Summary
Arginase 1 (Arg1) and indoleamine 2,3-dioxygenase 1 (IDO1) are immunoregulatory enzymes catalyzing the degradation of l-arginine and l-tryptophan, respectively, resulting in local amino acid deprivation. In addition, unlike Arg1, IDO1 is also endowed with non-enzymatic signaling activity in dendritic cells (DCs). Despite considerable knowledge of their individual biology, no integrated functions of Arg1 and IDO1 have been reported yet. We found that IDO1 phosphorylation and consequent activation of IDO1 signaling in DCs was strictly dependent on prior expression of Arg1 and Arg1-dependent production of polyamines. Polyamines, either produced by DCs or released by bystander Arg1+ myeloid-derived suppressor cells, conditioned DCs toward an IDO1-dependent, immunosuppressive phenotype via activation of the Src kinase, which has IDO1-phosphorylating activity. Thus our data indicate that Arg1 and IDO1 are linked by an entwined pathway in immunometabolism and that their joint modulation could represent an important target for effective immunotherapy in several disease settings.
Keywords: TGF-β, Arg1, IDO1, dendritic cell, amino acid metabolism, immune regulation, polyamine, ornithine
Highlights
-
•
Dendritic cells (DCs) can co-express Arg1 and IDO1 immunosuppressive enzymes
-
•
Arg1 activity is required for IDO1 induction by TGF-β in DCs
-
•
Spermidine, a downstream Arg1 product, but not arginine starvation, induces IDO1 in DCs
-
•
Arg1+ myeloid derived suppressor cells (MDSCs) can render DCs immunosuppressive via IDO1
Arginase 1 (Arg1) and indoleamine 2,3-dioxygenase 1 (IDO1) are immunosuppressive enzymes known to operate in distinct immune cells. Mondanelli and colleagues demonstrate that Arg1 and IDO1 cooperate in conferring long-term, immunosuppressive effects to dendritic cells.
Introduction
Over the course of evolution, the catabolic pathways of l-tryptophan (Trp) and of l-arginine (Arg) have evolved to be primary regulatory nodes in the control of immune responses (Grohmann and Bronte, 2010, Murray, 2016). Trp, an essential amino acid for mammals, is a substrate for indoleamine 2,3-dioxygenase 1 (IDO1), which catalyzes the first, rate-limiting step in the kynurenine pathway, leading to Trp depletion and the production of a series of immunoregulatory molecules collectively known as kynurenines (Grohmann et al., 2003b, Mellor and Munn, 2004, Puccetti and Grohmann, 2007a). Both effects—namely, Trp starvation and kynurenine production—are involved in the conversion of naive CD4+ T cells into Foxp3+ regulatory T (Treg) cells (Fallarino et al., 2006, Puccetti and Grohmann, 2007a). Moreover, the main IDO1 catalytic product, l-kynurenine (Kyn), has immunoregulatory effects in the absence of Trp starvation, via activation of the Aryl hydrocarbon Receptor (AhR) (Grohmann and Puccetti, 2015, Platten et al., 2012).
High IDO1 expression and catalytic activity occur in dendritic cells (DCs) in response to the cytokine interferon-γ (IFN-γ) (Grohmann et al., 2003b). In DCs stimulated with transforming growth factor β (TGF-β), IDO1 becomes instead phosphorylated in its immune-based inhibitory tyrosine motifs (ITIMs), so to mediate intracellular signaling events in a self-sustaining feedforward loop that leads to durable immunoregulatory effects (Bessede et al., 2014, Pallotta et al., 2014, Pallotta et al., 2011, Volpi et al., 2016). The composite mode of action of IDO1 may well explain its being recognized as an authentic regulator of immunity under several physiopathologic conditions (Orabona and Grohmann, 2011, Puccetti and Grohmann, 2007a).
Arg is a semi-essential amino acid, i.e., it is required by mammals only under special circumstances, such as immune responses. In immune cells, Arg is actively metabolized by arginase 1 (Arg1) or Arg2 to produce urea and l-ornithine (Orn) or it is used for protein biosynthesis (Wu and Morris, 1998). Moreover, nitric oxide synthases use Arg to make nitric oxide—a key anti-microbial gas and signaling molecule—and l-citrulline (Bronte and Zanovello, 2005). Arg consumption by Arg1, rather than Arg2 or nitric oxide synthases, represents a well-known immunoregulatory mechanism exploited by M2 macrophages (Murray, 2016, Sica and Mantovani, 2012), as well as myeloid-derived suppressor cells (MDSCs) in tumoral settings (Gabrilovich and Nagaraj, 2009, Marigo et al., 2008). Conversely, the immunoregulatory role of catalytic products via Arg1 activity (urea and Orn) has been unclear (Grohmann and Bronte, 2010). T helper 2 (Th2) cytokines, such as interleukin-4 (IL-4) and IL-13, represent efficient inducers of Arg1 expression (Gabrilovich and Nagaraj, 2009, Marigo et al., 2008, Sica and Mantovani, 2012), although TGF-β has also been shown to upregulate the enzyme, at least in rat peritoneal macrophages (Boutard et al., 1995). Besides its involvement in mouse dendritic cell (DC) differentiation (Yang et al., 2015), the biology of Arg1 in DCs is still obscure. As a whole, the bulk of the data on Arg1 and IDO1 would suggest that the two amino-acid metabolic enzymes might operate in quite distinct spatial (i.e., cells) and mechanistic modes, namely via either amino acid starvation itself (as is the case for Arg1) or via the combined effects of immunoregulatory Kyn and signaling activity (IDO1). This suggests that the enzymes have separate functions (to possibly cope with distinct environmental needs) and/or have evolved to complement, rather than integrate, each other.
DCs integrate multiple signals and mechanisms in order to promote either adaptive immunity or immune tolerance in response to specific conditions, such as distinct types of cytokinic milieu (Banchereau and Steinman, 1998, Macagno et al., 2007). By using the main cytokines capable of inducing those metabolic enzymes (i.e., IFN-γ, IL-4, and TGF-β), we here investigated whether Arg1 and IDO1 were co-expressed in DCs and what the functional meaning—if any—of their co-expression would be. We found that (1) TGF-β, but neither IL-4, IFN-γ, nor combination thereof, will induce both Arg1 and IDO1 in DCs, with Arg1 being upregulated before IDO1; (2) Arg1 activity is absolutely required for IDO1-dependent signaling events as initiated by TGF-β; (3) a polyamine—spermidine, resulting from Orn decarboxylation—can replace TGF-β in the activation of the Src kinase that phosphorylates IDO1 and trigger immunosuppressive IDO1 signaling; and (4) DCs can be conditioned by Arg1+ MDSCs to express an IDO1-dependent immunosuppressive phenotype.
Results
TGF-β Induces Co-expression of Arg1 and IDO1 in DCs
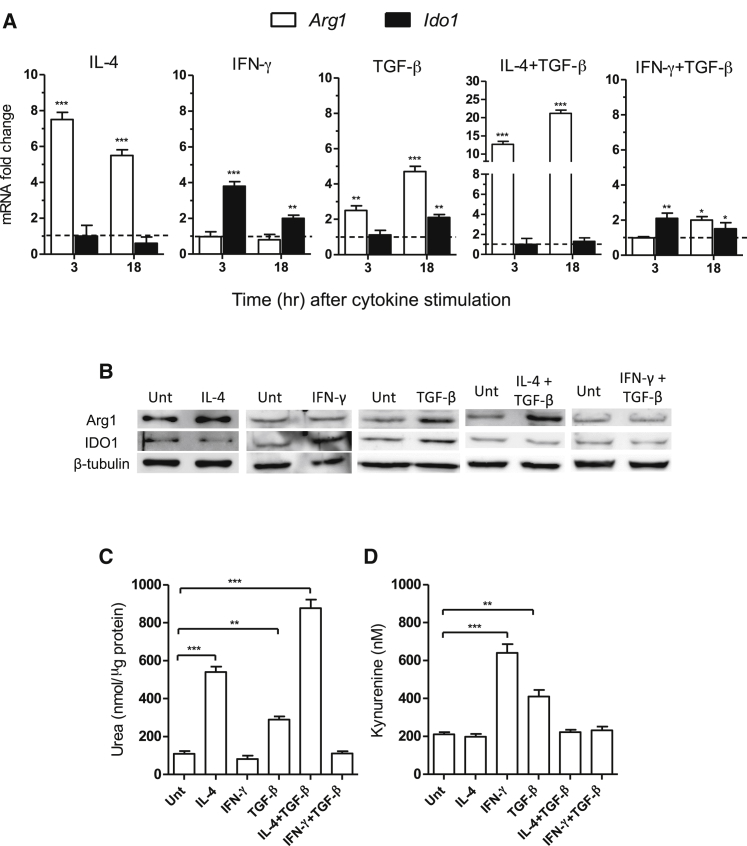
We investigated the expression and catalytic activity of Arg1 and IDO1 in splenic CD11c+ DCs stimulated with IL-4 (i.e., the main Arg1 inducer), IFN-γ, or TGF-β, both of which are IDO1 inducers. Combinations of TGF-β and IL-4 or IFN-γ were also tested. In accordance with previous data (Pallotta et al., 2011), transcript analysis at 3 and 18 hr of cytokine incubation revealed that both IFN-γ and TGF-β upregulated Ido1 expression, with IFN-γ inducing the IDO1-encoding gene to a higher extent and more rapidly than TGF-β (Figure 1A). By contrast, IL-4 did not increase Ido1, but it did induce Arg1 expression (>7-fold increase at 3 hr of stimulation). No modulatory effect could be observed for IFN-γ on Arg1 expression. In contrast, TGF-β upregulated Arg1 in a more intense fashion and with faster kinetics than Ido1. Negligible transcript expression for Arg2 and Nos2 (coding for the inducible nitric oxide synthase) could be observed in either unstimulated or TGF-β-stimulated DCs (data not shown). DC incubation with TGF-β plus IL-4 resulted in a synergistic effect on Arg1 upregulation (>12- and 20-fold increase at 3 and 18 hr, respectively) but in no induction of Ido1. On combining TGF-β with IFN-γ, no incremental effect over that of IFN-γ alone was observed at 3 hr and impaired upregulation of Ido1 expression by TGF-β would instead occur at 18 hr.
Figure 1.
Arg1 and IDO1 Are Co-expressed in DCs Stimulated with TGF-β
(A) Real-time PCR analysis of Arg1 and Ido1 transcripts in DCs stimulated for 3 or 18 hr with IL-4, IFN-γ, TGF-β, IL-4 plus TGF-β, or IFN-γ plus TGF-β normalized to the expression of Gapdh (encoding glyceraldehyde phosphate dehydrogenase) and presented relative to results in untreated cells (dotted line, 1-fold).
(B) Arg1 and IDO1 immunoblot analysis of cell lysates from DCs incubated with IL-4, IFN-γ, TGF-β, IL-4 plus TGF-β, IFN-γ plus TGF-β, or medium alone (untreated, unt) for 24 hr.
(C) Arg1 and (D) IDO1 activity measured in terms of urea in cell lysates and l-kynurenine in cell supernatants, respectively, of DCs incubated as in (B).
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (Student’s t test; cytokine-treated versus untreated samples). All data are from one experiment representative of three (A–D; means ± SD of triplicates in A, C, and D).
We next evaluated whether the differential modulation of Arg1 and IDO1 could also be observed at the level of protein expression and catalytic activity. Immunoblot analyses with anti-Arg1 and anti-IDO1 antibodies on lysates of DCs—subjected to the same treatments as in Figure 1A but for 24 hr—confirmed the real-time PCR experiments, i.e., IL-4 and IFN-γ induce only Arg1 and IDO1, respectively, whereas TGF-β alone, but not in combination with IL-4 or IFN-γ, upregulated protein expression of both enzymes (Figure 1B). Similarly, the enzymic activities of both Arg1 (measured in terms of urea contents in cell lysates; Figure 1C) and IDO1 (in terms of Kyn concentrations in culture supernatants; Figure 1D) were significantly upregulated at 24 hr by TGF-β but not IL-4, IFN-γ, or combinations thereof.
Therefore, these data showed that Arg1 can be expressed in DCs, specifically in response to IL-4 and/or TGF-β, and that Arg1 expression precedes that of IDO1 in DCs exposed to TGF-β alone. Perhaps more importantly, these data unveiled that the Arg and Trp catabolic pathways can be co-activated in DCs.
Arg1 Expression Is Required for IDO1 Induction by TGF-β in DCs
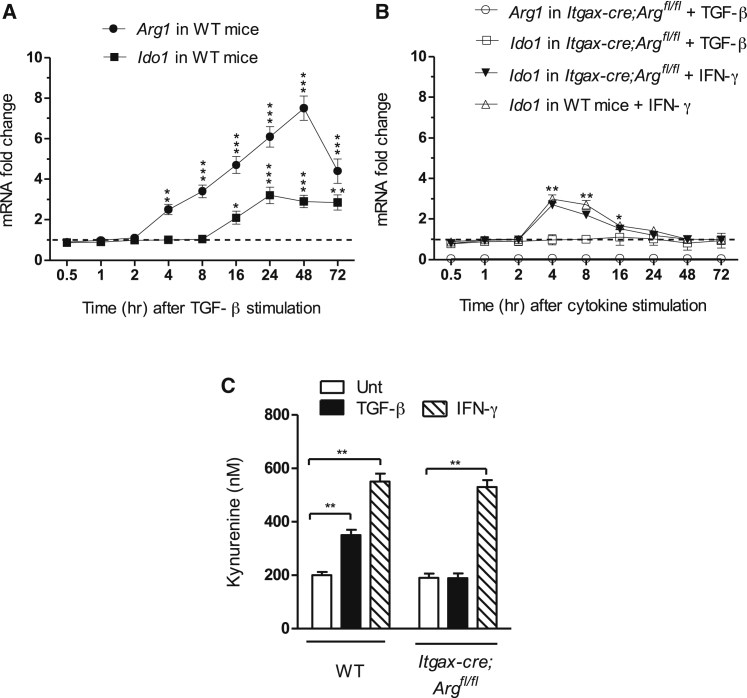
To better appreciate the temporal relationship between Arg1 and IDO1 expressions in TGF-β-stimulated DCs, we performed a more comprehensive kinetic analysis—i.e., from 0.5 to 72 hr of cytokine incubation—of transcripts coding for the two amino-acid catabolic enzymes. The results confirmed the earlier induction of Arg1 (3−4 hr; Figures 1A and 2A) as compared to Ido1 transcripts (16−18 hr; Figures 1A and 2A), as well as a higher extent of Arg1 expression that peaked at 24−48 hr (>7 fold increase as compared to unstimulated cells) of TGF-β stimulation. Ido1 expression did not increase more than 3-fold (Figure 2A). Nevertheless, in agreement with our previous data (Pallotta et al., 2011), the extent of Ido1 expression remained stable throughout the experiment, i.e., up to 72 hr, after which DCs could no longer be analyzed because of poor viability. Conversely, Arg1 transcripts appeared to decrease from 72 hr onward.
Figure 2.
Arg1 Expression Is Necessary for IDO1 Induction by TGF-β in DCs
(A) Kinetic analysis of Arg1 and Ido1 transcripts in DCs from wild-type (WT) mice after incubation with TGF-β for different times (indicated).
(B) Kinetic analysis of Ido1 and Arg1 transcripts in DCs from Itgax-cre;Argfl/fl animals prior to incubation with TGF-β or IFN-γ for different times. DCs from WT animals were used as control for IFN-γ stimulation. In (A) and (B), data are normalized and presented as in Figure 1A.
(C) Kynurenine concentrations in supernatants of DCs from Itgax-cre;Arg1fl/fl and WT animals incubated with TGF-β, IFN-γ, or medium alone for 24 hr.
∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001 (Student’s t test; cytokine-treated versus untreated samples). All data are from one experiment representative of three (means ± SD of triplicates). Please see also Figure S1.
The sequential expressions of Arg1 and Ido1 prompted us to investigate whether Arg1 was required for IDO1 induction by TGF-β in DCs. We examined Ido1 transcript amounts over time using DCs purified from the spleens of Itgax-cre;Arg1fl/fl mice, lacking Arg1 expression in CD11c+ (i.e., DCs) but not in CD11b+ cells (Figure S1). We found that Arg1 deficiency abrogated Ido1 induction by TGF-β but not by IFN-γ in DCs (Figure 2B). Analysis of IDO1 activity (Figure 2C) at 24 hr of TGF-β or IFN-γ stimulation also indicated that lack of Arg1 expression in DCs allowed IFN-γ but not TGF-β to upregulate IDO1. Thus these data identified a cytokine milieu whereby DCs exploit Arg1 expression to increase IDO1.
Orn Upregulates IDO1 in the Absence of Externally Added TGF-β
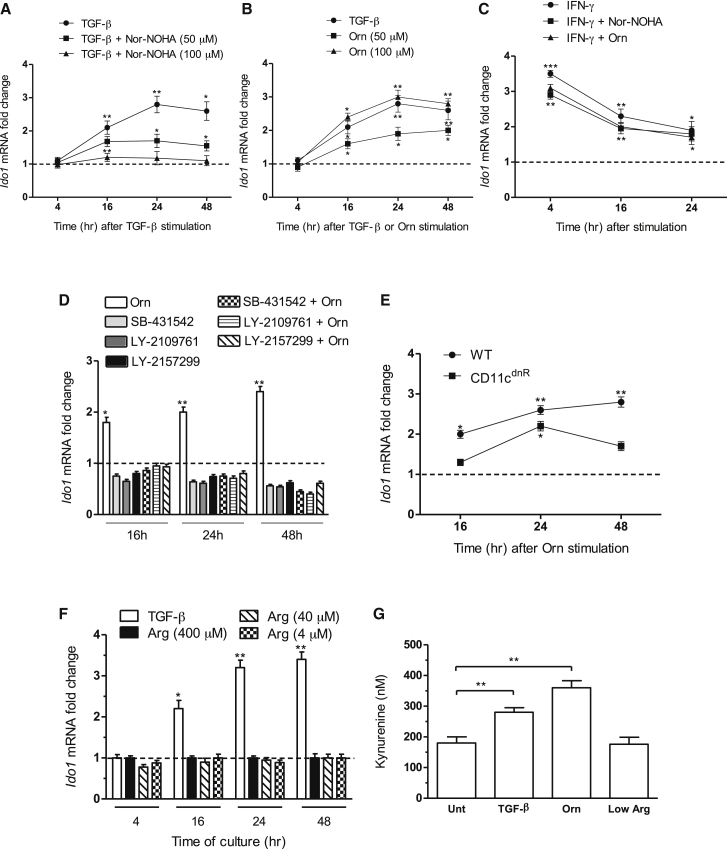
At variance with IDO1, which exerts immunoregulatory additional effects also via signaling activity (Pallotta et al., 2014, Pallotta et al., 2011, Volpi et al., 2016), Arg1-mediated mechanisms have been shown to occur only via Arg catalytic degradation. We thus investigated whether upregulation of Ido1 transcripts in DCs by TGF-β could be modulated by Nω-hydroxy-nor-Arg (nor-NOHA), an inhibitor of arginases. Pre-incubation with 50 μM or 100 μM nor-NOHA for 1 hr prior to the addition of TGF-β significantly impaired the Ido1-inducing ability of the cytokine (Figure 3A), indicating a functional role for Arg1 catalytic activity in IDO1 modulation.
Figure 3.
Orn Induces IDO1 Expression and Activity in DCs
Kinetic analysis of Ido1 transcripts in WT DCs stimulated with TGF-β (A and B), Orn (B and C), and/or IFN-γ (C) for different times (indicated) in the presence or absence of nor-NOHA (100 μM for IFN-γ), or medium with standard (i.e., 400 μM) or low (4–40 μM) Arg concentration (F).
Kinetic analysis of Ido1 transcripts in WT (D, E) or CD11cdnR (E) DCs stimulated with Orn in the presence (D) or absence (E) of TGF-β receptor inhibitors. No toxic effects of inhibitors could be observed in DCs at the used concentration of 10 μM (data not shown).
In (A)–(F), data are normalized and presented as in Figure 1A.
(G) Kynurenine concentrations in supernatants of WT DCs stimulated with TGF-β in a standard medium (where not specified) or a medium with low Arg (4 μM), in the presence or absence of nor-NOHA (100 μM) or Orn (100 μM).
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (stimulated samples versus untreated controls). All data are from one experiment representative of three (means ± SD of triplicates).
We next investigated whether Orn, an Arg1 product, was involved in Ido1 upregulation. Incubation of DCs with Orn at 50 or 100 μM (Figure 3B) significantly increased Ido1 expression, similar to what observed with the cytokine alone (Figure 3B). In contrast, no modulatory effect in Ido1 expression could be observed in DCs stimulated with IFN-γ in the presence of the Arg1 inhibitor or Orn, both at 100 μM (Figure 3C). To evaluate whether autocrine and/or paracrine TGF-β could be involved in the Ido1-upregulating effects of Orn, WT DCs were incubated with the Arg metabolite for different times after a 1 hr pretreatment with an inhibitor of TGF-β receptor signaling, namely SB-431542, LY2109761, or LY2157299 (also known as galunisertib) (Figure 3D). DCs purified from transgenic CD11cdnR mice (expressing a truncated form of TGF-β receptor II subunit in CD11c+ cells) (Laouar et al., 2005) were also assayed (Figure 3E). Both pharmacologic and genetic means of TGF-β signaling inhibition indicated that Orn effects do require autocrine and/or paracrine effects of TGF-β in both early inducing and late maintaining phase of Ido1 expression (see also Supplemental Text) and that the cytokine could be produced constitutively by DCs in basal conditions, in accordance to our previous data (Belladonna et al., 2008).
In contrast to Orn, media deficient in Arg (4 and 40 μM in the place of standard 400 μM; Figure 3F) did not significantly increased Ido1 expression. Moreover, Orn at 100 μM but not the use of an Arg-deficient medium (i.e., Arg at 4 μM) significantly increased IDO1 activity to an extent comparable to that observed with TGF-β (Figure 3G). Therefore, our data indicated that Orn, a main Arg1 product, but not Arg starvation, can condition DCs to upregulate IDO1 expression and activity if autocrine and/or paracrine TGF-β is present.
Orn Confers Immunosuppressive Properties on DCs via IDO1 Signaling
IDO1 immunosuppressive effects include non-enzymic functions, namely intracellular signaling events that, initiated by ITIM phosphorylation in the enzyme, are involved in reprogramming gene expression and in the induction of a stably regulatory phenotype in DCs (Orabona et al., 2012, Pallotta et al., 2011), capable of controlling T cell-mediated autoimmune responses (Pallotta et al., 2014, Volpi et al., 2016). In particular, IDO1’s ITIM phosphorylation is triggered in DCs by TGF-β via a pathway that requires phosphatidylinositide 3-kinase (PI3K) and a tyrosine kinase of the Src family (Bessede et al., 2014, Volpi et al., 2016), which phosphorylates IDO1 ITIMs. In turn, phosphorylated IDO1 ITIMs work as docking sites for tyrosine protein phosphatases, such as SHP-1 and SHP-2, which are concomitantly upregulated by TGF-β (Orabona et al., 2012, Pallotta et al., 2011). These events lead to the activation of an immunoregulatory signaling pathway in DCs that promotes endogenous production of TGF-β and induction of the Ido1 gene, perpetuating IDO1 signaling events and associated immunosuppressive effects over the long term.
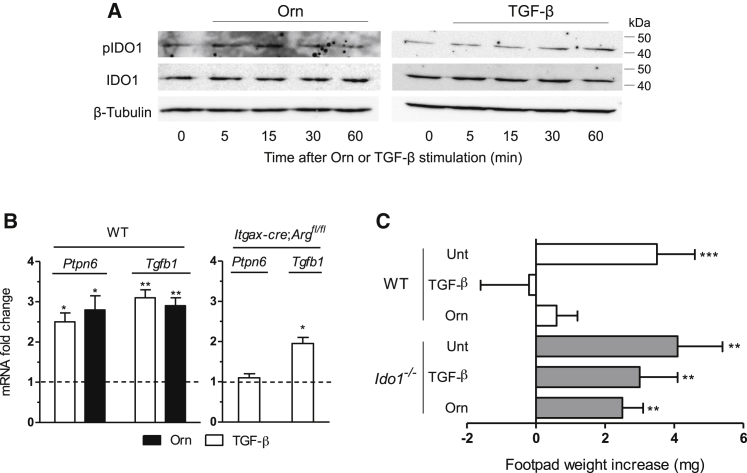
Because Orn was capable of upregulating Ido1 in DCs in a sustained fashion comparable to that of TGF-β (Figure 3), we investigated whether Orn could replace the cytokine in activating the IDO1’s signaling and immunosuppressive effects. By means of an antibody specific for the phosphorylated form of the enzyme (Bessede et al., 2014, Pallotta et al., 2011, Volpi et al., 2016), we found that the DC incubation with Orn did induce IDO1 phosphorylation, with a peak at 15 min, i.e., an effect earlier than that triggered by TGF-β (at 30–60 min) (Figure 4A). Moreover, similarly to TGF-β, stimulation with the Arg1 product upregulated transcript expression of Ptpn6 (coding for SHP-1) and Tgfb1 at 18 hr in WT DCs. In contrast, no or less upregulation of Ptpn6 and Tgfb1, respectively, was induced by TGF-β in Itgax-cre;Arg1fl/fl DCs (Figure 4B), further confirming the important role of Arg1 in triggering and maintaining the IDO1 signaling.
Figure 4.
Orn Activates IDO1 Signaling and Confers IDO1-Dependent Immunosuppressive Properties in DCs
(A) Immunoblot analysis of phosphorylated IDO1 (pIDO1) and total IDO1 in cell lysates of DCs incubated for different times with TGF-β or Orn (100 μM).
(B) Real-time PCR analysis of Ptpn6 and Tgfb1 transcripts in WT DCs stimulated with TGF-β or Orn (100 μM) or in Itgax-cre;Argfl/fl DCs stimulated with TGF-β for 18 hr. Data are normalized and presented as in Figure 1A. ∗p < 0.05 and ∗∗p < 0.01 (unpaired Student’s t test; cytokine- or Orn-treated versus untreated samples).
(C) In vivo suppression of the activity of HY-pulsed WT CD8− DCs into WT recipient mice, in combination with a minority fraction (5%) of CD8− DCs from either WT or Ido1−/− mice with no conditioning (unt, untreated) or conditioned in vitro with TGF-β or Orn (100 μM) for 24 hr; analysis of skin reactivity of recipient mice to the eliciting peptide at 15 days is presented as change in footpad weight. ∗∗p < 0.01 and ∗∗∗p < 0.001 (paired Student’s t test; mean weight of experimental versus control footpads). Data are from one experiment representative of two (A) or three (B and C; means ± SD of triplicates in B and six samples in C).
To appreciate the immunosuppressive potential of DCs conditioned by Orn, we used the skin test assay, an established protocol for measuring the in vivo induction of antigen-specific immunoreactivity versus tolerance in DCs (Grohmann et al., 2002, Grohmann et al., 2007, Pallotta et al., 2011, Puccetti et al., 1994). To this purpose, we sensitized wild-type (WT) mice with the HY peptide (containing the H-2Db epitope of male minor transplantation antigen) presented by WT CD8− DCs (constituting an immunostimulatory, splenic DC subset) (Grohmann et al., 2003a) administered alone or in combination with a minority fraction of the same cells (5%) purified from either WT or Ido1−/− animals after conditioning with TGF-β, Orn, or medium alone for 24 hr. After priming the mice, we assessed immune reactivity at two weeks by intrafootpad challenge with the HY peptide in the absence of DCs, as described (Grohmann et al., 2002, Grohmann et al., 2007, Pallotta et al., 2011, Puccetti et al., 1994). As expected, the default priming ability of immunostimulatory DCs was not affected by the presence of untreated cells. Yet, sensitization together with TGF-β- but also Orn-pretreated WT DCs caused suppression of HY-specific reactivity, an effect not detectable in mice sensitized with Ido1−/− DCs, regardless of whichever type of conditioning molecule had been used (Figure 4C).
As a whole, our data indicated that the mere incubation of DCs with an Arg metabolite can activate molecular events traceable to IDO1 signaling as well as IDO1-dependent immunosuppressive outcomes detectable in vivo.
Orn Decarboxylation Is Required for Orn Effects Mediated by IDO1 in DCs
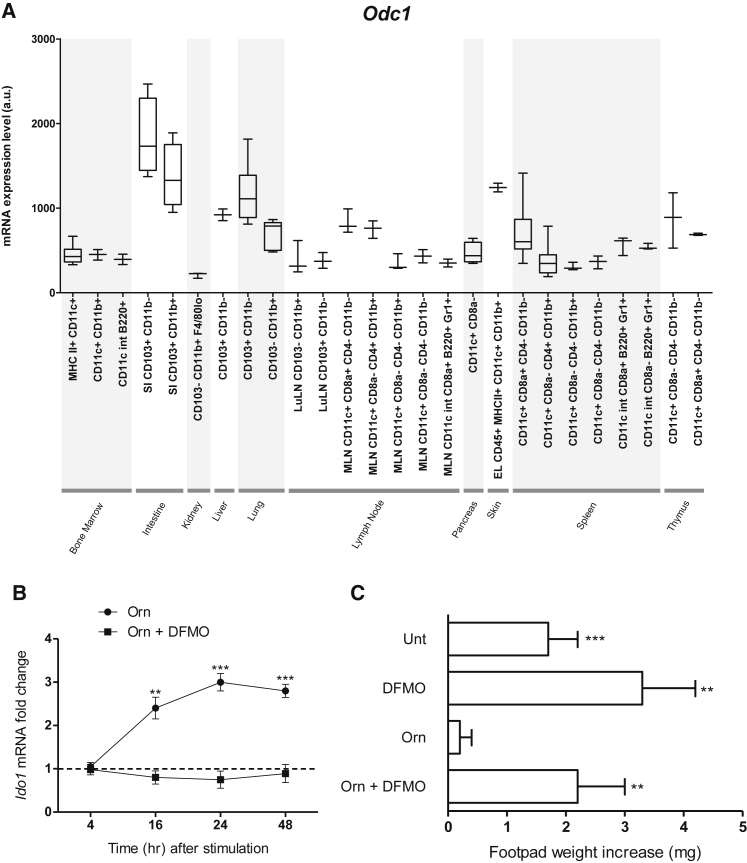
Orn can be further metabolized into putrescine, a polyamine that, in turn, can be transformed into spermidine and then spermine (Figure S2), by Orn decarboxylase (ODC). In tumor cells, where it is often highly expressed, ODC favors proliferative events. More recently, ODC has been shown to be likewise involved in immunoregulatory mechanisms that oppose anti-tumor immunity (Hayes et al., 2014). As a matter of fact, administration of α-difluoromethylornithine (DFMO), an ODC inhibitor, will inhibit tumor growth via impairment of MDSC-mediated suppressive effects (Ye et al., 2016). Whether DFMO-promoted immunity might also depend on impairment of IDO1 activity in DCs is currently unknown.
To evaluate ODC expression in DCs, we conducted a meta-analysis of public microarray data restricted to the expression of the Odc1 gene in several mouse DC subsets purified from both lymphoid and nonlymphoid organs. The results showed that high amounts of Odc1 transcripts were detectable in all DC subsets analyzed so far, including splenic, conventional CD11c+ DCs, the subject of the current study (Figure 5A). Prompted by these data, we investigated the effects of ODC inhibition on Orn actions in DCs. Because of the obligate requirement for ODC in many cell types (Cervelli et al., 2014, Guo et al., 2005), we addressed this issue by using DFMO. We found that co-incubation of splenic DCs with Orn and DFMO abrogated the capacity of Orn alone to upregulate Ido1 expression (Figure 5B). Moreover, by using the skin test assay as in Figure 4C, we found that no immunosuppressive effects could be conferred by Orn on WT DCs upon co-incubation of cells with DFMO (Figure 5C).
Figure 5.
Inhibition of Orn Decarboxylation Abrogates Orn Effects in DCs
(A) Expression of Odc1 transcripts in sub-populations of DCs derived from different tissues.
(B) Kinetic analysis of Ido1 transcripts in WT DCs stimulated with Orn (100 μM) in the presence or absence of 1 mM DFMO for different times (indicated). Data are normalized and presented as in Figure 1A. ∗∗p < 0.01 and ∗∗∗p < 0.001 (unpaired Student’s t test; Orn- or Orn plus DFMO-treated versus untreated samples).
(C) In vivo suppression of the activity of HY-pulsed WT CD8− DCs in combination with a minority fraction (5%) of the same cells with no conditioning (untreated, unt) or conditioned in vitro with Orn as in Figure 4C in the presence or absence of DFMO (1 mM); analysis of skin reactivity is as in Figure 4C. ∗∗p < 0.01 and ∗∗∗p < 0.001 (paired Student’s t test as in Figure 4C). Data are from one experiment representative of three (B and C; means ± SD of triplicates in B and six samples in C; means ± SD of samples indicated in Table S3 for each DC subset in A). Please see also Figures S2 and S3.
Because stimulation with Orn induced significant upregulation of the IDO1 gene in human DCs as well, and DFMO negated this effect (Figure S3), our data suggest that the ODC catalytic activity basally expressed in DCs exerts important immunoregulatory effects and that metabolites downstream of ODC function might represent the most proximal inducers of IDO1 signaling and IDO1-mediated immunosuppression in DCs.
Spermidine Activates the Src Kinase and Confers IDO1-Dependent, Immunosuppressive Properties in DCs
Polyamines, i.e., the diamine putrescine, the triamine spermidine, and the tetra-amine spermine, are highly bioactive polycations capable of binding nucleic acids and proteins and of modulating several signaling pathways. Polyamine functions have been studied most extensively in tumors, where they are often required for cell transformation and proliferation (Gerner and Meyskens, 2004). Whether polyamines can be produced by, or exert effects on, DCs is currently unknown.
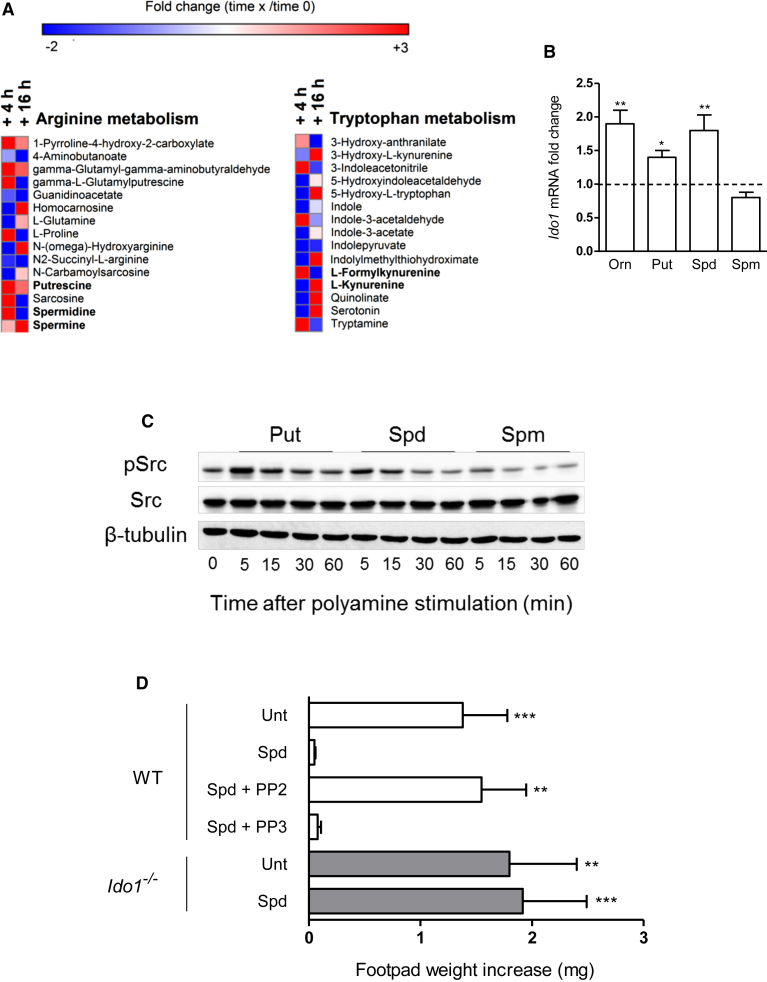
Real-time PCR analyses revealed that DCs expressed high amounts of Srm and Sms genes, coding for spermidine and spermine synthase, respectively, and that such expression was not modulated by either TGF-β or IL-4 (Figure S4A). To evaluate the effective production of polyamines by DCs, we assessed the profile of Arg metabolites along with that of Trp over time in culture supernatants of cells incubated with Orn (Figure 6A). Following Orn incubation, an increase was observed in DC release of all polyamines, with putrescine and spermidine production being evident at 4 hr and spermine at 16 hr. Moreover, Orn incubation also led to upregulation of IDO1 products, i.e., l-formylkynurenine at 4 hr, followed by l-kynurenine at 16 hr. An increase in other Orn metabolites (i.e., proline, 1-pyrroline 4-hydroxy-2-carboxylate, γ-l-glutamyl-putrescine, and γ-l-glutamyl-amino-butyraldehyde) and Trp metabolites (serotonin, tryptamine, 5-hydroxy-l-tryptophan, and indole-3-acetaldehyde) could also be observed upon DC incubation with Orn.
Figure 6.
Spermidine Activates IDO1 Immunosuppressive Signaling
(A) Analysis of Arg and Trp metabolite profiles in culture supernatants from DCs incubated with Orn (100 μM) for 4 or 16 hr. Results are expressed in terms of heatmap, in which each square represents the fold change in mass to charge ratio mean value of the relative metabolite as compared to time 0. Annotated targeted metabolites were derived from nontarget raw date and only represent signals of established chemical identities with 5 ppm.
(B) Real-time PCR analysis of Ido1 transcripts in DCs incubated with Orn, putrescine (Put), spermidine (Spd), or spermine (Spm; all at 20 μM) or medium alone for 24 hr. Data are normalized and presented as in Figure 1A. ∗p < 0.05 and ∗∗p < 0.01 (unpaired Student’s t test; Orn- or polyamine-treated versus untreated samples).
(C) Kinetics of Src phosphorylation in WT DCs incubated with Put, Spd, or Spm at 20 μM for different times. Cell lysates were analyzed by sequential immunoblotting with antibody to phosphorylated Src (pSrc), anti-Src, and β-tubulin.
(D) In vivo suppression assay with a minority fraction (5%) of WT or Ido1−/− CD8− DCs with no conditioning (Unt) or conditioned in vitro with 20 μM spermidine in the presence or absence of PP2 or PP3 at 5 μM; analysis of skin reactivity is as in Figure 4C. ∗∗p < 0.01 and ∗∗∗p < 0.001 (paired Student’s t test as in Figure 4C). Data are from one experiment representative of two (A) and three (B–D; means ± SD of triplicates in B and six samples in D). Please see also Figure S4.
We then compared the IDO1-inducing ability of polyamines with that of Orn in DCs. The results showed that DC incubation with either putrescine or spermidine for 24 hr significantly increased expression of the Ido1 gene to an extent comparable to those induced by Orn (Figure 6B), whereas induction of the IDO1 protein could be observed only for spermidine (Figure S4B). In contrast, spermine did not upregulate IDO1 but rather showed a tendency to downregulate the enzyme expression (Figure 6B and S4). In pro-inflammatory microenvironments, IDO1 is subjected to regulatory proteolysis mediated by the immunoproteasome in DCs (Orabona et al., 2008). We therefore investigated whether lack of IDO1 protein upregulation by putrescine could be due to a concomitant increase in immunoproteasome activity. We found that putrescine but not spermidine significantly upregulated Psmb8, Psmb9, and Psmb10 transcripts, coding for β5i, β1i, and β2i immunoproteasome subunits, respectively, in DCs (Figure S4C). Moreover, we analyzed IDO1 protein expression in lysates from DCs exposed to cycloheximide (an inhibitor of protein synthesis) prior to incubation with putrescine alone or in combination with MG132, a proteasome inhibitor. These results revealed that, in the presence of cycloheximide, putrescine alone rather promoted a reduction in IDO1 protein expression, which was opposed by the co-presence of MG132 (Figure S4D). The combination of MG132 and putrescine was accompanied by the appearance of proteins of higher molecular weight than 42 kDa, possibly representing polyubiquitinated IDO1 species, as described (Orabona et al., 2008).
In tumor cells, putrescine and spermidine but not spermine promote the phosphorylation and consequent activation of MAPK, Src, and PI3K (normally followed by Akt) kinases, via still undefined mechanisms (Hölttä et al., 1993). Conversely, spermine has been shown to restrain immune responses in activated macrophages by inhibiting gene expression of NOS2 (Zhang et al., 1997). In intestinal epithelial cells, spermine negatively modulates the activity of Src kinase via direct binding of the Src SH2 domain (Ray et al., 2012). Because Src is involved in the activation of IDO1 signaling in DCs (Bessede et al., 2014, Pallotta et al., 2011, Volpi et al., 2016), we evaluated the ability of polyamines in modulating Src phosphorylation over time. Putrescine and spermidine, but not spermine, increased Src phosphorylation in DCs, as soon as at 5 min of cell incubation (Figure 6C). To conduct a more comprehensive evaluation of the potential of putrescine and spermidine in modulating signaling pathways in DCs, we performed a kinomic analysis using a microarray of peptides phosphorylable by tyrosine, serine, and threonine kinases (van Baal et al., 2006) and cell extracts from DCs incubated with the polyamines for 15 or 30 min. We found that putrescine is a potent activator of several kinases in DCs (Figure S4E), including Src, Fyn, Hck, and Lck tyrosine kinases (Table S1), and that it also promotes phosphorylation of a peptide from IKKβ, a kinase involved in the activation of the pro-inflammatory NF-κB pathway (Table S4). Although less potent when considering the overall effect (Figure S4E), spermidine did promote tyrosine phosphorylation of peptides from protein kinase C (ι type) and β-catenin, known substrates of Src and Fyn kinases (Table S1), respectively, but not from IKKβ (Table S4).
In a skin test assay, DC incubation with spermidine (Figure 6D) led to immunosuppressive effects detectable in vivo. Moreover, as observed for Orn (Figure 4C), spermidine effects were lost in Ido1−/− DCs (Figure 6D). Importantly, the immunosuppressive effects were also impaired by coincubating WT DCs with spermidine and PP2 (a Src kinase inhibitor) but not PP3 (a negative control) (Figure 6D).
Overall, our data suggest that spermidine and its metabolic precursor putrescine are produced by DCs and represent major modulators of their own signaling pathways and function, including the activation of Src kinase. However, only spermidine triggered immunosuppressive IDO1 signaling, an effect possibly lost when using putrescine for DC conditioning because of the concomitant activation of several kinases other than those belonging in the Src family, including proinflammatory IKKβ, and because of the increased expression of proteasome subunits possibly degrading the IDO1 protein.
Arg1+ MDSCs Condition DCs toward an IDO1-Dependent, Immunosuppressive Phenotype
As evident from their name, the defining feature of MDSCs is their ability to suppress immune cell functions. Main factors implicated in MDSC-mediated immune suppression include high expression of Arg1, among others (Marvel and Gabrilovich, 2015). In addition to their inherent immunosuppressive activity, MDSCs might amplify regulatory properties of other immune cells, particularly in tumor microenvironments. Although some mechanisms underlying MDSC-macrophage interaction have been established (Ugel et al., 2015), the cross-talk between MDSCs and DCs is still unclear (Ostrand-Rosenberg et al., 2012).
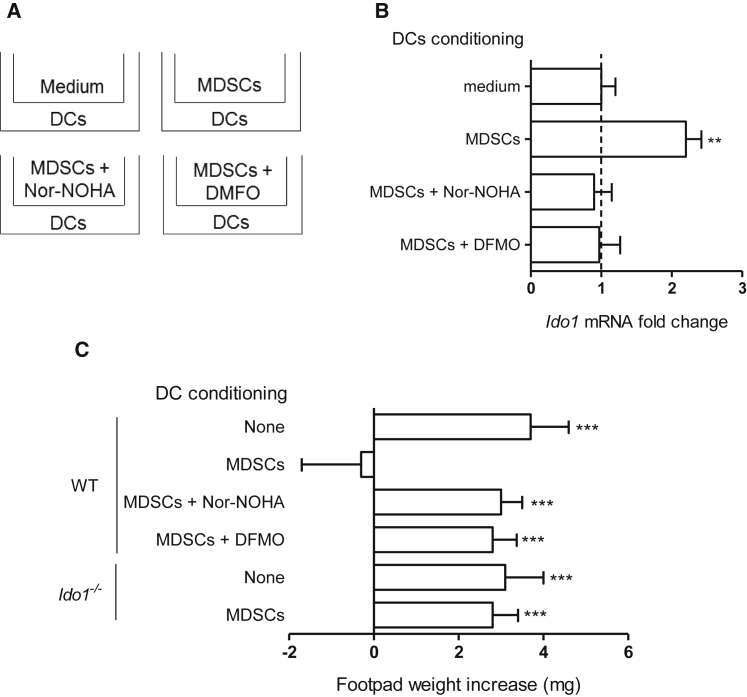
Prompted by the finding that an extracellular polyamine can condition DCs toward an IDO1-dependent, immunosuppressive phenotype, we investigated whether MDSCs could also exert similar effects on DCs via the Arg1 pathway. Arg1+ MDSCs were obtained from the bone marrow of WT mice as described (Youn et al., 2008), pre-treated with nor-NOHA, DFMO (100 μM and 1 mM, respectively), or medium alone for 1 hr, extensively washed, and cocultured with DCs in separate chambers of transwells (Figure 7A). After 24 hr, DCs were recovered and analyzed for Ido1 transcript expression by real-time PCR as well as immunosuppressive potential in vivo by skin test assay as in Figure 4C and Figure 6D. Controls included DCs cultured in transwells without MDSCs (medium; Figure 7A). We found that the separate coculture with untreated MDSCs significantly upregulated Ido1 expression (Figure 7B) and conferred in vivo immunosuppressive properties on DCs (Figure 7C). However, both effects were lost when either Arg1 or ODC catalytic activity had been inhibited in MDSCs before their co-culturing with DCs. Moreover, no in vivo immunosuppressive phenotype could be acquired by Ido1−/− DCs by conditioning with MDSCs not subjected to Arg1 or ODC inhibitor treatment (Figure 7C).
Figure 7.
MDSCs Confer DCs an IDO1-Dependent, Immunosuppressive Phenotype via Arg1 Metabolites
(A) General scheme of transwell experiments. Purified MDSCs, expressing high amounts of Arg1 (data not shown), were pre-incubated for 1 hr with medium alone, nor-NOHA (100 μM), or DMFO (1 mM), prior to incubation with CD8− DCs for 24 hr.
(B) Ido1 transcript expression in DCs conditioned as in (A). Data are normalized and presented as in Figure 1A. ∗∗p < 0.01 (unpaired Student’s t test; DCs conditioned with MDSCs alone or pretreated with enzyme inhibitors versus unconditioned DCs).
(C) In vivo suppression as in Figure 4C with a minority fraction (5%) of WT or Ido1−/− CD8− DCs preconditioned in vitro in transwells for 24 hr with MDSCs preincubated as in (A); analysis of skin reactivity is as in Figure 4C. ∗∗∗p < 0.001 (paired Student’s t test as in Figure 4C). Data are from one experiment representative of three (B and C; means ± SD of triplicates in B and six samples in C). Please see also Figure S5.
Thus our data confirmed that Arg and Trp immunoregulatory pathways are functionally integrated and that this integration can occur both intra- (i.e., DCs) and inter-cellularly (MDSCs and DCs) (Figure S5A and Figure 7).
Discussion
An effective communication networking represents the basis of life of evolved multicellular organisms. As a matter of fact, evidence for critical cross-talk mechanisms between metabolism and immunity—the main systems involved in maintaining and defending a constant internal milieu (Odegaard and Chawla, 2013, Pearce and Pearce, 2013)—are emerging. In establishing adaptive immunometabolic networks over evolution, functional repurposing of ancestral proteins (originally only metabolic or immune in nature) may have represented a powerful strategy. Such evolved structures—defined as “moonlighting proteins” when they maintain their original function—often derive from gene duplication, a major driving force that renders biological systems more robust to environmental perturbations (Espinosa-Cantú et al., 2015).
Immune regulation is a highly evolved form of biologic response that controls immunity to self but also dampens exaggerated inflammation (Fazekas de St Groth, 1998, Flajnik and Kasahara, 2010, Grohmann and Bronte, 2010). Metabolism of Arg and Trp and their consequent starvation in cell microenvironments still represent a survival strategy in phylogenetically ancient organisms. Yet, Arg and Trp catabolisms have been co-opted by immune regulation in mammals (Murray, 2016). In this regard, the bulk of available information would suggest that the Arg1 enzyme, known to catabolize Arg mainly in immune cells such as MDSCs and macrophages, might have acquired immunoregulatory functions by “simply” extending the ancient mechanism of starvation to the control of activation and proliferation of T lymphocytes. This is even more true if one considers that, for instance, mouse macrophages stimulated with IL-4 will upregulate Arg1 expression by 100- to 1,000-fold (Pauleau et al., 2004). In contrast, in our study, the increase in Arg1 transcripts in DCs incubated with IL-4 was no higher than 8-fold, suggesting that the Arg1 function in DCs might be considerably different as compared to the profound Arg starvation determined by Arg1+ macrophages. Nevertheless, the co-presence of IL-4 and TGF-β further upregulated Arg1 transcripts more than 20-fold, suggesting that maximal expression of Arg catabolism would require multiple signals in DCs. In contrast, IDO1, the Trp catabolizing enzyme mainly operating in immune cells such as DCs, has been shown to exert immunoregulatory effects via Trp starvation but also via its catalytic products, i.e., kynurenines, and, perhaps most importantly, via a non-enzymic signaling activity (Chen, 2011, Grohmann et al., 2003b, Puccetti and Grohmann, 2007a). In fact, by means of its ITIM domains (phosphorylable by Src tyrosine kinases activated in the presence of TGF-β), IDO1 establishes an intracellular signaling network in DCs that leads to the long-term expression of Ido1 itself and subsequent, sustained control of adaptive immunity. At variance with Arg1, mouse Ido1, and human IDO1 as well, might be the result of duplication of the more ancient Ido2 (IDO2 in humans) gene, an Ido1 (IDO1) paralog characterized by low efficiency in Trp degradation and inability to act as a signaling molecule (Orabona et al., 2012, Yuasa et al., 2007). This represents the successful repurposing of a very ancient enzyme. Tryptophan 2,3-dioxygenase (TDO), another ancient enzyme mainly expressed in the liver and responsible for the degradation of Trp entered by diet, does not contain ITIMs.
We here showed that, in the presence of TGF-β but not IL-4, IFN-γ, or combinations thereof, Arg1 and IDO1 expressions co-exist in DCs. If anything, the co-presence of other cytokines rather impaired the TGF-β upregulating action on both amino-acid catabolizing enzymes, an effect possibly due the antagonistic effects of IL-4 (Musso et al., 1994) and IFN-γ (Bronte and Zanovello, 2005) (G. Natoli, personal communication) on Ido1 and Arg1 gene expressions, respectively. The apparently transient, yet intense, induction of Arg1 enzymatic activity by TGF-β was mandatory for the subsequent IDO1 upregulation in terms of both catalytic and signaling mechanisms. These Arg1 effects were not mediated by Arg deprivation (as one might have expected, due to the poor or absent proliferative capacity of DCs) but rather by its downstream enzymatic catabolites, namely, the polyamine spermidine, generated downstream of decarboxylation of Orn, one of the direct Arg1 catalytic products. Moreover, our data indicated that this polyamine can promote IDO1 phosphorylation and signaling events in DCs (Bessede et al., 2014, Pallotta et al., 2011, Volpi et al., 2016), possibly via direct activation of the Src kinase. Spermidine might therefore represent a two-sided node responsible for an intersection between the immunometabolic pathways of Arg1 and IDO1. Perhaps most importantly, this relay pathway would allow the immune system to translate an initial short-term (Arg1-mediated; typical of early-acting MDSCs and regulatory macrophages; Goldszmid et al., 2014) into a sustained regulatory response (via IDO1; typical of long-term acting tolerogenic DCs; Morelli and Thomson, 2007).
TGF-β is a cytokine that appeared in metazoans and, by virtue of its marked pleiotropy and multiple effects, plays a prominent role in the logic of communicative networks in multicellular organisms (Massagué and Gomis, 2006). Although the output of a TGF-β response is highly contextual, the presence of this cytokine in microenvironments often favors local immunosuppression, inhibiting anti-tumor immunity (Gorelik and Flavell, 2002, Pickup et al., 2013, Tu et al., 2014). The TGF-β potential for inducing both enzymes, i.e., Arg1 in macrophages (Boutard et al., 1995) and IDO1 in DCs (Belladonna et al., 2009, Pallotta et al., 2011) was already known. However, we here demonstrated that the immunoregulatory effects of TGF-β in DCs would go beyond the mutually exclusive upregulation of the Arg1 and IDO1 enzymes (observable in the presence of IL-4 and IFN-γ, respectively) by allowing the establishment of a network involving both Arg1 and IDO1 in DCs, further underlining the functional plasticity of these cells. Moreover, because MDSCs themselves are an abundant source of TGF-β production (Bierie and Moses, 2010), the network established by the triad constituted by TGF-β, Arg1, and IDO1 might be highly relevant to establishing potent immunosuppressive environments if also DCs are present as well.
In conclusion, our data, besides further underlining the importance of critical pathways linking metabolism and immunity in multicellular animals, suggest that the appearance of a highly evolved form of biologic response such as immune regulation would rely on a network based on the co-option of two ancient pathways, i.e., Arg and Trp catabolisms (Anderson et al., 2016) as reinforced by TGF-β. A consequence of this could be that tumors, considered to be the result of an evolutionary process (Billaud and Santoro, 2011), have become particularly apt to co-opt metabolic and immunosuppressive networks to propel their generation and progression. Thus our data might predict that the simultaneous inhibition of two immune checkpoints such as Arg1 (https://clinicaltrials.gov/show/NCT02903914) and IDO1 (Buqué et al., 2016) could represent a successful strategy in tumor immunotherapy.
Experimental Procedures
Mice
Eight- to ten-week-old female C57BL/6 mice were obtained from Charles River Breeding Laboratories. Ido1−/− mice were purchased from The Jackson Laboratory and bred at Charles River. To obtain Itgax-cre;Argfl/fl mice (i.e., lacking Arg1 expression in CD11c+ cells), C57BL/6-Arg1tm1Pmu/J were bred to the strain C57BL/6J-Tg (Itgax-cre,-EGFP)4097Ach/J (The Jackson Laboratory), with inducible Cre recombinase expression in the CD11c+ cells (Figure S1). CD11cdnR mice on C57/BL6 background (Laouar et al., 2005) were bred and maintained at the animal facility of the University of Michigan School of Medicine.
Isolation and Treatments of DCs and MDSCs
Splenic DCs were purified using CD11c MicroBeads (Miltenyi Biotec), as described (Grohmann et al., 2002). Purity of DCs is detailed in Supplemental Experimental Procedures. MDSCs were obtained from bone marrow cells, as previously described (Youn et al., 2008). Details of MDSC purification and treatments can be found in Supplemental Experimental Procedures. For all in vitro studies, CD11c+ or CD8− DCs were cultured at 1 × 106 cells per well in 24-well plates in Iscove’s Modified Dulbecco’s medium (IMDM, Thermo Fisher Scientific) or, in selected experiments, in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific), containing low or standard Arg levels (4, 40, or standard 400 μM) and completed by adding l-asparagine (120 μM) and l-lysine (790 μM). Recombinant human TGF-β (R&D System), Orn, and polyamines were used at the final concentration of 20 ng/ml, 50–100 μM, and 20 μM, respectively. In specific experiments, DCs were conditioned by co-culture for 24 hr with MDSCs (either such or pretreated for 1 hr with inhibitors) using transwell cell culture inserts (Nunc).
Real-Time RT-PCR, Western Blotting, and Determination of Arg1 and IDO1 Catalytic Activity
All these procedures are detailed in Supplemental Experimental Procedures. Arg1 protein expression was investigated in DCs by immunoblot with a goat polyclonal anti-mouse Arg1 antibody (Abcam). IDO1 and pIDO1 expressions were investigated with a rabbit monoclonal anti-mouse IDO1 antibody (cv152) (Romani et al., 2008) or a rabbit polyclonal antibody to the phosphorylated ITIM1 motif of IDO1 (Pallotta et al., 2011), respectively.
Immunization and Skin Test Assay
A skin test assay was used for measurements of major histocompatibility complex class I–restricted delayed-type hypersensitivity (DTH) responses to the HY peptide (WMHHNMDLI) in C57BL/6 female recipient mice, as described (Pallotta et al., 2011). For in vivo immunization, 3 × 105 peptide-loaded CD8− DCs, combined with a minority fraction (5%) of peptide-loaded C57BL/6 or Ido1−/− CD8− DCs, were injected subcutaneously into recipient mice. Two weeks later, a DTH response was measured to intrafootpad challenge with the eliciting peptide, and results were expressed as the increase in footpad weight of peptide-injected footpads over that of vehicle-injected (internal control) counterparts. The minority cell fraction, constituted by WT or Ido1−/− CD8− DCs, was left untreated or treated overnight with specific reagents as above.
Meta-analysis of DC Gene Expression Data, Metabolomic Analyses, and Kinome Profiling Analyses
All these procedures are detailed in Supplemental Experimental Procedures.
Statistical Analyses
Unpaired Student’s t test was used for in vitro analyses, using at least three values from 2–3 experiments per group, whereas for the skin test assay paired Student’s t test was used (using at least six mice per group). Differences were considered significant with p < 0.05.
Author Contributions
G.M., C. Volpi, and U.G. designed the study. P.P., C. Volpi, and U.G. supervised the study as a whole. G.M. performed the majority of in vitro experiments and prepared most of the figures. R.B. and C.O. performed in vivo experiments. M.T.P. and A.M. supervised IDO1 signaling experiments. E.A. performed measurements of l-kynurenine. C. Vacca, M.L.B., A.I., F.F., Y.L., and L.S. helped with some experiments. S.U. and V.B. performed measurements of urea and supervised experiments with MDSCs. F.G. and L.Z. performed and supervised, respectively, metabolomics analyses. A.V. and M.P. performed and supervised, respectively, kinomic analyses. E.M.C.M. and S.B. performed and supervised, respectively, meta-analyses of Odc1 expression. U.G. wrote the manuscript.
Acknowledgments
We thank Ioana M. Iamandii for statistical analyses. This work was supported by the European Research Council (338954-DIDO; to U.G. and A.M.) and in part by Ministero dell’Istruzione, Università e Ricerca, Italy (FIRB RBAP11T3WB; to U.G., V.B., and S.B.).
Published: February 14, 2017
Footnotes
Supplemental Information includes five figures, four tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2017.01.005.
Contributor Information
Claudia Volpi, Email: claudia.volpi@unipg.it.
Ursula Grohmann, Email: ursula.grohmann@unipg.it.
Supplemental Information
References
- Anderson D.P., Whitney D.S., Hanson-Smith V., Woznica A., Campodonico-Burnett W., Volkman B.F., King N., Thornton J.W., Prehoda K.E. Evolution of an ancient protein function involved in organized multicellularity in animals. eLife. 2016;5:e10147. doi: 10.7554/eLife.10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Belladonna M.L., Volpi C., Bianchi R., Vacca C., Orabona C., Pallotta M.T., Boon L., Gizzi S., Fioretti M.C., Grohmann U., Puccetti P. Cutting edge: Autocrine TGF-beta sustains default tolerogenesis by IDO-competent dendritic cells. J. Immunol. 2008;181:5194–5198. doi: 10.4049/jimmunol.181.8.5194. [DOI] [PubMed] [Google Scholar]
- Belladonna M.L., Orabona C., Grohmann U., Puccetti P. TGF-beta and kynurenines as the key to infectious tolerance. Trends Mol. Med. 2009;15:41–49. doi: 10.1016/j.molmed.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Bessede A., Gargaro M., Pallotta M.T., Matino D., Servillo G., Brunacci C., Bicciato S., Mazza E.M., Macchiarulo A., Vacca C. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B., Moses H.L. Transforming growth factor beta (TGF-beta) and inflammation in cancer. Cytokine Growth Factor Rev. 2010;21:49–59. doi: 10.1016/j.cytogfr.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaud M., Santoro M. Is Co-option a prevailing mechanism during cancer progression? Cancer Res. 2011;71:6572–6575. doi: 10.1158/0008-5472.CAN-11-2158. [DOI] [PubMed] [Google Scholar]
- Boutard V., Havouis R., Fouqueray B., Philippe C., Moulinoux J.P., Baud L. Transforming growth factor-beta stimulates arginase activity in macrophages. Implications for the regulation of macrophage cytotoxicity. J. Immunol. 1995;155:2077–2084. [PubMed] [Google Scholar]
- Bronte V., Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Buqué A., Bloy N., Aranda F., Cremer I., Eggermont A., Fridman W.H., Fucikova J., Galon J., Spisek R., Tartour E. Trial Watch-Small molecules targeting the immunological tumor microenvironment for cancer therapy. OncoImmunology. 2016;5:e1149674. doi: 10.1080/2162402X.2016.1149674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervelli M., Angelucci E., Germani F., Amendola R., Mariottini P. Inflammation, carcinogenesis and neurodegeneration studies in transgenic animal models for polyamine research. Amino Acids. 2014;46:521–530. doi: 10.1007/s00726-013-1572-3. [DOI] [PubMed] [Google Scholar]
- Chen W. IDO: more than an enzyme. Nat. Immunol. 2011;12:809–811. doi: 10.1038/ni.2088. [DOI] [PubMed] [Google Scholar]
- Espinosa-Cantú A., Ascencio D., Barona-Gómez F., DeLuna A. Gene duplication and the evolution of moonlighting proteins. Front. Genet. 2015;6:227. doi: 10.3389/fgene.2015.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F., Grohmann U., You S., McGrath B.C., Cavener D.R., Vacca C., Orabona C., Bianchi R., Belladonna M.L., Volpi C. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J. Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- Fazekas de St Groth B. The evolution of self-tolerance: a new cell arises to meet the challenge of self-reactivity. Immunol. Today. 1998;19:448–454. doi: 10.1016/s0167-5699(98)01328-0. [DOI] [PubMed] [Google Scholar]
- Flajnik M.F., Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner E.W., Meyskens F.L., Jr. Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Goldszmid R.S., Dzutsev A., Trinchieri G. Host immune response to infection and cancer: unexpected commonalities. Cell Host Microbe. 2014;15:295–305. doi: 10.1016/j.chom.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik L., Flavell R.A. Transforming growth factor-beta in T-cell biology. Nat. Rev. Immunol. 2002;2:46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Bronte V. Control of immune response by amino acid metabolism. Immunol. Rev. 2010;236:243–264. doi: 10.1111/j.1600-065X.2010.00915.x. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Puccetti P. The Coevolution of IDO1 and AhR in the Emergence of Regulatory T-Cells in Mammals. Front. Immunol. 2015;6:58. doi: 10.3389/fimmu.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U., Orabona C., Fallarino F., Vacca C., Calcinaro F., Falorni A., Candeloro P., Belladonna M.L., Bianchi R., Fioretti M.C., Puccetti P. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat. Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Bianchi R., Orabona C., Fallarino F., Vacca C., Micheletti A., Fioretti M.C., Puccetti P. Functional plasticity of dendritic cell subsets as mediated by CD40 versus B7 activation. J. Immunol. 2003;171:2581–2587. doi: 10.4049/jimmunol.171.5.2581. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Fallarino F., Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Grohmann U., Volpi C., Fallarino F., Bozza S., Bianchi R., Vacca C., Orabona C., Belladonna M.L., Ayroldi E., Nocentini G. Reverse signaling through GITR ligand enables dexamethasone to activate IDO in allergy. Nat. Med. 2007;13:579–586. doi: 10.1038/nm1563. [DOI] [PubMed] [Google Scholar]
- Guo Y., Cleveland J.L., O’Brien T.G. Haploinsufficiency for odc modifies mouse skin tumor susceptibility. Cancer Res. 2005;65:1146–1149. doi: 10.1158/0008-5472.CAN-04-3244. [DOI] [PubMed] [Google Scholar]
- Hayes C.S., Burns M.R., Gilmour S.K. Polyamine blockade promotes antitumor immunity. OncoImmunology. 2014;3:e27360. doi: 10.4161/onci.27360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölttä E., Auvinen M., Andersson L.C. Polyamines are essential for cell transformation by pp60v-src: delineation of molecular events relevant for the transformed phenotype. J. Cell Biol. 1993;122:903–914. doi: 10.1083/jcb.122.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouar Y., Sutterwala F.S., Gorelik L., Flavell R.A. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- Macagno A., Napolitani G., Lanzavecchia A., Sallusto F. Duration, combination and timing: the signal integration model of dendritic cell activation. Trends Immunol. 2007;28:227–233. doi: 10.1016/j.it.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Marigo I., Dolcetti L., Serafini P., Zanovello P., Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol. Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- Marvel D., Gabrilovich D.I. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest. 2015;125:3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., Gomis R.R. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Mellor A.L., Munn D.H. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- Morelli A.E., Thomson A.W. Tolerogenic dendritic cells and the quest for transplant tolerance. Nat. Rev. Immunol. 2007;7:610–621. doi: 10.1038/nri2132. [DOI] [PubMed] [Google Scholar]
- Murray P.J. Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol. 2016;17:132–139. doi: 10.1038/ni.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso T., Gusella G.L., Brooks A., Longo D.L., Varesio L. Interleukin-4 inhibits indoleamine 2,3-dioxygenase expression in human monocytes. Blood. 1994;83:1408–1411. [PubMed] [Google Scholar]
- Odegaard J.I., Chawla A. The immune system as a sensor of the metabolic state. Immunity. 2013;38:644–654. doi: 10.1016/j.immuni.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orabona C., Grohmann U. Indoleamine 2,3-dioxygenase and regulatory function: tryptophan starvation and beyond. Methods Mol. Biol. 2011;677:269–280. doi: 10.1007/978-1-60761-869-0_19. [DOI] [PubMed] [Google Scholar]
- Orabona C., Pallotta M.T., Volpi C., Fallarino F., Vacca C., Bianchi R., Belladonna M.L., Fioretti M.C., Grohmann U., Puccetti P. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:20828–20833. doi: 10.1073/pnas.0810278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orabona C., Pallotta M.T., Grohmann U. Different partners, opposite outcomes: a new perspective of the immunobiology of indoleamine 2,3-dioxygenase. Mol. Med. 2012;18:834–842. doi: 10.2119/molmed.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S., Sinha P., Beury D.W., Clements V.K. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin. Cancer Biol. 2012;22:275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta M.T., Orabona C., Volpi C., Vacca C., Belladonna M.L., Bianchi R., Servillo G., Brunacci C., Calvitti M., Bicciato S. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011;12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- Pallotta M.T., Orabona C., Bianchi R., Vacca C., Fallarino F., Belladonna M.L., Volpi C., Mondanelli G., Gargaro M., Allegrucci M. Forced IDO1 expression in dendritic cells restores immunoregulatory signalling in autoimmune diabetes. J. Cell. Mol. Med. 2014;18:2082–2091. doi: 10.1111/jcmm.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauleau A.L., Rutschman R., Lang R., Pernis A., Watowich S.S., Murray P.J. Enhancer-mediated control of macrophage-specific arginase I expression. J. Immunol. 2004;172:7565–7573. doi: 10.4049/jimmunol.172.12.7565. [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup M., Novitskiy S., Moses H.L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M., Wick W., Van den Eynde B.J. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72:5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- Puccetti P., Grohmann U. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-kappaB activation. Nat. Rev. Immunol. 2007;7:817–823. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- Puccetti P., Bianchi R., Fioretti M.C., Ayroldi E., Uyttenhove C., Van Pel A., Boon T., Grohmann U. Use of a skin test assay to determine tumor-specific CD8+ T cell reactivity. Eur. J. Immunol. 1994;24:1446–1452. doi: 10.1002/eji.1830240631. [DOI] [PubMed] [Google Scholar]
- Ray R.M., Li C., Bhattacharya S., Naren A.P., Johnson L.R. Spermine, a molecular switch regulating EGFR, integrin β3, Src, and FAK scaffolding. Cell. Signal. 2012;24:931–942. doi: 10.1016/j.cellsig.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L., Fallarino F., De Luca A., Montagnoli C., D’Angelo C., Zelante T., Vacca C., Bistoni F., Fioretti M.C., Grohmann U. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–215. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu E., Chia P.Z., Chen W. TGFβ in T cell biology and tumor immunity: Angel or devil? Cytokine Growth Factor Rev. 2014;25:423–435. doi: 10.1016/j.cytogfr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugel S., De Sanctis F., Mandruzzato S., Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J. Clin. Invest. 2015;125:3365–3376. doi: 10.1172/JCI80006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baal J.W., Diks S.H., Wanders R.J., Rygiel A.M., Milano F., Joore J., Bergman J.J., Peppelenbosch M.P., Krishnadath K.K. Comparison of kinome profiles of Barrett’s esophagus with normal squamous esophagus and normal gastric cardia. Cancer Res. 2006;66:11605–11612. doi: 10.1158/0008-5472.CAN-06-1370. [DOI] [PubMed] [Google Scholar]
- Volpi C., Mondanelli G., Pallotta M.T., Vacca C., Iacono A., Gargaro M., Albini E., Bianchi R., Belladonna M.L., Celanire S. Allosteric modulation of metabotropic glutamate receptor 4 activates IDO1-dependent, immunoregulatory signaling in dendritic cells. Neuropharmacology. 2016;102:59–71. doi: 10.1016/j.neuropharm.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Morris S.M., Jr. Arginine metabolism: nitric oxide and beyond. Biochem. J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Wei J., Zhong L., Shi M., Zhou P., Zuo S., Wu K., Zhu M., Huang X., Yu Y. Cross talk between histone deacetylase 4 and STAT6 in the transcriptional regulation of arginase 1 during mouse dendritic cell differentiation. Mol. Cell. Biol. 2015;35:63–75. doi: 10.1128/MCB.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Geng Z., Dominguez D., Chen S., Fan J., Qin L., Long A., Zhang Y., Kuzel T.M., Zhang B. Targeting Ornithine Decarboxylase by α-Difluoromethylornithine Inhibits Tumor Growth by Impairing Myeloid-Derived Suppressor Cells. J. Immunol. 2016;196:915–923. doi: 10.4049/jimmunol.1500729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Nagaraj S., Collazo M., Gabrilovich D.I. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa H.J., Takubo M., Takahashi A., Hasegawa T., Noma H., Suzuki T. Evolution of vertebrate indoleamine 2,3-dioxygenases. J. Mol. Evol. 2007;65:705–714. doi: 10.1007/s00239-007-9049-1. [DOI] [PubMed] [Google Scholar]
- Zhang M., Caragine T., Wang H., Cohen P.S., Botchkina G., Soda K., Bianchi M., Ulrich P., Cerami A., Sherry B., Tracey K.J. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J. Exp. Med. 1997;185:1759–1768. doi: 10.1084/jem.185.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.