Summary
Tissue-resident memory T (Trm) cells form a heterogeneous population that provides localized protection against pathogens. Here, we identify CD49a as a marker that differentiates CD8+ Trm cells on a compartmental and functional basis. In human skin epithelia, CD8+CD49a+ Trm cells produced interferon-γ, whereas CD8+CD49a− Trm cells produced interleukin-17 (IL-17). In addition, CD8+CD49a+ Trm cells from healthy skin rapidly induced the expression of the effector molecules perforin and granzyme B when stimulated with IL-15, thereby promoting a strong cytotoxic response. In skin from patients with vitiligo, where melanocytes are eradicated locally, CD8+CD49a+ Trm cells that constitutively expressed perforin and granzyme B accumulated both in the epidermis and dermis. Conversely, CD8+CD49a– Trm cells from psoriasis lesions predominantly generated IL-17 responses that promote local inflammation in this skin disease. Overall, CD49a expression delineates CD8+ Trm cell specialization in human epithelial barriers and correlates with the effector cell balance found in distinct inflammatory skin diseases.
Keywords: Skin, tissue resident T cells, cytotoxicity, CD49a, psoriasis, vitiligo, immunopathology, perforin, granzyme B, IL-15
Graphical Abstract
Highlights
-
•
CD49a expression marks CD8+ Trm cells poised for IFN-γ production in human skin
-
•
IL-15 drives potent cytotoxic capacity in CD49a+ Trm cells
-
•
IL-17 is preferentially produced by CD49a− CD8+ Trm cells in the skin
-
•
CD49a+ versus CD49- Trm cell functional dichotomy is preserved in vitiligo and psoriasis
Tissue-resident memory T (Trm) cells provide localized adaptive immunity in peripheral tissues. Cheuk et al. identify cytotoxic CD49a+CD8+ Trm cells and IL-17-producing CD49a−CD8+ Trm cells in healthy human skin. The functional dichotomy of pathogenic Trm cells based on CD49a expression is preserved in focal skin diseases vitiligo and psoriasis.
Introduction
The human skin forms a barrier to the external environment that is constantly exposed to colonizing microbiota, invasive pathogens, and allergens (Belkaid and Segre, 2014, Pasparakis et al., 2014). These encounters drive differentiation and colonization of tissue-resident memory T (Trm) cells (Gaide et al., 2015, Gebhardt et al., 2009, Naik et al., 2015, Naik et al., 2012), originally characterized in barrier tissues such as skin, gut, lung, and the female genital tract (Gaide et al., 2015, Schenkel and Masopust, 2014, Thome et al., 2014, Watanabe et al., 2015). Of the 20 billion T cells contained in human skin (Clark et al., 2006), 50%–70% express the Trm cell markers CD103 (α-subunit of the αEβ7 integrin receptor) and CD69 (Watanabe et al., 2015), both implicated in lodging Trm cells in peripheral tissues. CD103 binds E-cadherin, which is highly expressed on epithelia, whereas CD69 antagonizes sphingosine 1-phosphate receptor 1 (S1PR1)-mediated egress from tissues (Mackay et al., 2015, Skon et al., 2013).
Following in situ activation, Trm cells provide strong defense against recurrent infections (Ariotti et al., 2014, Schenkel et al., 2014). While such local immune responses contribute to immunity (Gebhardt et al., 2009, Glennie et al., 2015, Jiang et al., 2012, Naik et al., 2012), aberrant activation might cause disease (Hondowicz et al., 2016, Jabri and Abadie, 2015, Park and Kupper, 2015). In the skin, the patchy appearance of several T cell-mediated diseases, such as psoriasis and vitiligo (Boehncke and Schön, 2015, Ezzedine et al., 2015), suggests that tissue-resident rather than circulating cells drive immunopathology (Cheuk et al., 2014, Gaide et al., 2015, Kirsch et al., 2015). Demarcated, inflamed and hyperproliferative plaques are maintained by interleukin-23 (IL-23) and IL-17 in psoriasis (Hueber et al., 2010, Leonardi et al., 2008), whereas vitiligo presents with persistent depigmentations attributed to local interferon-γ (IFN-γ) production and T cell-mediated killing of melanocytes (Harris et al., 2012, Rashighi et al., 2014, van den Boorn et al., 2009). Thus, if Trm cells drive these diseases, then vitiligo and psoriasis would be expected to involve the contribution of functionally different subsets.
Trm cell subsets with distinct cytokine profiles have been reported in skin (Naik et al., 2015, Sanchez Rodriguez et al., 2014, Watanabe et al., 2015), possibly reflecting a diversity of immune functionalities and responses to pathogenic and commensal microbiota. Nonetheless, phenotypic markers of Trm cell subsets with specialized effector functions have not been established. In addition to CD103 and CD69, murine Trm cells that create local immunity to recurrent viral infection homogenously express CD49a (Gebhardt et al., 2009, Ray et al., 2004, Zhang and Bevan, 2013). CD49a constitutes the α-subunit of the α1β1 integrin receptor, also known as very late antigen 1 (VLA-1), and binds collagen IV enriched in the basement membrane separating epidermis and dermis. As only 15% of human skin-derived T cells express CD49a (Purwar et al., 2011), CD49a expression might distinguish Trm cell subsets with distinct effector functions and putative roles in immunopathology.
Here, we determined the anatomical localization, transcriptional profiles, and functional properties of Trm cell subsets in human skin with respect to CD49a, CD69, and CD103. In healthy individuals, CD8+CD103+CD49a– Trm cells were present in both the dermis and epidermis, whereas CD8+CD103+CD49a+ Trm cells specifically localized to the epidermis. Epidermal CD8+CD103+CD49a– Trm cells were the predominant IL-17-producers, whereas CD8+CD103+CD49a+ Trm cells excelled at IFN-γ production and rapidly gained a cytotoxic capacity following IL-15 stimulation. This functional dichotomy was evident in the comparison of distinct immune-mediated skin diseases, with skin biopsies from vitiligo patients showing a predominance of cytotoxic CD8+CD103+CD49a+ Trm cells while skin biopsies from psoriasis patients featured the accumulation of the IL-17 producing CD8+CD103+CD49a– counterparts. Together, our results reveal a functional specialization of distinct skin Trm cell subsets in health and disease.
Results
CD49a Expression Marks Epidermal, Oligoclonal CD8+CD103+ Trm Cells
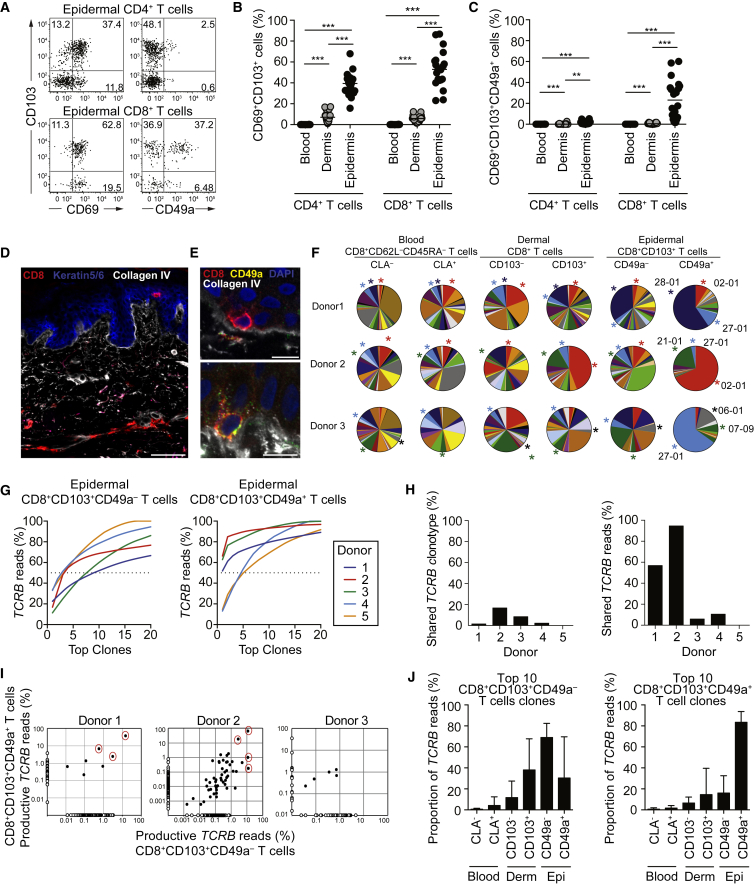
To gain insights into the diversity among Trm cells with respect to spatial niches, we determined the relative expression of tissue-residency markers CD69, CD103, and CD49a on CD4+ and CD8+ T cells from healthy human skin (Figure S1A). Compared to the dermis, an increased proportion of epidermal CD4+ and CD8+ T cells co-expressed CD69 and CD103 (Figures 1A and 1B) as previously reported (Watanabe et al., 2015). In these healthy skin samples, CD49a expression was restricted to CD8+CD69+CD103+ T cells that were located in the epidermis (Figures 1A and 1C). Considerable variation in the frequencies of CD8+CD69+CD103+CD49a+ T cells was apparent among (Figure 1C) and within different donors (Figures S1B and S1C), indicating high inter- and intra-individual variation of Trm cell compositions within the epidermal compartment.
Figure 1.
Healthy Human Skin Contains a Population of Clonally Enriched CD49a+ Trm Cells in Epidermis
(A–C) Phenotypic analysis of live CD3+TCRγδ− T cells from blood, dermis, and epidermis from healthy donors (n = 20). (A) Representative FACS plots of CD103, CD69, and CD49a expression in epidermal CD4 or CD8 T cells. (B and C) Frequencies of CD103+CD69+ (B) and CD103+CD69+CD49a+ (C) in CD4+ or CD8+ T cells. Each point represents data from an individual donor. Median depicted. Two-tailed Wilcoxon tests, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Confocal image of cryopreserved healthy skin depicting CD8 (red), Keratin5/6 (blue), and collagen IV (white). Scale bar, 100 μm.
(E) Confocal images of epidermal CD8+CD49− (upper panel) and CD8+CD49+ (lower panel) cells relative to the collagen IV-rich basement membrane. CD8 (red), CD49a (green) DAPI (blue), collagen IV (white) are depicted. Scale bars, 10 μm. Images are representative of ten healthy donors.
(F–J) Analysis of TCRβ CDR3 region in sorted CD8+ T cells from blood, dermis, and epidermis of healthy donors (n = 5). (F) Pie charts show the frequency of TCR Vβ families in T cell subsets from three donors. The top three Vβ families in epidermal CD8+CD103+CD49a+ are marked by color-coded asterisks. (G) The cumulative frequency of reads among top 20 clonotypes from epidermal CD8+CD103+CD49a– (left) and CD8+CD103+CD49a+ (right) Trm cells. (H) Proportion of shared clonotypes (left) and cumulative frequency of shared reads (right) between CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells. (I) Log scatterplot from donor 1–3 showing proportion of reads for each unique clonotype in CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells. Major shared clones are highlighted with red circles. (J) Total frequency of productive reads of the top ten clonotypes of CD8+CD103+CD49a– (left) and CD8+CD103+CD49a+ (right) Trm cells in the total reads of CD8+ T cell subsets in blood, dermis and epidermis. Mean ± SD is depicted. See also Figure S1.
Skin CD4+ Trm cells were predominately CD62L–CD45RA–CD28+, whereas CD8+ Trm cells were CD62L– but heterogeneous with respect to expression of CD27, CD28, and CD45RA (Figures S1D and S1E). To delineate the microanatomical localization of skin CD8+ T cell subsets, we performed confocal imaging of healthy skin. Regardless of CD49a expression, epidermal CD8+ T cells were juxtaposed to the collagen IV-rich basement membrane, embedded among basal keratinocytes (Figures 1D and 1E). Perivascular dermal CD8+ T cells were predominately detected in papillary dermis well separated from epidermis (Figure 1D). In total, 24% (6/33) of epidermal and 6% (3/53) of dermal CD8+ T cells expressed CD49a in direct contact with the basement membrane (Figure 1E; data not shown).
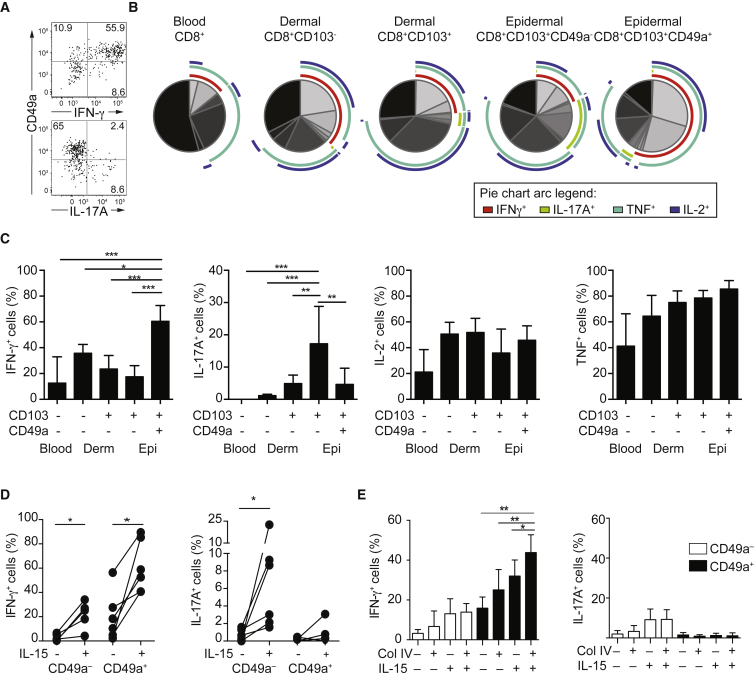
Bulk T cells in full thickness human skin display a diverse T cell receptor (TCR) repertoire that is in equilibrium with that of circulating T cells (Clark et al., 2006, Harden et al., 2015). To determine whether epidermal CD8+CD103+CD49a+ Trm cells shared a clonal origin with circulating effector memory CD8+ T cells or skin T cell subsets, CD8+ T cell subsets from five healthy donors were sorted (Figures S1F and S1G) and analyzed by targeted, deep, high-throughput genomic DNA sequencing of the TCRβ complementarity-determining region 3 (CDR3). A single Vβ family dominated the epidermal CD8+CD103+CD49a+ Trm cell population in three donors, with CDR3 diversity more restricted in CD8+CD103+CD49a+ versus CD8+CD103+CD49a– Trm cells (Figures 1F and 1G). Although only a small proportion of the detected clonotypes overlapped between epidermal CD8+CD103+CD49a+ and CD8+CD103+CD49a– Trm cells (Figure 1H), a few dominant clones were shared and comprised the majority of total reads in two donors (Figures 1H and 1I). Furthermore, whereas the most prevalent CD8+CD103+CD49a– Trm clonotypes showed some overlap with other epidermal and dermal CD8+ T cell subsets (Figure 1J), Vβ family distribution differed in epidermal and dermal CD8+CD103+CD49a– Trm cells (Figure 1F). The ten largest CD8+CD103+CD49a+ Trm cell clones were highly enriched in the epidermis (Figure 1J). Although the mechanical procedure of dermal and epidermal preparations might result in some degree of cellular cross-contamination, these results together demonstrate the epidermal localization of expanded, oligoclonal CD8+CD103+CD49a+ Trm cells in human skin.
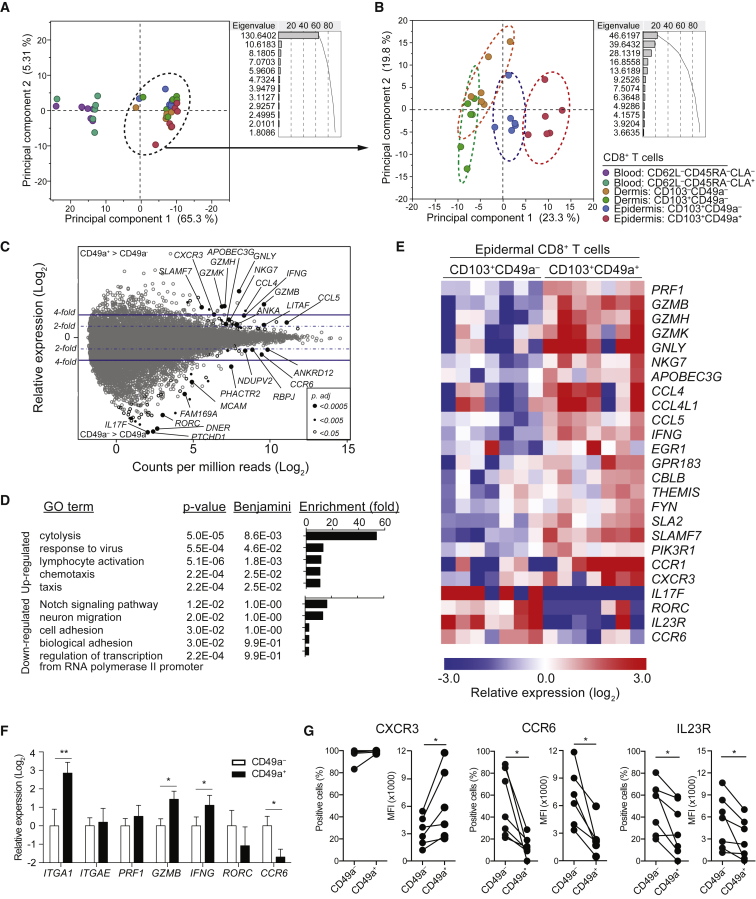
Transcriptional Profiles of Epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm Cells Indicate a Functional Dichotomy
To further interrogate the functional properties of epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cell populations, we determined their transcriptional profiles, along with those of dermal CD8+CD103–CD49a– and CD8+CD103+CD49a– T cells as well as peripheral blood CD8+CD62L–CD45RA– T cell populations expressing or lacking the cutaneous leukocyte antigen (CLA). Cells were sorted from healthy donors (Figures S1F and S1G) and analyzed by RNA-sequencing (Ramsköld et al., 2012). Principal component analysis on all sorted T cells subsets from six donors distinguished circulating and skin T cell populations (Figure 2A). Among skin T cells, principal component analysis discriminated epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells from dermal populations (Figure 2B). Focusing on the epidermal Trm cells, 92 genes were differentially expressed between CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells (Figure 2C, Tables S1 and S2). Gene enrichment analysis (DAVID) indicated acquisition of cytotoxicity-related function as the most compelling difference between the populations (Figure 2D). Specifically, expression of PRF1, GZMB, GZMH, GZMK, GNLY, and NKG7 transcripts, all encoding cytotoxic granule components, were elevated in epidermal CD8+CD103+CD49a+ Trm cells (Figure 2E), indicating that this subset might mediate cellular cytotoxicity. Moreover, genes mediating response to viruses, lymphocyte activation, and chemotaxis were also upregulated in CD8+CD103+CD49a+ Trm cells (Figure 2D). Conversely, gene enrichment analyses did not identify any significantly downregulated gene programs (Figure 2D). Nonetheless, transcripts of genes associated with IL-17 production, such as IL17F, RORC, IL23R, and CCR6, were significantly decreased in CD8+CD103+CD49a+ relative to CD8+CD103+CD49a– Trm cells, whereas transcripts for IFN-γ were elevated (Figures 2D-E). Transcription of ITGA1, encoding CD49a, was not significantly different following correction for multiple comparisons (uncorrected p = 0.0007, adjusted p = 0.12). Quantitative PCR (qPCR) analysis demonstrated significantly higher ITGA1 transcription in CD8+CD103+CD49a+ versus CD8+CD103+CD49a– Trm cell subsets (Figure 2F; p = 0.002). Moreover, regulation of GZMB, IFNG, and CCR6 was confirmed by qPCR, whereas no difference in ITGAE, encoding CD103, was found (Figure 2F). Further validating transcriptional data, CXCR3 expression was higher on CD8+CD103+CD49a+ Trm cells, whereas IL-23R and CCR6 were preferentially expressed by CD8+CD103+CD49a– Trm cells (Figure 2G). As such, transcriptional profiles as well as surface receptor expression suggested functional differences between CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells.
Figure 2.
Distinct Transcriptional Profiles of CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm Cells
(A–E) RNA-seq analyses of CD8+ T cell subpopulations sorted from blood, dermis, or epidermis of healthy individuals. (A and B) Principal component analysis of the top 200 differentially regulated genes among (A) all six subpopulations or (B) four skin-derived subpopulations after mean centering to correct for batch effects. Dots indicate samples of different subpopulations from a total of 6 donors, as indicated. Principal component 1 and 2 represents the largest source of variation, combined accounting for (A) 70.6% and (B) 43.1% of the total variation, respectively. Color-coded eclipses indicate the 95% confident area of particular T cell subpopulations. (C–E) Transcriptome analysis of CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells sorted from healthy epidermis (n = 7). (C) MA-plot (Log2 fold change against Log2 count per million) showing differential gene expression between epidermal CD8+CD103+CD49a− and CD8+CD103+CD49a+ Trm cells. Each gene was symbol-coded as indicated according to their adjusted p values generated using EdgeR with FDR correction. (D) Functional annotation analysis using DAVID tool listing gene sets upregulated (upper) or downregulated (lower) of CD8+CD103+CD49a+ Trm cells as compared to CD8+CD103+CD49a– Trm cells. (E) Heatmap showing gene expression of selected genes associated with functional annotations. Each column represents an individual donor. Row-mean centered relative expression values are shown.
(F) Quantitative real-time PCR measurements of transcript levels of specific genes, as indicated, in epidermal CD8+CD103+CD49a– relative to CD8+CD103+CD49a+ Trm cell subsets. Fold-change was calculated against the mean expression of the CD8+CD103+CD49a– Trm cells. Mean ± SD depicted.
(G) Surface expression of CXCR3, CCR6 and IL-23 receptor (IL23R) in CD8+CD103+CD49a– or CD8+CD103+CD49a+ Trm cells, as assessed by flow cytometry. Data are presented with respect to frequency of positive cells and their respective median florescence intensity (MFI) in subsets, as indicated. Two-tailed Wilcoxon tests. ∗p < 0.05, ∗∗p < 0.01. See also Figure S1.
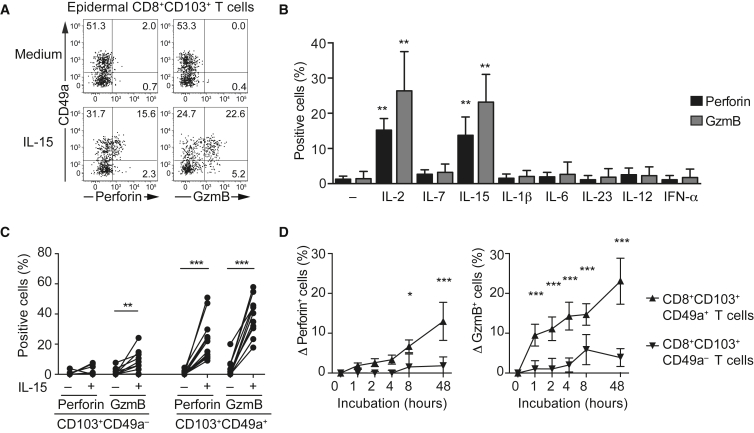
IL-2 and IL-15 Induce Cytotoxic Effector Protein Expression in Epidermal CD8+CD103+CD49a+ Trm Cells
Despite enrichment of transcripts for cytotoxic granule constituents in epidermal CD8+CD103+CD49a+ Trm cells, perforin, and granzyme B protein expression was virtually undetectable when examined ex vivo (Figures 3A and S2A and S2B). We reasoned that in situ inflammatory stimuli might be required to elicit expression of cytotoxic granule constituents in Trm cell subsets, as recently proposed for IL-15 (Jabri and Abadie, 2015). Following incubation of epidermal cell suspensions with a variety of proinflammatory cytokines relevant to skin inflammation, intracellular protein expression of perforin and granzyme B was detected in CD8+CD103+CD49a+ Trm cells following IL-2 and IL-15 stimulation (Figures 3A–3C). Granzyme B expression was upregulated as early as 1 hr after IL-15 stimulation, whereas perforin expression was detected at later time points (Figure 3D). Antibody-mediated blockade of MHC class I did not diminish IL-15-mediated granzyme B upregulation (data not shown) and other inflammatory cytokines, including IL-1β, IL-6, IL-7, IL-12, IL-23, and IFN-α, did not elicit expression of cytotoxic granule constituents (Figure 3G). In an inflammatory milieu, several cytokines might interact for induction of cytotoxic granule constituent expression. Accordingly, IL-15-dependent expression of perforin and granzyme B was augmented by IL-6, but not other cytokine combinations tested (Figures S2C–S2E). Differential expression of IL-2 and IL-15 receptor components could have explained the preferential responses of epidermal CD8+CD103+CD49a+ Trm cells. However, the IL-2 receptor β and γ chains were expressed in both epidermal subsets (Figures S2F and S2G). Thus, epidermal CD8+CD103+CD49a+ Trm cells appeared quiescent in healthy skin, but IL-2 from activated immune cells or IL-15 from keratinocytes (Loser et al., 2004) might prime their cytotoxic potential.
Figure 3.
IL-2 and IL-15 Induce Perforin and Granzyme Expression Specifically in Epidermal CD8+CD103+CD49a+ T Cells
(A) Representative FACS plot of perforin and granzyme B expression in relation to CD49a expression in epidermal CD8+CD103+ Trm cells with or without IL-15 (20 ng/mL) stimulation for 48 hr.
(B) Perforin (black) and granzyme B (gray) expressing cells among epidermal CD8+CD103+ Trm cells following 48 hr incubation with medium, IL-2 (20 ng/ml), IL-7 (20 ng/ml), IL-15 (20 ng/ml), IL-1β (20 ng/ml), IL-6 (20 ng/ml), IL-23 (20 ng/ml), IL-12, (50 ng/ml), IFN-α (2,000 U/ml). Mean ± SD is depicted. Kruskal-Wallis with Dunn’s multiple comparison tests.
(C) Proportion of perforin and granzyme B expressing cells among epidermal CD8+CD103+CD49a– or CD8+CD103+CD49a+ Trm cells upon IL-15 stimulation (n = 12). Two-tailed Wilcoxon test.
(D) CD8+CD103+ Trm cells were sorted and stimulated with IL-15 (20 ng/ml) for 1, 2, 4, 8, or 48 hr. Mean ± SD of the increase (Δ) in perforin (left) and granzyme B (right) expressing cells among CD8+CD103+CD49a– (inverted triangle) or CD8+CD103+CD49a+ (upright triangle) Trm cells as compared to unstimulated (n = 4). Two-tailed paired t test with Holm-Sidak correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S2.
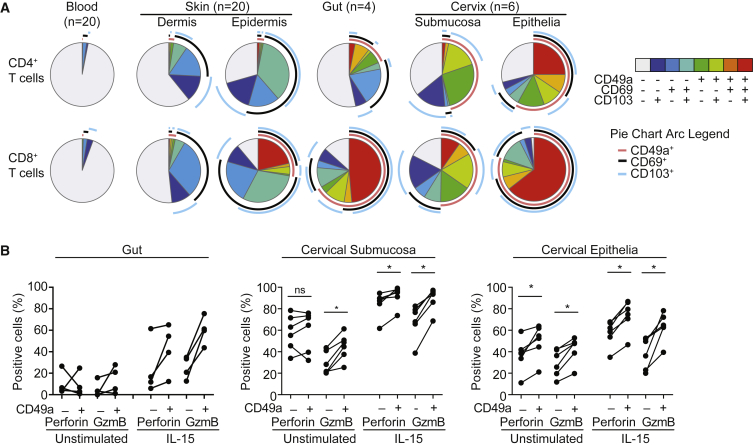
CD49a Expression Broadly Identifies Trm Cells with Cytotoxic Potential in Mucosa
Besides the skin, Trm cells are widely distributed in other barrier tissues. To determine whether CD49a expression more broadly defined Trm cells with a cytotoxic potential, we examined two mucosal barriers, gut and cervix (Figure S3A). CD8+CD103+CD69+CD49a+ Trm cells were abundant in these tissues, with an overall higher frequency of CD49a+ T cells (Figure 4A). Unlike skin, substantial proportions of CD8+CD49a+ T cells lacking CD69 or CD103 expression were present in both gut and cervix and a population of CD4+CD103+CD69+CD49a+ Trm cells existed in cervical epithelium (Figure 4A). With respect to expression of cytotoxic granule constituents, gut, and particularly cervical Trm cells displayed higher basal expression of perforin and granzyme B relative to skin (Figures 3A–3C, 4B and S3B). Perforin and granzyme B were more highly expressed in CD8+CD103+CD69+CD49a+ compared to CD8+CD103+CD69+CD49a– gut and cervical Trm cells, being further augmented by IL-15 stimulation (Figure 4B). These results indicate that CD49a expression defines Trm cell subsets with cytotoxic potential in a variety of epithelial tissues.
Figure 4.
CD49a Expression Identifies Trm Cells with Cytotoxic Potential in Diverse Epithelial Tissues
(A) SPICE charts of CD103, CD69, and CD49a expression in CD4+ (upper) and CD8+ (lower) T cells from peripheral blood, skin dermis and epidermis, full thickness gut (ileum), and ectocervical submucosa and epithelia.
(B) Frequency of perforin and granzyme B-expressing cells epidermal Trm cells from gut, cervical submucosa and epithelium, as indicated, in the absence or presence of IL-15 stimulation (20 ng/ml) for 48 hr. Two-tailed Wilcoxon tests were used to test for statistical significance, ∗p < 0.05. See also Figure S3.
IL-15 Activated Epidermal CD8+CD103+CD49a+ Trm Cells Are Cytotoxic
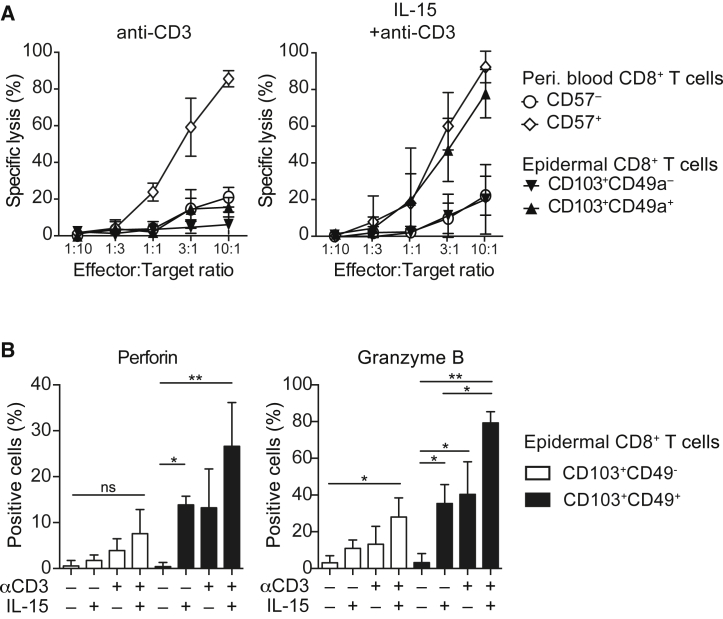
Having found that CD49a generally identifies Trm cells with cytotoxic potential and having elucidated inflammatory signals for their priming, we set out to determine their cytotoxic capacity. Sorted T cell subsets from blood and skin collected from healthy donors (Figure S4) were incubated with 51Cr-labeled target cells in 4 hr redirected antibody-dependent cellular cytotoxicity assays. The cytotoxic activity of epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cell subsets was compared to that of peripheral blood CD8+CD57+ T cells, a highly cytotoxic effector memory subset (Chiang et al., 2013), as well as less cytotoxic CD8+CD57– T cells, constituting naive and memory subsets. CD8+CD57+ T cells displayed strong cytotoxic activity, whereas skin-derived CD8+CD103+CD49a+ Trm cells mediated very poor activity following anti-CD3 antibody stimulation (Figure 5A). However, following 48 hr of IL-15 pre-stimulation, epidermal CD8+CD103+CD49a+ Trm cells manifested cytotoxic activity comparable to that of peripheral blood CD8+CD57+ T cells, whereas CD8+CD103+CD49a– Trm cells and CD8+CD57– T cells remained poor cytotoxic effectors (Figure 5A). Stimulation of epidermal T cells with anti-CD3 antibodies for 24 hr induced similar levels of CD8+CD103+CD49a+ Trm cell granzyme B and perforin expression as IL-15, whereas these stimulations in combination displayed an additive effect (Figure 5B). In summary, skin-derived CD8+CD103+CD49a+ Trm cells lack perforin expression, with short-term TCR stimulation alone being insufficient to induce cytotoxicity. Rather, IL-15-stimulation prompted strong TCR-dependent cellular cytotoxicity by CD8+CD103+CD49a+ Trm cells.
Figure 5.
IL-15 Stimulation Induces Strong Cytotoxic Function by CD8+CD103+CD49a+ Trm Cells
(A) Specific lysis by sorted effector cells of P815 target cells in the presence of anti-CD3 antibody. Unstimulated (n = 3, left) or IL-15 stimulated (n = 6, right) blood derived CD57+CD8+ or CD57−CD8+ T cells, epidermal CD8+CD103+CD49a+, or CD8+CD103+CD49a– Trm cells at effector to target ratios as displayed. Mean ± SD depicted.
(B) Perforin (left) and granzyme B (right) expressing Trm cells following stimulation with IL-15 (20 ng/ml), anti-CD3 (1 μg/ml), or IL-15 + anti-CD3 in the presence of P815 cells (n = 4). Mean ± SD depicted. RM-ANOVA test with Holm-Sidak’s multiple comparisons test was employed for statistical significance. ∗p < 0.05, ∗∗p < 0.01. See also Figure S4.
CD8+CD103+CD49a– Trm Cells Respond to Activation with IL-17 Whereas CD8+CD103+CD49a+ Trm Cells Excel at Production of IFN-γ
T cell-derived cytokines are crucial for defense against invading pathogens at barrier sites. Generally, IFN-γ contributes to immunity toward intracellular infections while IL-17 provides anti-fungal defense and both of these cytokines initiate inflammatory keratinocyte responses. Prompted by differences in transcription of IFNG as well as RORC, a key transcriptional regulator of IL-17 production, we determined whether CD49a expression also defined epidermal skin Trm cell subsets in regards to cytokine production. Corroborating transcriptional profiles, CD8+CD103+CD49a– Trm cells produced IL-17 while CD8+CD103+CD49a+ Trm cells excelled in IFN-γ production upon stimulation with phorbol 12-myristate 13-acetate and ionomycin (Figures 6A–6C). Relative to the epidermal CD8+CD103+CD49a– Trm cells, dermal counterparts produced 3.5-fold less IL-17. TNF and IL-2 were abundantly produced by dermal and epidermal Trm cell subsets (Figures 6B and 6C). TCR engagement using anti-CD3 antibodies also preferentially induced IFN-γ by epidermal CD8+CD103+CD49a+ Trm cells (Figure 6D). Moreover, IL-15 stimulation potentiated TCR-dependent expression of IL-17 and IFN-γ by epidermal CD8+CD103+CD49a– and IFN-γ by CD8+CD103+CD49a+ Trm cells, respectively (Figure 6D), substantiating effectual γ chain receptor signaling in both subsets. Coating of wells with collagen IV in addition to anti-CD3 antibodies specifically augmented production of IFN-γ by epidermal CD8+CD103+CD49a+ Trm cells (Figure 6E), indicating VLA-1-mediated regulation of cytokine production. Unconventional mucosal-associated invariant T (MAIT) cells represent a fraction of peripheral blood IL-17-producing T cells (Dusseaux et al., 2011). Although up to 30% of skin CD8+ T cells expressed CD161, CD8+TCR-Vα7.2+CD161+ MAIT cells comprised less than 25% of dermal or epidermal skin CD8+ Trm cell populations (Figures S5A and S5B) and made up less than 25% of the IL-17-producing CD8+CD103+ T cells (Figures S5C and S5D). Thus, CD49a expression delineated a dichotomy in Trm cell cytokine production, augmented by IL-15, with CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells preferentially producing IL-17 and IFN-γ, respectively.
Figure 6.
Specialization with Respect to IL-17 and IFN-γ Production between Epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm Cells
(A) Representative FACS plots of IFN-γ, IL-17A, and CD49a expression in epidermal CD8+CD103+ T cells.
(B) SPICE charts depict expression of IFN-γ, IL-17A, TNF, and IL-2 in PMA plus ionomycin stimulated blood-derived total CD8+ T cells, dermal CD8+CD103– and CD8+CD103+ T cells, as well as epidermal CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells (n = 6).
(C) Proportion of IFN-γ, IL-17A, IL-2, or TNF expressing cells among indicated T cell subpopulations (n = 6). ANOVA test with Holm-Sidak’s multiple comparisons. Mean ± SD depicted.
(D) Proportion of IFN-γ- or IL-17A-expressing CD8+CD103+CD49a– or CD8+CD103+CD49a+ Trm cells, as indicated, following sorting of epidermal explanted CD8+CD103+ Trm cells and combinations of stimulation with anti-CD3 antibody and IL-15 (20 ng/mL).
(E) Sorted CD8+CD103+ Trm cells were seeded in non-coated or collagen IV-coated wells and cultured in the presence or absence of IL-15 (20 ng/mL) for 20–24 hr before anti-CD3 stimulation. Bar charts show the frequency of IFN-γ- and IL-17A-expressing cells among CD8+CD103+CD49a– or CD8+CD103+CD49a+ T cells (n = 5), as indicated. Mean ± SD depicted. ANOVA test with Holm-Sidak’s multiple comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S5.
Functional Specialization According to CD49 Expression Is Preserved in Distinct Focal Skin Diseases
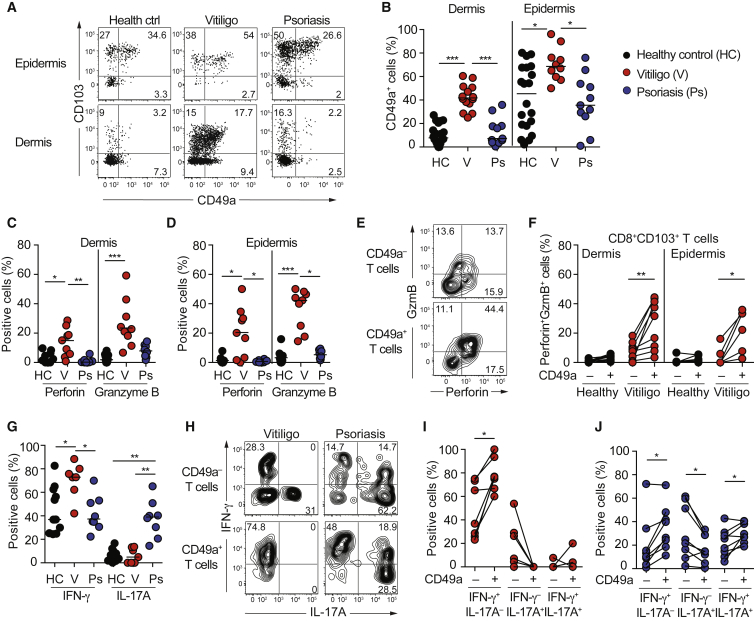
A population of epithelial Trm cells that upon controlled activation perform cytotoxic killing and IFN-γ production would be perfectly placed to safeguard against infections and malignant transformation in situ. Hypothetically, such specialized Trm cells might also participate in focal immune pathology in vitiligo (Figure S6A), a disease where cytotoxic T cells have been implicated in eradication of melanocytes. The proportion of epidermal CD8+CD103+CD49a+ Trm cells was increased in lesional vitiligo but not in psoriasis (Figures 7A and 7B). CD49a was expressed on almost half of dermal CD8+CD103+ Trm cells in lesional vitiligo (Figures 7A and 7B), adjacent to collagen IV-expressing vessels (Figure S6B), while healthy skin, non-lesional vitiligo and psoriasis was devoid of this population (Figures 7A and 7B, S6C and S6D). In contrast, CD4+CD103+ T cells in lesional vitiligo did not express CD49a (Figure S6E). In vitiligo, a substantial proportion of lesional CD8+CD103+CD49a+ Trm cells recognized melanocyte-derived antigens (Figures S6F and S6G). Perforin and granzyme B-expressing Trm cells were detected in vitiligo, but not in healthy skin or psoriasis (Figures 7C and 7D). Moreover, CD8+CD103+CD49a+ Trm cells co-expressed perforin and granzyme B in lesional vitiligo ex vivo (Figures 7E and 7F). In line with increased CD49a frequencies, IFN-γ producing Trm cells were enriched in vitiligo lesions (Figure 7G). Conversely, IL-17-producing Trm cells were enriched in psoriasis plaques (Figure 7G). As in skin from healthy volunteers, CD8+CD103+CD49a+ Trm cells thus preferentially expressed IFN-γ and CD8+CD103+CD49a– Trm cells IL-17 in both vitiligo and psoriasis (Figures 7H–7J). Similar to healthy skin and in agreement with a previous study (Teunissen et al., 2014), less than 5% of skin IL-17 producing cells were CD161highVα7.2+ MAIT cells (Figures S6H–S6J). Trm cells with the capacity to co-express IFN-γ and IL-17 were present in psoriasis plaques (Figure 7H and 7J), indicating a degree of functional plasticity in the context of chronic inflammation. However, constrained plasticity of healthy skin-derived Trm cells was indicated, as sorted CD8+CD103+CD49a+ Trm cells from healthy skin failed to produce IL-17 following 72 hr of stimulation with IL-1, IL-6, and IL-23 (Figures S6L and S6M). In resolved psoriasis, CD8+ Trm cells poised for IL-17 production accumulate (Cheuk et al., 2014), and IL-17 expression was highly enriched within the CD8+CD103+CD49a– Trm cell subset (Figure S6N). Thus, functional dichotomies associated with CD49a expression were generally preserved in disease-associated Trm cells arguing that CD49a marks subsets of Trm cells with imprinted effector profiles and distinct functions in inflammatory skin diseases.
Figure 7.
Functional Dichotomy of CD8+CD103+CD49a+ and CD8+CD103+CD49a– Trm Cells Is Preserved in Vitiligo and Psoriasis
(A) Representative FACS plots show CD103 and CD49a expression on dermal and epidermal CD8+ T cells from healthy skin, vitiligo, and psoriasis lesions.
(B) Frequency of CD49a+ cells among dermal or epidermal CD8+CD103+ Trm cells in skin from healthy donors (HC, n = 20), vitiligo (V, n = 13), or psoriasis lesions (Ps, n = 11).
(C and D) Proportion of perforin or granzyme B expressing cells among (C) dermal or (D) epidermal CD8+CD103+ Trm cells from healthy skin (n = 13), vitiligo (n = 9), or psoriasis lesions (n = 8).
(E) Representative contour plots of dermal CD8+CD103+CD49a− (upper) or CD8+CD103+CD49a+ (lower) Trm cells in vitiligo lesions.
(F) Proportion of perforin and granzyme B co-expressing cells among Trm cells from healthy skin or vitiligo lesions, as indicated.
(G) Proportion of IFN-γ or IL-17A expressing cells among PMA/ionomycin stimulated epidermal CD8+CD103+ T cells from healthy skin (n = 12), vitiligo (n = 7), or psoriasis lesions (n = 8).
(H) Representative contour plots showing IFN-γ/IL-17A expression in PMA/ionomycin stimulated epidermal CD8+CD103+CD49a− (upper) and CD8+CD103+CD49a+ T cells (lower) from vitiligo (left) and psoriasis (right) lesions.
(I and J) Proportion of IFN-γ and IL-17A expressing epidermal CD8+CD103+ T cells from (I) vitiligo or (J) psoriasis lesions. Two-tailed Wilcoxon tests in (G, J, and K) and Kruskal-Wallis tests with Dunn’s multiple comparison tests in (B–E and H). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.
Discussion
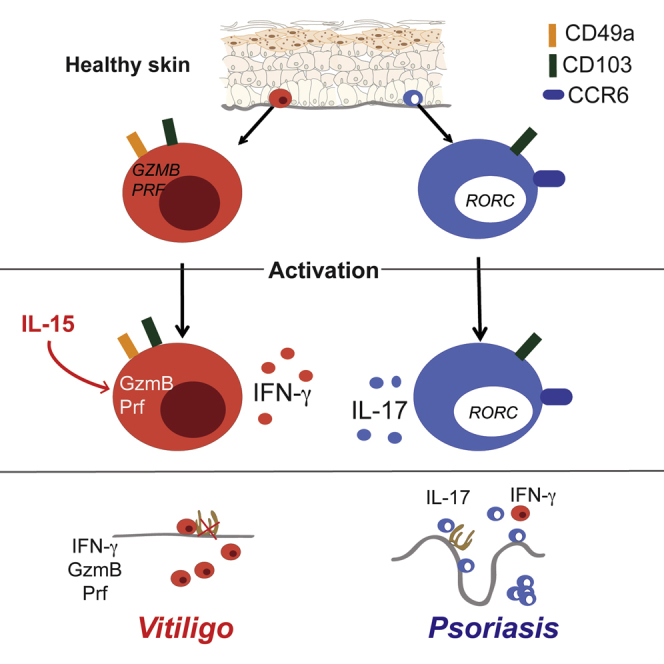
At epithelial boundaries, Trm cells differentiate and form a first line of adaptive defense against multiple pathogens, tailored to effectively control recurrent infections. Consequently, it would be expected that distinct subsets of Trm cells respond to activation with heterogeneous cytokine responses (Naik et al., 2015, Sanchez Rodriguez et al., 2014, Schlapbach et al., 2014, Watanabe et al., 2015) Despite their protective role in recurrent viral infections, direct proof of Trm cell-mediated cytotoxicity is lacking (Mueller and Mackay, 2016). In addition to anti-microbial defense, pathogenic Trm cells are implicated in several inflammatory diseases (Clark, 2015). Here, we identify CD49a expression as a marker delineating a subpopulation of CD8+ Trm cells in human skin that specifically localize to the basal layer of epidermis, preferentially produce IFN-γ, and display high cytotoxic capacity upon stimulation. We also find a high proportion of CD49a+ Trm cells poised for cellular cytotoxicity in other epithelial tissues. Conversely, CD49a– Trm cells excelled in IL-17 production. Lastly, we determined that this functional dichotomy among Trm cell subsets was preserved in the inflammatory skin diseases psoriasis or vitiligo.
In healthy skin, we found CD49a+ Trm cells confined to basal epidermis in contact with collagen IV expressed in the basement membrane. Mouse epidermis displays analogous spatial restrictions to the number of Trm cells during homeostasis (Zaid et al., 2014). Interactions with collagen IV might anchor CD49a+ Trm cells in the epidermis. In murine models, protective CD8+CD49a+ Trm cells develop in barrier tissues (Gebhardt et al., 2009, Ray et al., 2004, Zhang and Bevan, 2013) following viral infections. In line with our study of human skin, granzyme B-expressing Trm cells are retained in mouse epithelium following epidermotropic viral infections (Gebhardt et al., 2009, Schenkel et al., 2014). Epithelial tissues represent first sites of viral entry and with high cellular turnover of keratinocytes, offer an attractive niche for viral propagation and shedding. Thus, by expressing CD49a, Trm cells seem well-positioned for immunosurveillance of infected epithelial cells via production of IFN-γ as well as cellular cytotoxicity. Collagen IV-mediated engagement of CD49a enhanced IFN-γ production by CD8+CD103+CD49a+ Trm cells, possibly through stabilizing IFNG transcripts (Wang et al., 2006). Similar to mouse CD8+ Trm cells in the context of viral infection (Mackay et al., 2013, Wakim et al., 2013), the transcriptional profiles of human CD8+CD103+CD49a+ Trm cells suggested that anti-viral defense and target cell killing represent key functions of this subset. Such cells might also contribute to local surveillance and defense against malignancy (Jameson et al., 2002).
Freshly isolated human skin CD8+CD103+CD49a+ Trm cells displayed a transcriptional profile indicative of cytotoxic function, but did not express key mediators of cellular cytotoxicity. Rather, their cytotoxic capacity was primed through IL-2 and IL-15-mediated induction of perforin and granzyme B expression. IL-15 is essential for Trm development (Mackay et al., 2013) and entry into the epidermis (Adachi et al., 2015). Here we find that IL-15 additionally acts both to potentiate cytokine responses and as a key mediator in cytotoxic licensing of CD8+CD103+CD49a+ Trm cell. Our results suggest an important role for bystander keratinocytes or T cells in activation of Trm cell-mediated effector functions within the skin. These observations validate the hypothesis that IL-15 might generally act as a central danger signal regulating Trm cell responses (Jabri and Abadie, 2015) and extend knowledge by providing a cellular marker of specific Trm cell subsets capable of mediating cellular cytotoxicity. In the gut, IL-15 is implicated in driving cytotoxic lymphocyte responses that turn pathological in celiac disease (Meresse et al., 2004). In our analyses, CD49a was abundantly expressed on CD8+ Trm cells in gut and cervix, representing mucosal barrier tissues. In these tissues, a greater proportion of Trm cells expressed CD49a than in skin and displayed a more activated phenotype, with constitutive expression of perforin and granzyme B. It is possible that mucosa is a milieu more commonly challenged by pathogens, requiring activated, cytotoxic CD49a+ Trm cells that provide continuous surveillance. Nonetheless, IL-15-mediated potentiation of CD49a+ Trm cells might represent a mechanism to safeguard tissues against immunopathology.
Revealing functional specialization among epidermal Trm cells with respect to CD49a expression, CD8+CD103+CD49a– Trm cells preferentially produced IL-17, a cytokine required for control of bacterial and fungal infections. Epidermal CD8+CD103+CD49a– Trm cells excelled in IL-17 production relative to dermal counterparts as well as dermal CD8+CD103– T cells. This observation, combined with different gene-expression profiles, indicates distinct subsets of CD8+CD103+CD49a– Trm in dermis versus epidermis. To explore potential origins of epidermal CD8+CD103+CD49a+ Trm cells, we sorted dermal CD8+CD103– Trm cells and stimulated them with IL-15 and TGF-β, cytokines implicated in Trm cell differentiation (Mackay et al., 2013, Watanabe et al., 2015). A fraction of dermal CD8+CD103– Trm cells upregulated both CD103 and CD49a following stimulation (S.C. and L.E., unpublished observations), indicating that inflammatory responses might differentiate dermal T cells to functionally distinct Trm cells embedded in epidermis. Animal models provide further opportunities to dissect the molecular requirements for induction of CD49a and determine the role of CD49a in ensuring effective Trm cell-mediated immunity.
The patchy appearance and fixed recurrence of several inflammatory skin diseases implies the activity of Trm cells. In vitiligo, activated CD8+CD103+CD49a+ Trm cells expressing cytotoxic granule constituents accumulated and penetrated into the dermis. There, they are well-positioned to eradicate repopulating melanocytes. In contrast, in active psoriasis, with up to 100-fold more Trm cells within the hypertrophic epidermis compared to healthy skin (Cheuk et al., 2014), IL-17-producing Trm cells were profoundly expanded. Moreover, IL-17 or IFN-γ production by distinct Trm cells subsets was generally maintained even in the context of the vigorous tissue inflammation. Co-expression of IL-17 and IFN-γ was detected in some CD8+CD103+CD49a– and CD8+CD103+CD49a+ Trm cells. However, we could not induce IL-17 production by CD8+CD103+CD49a+ Trm cells sorted from healthy skin following IL-23, IL-6, and IL-1β stimulation. Thus, we speculate that during differentiation Trm cells require TCR engagement in addition to IL-17 polarizing inflammatory cues to display the level of functional plasticity we observe in psoriasis. In resolved psoriasis where numbers of Trm cells are similar to healthy skin, we previously described enrichment of CD8+CD103+CCR6+IL-23R+ Trm cells poised to IL-17 production (Cheuk et al., 2014). Congruently, preferential expression of IL-17 by CD8+CD103+CD49a– Trm cells was apparent. In contrast, IFN-γ-producing CD8+CD103+CD49a+ Trm cells were enriched in vitiligo, further supporting the notion of a dichotomy of Trm cell subsets according to CD49a expression even in the setting of skin inflammation and immunopathology (Figure S6O).
Given the role of IL-2 and IL-15 in potentiating both IL-17 and cytotoxic effector functions in Trm cells, blockade of the downstream JAK/STAT pathway represents a promising therapeutic target for skin diseases putatively caused by aberrant Trm cell activation. Interestingly, a case report suggests clinical efficacy of toficitinib, a small molecule inhibitor of JAK1 and JAK3, on vitiligo (Craiglow and King, 2015). Systemic administration of IL-15Rβ blocking antibodies prevented hair loss in a mouse model of alopecia areata (Xing et al., 2014), a disease characterized by aberrant killing of hair follicles. Most patients present with limited burden of disease that cause considerable suffering, yet do not require systemic treatment. Future development of topical treatments aiming at controlling pathogenic Trm cells in situ would offer an attractive therapeutic strategy, with our data highlighting γ chain signaling an attractive target.
In summary, our data reveal that human skin contains different subset of epidermal Trm cells poised toward cytotoxicity and IFN-γ or IL-17 production, respectively. Inflammatory cytokines unleashed TCR engagement-dependent cellular cytotoxicity by CD49a+ Trm cells from healthy skin, whereas activated, perforin- and granzyme-expressing CD49a+ Trm cells accumulated in vitiligo lesions. Conversely, CD49a– Trm cells accounted for the augmented IL-17 production in lesional psoriasis. Further insights into the dynamics of the composition, retention, and activation of expanded, functionally specialized Trm cell populations might contribute to improved management of both infections and chronic inflammatory skin diseases.
Experimental Procedures
Tissue Samples
Human peripheral blood and healthy skin samples from trunk were obtained from reconstructive skin surgery at AdVita Clinic, Stockholm, Sweden, and Karolinska University Hospital, Solna. Cervical tissue samples were obtained from non-malignant and non-inflammatory surgical specimens, St. Göran Hospital, Stockholm, Sweden. Gut biopsies were obtained from the ileum of healthy patients with hereditary predisposition for colorectal cancer (Lynch syndrome) at Karolinska University Hospital, Huddinge. Patients with non-segmental vitiligo, or plaque psoriasis were collected at the Swedish Psoriasis Association Clinic or the dermatology clinic at Karolinska University Hospital, Solna (Table S3). Resolved psoriasis was collected as previously described (Cheuk et al., 2014). Lesional vitiligo biopsies were sampled from within 1 cm to and non-lesional at least 10 cm away from lesional borders. All tissue samples were collected according to the Declaration of Helsinki with signed consent. Ethical permits 2012/50-31/2, 2015/0041-31, 2015/933-32, 2012/1900-31/1, 2013/1800-32, 2015/1078-32, regional ethical committee of Stockholm.
Confocal Microscopy
Skin biopsies were cryopreserved at −80°C. 8–10 μm thick sections were stained as previously described (Cheuk et al., 2014) with primary antibodies to collagen IV (clone COL-94, Abcam), MelanA (clone EP1422Y, Abcam), keratin 5/6 (cloneD516, DAKO), CD8 (AB4055, AbCAM), CD49a (clone 550594, BD Bioscience), and CD3 (clone CD3-12, Abcam). Images were acquired by Zeiss LSM700 and LSM800 (Zeiss) and analyzed with Fiji-ImageJ.
Cell Suspension and Culture
Whole-skin punch biopsies and cervix samples were incubated in 5U dispase (Life Technologies) overnight at 4°C followed by manual separation of epidermis and cervical epithelium from dermis or cervical submuocsa respectively followed by 90 min incubation in collagenase III (3 mg/ml; Worthington) with DNase (5 μg/ml; Roche) in RPMI 1640. Epidermal cell suspension was prepared by repeated pipetting. Dermis and submucosa were further processed by Medicon tissue disruptor (BD Biosciences) as previously described (Cheuk et al., 2014). Ileum biopsies were digested in collagenase II (0.25 mg/ml; Sigma-Aldrich) with DNase (0.2 mg/ml; Roche) in IMDM (Life Technologies) for 30–45 min. Complete RPMI medium was added and the cell suspension were subsequently passed through a 40 μm (gut) / 70 μm (skin or cervix) cell strainer (BD Bioscience). Peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll (GE Healthcare) density separation.
P815 cells were purchased from ATCC and maintained in complete medium (RPMI 1640 supplemented with 10% fetal bovine serum [FBS], L-glutamine; all Hyclone). Recombinant IL-15, IL-1β, IL-2, IL-6, IL-7, IL-23, IL-12 and IFN-α (all R&D Systems) were stored at −80°C. Human collagen IV, phorbol 12-myristate 13-acetate (PMA), and ionomycin were purchased from Sigma.
Flow Cytometry and Cell Sorting of Tissue-Derived Cells
Fluorochrome-conjugated antibodies are specified in the Table S4. Freshly isolated cell suspensions were stained and kept on ice before sorting. Sorting was performed within 2-4 hr using MoFlo XDP (Beckman Coulter) cell sorter or BD FACZJAZZ (BD). The purity of sorted cells was at least 90%. Sorted cells were maintained in complete medium overnight before further experiment. Details on flow cytometry and sorting experiments, as well as functional evaluations are provided in the Supplemental Experimental Procedures.
DNA Extraction and TCR Sequencing
DNA was extracted (Puregene, QIAGEN) and the TCR-β CDR3 regions were sequenced and mapped (ImmunoSEQ, Adaptive Biotech). Data from productive reads were extracted from ImmunoSEQ platform for further analysis. Raw data were uploaded on the immuneACCESS platform provided by AdaptiveBiotech (doi.org/10.21417/B76K56). Details on TCR clonality analysis are provided in the Supplemental Procedures.
RNA Extraction and Transcriptome Analyses
RNA was purified from cell-qiazol lysate (miRNeasy, QIAGEN) and quality was checked (Bioanalyzer RNA 6000, Agilent) before library construction (SMART-Seq, ClonTech) and sequencing (Illumina HiSeqTM 2000). Data were analyzed as detailed in the Supplemental Experimental Procedures.
Cytotoxicity Assay
The cytotoxicity assay was performed and analyzed as described previously (Chiang et al., 2013). In brief, sorted and rested cell populations were used as effector cells against 51Cr-labeled P815 target cells supplemented with 0.5 μg/ml of anti-CD3 antibody (clone S4.1, Invitrogen) with effector-to-target ratios between 10 and 0.3. Samples were run in duplicates for 4 hr at 37°C. 51Cr release in supernatant was measured on a γ-counter (Wizard2, PerkinElmer) and specific lysis was calculated as previously described (Chiang et al., 2013).
Statistics and Graphics
Statistical analysis and graphical illustration of numerical data was performed by using either PRISM (v6, GraphPad), JMP 13 or RStudio. SPICE charts were generated by SPICE (https://exon.niaid.nih.gov/spice/). Heatmaps were generated by MeV: MultiExperiment Viewer (http://mev.tm4.org/).
Author Contributions
Designed study: S.C., Y.T.B., and L.E. Performed experiments: S.C., H.S., I.G.S., S.C.C., E.M., E.D., N.M., A.G., M.F., A.I., and L.E. Provided reagents and clinical material: M.E., L.B., J. Mjösberg, J. Michaëlsson, A.T., C.H., M.S., and M.E. Performed analysis: S.C. and L.F., Wrote paper: S.C., Y.T.B., and L.E. L.E. supervised ex vivo preparation and analysis of human skin cells, Y.T.B. supervised cytotoxicity experiments.
Acknowledgments
We would like to thank Emma Wadman and Kerstin Bergh for excellent help in recruitment of patients and technical assistance and Frank Carbone for scientific advice and for critically reading the manuscript. This work was supported in part by funds from Novartis. We acknowledge funding from the Swedish Research Council (Y.T.B. and L.E.), Ragnar Söderberg Stiftelse, Svenska Läkaresällskapet, Stockholm County Council (ALF and Clinical Research Appointment), Psoriasisfonden, Hudfonden (L.E.), European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement no. 311335 (Y.T.B.), Swedish Cancer Foundation (Y.T.B.), Swedish Foundation for Strategic Research (Y.T.B.), and Stockholm City Council and Karolinska Institutet Center for Innovative Medicine (Y.T.B.), and Wallenberg Foundation (KAW for Y.T.B. and MMW for L.E.).
Published: February 14, 2017
Footnotes
Supplemental Information includes six figures, four tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2017.01.009.
Contributor Information
Yenan T. Bryceson, Email: yenan.bryceson@ki.se.
Liv Eidsmo, Email: liv.eidsmo@ki.se.
Accession Numbers
The accession number for RNA-seq data reported in this paper is GEO: GSE83637. The sequencing data for T cell receptor beta CDR3 region is available at immuneACCESS: http://dx.doi.org/10.21417/B76K56.
Supplemental Information
References
- Adachi T., Kobayashi T., Sugihara E., Yamada T., Ikuta K., Pittaluga S., Saya H., Amagai M., Nagao K. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 2015;21:1272–1279. doi: 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti S., Hogenbirk M.A., Dijkgraaf F.E., Visser L.L., Hoekstra M.E., Song J.Y., Jacobs H., Haanen J.B., Schumacher T.N. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346:101–105. doi: 10.1126/science.1254803. [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Segre J.A. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Boehncke W.H., Schön M.P. Psoriasis. Lancet. 2015;386:983–994. doi: 10.1016/S0140-6736(14)61909-7. [DOI] [PubMed] [Google Scholar]
- Cheuk S., Wikén M., Blomqvist L., Nylén S., Talme T., Ståhle M., Eidsmo L. Epidermal Th22 and Tc17 cells form a localized disease memory in clinically healed psoriasis. J. Immunol. 2014;192:3111–3120. doi: 10.4049/jimmunol.1302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S.C., Theorell J., Entesarian M., Meeths M., Mastafa M., Al-Herz W., Frisk P., Gilmour K.C., Ifversen M., Langenskiöld C. Comparison of primary human cytotoxic T-cell and natural killer cell responses reveal similar molecular requirements for lytic granule exocytosis but differences in cytokine production. Blood. 2013;121:1345–1356. doi: 10.1182/blood-2012-07-442558. [DOI] [PubMed] [Google Scholar]
- Clark R.A. Resident memory T cells in human health and disease. Sci. Transl. Med. 2015;7:269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K., Dowgiert R.K., Kupper T.S. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- Craiglow B.G., King B.A. Tofacitinib Citrate for the Treatment of Vitiligo: A Pathogenesis-Directed Therapy. JAMA Dermatol. 2015;151:1110–1112. doi: 10.1001/jamadermatol.2015.1520. [DOI] [PubMed] [Google Scholar]
- Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., Lantz O. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- Ezzedine K., Eleftheriadou V., Whitton M., van Geel N. Vitiligo. Lancet. 2015;386:74–84. doi: 10.1016/S0140-6736(14)60763-7. [DOI] [PubMed] [Google Scholar]
- Gaide O., Emerson R.O., Jiang X., Gulati N., Nizza S., Desmarais C., Robins H., Krueger J.G., Clark R.A., Kupper T.S. Common clonal origin of central and resident memory T cells following skin immunization. Nat. Med. 2015;21:647–653. doi: 10.1038/nm.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T., Wakim L.M., Eidsmo L., Reading P.C., Heath W.R., Carbone F.R. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- Glennie N.D., Yeramilli V.A., Beiting D.P., Volk S.W., Weaver C.T., Scott P. Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J. Exp. Med. 2015;212:1405–1414. doi: 10.1084/jem.20142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden J.L., Hamm D., Gulati N., Lowes M.A., Krueger J.G. Deep Sequencing of the T-cell Receptor Repertoire Demonstrates Polyclonal T-cell Infiltrates in Psoriasis. F1000Res. 2015;4:460. doi: 10.12688/f1000research.6756.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.E., Harris T.H., Weninger W., Wherry E.J., Hunter C.A., Turka L.A. A mouse model of vitiligo with focused epidermal depigmentation requires IFN-γ for autoreactive CD8+ T-cell accumulation in the skin. J. Invest. Dermatol. 2012;132:1869–1876. doi: 10.1038/jid.2011.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondowicz B.D., An D., Schenkel J.M., Kim K.S., Steach H.R., Krishnamurty A.T., Keitany G.J., Garza E.N., Fraser K.A., Moon J.J. Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity. 2016;44:155–166. doi: 10.1016/j.immuni.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber W., Patel D.D., Dryja T., Wright A.M., Koroleva I., Bruin G., Antoni C., Draelos Z., Gold M.H., Durez P., Psoriasis Study Group. Rheumatoid Arthritis Study Group. Uveitis Study Group Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- Jabri B., Abadie V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 2015;15:771–783. doi: 10.1038/nri3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson J., Ugarte K., Chen N., Yachi P., Fuchs E., Boismenu R., Havran W.L. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- Jiang X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., Kupper T.S. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch I.R., Watanabe R., O’Malley J.T., Williamson D.W., Scott L.L., Elco C.P., Teague J.E., Gehad A., Lowry E.L., LeBoeuf N.R. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci. Transl. Med. 2015;7:308ra158. doi: 10.1126/scitranslmed.aaa9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi C.L., Kimball A.B., Papp K.A., Yeilding N., Guzzo C., Wang Y., Li S., Dooley L.T., Gordon K.B., PHOENIX 1 study investigators Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1) Lancet. 2008;371:1665–1674. doi: 10.1016/S0140-6736(08)60725-4. [DOI] [PubMed] [Google Scholar]
- Loser K., Mehling A., Apelt J., Ständer S., Andres P.G., Reinecker H.C., Eing B.R., Skryabin B.V., Varga G., Schwarz T., Beissert S. Enhanced contact hypersensitivity and antiviral immune responses in vivo by keratinocyte-targeted overexpression of IL-15. Eur. J. Immunol. 2004;34:2022–2031. doi: 10.1002/eji.200324785. [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Braun A., Macleod B.L., Collins N., Tebartz C., Bedoui S., Carbone F.R., Gebhardt T. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 2015;194:2059–2063. doi: 10.4049/jimmunol.1402256. [DOI] [PubMed] [Google Scholar]
- Meresse B., Chen Z., Ciszewski C., Tretiakova M., Bhagat G., Krausz T.N., Raulet D.H., Lanier L.L., Groh V., Spies T. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Mueller S.N., Mackay L.K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Wilhelm C., Molloy M.J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.O., Kupper T.S. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 2015;21:688–697. doi: 10.1038/nm.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Haase I., Nestle F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014;14:289–301. doi: 10.1038/nri3646. [DOI] [PubMed] [Google Scholar]
- Purwar R., Campbell J., Murphy G., Richards W.G., Clark R.A., Kupper T.S. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS ONE. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D., Luo S., Wang Y.C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashighi M., Agarwal P., Richmond J.M., Harris T.H., Dresser K., Su M.W., Zhou Y., Deng A., Hunter C.A., Luster A.D., Harris J.E. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci. Transl. Med. 2014;6:223ra23. doi: 10.1126/scitranslmed.3007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S.J., Franki S.N., Pierce R.H., Dimitrova S., Koteliansky V., Sprague A.G., Doherty P.C., de Fougerolles A.R., Topham D.J. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity. 2004;20:167–179. doi: 10.1016/s1074-7613(04)00021-4. [DOI] [PubMed] [Google Scholar]
- Sanchez Rodriguez R., Pauli M.L., Neuhaus I.M., Yu S.S., Arron S.T., Harris H.W., Yang S.H., Anthony B.A., Sverdrup F.M., Krow-Lucal E. Memory regulatory T cells reside in human skin. J. Clin. Invest. 2014;124:1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., Masopust D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98–101. doi: 10.1126/science.1254536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapbach C., Gehad A., Yang C., Watanabe R., Guenova E., Teague J.E., Campbell L., Yawalkar N., Kupper T.S., Clark R.A. Human TH9 cells are skin-tropic and have autocrine and paracrine proinflammatory capacity. Sci. Transl. Med. 2014;6:219ra8. doi: 10.1126/scitranslmed.3007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skon C.N., Lee J.Y., Anderson K.G., Masopust D., Hogquist K.A., Jameson S.C. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen M.B., Yeremenko N.G., Baeten D.L., Chielie S., Spuls P.I., de Rie M.A., Lantz O., Res P.C. The IL-17A-producing CD8+ T-cell population in psoriatic lesional skin comprises mucosa-associated invariant T cells and conventional T cells. J. Invest. Dermatol. 2014;134:2898–2907. doi: 10.1038/jid.2014.261. [DOI] [PubMed] [Google Scholar]
- Thome J.J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., Farber D.L. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boorn J.G., Konijnenberg D., Dellemijn T.A., van der Veen J.P., Bos J.D., Melief C.J., Vyth-Dreese F.A., Luiten R.M. Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J. Invest. Dermatol. 2009;129:2220–2232. doi: 10.1038/jid.2009.32. [DOI] [PubMed] [Google Scholar]
- Wakim L.M., Gupta N., Mintern J.D., Villadangos J.A. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 2013;14:238–245. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- Wang J.G., Collinge M., Ramgolam V., Ayalon O., Fan X.C., Pardi R., Bender J.R. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J. Immunol. 2006;176:2105–2113. doi: 10.4049/jimmunol.176.4.2105. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., Clark R.A. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 2015;7:279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Dai Z., Jabbari A., Cerise J.E., Higgins C.A., Gong W., de Jong A., Harel S., DeStefano G.M., Rothman L. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 2014;20:1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid A., Mackay L.K., Rahimpour A., Braun A., Veldhoen M., Carbone F.R., Manton J.H., Heath W.R., Mueller S.N. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl. Acad. Sci. USA. 2014;111:5307–5312. doi: 10.1073/pnas.1322292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bevan M.J. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.