Abstract
Background:
The canonical Wnt signal transduction (or the Wnt/β-catenin pathway) plays a crucial role in the development of animals and in carcinogenesis. Beta-catenin is the central component of this signaling pathway. The activation of Wnt/β-catenin signaling results in the cytoplasmic and nuclear accumulation of β-catenin. In the nucleus, β-catenin interacts with the TCF/LEF transcription factors and, therefore, participates in the upregulation or downregulation of some important genes involved in diverse cellular activities. In addition, β-catenin is a critical component of the cadherin-mediated cell adherens junction. We had previously noticed that very high cellular concentrations of β-catenin had a negative effect on the transcriptional activity of this protein and, therefore, the aim of this study was to find a mechanism for this negative interaction.
Methods:
Cell fractionation, western blotting, and immunofluorescence microscopy experiments were performed to measure β-catenin protein levels and β-catenin cellular localization in HEK293Tcells transfected with various amounts of a β-catenin-encoding plasmid. Also, total RNA was extracted from the cells and used for reverse transcriptase-PCR experiments to measure the expression of the β-catenin target genes. SPSS, version 16, was used to analyze the results statistically.
Results:
We demonstrated that overexpression of β-catenin led to the formation of rod-shaped protein aggregates. The aggregate structures were mainly formed in the cell nucleus and were heavy enough to be isolated by centrifugation. Beta-catenin aggregate formation was accompanied by a decrease in the expression of the β-catenin target genes used in this study.
Conclusion:
Since deregulation of β-catenin function occurs in several human diseases, including cancer and neurological disorders, the results of this paper further support the possible biological and clinical significance of β-catenin aggregate formation.
Keywords: HEK293Tcells, Beta catenin, Cell aggregation factors, Nuclear localization signals
What’s Known
Wnt/β-catenin pathway plays a crucial role in animal development and several human diseases. β-catenin is the central component of this signaling pathway. Upregulation of β-catenin activity has been reported in several human cancers. Furthermore, downregulation of β-catenin function is the cause of some neurological diseases.
What’s New
It is demonstrated that in human HEK-293T cells, higher cellular concentrations of the β-catenin protein may lead to the formation of its aggregate structures in the cell nuclei accompanied by a decrease in transcriptional activity of this protein. Our results further support the biological relevance of the β-catenin aggregate formation.
Introduction
Beta-catenin-dependent signaling pathways play essential roles in various cellular processes and in normal tissue homeostasis.1-4 Although β-catenin was primarily known as a central component of the canonical Wnt signaling, several other signaling pathways like those mediated by tyrosine kinases, PI3-kinases, and heterotrimeric G proteins can also regulate β-catenin function. The regulation of β-catenin by these signaling pathways is probably through the inhibition of the enzymatic activity of GSK-3β.5-9 The proposed mechanism is that the activation of the above signaling pathways leads to phosphorylation (at serine 9) and, therefore, inactivation of GSK-3β.5-7 The inhibition of GSK-3β may result in protein stabilization and, thus, cellular accumulation of β-catenin. For example, we have reported that the activation of the Gq class of heterotrimeric G-proteins in Xenopus oocytes or in HEK293Tcells leads to the inhibition of GSK-3β and cellular accumulation of β-catenin.8,9
The cellular accumulation of β-catenin can result in the translocation of this protein into the nucleus. In the nucleus, β-catenin interacts with the TCF/LEF transcription factors and, therefore, regulates the transcription of many genes involved in diverse cellular processes, including proliferation, differentiation, migration, and apoptosis.2,4 In addition, β-catenin has an important role in maintaining epithelial tissues by interacting with the E-cadherin cell-cell adhesion protein.10,11
Given the critical role of β-catenin in different cellular processes, the abnormal function of this protein has been observed in animal developmental disorders and also in several human diseases, including human malignancies.1-4 The role of β-catenin-mediated signaling in colon cancer has been intensively investigated, and it has been well known that upregulation of β-catenin oncogenic activities occurs in more than 85% of the sporadic forms of colon cancer and in almost all patients with familial adenomatous polyposis.1-4,12,13 In patients with colon cancer, the upregulation of β-catenin is mainly due to the truncating mutations of the tumor suppressor, the APCgene,12,13 although in some patients stabilizing mutations of β-catenin itself or inactivating mutations of Axin have been reported.14,15 Since the upregulation of the β-catenin protein is an early event in colon cancer, the functional blockade of this protein for the prevention or treatment of colon cancer is a valuable clinical approach.
We had previously observed a reduction in β-catenin-mediated gene expression when the protein was produced at very high cellular concentrations. To find out the reason, we primarily used immunostaining methods; and during our immunofluorescence microscopy experiments in HEK293T cells, we noticed that the transfection of the cells with larger amounts of the β-catenin plasmid resulted in the formation of rod-shaped β-catenin protein aggregates. In summary, in this paper, for the first time, we report that β-catenin can form visible protein aggregates when expressed at high levels in HEK293T cells. The β-catenin aggregate formation in the HEK293T cells was predominantly observed in the cell nucleus, suggesting that aggregate formation can negatively affect the transcriptional activity of β-catenin. Our gene expression results, for the first time, support this suggestion.
Materials and Methods
Cell Culture and Transfection
In total, 2 × 105 HEK293T cells were seeded in each well of 6-well plates and grown at 37°C, 5% CO2 in DMEM supplemented with 10% FBS and antibiotics (100 μg/mL of streptomycin and 100 units/mL of penicillin). At 60% confluency, the medium was replaced with a fresh medium containing 25 μg/mL of chloroquine phosphate (Ipca Laboratories, India); and 2 hours later, the cells were transfected with various amounts of the β-catenin expression plasmid. (See the figure legends.) A standard calcium phosphate protocol was used for transfection.9 Six hours after transfection, the medium was changed to a medium lacking chloroquine phosphate; and 48 hours later, the cells were harvested. The cell pellets were used directly or stored at -70°C until use.
Cell Fractionation and Western Blotting
Cell fractionation of theHEK293Tcells was performed using the method described by Holden and Horton.16 Protein concentration and western blotting experiments were performed as described previously.9
Indirect Immunofluorescence
In order to study the quantity and cellular localization of the β-catenin protein, we used indirect immunofluorescence microscopy. The experiments were performed as described previously.9
RNA Extraction and Reverse Transcriptase-PCR (RT-PCR)
For the measurement of the expression of β-catenin target genes, the cells were harvested 48 hours post-transfection with Trypsin/EDTA (0.53 mM of EDTA and 0.05% [w/v] Trypsin in PBS) and washed twice with cold PBS. Total RNA was extracted from the cell pellets using the RNX-plus kit (CinnaGen, Tehran, Iran) as described by the supplier. Thereafter, 2 μg of RNA was treated with 1 U of DNase I in a total volume of 10 μL at 37°C for 30 minutes. The DNase enzyme was inactivated in 2.5 mM of EDTA at 65°C for 10 minutes, and then the reaction was used for reverse transcription by adding 200 U of reverse transcriptase enzyme (Fermentas), 1X RT buffer, 20 U of RiboLock RNase inhibitor, 1 mM of dNTP mix, and 0.2 μg of a random hexamer primer, in a total volume of 25 μL. PCR amplification was performed on 1 μL of the reverse transcription reaction using 30 pmol of each primer and 2.5 U of Taq polymerase in a total volume of 25 μL. The amplification protocol involved denaturation at 95°C for 60 seconds, annealing (GAPDH at 59°C, cyclin D1 at 57°C, and luciferase at 62°C) for 60 seconds, and extension at 72°C for 60 seconds. The 30 cycles of PCR were followed by a final extension at 72°C for 10 minutes. The PCR products were separated on a 1% agarose gel and visualized by ethidium bromide. The results of the RT-PCR experiments were then quantified with the ImageJ software. The PCR primers are listed in table 1.
Table 1.
Oligonucleotide primers for reverse transcriptase-PCR reactions
| Gene | Primers |
|---|---|
| GAPDH | F: 5’ CCA GGT GGT CTC CTC TGA CTT CAA CAG 3’ |
| R: 5’ AGG GTC TCT CTC TTC TTC CTC TTG TGC TGC 3’ | |
| Cyclin D1 | F: 5’ TTC CTC TCC AAA ATG CCA G 3 ’ |
| R: 5’ AGA GAT GGA AGG GGG AAA GA 3 ’ | |
| Luciferase | F: 5’ CTC ATA GAA CTG CCT GCG TG 3’ |
| R: 5’ GGC GAA GAA GGA GAA TAG GG 3’ |
F: Forward primer; R: Reverse primer
Statistical Analysis
The gene expression results are presented as mean ± standard error. SPSS, version 16, was used to analyze the results statistically. The analysis of variance (ANOVA) test was used to compare the means of the gene expression levels between the different treated groups. All the experiments were carried out in triplicate. A P>0.05 was considered significant.
Results
Formation of Protein Aggregates by β-Catenin at High Concentrations
We had previously noticed that the overexpression and production of the β-catenin protein at very high cellular levels leads to a decrease in the expression of the β-catenin target genes (Saghaeian Jazi and A. Najafi, unpublished results). To find out the reason, we first studied the cellular β-catenin protein by immunostaining methods. HEK293Tcells were transfected with increasing amounts of an expression plasmid harboring the β-catenin cDNA, and then the cellular levels of β-catenin were measured by western blotting. As was expected, the cells produced more β-catenin protein when transfected with greater amounts of the plasmid (figure 1A). When immunofluorescence microscopy was used, fluorescent rod-like bodies were clearly seen in some cells (figure 1B). The aggregate structures were not observed in the absence of the β-catenin antibody, suggesting that β-catenin forms protein aggregates at high concentrations (figure 1B).
Figure 1.
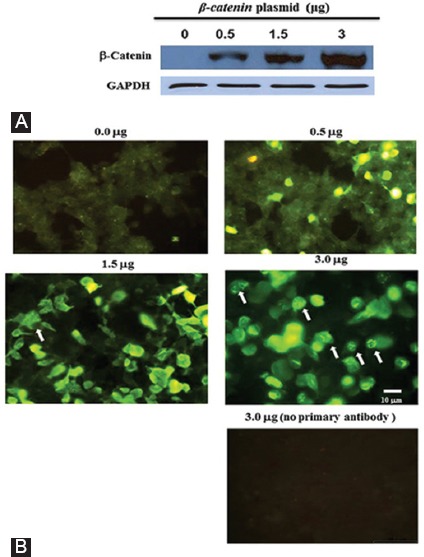
HEK293T cells were seeded in duplicate and transfected with different amounts of the β-catenin plasmid (the number on top of each panel). One group of cells was used for western blotting experiments to measure β-catenin protein levels (A) and the other group was used for immunofluorescence staining of β-catenin (B). The cells harboring β-catenin protein aggregates are indicated by arrows. The lowest panel represents the cells transfected with 3 μg of the β-catenin plasmid, but the primary antibody was omitted from the staining protocol to test the specificity of the β-catenin antibody. The expression of the GAPDH protein was used as a loading control for the blot shown in figure 1A.
The rod bodies varied in length, and they were mainly seen in the cell nucleus (figure 2A). It was also observed that the precipitates of the β-catenin aggregates could be recovered by centrifugation. Nuclear extracts were isolated from the cells and centrifuged at 63000 g to separate soluble and insoluble forms of the β-catenin protein. The 2fractions, after protein measurement, were used for western blotting experiments with the β-catenin antibody (figure 2B). As is shown in this figure, the amount of both soluble and insoluble forms of β-catenin increased when higher concentrations of the β-catenin plasmid were used for cell transfection. It is likely that these β-catenin protein aggregates had different sizes and only the heavier ones were precipitated during centrifugation.
Figure 2.
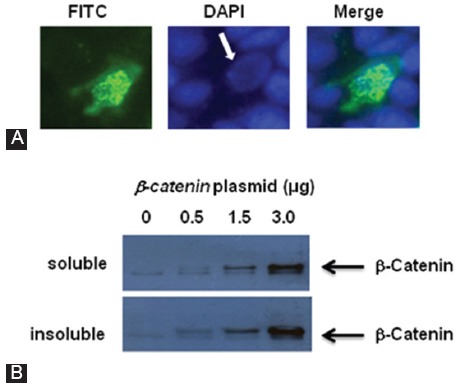
(A) FITC (left) and DAPI (middle) staining of a HEK293T cell, overexpressing β-catenin. The figure shows that the β-catenin protein aggregates predominantly formed in the cell nucleus (the arrow). The right hand panel is the merged image. (B) HEK293T cells were transfected with increasing amounts of the β-catenin plasmid; and 48hours after transfection, the cells were harvested and fractionated as described in Reference 16. Nuclear soluble and insoluble proteins were utilized for immunoblotting experiments using β-catenin antibody.
We then considered whether β-catenin aggregates were present in physiological conditions or whether they were only the result of overexpression experiments. SW480 colon cancer cells have naturally high protein levels of β-catenin. Therefore, the β-catenin proteins in these cells were carefully examined by immunofluorescence microscopy, but there was no indication of protein aggregation (figure 3).
Figure 3.
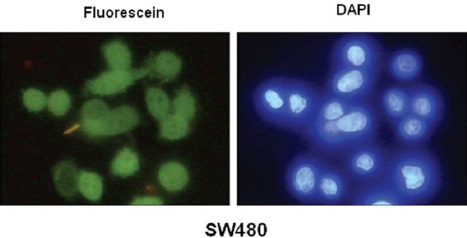
Immunofluorescence staining of SW480 colon cancer cells for the β-catenin protein. The DAPI staining is shown in the right hand panel.
Transcriptional Activity of β-Catenin Decreases at Higher Concentrations
Since the β-catenin aggregate structures were mainly formed in the cell nucleus, we asked whether this could have any effect on the expression of β-catenin target genes. The upregulation of cyclin D1 in response to the activation of the canonical Wnt/β-catenin signaling has been considered by many laboratories, including ours.17,18 Therefore, we chose cyclin D1 as a native cellular β-catenin-responsive gene. We also used luciferase under the control of β-catenin-TCF/LEF binding regulatory elements (pTOPFlash), as a β-catenin-responsive reporter gene.14 As we intended to test the β-catenin transcriptional activity more directly, we used RT-PCR experiments to measure the luciferase gene expression. The transcription of the above genes in the cells transfected with different amounts of the β-catenin-encoding plasmid was measured. As is shown in figure 4, compared to that of the cells transfected with an empty vector, the transcription of the cyclin D1 and luciferase genes was upregulated (1.5 to 2-fold) in the cells transfected with 0.5 μg of the b-catenin plasmid. Interestingly, the transfection of the cells with 1.5 μg of the plasmid either did not change or led to a decrease in the expression of the cyclin D1 and luciferase genes and the use of 3.0 μg of the β-catenin plasmid for cell transfection generally resulted in a further decrease in gene expression (figure 4). Interestingly, in some experiments, the transfection of the cells with 3.0 μg of the β-catenin plasmid lowered the transcription of the cyclin D1 and luciferase genes below the basal level (figure 4). It is worth mentioning that we observed some degree of variability in the gene expression results, which may have been due to the difference in the efficiency of cell transfection from one experiment to another. Collectively, the results of these experiments suggest that β-catenin is transcriptionally active only up to certain cellular levels and higher concentrations of this protein have a negative effect on the transcription of the target genes, perhaps due to the formation of inactive protein aggregate structures.
Figure 4.
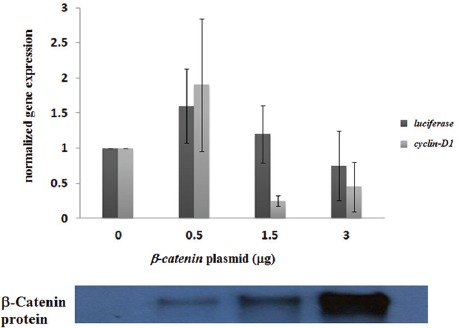
RT-PCR experiments were performed to measure the expression of the cyclin D1 and luciferase genes in HEK293T cells transfected with varying amounts of the β-catenin plasmid. To measure the luciferase gene, we also transfected the cells with 0.5 μg of the reporter, TOP Flash plasmid. The chart represents the average of gene expression for 3 independent experiments. The gene expression level was significantly different between the groups evaluated by the ANOVA test (P=0.01). The result of a western blot experiment measuring the corresponding β-catenin protein levels is shown below the chart.
Discussion
Beta-catenin is a multifunctional protein which regulates different cellular activities.1-4 As a component of cell adherens junctions, β-catenin physically interacts with the cell membrane protein, E-cadherin, to stabilize the epithelial tissues.10,11 In addition, as a signaling protein, nuclear β-catenin interacts with the TCF/LEF transcription factors to regulate the expression of many cellular genes encoding very important proteins, including growth factors, transmembrane receptors, and transcription factors. A list of these genes is available at the Wnt-signaling homepage (www.stanford.edu/~rnusse/wntwindow.html). For years, β-catenin has been the focus of research by scientists in the field of developmental biology, cancer biology, and cell and molecular biology. Beta-catenin was primarily known as a central component of the canonical Wnt signaling.10,11 Upon interaction between some Wnt ligands and their cognate receptors, the canonical Wnt signaling becomes activated and leads to the chemical and functional modification of a protein complex (destruction complex), which is involved in the regulation of β-catenin cellular stabilization.1-4 The activation of the canonical Wnt signaling blocks GSK-3β-mediated β-catenin phosphorylation at a few serine or threonine residues located at the N-terminal segment of the protein.5-7 The phosphorylation of β-catenin at these sites makes this protein susceptible to the proteasome degradation system, thereby decreasing the stabilization and cellular accumulation of β-catenin. Interestingly, the natural oncogenic mutants of β-catenin carry mutations in the GSK-3β phosphorylation sites and these mutants are much more resistant to proteolysis and are, thus, dominantly active.14 The growth, proliferation, and survival of some human cancer cells are dependent on β-catenin function, and the deregulation of this protein has been observed in some human cancers.1-4 The oncogenic function of β-catenin has been highlighted in colorectal cancers since most patients carry genetic and epigenetic alterations leading to the upregulation of β-catenin function.1-4
In this paper, we showed that the overexpression of β-catenin in HEK293T cells might cause this protein to form aggregate structures. We also showed that the β-catenin aggregate structures were mainly formed in the cell nucleus (figure 2). This was an interesting observation because it could be a mechanism for our previous finding that β-catenin had a lower transcriptional activity at very high cellular concentrations. We repeated gene expression experiments and, consistent with previous results, observed that the transcription of cyclin D1 was decreased when the cells were transfected with a higher amount of the β-catenin-encoding plasmid (e.g. 3.0 µg for each well of a 6-well plate) (figure 4). The decrease was also observed for the luciferase gene in the TOPFlash plasmid (figure 4). In this construct, the luciferase gene is under the control of 3repeats of a β-catenin-TCF/LEF-binding element.14 These results suggest that β-catenin aggregate formation could functionally block β-catenin-dependent gene expression.
At least 2 other research groups have also noticed β-catenin aggregates in cells having large amounts of this protein,19,20 and the protein regions necessary for the formation of such aggregates have been mapped.20 The most important region appears to be the armadillo repeat domain of β-catenin since the expression of the proteins that bind to this domain (e.g. N-cadherin, Tcf-4, and APC) can block the formation of aggregates.20 The formation of β-catenin aggregates has been reported in several cell lines, including PC12, MDCK, and NIH3T3.19,20 Here, we showed that β-catenin aggregates were also able to form inHEK293Tcells, suggesting that the formation of these aggregate structures could occur in many cells. Other laboratories have also discovered that β-catenin aggregates are mainly found in cell nucleus,19,20 and it has been suggested that vinculin and LEF-1 are also present with β-catenin in the aggregates.20
An important question is whether β-catenin can form aggregate structures at normal and physiological concentrations. In normal cells, β-catenin is mainly found at the cell membrane (as a component of cell adherens junctions) and its intracellular concentrations is very low.1-4 However, some cancer cells have increased the intracellular protein levels of β-catenin which provide them with growth and proliferative advantages.12,13 An appropriate example is the invasive SW480 colon cancer cell, which carries easily detectable amounts of intracellular β-catenin (figure 3). However, we did not find any indication of β-catenin aggregate formation in SW480 cells (figure 3). This was expectable, because if β-catenin formed aggregate structures in the SW480 cells, this protein could not function as a potent oncogene in these cells. It is very interesting to know whether the cells like SW480 have a mechanism to block β-catenin aggregates. It is also likely that β-catenin protein levels in SW480 cells do not reach those levels to form protein aggregates. Among the HEK293T cells transfected with 0.5 µg of β-catenin encoding plasmid, we found cells which had β-catenin protein levels apparently higher than those of the SW480 cells, but these cells did not have visible aggregate structures (figure 1B). The results shown in this paper only demonstrate that β-catenin forms protein aggregation at very high cellular concentrations, but they do not measure the cellular levels of this protein which initiate protein aggregation. A stable transfection of HEK293T cells with β-catenin, followed by a selection of different clones expressing different amounts of β-catenin, might create a more defined direction toward estimating the β-catenin concentrations required for aggregate formation.
The formation of protein aggregates has been observed in several neurodegenerative diseases.21,22 In general, protein aggregation is toxic to cells because the aggregate structures can functionally block important cellular proteins. There are several studies indicating that the downregulation of the Wnt/β-catenin signaling pathway is involved in the pathogenesis of some neurodegenerative disorders, including amyotrophic lateral sclerosis, Alzheimer’s disease, and Huntington’s diseases.23-25 Beta-catenin aggregate formation might be one of the mechanisms for the Wnt/β-catenin signaling deficiency in neurodegenerative diseases. Interestingly, Pinto and colleagues23 used NSC34 cells stably expressing the enzyme superoxide dismutase-1 (as an in vitro model) to study Wnt/β-catenin signaling in the neurodegenerative disease, amyotrophic lateral sclerosis. The authors suggested that the decrease in the Wnt/β-catenin pathway-dependent gene expression in the NSC34 cells was probably due to the cellular aggregation of β-catenin.
Beta-catenin is a critical protein with at least 2 important biological functions.1-4 As was mentioned above, deregulation of β-catenin activity has been observed in several human cancers and also in other human diseases, including neurodegenerative disorders. Accordingly, further research is warranted with a view to establishing whether the aggregation of this protein is biologically relevant.
Conclusion
In summary, in this paper, we report that β-catenin can form visible protein aggregates when expressed at high levels in HEK293T cells, re-emphasizing the possible biological significance of this phenomenon. The β-catenin aggregate formation in the HEK293Tcells was predominantly observed in the cell nucleus, suggesting that aggregate formation can negatively affect the transcriptional activity of β-catenin.
Acknowledgement
This work was supported in part by a grant (# 86103/31) from Iran National Science Foundation to S. Mahmoud A. Najafi and also by the Research Department of Tehran University.
Conflict of Interest: None declared.
References
- 1.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell2012.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Peak M, Rochford JJ, Borthwick AC, Yeaman SJ, Agius L. Signalling pathways involved in the stimulation of glycogen synthesis by insulin in rat hepatocytes. Diabetologia. 1998;41:16–25. doi: 10.1007/s001250050861. [DOI] [PubMed] [Google Scholar]
- 6.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–32. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 7.Halse R, Rochford JJ, McCormack JG, Vandenheede JR, Hemmings BA, Yeaman SJ. Control of glycogen synthesis in cultured human muscle cells. J Biol Chem. 1999;274:776–80. doi: 10.1074/jbc.274.2.776. [DOI] [PubMed] [Google Scholar]
- 8.Najafi SM. Activators of G proteins inhibit GSK-3beta and stabilize beta-Catenin in Xenopus oocytes. Biochem Biophys Res Commun. 2009;382:365–9. doi: 10.1016/j.bbrc.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Salmanian S, Najafi SM, Rafipour M, Arjomand MR, Shahheydari H, Ansari S, et al. Regulation of GSK-3beta and beta-Catenin by Galphaq in HEK293T cells. Biochem Biophys Res Commun. 2010;395:577–82. doi: 10.1016/j.bbrc2010.04.087. [DOI] [PubMed] [Google Scholar]
- 10.McCrea PD, Turck CW, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–61. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 11.Peifer M, Pai LM, Casey M. Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev Biol. 1994;166:543–56. doi: 10.1006/dbio1994.1336. [DOI] [PubMed] [Google Scholar]
- 12.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 13.Senda T, Iizuka-Kogo A, Onouchi T, Shimomura A. Adenomatous polyposis coli (APC) plays multiple roles in the intestinal and colorectal epithelia. Med Mol Morphol. 2007;40:68–81. doi: 10.1007/s00795-006-0352-5. [DOI] [PubMed] [Google Scholar]
- 14.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 15.Jin LH, Shao QJ, Luo W, Ye ZY, Li Q, Lin SC. Detection of point mutations of the Axin1 gene in colorectal cancers. Int J Cancer. 2003;107:696–9. doi: 10.1002/ijc.11435. [DOI] [PubMed] [Google Scholar]
- 16.Holden P, Horton WA. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res Notes. 2009;2:243. doi: 10.1186/1756-0500-2-243. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–7. doi: 10.1073/pnas.96.10.5522. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 19.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–48. doi: 10.1083/jcb.141.6.1433. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giannini AL, Vivanco MM, Kypta RM. Analysis of beta-catenin aggregation and localization using GFP fusion proteins: nuclear import of alpha-catenin by the beta-catenin/Tcf complex. Exp Cell Res. 2000;255:207–20. doi: 10.1006/excr1999.4785. [DOI] [PubMed] [Google Scholar]
- 21.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 22.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501:45–51. doi: 10.1038/nature12481. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto C, Cardenas P, Osses N, Henriquez JP. Characterization of Wnt/beta-catenin and BMP/Smad signaling pathways in an in vitro model of amyotrophic lateral sclerosis. Front Cell Neurosci. 2013;7:239. doi: 10.3389/fncel.2013.00239. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 25.L’Episcopo F, Serapide MF, Tirolo C, Testa N, Caniglia S, Morale MC, et al. A Wnt1 regulated Frizzled-1/beta-Catenin signaling pathway as a candidate regulatory circuit controlling mesencephalic dopaminergic neuron-astrocyte crosstalk: Therapeutical relevance for neuron survival and neuroprotection. Mol Neurodegener. 2011;6:49. doi: 10.1186/1750-1326-6-49. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]


