ramoplanin | |
| F-151,217 | F-151,217-C02-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 0 | 27|D(2) | 26|D | Related to ramoplanin | |
| F-151,222 | F-151,222-C01-C01 | FR23(13) | 0.015 | 0.03 | (−) | 0.015 | (−) | 0 | 16|A(3) | 18|A | A2 | |
| F-151,222-C02-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 0 | 16|A(3) | 15|A | A2 | ||
| F-152,578 | F-152,578-C01-C01 | DNPM(7) | (−) | (−) | (−) | (−) | (−) | 0 | 7|C(2) | 7|C | Related to ramoplanin | |
| F-161,197 | F-161,197-C01-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 27|E | 27|D(2) | 27|D | Related to ramoplanin | |
| F-161,197-C03-C01 | RAM2(13) | (−) | (−) | (−) | (−) | (−) | 0 | 7|E(2) | 7|E | Related to ramoplanin | ||
| F-161,198 | F-161,198-C02-C01 | MPG(13) | 0.06 | 0.06 | (−) | 0.06 | (−) | 0 | 10|A(2) | 10|A | Related to ramoplanin | |
| F-161,203 | F-161,203-C01-C01 | GOT(13) | 0.06 | 0.06 | (−) | (−) | (−) | 0 | 11|A(2) | 10|A | Related to ramoplanin | |
| F-161,233 | F-161,233-C01-C01 | GOT(13) | 0.06 | 0.06 | (−) | 0.03 | (−) | 0 | 14|A(3) | 16|A | Related to ramoplanin | |
| F-161,272 | F-161,272-C01-C01 | GOT(13) | (−) | 0.06 | (−) | 0.06 | (−) | 0 | 8|B(2) | 8|B | Related to ramoplanin | |
| F-162,175 | F-162,175-C03-C01 | MPG(13) | 0.06 | 0.015 | (−) | 0.03 | (−) | 0 | 15|A(3) | 16|A | Related to ramoplanin | |
| F-168,651 | F-168,651-C02-C01 | GOT(13) | 0.06 | 0.03 | (−) | 0.06 | (−) | 0 | 16|A(3) | 18|A | Related to ramoplanin | |
| F-168,651-C03-C01 | MPG(13) | (−) | 0.06 | (−) | 0.03 | (−) | 0 | 15|A(3) | 15|A | Related to ramoplanin | ||
| F-169,254 | F-169,254-C02-C01 | GOT(13) | (−) | (−) | (−) | 0.06 | (−) | 0 | 16|A(3) | 17|A | Related to ramoplanin | |
| F-169,254-C03-C01 | MPG(13) | (−) | (−) | (−) | (−) | (−) | 0 | 11|A(3) | 11|A | Related to ramoplanin | ||
| F-169,257 | F-169,257-C02-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 0 | 12|A(3) | 13|A | Related to ramoplanin | |
| F-169,257-C03-C01 | MPG(13) | 0.06 | 0.06 | (−) | 0.06 | (−) | 0 | 13|A(3) | 13|A | Related to ramoplanin | ||
| F-169,627 | F-169,627-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | 0 | 14|C(2) | 13|C | Related to ramoplanin | |
| F-170,297 | F-170,297-C02-C01 | GOT(13) | 0.06 | 0.06 | (−) | (−) | (−) | 0 | 12|A(2) | 11|A | Related to ramoplanin | |
| F-170,386 | F-170,386-C01-C01 | FR23(13) | (−) | 0.03 | (−) | 0.06 | (−) | 0 | 13|A(3) | 13|A | Related to ramoplanin | |
| F-170,386-C02-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 0 | 17|A(3) | 19|A | Related to ramoplanin | ||
| F-170,386-C03-C01 | MPG(13) | (−) | 0.06 | (−) | 0.03 | (−) | 0 | 14|A(3) | 16|A | Related to ramoplanin | ||
| F-171,173 | F-171,173-C02-C01 | GOT(13) | 0.06 | 0.06 | (−) | 0.06 | (−) | 0 | 9|B(2) | 9B | Related to ramoplanin | |
| F-171,509 | F-171,509-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | 0 | 12|A(3) | 13|A | Related to ramoplanin | |
| F-171,509-C02-C01 | GOT(13) | (−) | (−) | (−) | (−) | (−) | 0 | 11|C(2) | 11|C | Related to ramoplanin | ||
| F-175,357 | F-175,357-C03-C01 | SOTM(13) | 0.0075 | 0.001875 | 0.06 | 0.0075 | (−) | 0 | 13|A(2) | 12A | Related to ramoplanin | |
| F-175,914 | F-175,914-C02-C01 | FR23(13) | (−) | 0.06 | (−) | (−) | (−) | 0 | 7|C(2) | 7|C | Related to ramoplanin | |
| F-175,917 | F-175,917-C02-C01 | FR23(13) | (−) | 0.06 | (−) | (−) | (−) | 0 | 7|C(2) | 7|C | Related to ramoplanin | |
| F-177,328 | F-177,328-C02-C01 | MPG(7) | 0.03 | 0.015 | (−) | 0.03 | (−) | 0 | 17|A(3) | 20|A | Related to ramoplanin | |
| F-177,430 | F-177,430-C02-C01 | MPG(7) | 0.0075 | 0.00375 | (−) | 0.015 | (−) | 0 | 19|A(3) | 21|A | Related to ramoplanin | |
| F-177,720 | F-177,720-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | 0 | 12|A(3) | 13|A | Related to ramoplanin | |
| F-177,720-C02-C01 | FR23(13) | (−) | 0.03 | (−) | 0.06 | (−) | 0 | 14|A(3) | 13|A | A2/A1 | ||
| F-177,770 | F-177,770-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 14|A(3) | 16|A | A2/A1 |
| F-177,770-C02-C01 | FR23(13) | 0.001875 | 0.001875 | 0.06 | 0.00375 | (−) | (−) | 0 | 14|A(2) | 14|A | related to ramoplanin | |
| F-177,777 | F-177,777-C02-C01 | FR23(13) | 0.015 | 0.015 | (−) | 0.015 | (−) | (−) | 0 | 14|A(3) | 14|A | Related to ramoplanin |
| F-178,247 | F-178,247-C02-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 6|B(2) | 6|B | Related to ramoplanin |
| F-178,252 | F-178,252-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 12|A(2) | 13|A | Related to ramoplanin |
| F-178,253 | F-178,253-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 14|E(2) | 14|E | Related to ramoplanin |
| F-178,645 | F-178,645-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 13|A(3) | 14|A | Related to ramoplanin |
| F-178,645-C02-C01 | FR23(13) | 0.03 | 0.03 | (−) | 0.06 | (−) | (−) | 0 | 13|A(3) | 14|A | Related to ramoplanin | |
| F-178,647 | F-178,647-C01-C01 | CLA(13) | 0.06 | 0.015 | (−) | 0.06 | (−) | (−) | 0 | 13|A(3) | 15|A | A2 |
| F-178,647-C02-C01 | FR23(13) | 0.015 | 0.015 | (−) | 0.03 | (−) | (−) | 0 | 16|A(3) | 17|A | A1/A2 | |
| F-179,400 | F-179,400-C01-C01 | CLA(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 10|A(3) | 11|A | Related to ramoplanin |
| F-179,413 | F-179,413-C01-C01 | CLA(13) | (−) | (−) | (−) | 0.06 | (−) | (−) | 0 | 18|A(3) | 19|A | Related to ramoplanin |
| F-179,413-C02-C01 | FR23(13) | (−) | 0.06 | (−) | 0.06 | (−) | (−) | 0 | 15|A(3) | 16|A | Related to ramoplanin | |
| F-179,454 | F-179,454-C02-C01 | FR23(13) | 0.015 | 0.0075 | (−) | 0.03 | (−) | (−) | 0 | 13|A(3) | 13|A | Related to ramoplanin |
| F-182,202 | F-182,202-C03-C01 | MPG(13) | 0.03 | 0.03 | (−) | 0.06 | (−) | (−) | 0 | 11|A(2) | 11|A | Related to ramoplanin |
| F-182,239 | F-182,239-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 12|A(2) | 12|A | Related to ramoplanin |
| F-182,241 | F-182,241-C01-C01 | FR23(13) | (−) | 0.06 | (−) | 0.06 | (−) | (−) | 0 | 13|A(3) | 13|A | Related to ramoplanin |
| F-182,241-C03-C01 | MPG(13) | (−) | 0.06 | (−) | 0.06 | (−) | (−) | 0 | 11|A(3) | 13|A | Related to ramoplanin | |
| F-186,787 | F-186,787-C01-C01 | CLA(7) | (−) | 0.06 | (−) | 0.06 | (−) | (−) | 0 | 6|B(2) | 6|B | Related to ramoplanin |
| F-188,416 | F-188,416-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 9|C(2) | 9|C | Related to ramoplanin |
| F-188,417 | F-188,417-C01-C01 | FR23(13) | 0.03 | 0.015 | (−) | 0.015 | (−) | (−) | 0 | 14|A(3) | 14|A | Related to ramoplanin |
| F-188,442 | F-188,442-C01-C01 | FR23(13) | (−) | (−) | (−) | (−) | (−) | (−) | 0 | 9|C(2) | 9|A | Related to ramoplanin |
ZOI (mm), Halo of inhibition.
Q, Qualifier (A or B-clear, C or D-hazy, E-very hazy).
(#) β-galactosidase intensity (blue ring).
Ramoplanin analogs: A1 MW = 2538; A2 MW = 2552; A3 MW = 2566.
IC90 > 0.06: (−).
Media and days of cultivation. Media composition are indicated in Supplementary Data.
These results indicate that the extracts keep similar antibacterial spectrum to ramoplanins.
Inhibition peptidoglycan biosynthesis
All extracts tested for inhibition of macromolecular labeling exhibited specific activity by inhibiting peptidoglycan synthesis (PG) above 80%. Figure 3 illustrates an example of the labeling test of one of the active extracts from the NP screening in a two-fold serial dilution series for inhibition of DNA and peptidoglycan syntheses in B. subtilis HB0950. This sample inhibited PG to a greater extent than they inhibited protein or DNA syntheses, indicating that it is a selective inhibitor of PG synthesis.
Figure 3.

Dose-Response curves from the Labeling test of one of the active extracts from the NP screening in B. subtilis HB0950.
Chemical analysis of LiaRS positive extracts
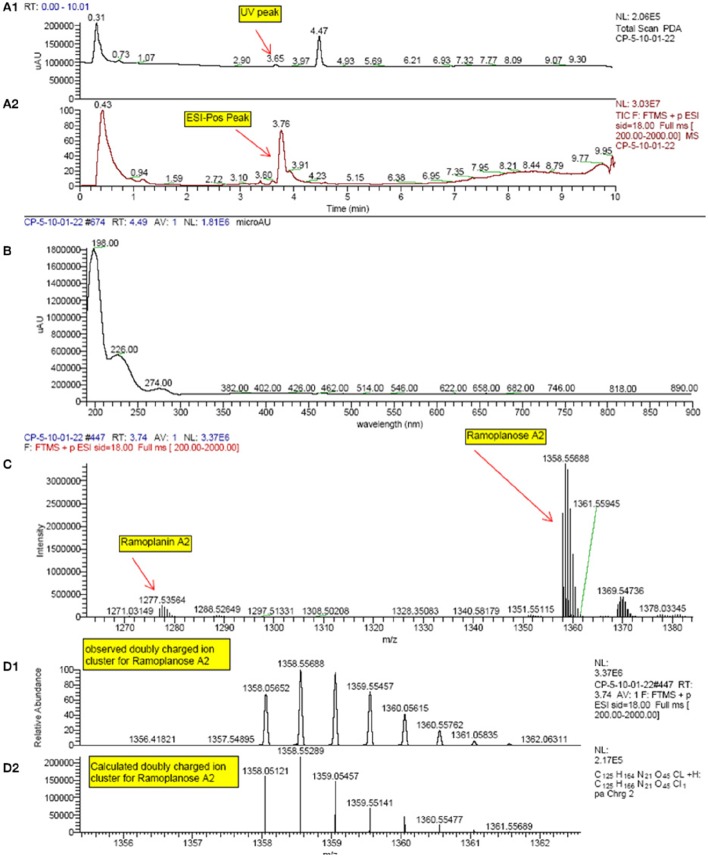
All extracts that were confirmed hits in the LiaRS screen were analyzed by HR-LCMS. Since ramoplanin was a known compound active in this assay, sample dereplication was performed in order to eliminate those samples containing this molecule from undergoing further isolation efforts. High resolution mass spectrometry signatures of each extract were evaluated and scanned for single ions corresponding to doubly charged species of ramoplanin A1, A2, and A3. In addition, the presence of ramoplanoses A1, A2, and A3, glycosylated versions of the ramoplanins, was also observed (Figure 4). Further, some additional ramoplanin analogs were also identified. These analogs corresponded to reduced versions (+4H and +O,4H) of ramoplanins, with the modifications presumably occurring in the N-acyl hydrophobic sidechain. All extracts showed mass spectrometry signatures (range of molecular weights and isotopic distributions) that confirmed the presence of compounds related to ramoplanin structural family and within them new analog compounds.
Figure 4.
HPLC and High Resolution LC-MS spectrum profile of some of the actives from the natural products screening which matched with Ramoplanin A2 and Ramoplanose. (Extract example: F-151,222-C01-C01). (A1): UV-visible trace; (A2): TIC (Total ion chromatogram); (B): UV-visible spectra at 4.49 min; (C): Mass spectra at 3.74 min from FTMS positive ion; (D1): Mass spectra at 3.74 min. (D2): Theoretical isotopic distribution of Ramoplanose A2.
Discussion
Ramoplanin is a suitable clinical candidate for the treatment of infections caused by aerobic or anaerobic Gram-positive pathogens (Farver et al., 2005). The mechanism of action of ramoplanin makes it unlikely that it will develop high levels of resistance since this compound acts on the second phase of peptidoglycan biosynthesis, capturing the Lipid intermediate II (Somner and Reynolds, 1990).
Thus, far, ramoplanin was only isolated from the fermentation of a strain of Actinoplanes sp. ATCC 33076 (Farver et al., 2005). However, in this study we have found 49 distinct actinomycete strains producing compounds related to this antibiotic. Further, our findings demonstrate that the extracts prepared from this group of strains display antibacterial profiles similar to ramoplanin.
These extracts revealed a positive response to the agar diffusion assay developed from studies in Cornell University (Mascher et al., 2003, 2004) showing activity against B. subtilis strains HB0950 and HB0953, being that they revealed a blue ring (X-Gal hydrolysis by the presence of β-galactosidase) in the strain containing the P liaI-lacZ fusion and they did not show such a response in the strain that additionally contained a gene insertion of kanamycin resistance in the liaR gene. Extracts positive in this test proved to be compounds that specifically interfere with the Lipid II as ramoplanin does (Mascher et al., 2004).
Moreover, most of them exhibited specific activities against Gram-positive bacteria (S. aureus, E. faecalis), as shown in Table 3. Also, all the extracts completely and selectively inhibited peptidoglycan biosynthesis, as demonstrated by the results obtained in a macromolecular labeling assay.
Analysis of the extracts by HR-LCMS indicated that they all contained compounds that belong to ramoplanin family. Dereplication was able to quickly identify these components and eliminated the need for extensive isolation efforts. Scale-up of selected cultures and isolation of novel ramoplanin analogs identified is in progress.
The point that most of the producing organisms belong to the Micromonosporaceae family is consistent with the taxonomy of the original organism producer of ramoplanin (Farver et al., 2005). Our results additionally suggest that the production of ramoplanin analogs is widespread within the actinomycetes. Geographically, the producers of these compounds were dispersed throughout the whole world based on the different origin of the samples from which the strains of our study came from.
The results obtained in this study support the fact that natural products are an unlimited source of potential drugs, in particular of antibiotics (Peláez, 2006; Chopra, 2013; Lacret et al., 2015; Ling et al., 2015; Crespo et al., 2016). They also demonstrate the power of using novel screening strategies that combine new knowledge in biotechnology with libraries of natural products in order to find new drug candidates against multi-drug resistance pathogens.
Our data strongly suggest the presence of new ramoplanin-analogs among the actinomycete strains of this study, so large scale fermentations and purifications of selected strains are being currently performed in order to identify these new antimicrobials which perhaps could overcome the low local tolerability of ramoplanin when injected intravenously.
Authors contributions
MD performed the HTS assay, collected, and analyzed data; IG performed the fermentation of microorganisms and taxonomic identification of the microbial strains. OG supervised the microbial work; CP, DZ, JM performed the chemical work; RO performed macromolecular labeling assay; JT, NE generated the extracts; FP general supervision; MD, CP, FR, FV, and OG wrote the manuscript which was revised and approved by all the authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the contribution of all CIBE (Centro de Investigación Básica de España) employees during the years of operation of the research center, and the support of many others at MRL (Merck Research Laboratories).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00343/full#supplementary-material
References
- Bozdogan B., Esel D., Whitener C., Browne F. A., Appelbaum P. C. (2003). Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52, 864–868. 10.1093/jac/dkg457 [DOI] [PubMed] [Google Scholar]
- Cao M., Wang T., Ye R., Helmann J. D. (2002). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45, 1267–1276. 10.1046/j.1365-2958.2002.03050.x [DOI] [PubMed] [Google Scholar]
- Cavalleri B., Hermes P., Giancarlo V., Enrico S., Franco P. (1984). A-16686, a new antibiotic from Actinoplanes. I. Fermentation, isolation and preliminary physico-chemical characteristics. J. Antibiot. 37, 309–317. 10.7164/antibiotics.37.309 [DOI] [PubMed] [Google Scholar]
- Chopra I. (2013). The 2012 garrod lecture: discovery of antibacterial drugs in the 21st century. J. Antimicrob. Chemother. 68, 496–505. 10.1093/jac/dks436 [DOI] [PubMed] [Google Scholar]
- Chun J., Int J. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Syst. Evol. Microbiol. 57, 2259–2261. 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- Crespo G., Gonzalez-Menendez V., de la Cruz M., Martin J., Cautain B., Sanchez P., et al. (2016). Antifungal long-chain alkenyl sulphates isolated from culture broths of the fungus Chaetopsina sp. Planta Med. [Epub ahead of print]. 10.1055/s-0042-118190 [DOI] [PubMed] [Google Scholar]
- Donadio S., Carrano L., Brandi L., Serina S., Soffientini A., Raimondi E., et al. (2002). Targets and assays for discovering novel antibacterial agents. J. Biotechnol. 99, 175–185. 10.1016/S0168-1656(02)00208-0 [DOI] [PubMed] [Google Scholar]
- Farver D. K., Hedge D. D., Lee S. C. (2005). Ramoplanin: a lipoglycodepsipeptide antibiotic. Ann. Pharmacother. 39, 863–868. 10.1345/aph.1E397 [DOI] [PubMed] [Google Scholar]
- Finegold S. M., John S. S., Vu A. W., Li C. M., Molitoris D., Song Y., et al. (2004). In vitro activity of ramoplanin and comparator drugs against anaerobic intestinal bacteria from the perspective of potential utility in pathology involving bowel flora. Anaerobe 10, 205–211. 10.1016/j.anaerobe.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Fischer H. P., Brunner N. A., Wieland B., Paquette J., Macko L., Ziegelbauer K., et al. (2004). Identification of antibiotic stress-inducible promoters: a systematic approach to novel pathway-specific reporter assays for antibacterial drug discovery. Genome Res. 14, 90–98. 10.1101/gr.1275704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits E., Blommaert E., Lippell A., O'Neill A. J., Weytjens B., De Maeyer D., et al. (2016). Elucidation of the mode of action of a new antibacterial compound active against Staphylococcus aureusand Pseudomonas aeruginosa. PLoS ONE 11:e0155139. 10.1371/journal.pone.0155139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M., Kämpfer P., Busse H.-J., Trujillo M. E., Susuki K.-I., Ludwig W., et al. (2012). Bergey's Manual of Systematic Bacteriology, 2nd Edn. The Actinobacteria, Vol. 5 New York, NY: Springer. [Google Scholar]
- Goodfellow M., Mordarki M., Williams S. T. (1984). The Biology of Actinomycetes. New York, NY: Academic Press. [Google Scholar]
- Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (1990). PCR Protocols. A Guide to Methods and Amplifications. London: Academic Press. [Google Scholar]
- Jabes D., Brunati C., Candiani G., Riva S., Romano G., Maffioli S., et al. (2014). Pharmacological Properties of NAI-603, a well-tolerated semisynthetic derivative of ramoplanin. Antimicrob. Agents Chemother. 58, 1922–1929. 10.1128/AAC.01620-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Lacret R., Oves-Costales D., Gomez C., Diaz C., de la Cruz M., Perez-Victoria I., et al. (2015). New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces zhaozhouensis. Mar. Drugs 13, 128–140. 10.3390/md13010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. (1991). Chapter 6. 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: Wiley; ), 115–175. [Google Scholar]
- Ling L. L., Schneider T., Peoples A. J., Spoering A. L., Engels I., Conlon B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T., Margulis N. G., Wang T., Ye R. W., Helmann J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50, 1591–1604. 10.1046/j.1365-2958.2003.03786.x [DOI] [PubMed] [Google Scholar]
- Mascher T., Zimmer S. L., Smith T. A., Helmann J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. 10.1128/AAC.48.8.2888-2896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCLS (2007). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standars Institute - NCCLS. [Google Scholar]
- Peláez F. (2006). The historical delivery of antibiotics from microbial natural products—can history repeat? Biochem. Pharmacol. 71, 981–990. 10.1016/j.bcp.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Peláez T., Alcalá L., Alonso R., Martín-López A., García-Arias V., Marín M., et al. (2005). In vitro activity of ramoplanin against clostridium difficile, including strains with reduced susceptibility to vancomycin or with resistance to metronidazole. Antimicrob. Agents Chemother. 49, 1157–1159. 10.1128/A.C.49.3.1157-1159.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R. Tormo J., García J., DeAntonio M., Feliz J., Mira A., Peláez F., et al. (2003). A method for the selection of production media for actinomycete strains based on their metabolite HPLC profiles. J. Ind. Microbiol. Biotechnol. 30, 582–588. 10.1007/s10295-003-0084-7 [DOI] [PubMed] [Google Scholar]
- Schmidt J. W., Greenough A., Burns M., Luteran A. E., McCafferty D. G. (2010). Generation of ramoplanin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 310, 104–111. 10.1111/j.1574-6968.2010.02051.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E., Baneyx F. (2002). Stress-based identification and classification of antibacterial agents: second-generation Escherichia coli reporter strains and optimization of detection. Antimicrob. Agents Chemother. 46, 2490–2497. 10.1128/AAC.46.8.2490-2497.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somner E. A., Reynolds P. E. (1990). Inhibition of peptidoglycan biosynthesis by ramoplanin. Antimicrob. Agents Chemother. 34, 413–419. 10.1128/AAC.34.3.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Cohen S., Mani N., Murphy C., Rothstein D. M. (2002). A pathway-specific cell based screening system to detect bacterial cell wall inhibitors. J. Antibiot. 55, 279–287. 10.7164/antibiotics.55.279 [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. 10.1128/jb.173.2.697-703.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.