Abstract
Idiopathic pulmonary fibrosis (IPF) is a disease characterized by the accumulation of apoptosis-resistant fibroblasts in the lung. We have previously shown that high expression of the transcription factor Twist1 may explain this prosurvival phenotype in vitro. However, this observation has never been tested in vivo. We found that loss of Twist1 in COL1A2+ cells led to increased fibrosis characterized by very significant accumulation of T cells and bone marrow–derived matrix-producing cells. We found that Twist1-null cells expressed high levels of the T cell chemoattractant CXCL12. In vitro, we found that the loss of Twist1 in IPF lung fibroblasts increased expression of CXCL12 downstream of increased expression of the noncanonical NF-κB transcription factor RelB. Finally, blockade of CXCL12 with AMD3100 attenuated the exaggerated fibrosis observed in Twist1-null mice. Transcriptomic analysis of 134 IPF patients revealed that low expression of Twist1 was characterized by enrichment of T cell pathways. In conclusion, loss of Twist1 in collagen-producing cells led to increased bleomycin-induced pulmonary fibrosis, which is mediated by increased expression of CXCL12. Twist1 expression is associated with dysregulation of T cells in IPF patients. Twist1 may shape the IPF phenotype and regulate inflammation in fibrotic lung injury.
Introduction
One theory of the pathogenesis of idiopathic pulmonary fibrosis (IPF) suggests that a fibroblast of a very distinct phenotype, following an unknown injury, accumulates in the lung—a fibroblast that paradoxically demonstrates little apoptosis (1) despite a microenvironment in the lung that is hostile to cell survival (2, 3). Evidence for this theory comes from ex vivo fibroblasts derived from IPF lungs that demonstrate resistance to proapoptotic stimuli compared with normal human fibroblasts (4). Recent data suggest that signals derived from the extracellular matrix (5) drive the so-called IPF phenotype. Persistence of fibroblasts in fibrotic lungs may be due to soluble survival factors (6) or may result from cell-autonomous prosurvival signaling, possibly through Akt-related pathways (7, 8). We have previously shown that the transcription factor Twist1 is one of the most highly expressed transcription factors by gene expression profiling of IPF lungs and may mediate the pathologic prosurvival phenotype of fibroblasts in vitro (3). However, in vivo data demonstrating the relevance of Twist1 to human or experimental pulmonary fibrosis have been lacking.
Twist1 is a member of the basic helix–loop–helix family of transcription factors and is critical for mesoderm differentiation in Drosophila (9). Twist1 mutations in humans lead to Saethre–Chotzen syndrome (10), and Twist1 knockout mice die in utero due to incomplete neural tube closure (11). Twist1 has multiple functions that may be relevant to the pathogenesis of pulmonary fibrosis, including inhibition of NF-κB inflammatory cells (12, 13). In cancer, Twist1 expression is a potent prosurvival factor portending a poor prognosis (14) and contributes to the resistance of cancers to chemotherapy-induced apoptosis (14). Related to the cancer phenotype, and perhaps most relevant to fibrosis is the role of Twist1 in epithelial–mesenchymal transition (15, 16), a process that has been suggested as a source of matrix-producing cells in fibrosis. We hypothesized that a strategy to silence expression of Twist1 in fibroblasts would block fibrosis by counteracting the prosurvival phenotype and thus promote a fibroblast that is susceptible to apoptosis. For this study, we tested the hypothesis that transgenic mice with loss of Twist1 in the mesenchymal compartment would be protected from experimental lung fibrosis. These animals, in the presence of tamoxifen, are engineered to express Cre recombinase in collagen-expressing cells (COL1A2) (17) and to excise the Twist1 locus flanked by loxP sites (Twist1 FL) (18).
Materials and Methods
Ethical statement
This work was approved by the Institutional Review Board and the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Reagents
mAb against Twist (clone 2C1a) was purchased from Abcam (Cambridge, MA). Abs to RelB (clone C1E4) and RelA (clone C22B4) were from Cell Signaling Technology (Danvers, MA). Anti-CXCL12 (clone K15C) was from EMD Millipore (Billerica, MA). Collagen type I and CXCR4 (immunoblotting) Abs were obtained from Rockland Immunochemicals (Limerick, PA). An Ab against β-actin was purchased from Santa Cruz Biotechnology (Dallas, TX). A Quantikine ELISA kit for mouse CXCL12/SDF-1α was from R&D Systems (Minneapolis, MN). All secondary Abs were from Jackson ImmunoResearch Laboratories (West Grove, PA). Bleomycin, tamoxifen, AMD3100, collagenase type I, DNase I, brefeldin A, sodium orthovanadate, and a mAb against α-smooth muscle actin (α-SMA; clone 1A4) were purchased from Sigma-Aldrich (St. Louis, MO). Wortmannin was obtained from Enzo (Farmingdale, NY). A Sircol collagen assay kit was purchased from Biocolor (Belfast, U.K.). Short interfering RNA (siRNA) oligonucleotides and nontargeting controls (SMARTpool) were purchased from Dharmacon (Lafayette, CO). HiPerFect transfection reagent, an RNeasy Plus mini kit, and all primers for RT-PCR were from Qiagen. RT-PCR primers specific for mouse Twist1, Collagen1, Fibronectin1, Acta2, macrophage migration inhibitory factor (MIF), CXCL9, CXCL10, CXCL11, CXCL12, CCL19, and CCL21, as well as human Twist1, CXCL12, and RelB and the endogenous mouse/human housekeeping gene PPIA, were obtained from Qiagen (Valencia, CA). A chemiluminescence plus kit was purchased from PerkinElmer (Boston, MA). Power SYBR Green and high-capacity cDNA reverse transcription kit were from Applied Biosystems (Foster City, CA).
Isolation of human lung tissue
Human lung tissues were obtained from excess pathologic tissue after lung transplantation and organ donation, under a protocol approved by the University of Pittsburgh Institutional Review Board (19). IPF tissues were obtained from explanted lungs of subjects with advanced disease, and normal lungs were donated organs not suitable for transplantation from the Center for Organ Recovery and Education. Lung tissues were stored at −80°C.
Primary human lung fibroblast culture
Human primary lung fibroblasts were cultured from the explanted normal lungs from organ donors or from the lungs of patients with IPF undergoing lung transplant surgery as previously described (20). All primary lung fibroblasts were maintained in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic and were used between passages 3 and 6.
Twist1 conditional knockout mice
ROSA26-STOP tdTomato+ mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice expressing Cre recombinase in collagen-expressing cells (COL1A2) in the presence of tamoxifen were provided by Dr. Benoit de Crombrugghe (MD Anderson Cancer Center) (17). Mice expressing Twist1 FL were provided by Dr. Gerard Karsenty (Columbia University) (18). The three strains were bred to generate triple transgenic mice. Control mice expressed Cre and tdTomato with two wild-type Twist1 alleles. All mice were subjected to i.p. injection of tamoxifen (80 mg/kg for 5 d) at 5 wk and six additional injections delivered three times per week beginning at 8 wk of age during bleomycin injury experiments.
Bleomycin-induced lung injury and treatment with AMD3100
Eight-week-old mice were anesthetized with isoflurane in an anesthesia chamber. Bleomycin was administered at 2 U/kg by inhalation (21). Beginning from day 1 after bleomycin injury, the antagonist of the CXCL12 receptor CXCR4, AMD3100, or the vehicle (sterile water) was s.c. injected at 5 mg/kg daily (22). Mice were sacrificed on day 14.
Immunofluorescence
Immunofluorescent staining of mouse lungs was performed as previously described (21). Briefly, mouse lungs were inflated to 25 cm H2O pressure with 4% paraformaldehyde, and the tissue was excised and submerged in paraformaldehyde for 1 h. The tissue was then cryoprotected overnight in 30% sucrose. Five-micrometer sections were cut and mounted. Tissue was then refixed and permeabilized with 0.2% Triton X-100. Tissue was stained for α-SMA and surfactant protein C (Seven Hills Bioreagents, Cincinnati, OH) and CD45 (eBioscience, San Diego, CA). Cy5-conjugated secondary Abs were from Jackson ImmunoResearch Laboratories. Tissue was mounted with DAPI and anti-fade solution.
Sircol assay for acid-soluble collagen
Mouse lungs were perfused through the right ventricle with PBS. The left lungs were excised and flash-frozen in liquid nitrogen, followed by lyophilization in preparation for collagen determination by the Sircol collagen assay. Lungs were homogenized in 0.5 M acetic acid with protease inhibitors (Sigma-Aldrich). The homogenate was pelleted, and the supernatant was run across a QIAshredder column (Qiagen). The lung–acetic acid mixture was incubated with Sircol Red reagent. The collagen-dye complex was pelleted, and the precipitated material was redissolved in NaOH. OD540 was recorded using a microplate reader.
Single-cell suspensions of mouse lungs for flow cytometry
Mouse lungs were minced into 1- to 2-mm pieces with sterile scissors and then incubated with 100 mg/ml collagenase I and 1 mg/ml DNase I in wash media (DMEM plus 5% FBS) at 37°C. Cells were dispersed by gentle pipetting and filtered through a cell strainer to eliminate debris. Cells were pelleted and resuspended in appropriate volume of flow cytometry staining buffer to achieve a final cell concentration of 2 × 107/ml. Sorting was performed in BD FACSAria II (BD Biosciences, San Jose, CA). Cells were stained with anti–CD45-FITC Ab (clone 30-F11; BioLegend, San Diego, CA). tdTomato samples were read in the PE channel with a 550LP mirror and 585/42 filter set.
RNA extraction, Nanostring analysis, and cDNA synthesis for RT-PCR of tdTomato+ cells isolated with flow sorting
Total RNA was extracted from tdTomato+ cells isolated from single-cell suspensions of mouse lungs with flow sorting according to the RNeasy Plus mini kit handbook. In one experiment, RNA was isolated and subjected to the NanoString nCounter codeset for mouse inflammation. cDNA was synthesized from the purified mRNA with a high-capacity cDNA reverse transcription kit according to the manufacturer’s protocol (Applied Biosystems).
Quantitative RT-PCR
Quantitative RT-PCR amplifications were performed with the Power SYBR Green PCR master mix and the ABI Prism 7900HT fast real-time PCR system (Applied Biosystems).
Bronchoalveolar lavage and detection of CXCL12 by ELISA
Immediately after euthanasia, mice were subjected to median sternotomy, and the trachea was exposed and intubated with an 18-gauge catheter. One microliter of sterile saline was instilled into the lung through the tracheal catheter at a pressure of 20 cm water, and cells were retrieved. Cells in the bronchoalveolar lavage (BAL) fluid were isolated by centrifugation. The supernatant was collected and processed to detect CXCL12 by the Quantikine ELISA kit according to the manufacturer’s instructions. Active TGF-β from BAL was measured using an ELISA kit from BioLegend. Additional cytokines were analyzed using the Luminex mouse 23-plex cytokine kit and a Bio-Plex 100 according to the manufacturer’s instructions (Bio-Rad Systems, Austin, TX).
Flow cytometry
BAL cells were counted by the trypan blue exclusion method followed by incubation with CD16/CD32 Ab (BD Pharmingen, San Diego, CA) for 30 min on ice. Mouse bone marrow cells were obtained as previously described (23). Cells were then stained with the following: Ly6G-FITC (clone IA8; BD Pharmingen), CD68-PE (clone FA-11; BioLegend), CD3-allophycocyanin (clone 145-2011; BD Pharmingen), B220-PeCy7 (clone RA3-6B2; BD Pharmingen), CD4-V450 (clone RM4-5; BioLegend), CD8-PerCP (clone 53-6.7; BD Pharmingen), and rat anti-mouse CD184/CXCR4 (clone 2B11; BD Pharmingen). FITC–anti-mouse/rat Ki67 and rat IgG2a κ isotype control was from eBioscience. Samples were acquired on a FACSAria II or LSRFortessa (BD Biosciences) and analyzed by FlowJo software (Tree Star, Ashland, OR). Intracellular cytokine staining of cells obtained from single-cell suspensions of mouse lungs was performed as described previously (24). Briefly, cells were ex vivo stimulated with PMA and ionomycin (Calbiochem, La Jolla, CA) in the presence of monensin (BD Pharmingen) for 4–6 h. Cells were stained for intracellular IFN-γ, IL-4, and IL-17A with anti–IFN-γ (clone XMG1.2; BD Pharmingen), anti–IL-4 (clone 11B11; BD Pharmingen), and anti–IL-17 (clone eBio1787; eBioscience) Abs. Intracellular staining was performed using the BD intracellular cytokine staining kit (BD Pharmingen), followed by flow cytometry. Staining of Foxp3 was performed with the mouse Foxp3 staining kit (eBioscience).
Gene silencing in human lung fibroblasts
Complexes with siRNA oligonucleotides or scrambled controls were transfected employing the HiPerFect transfection reagent according to the manufacturer’s recommendations (Dharmacon). Twist1 or control siRNA complexes were transfected on the day of plating (day 0) and the next day (day 1). Silencing of additional targets was performed consecutively on days 1 and 2.
Immunoblotting
After incubation with Twist1/control siRNA, IPF lung fibroblasts were lysed for immunoblotting to measure the protein expression of Twist1, CXCL12, collagen I, α-SMA, β-actin, RelA, and RelB. Immunoblotting was performed as previously described (25). Cells were treated with brefeldin A at 1 h prior to lysis to inhibit secretion of CXCL12.
Identification of E-box binding motifs in CXCL12 and RelB gene and chromatin immunoprecipitation assays
E-box binding motifs, CANNTG, were identified by simple sequence scanning for the 5′UTR and an additional 2 kb upstream sequences of both gene using Sequencher 5.0 (Genecode) for human CXCL12 and RelB. The identified E-box binding motifs were further characterized using chromatin immunoprecipitation (ChIP) assays. The ChIP assay was performed as described (26, 27). Chromatin was precipitated with Abs against Twist1, mouse IgG1, or rabbit IgG as controls. DNA from the pull-down reactions and the input DNA were used as a template for PCR employing primers designed to amplify the regions containing putative E-box binding motifs. A total of 5 and 12 potential E-box binding sites were identified in CXCL12 and the RelB promoter, respectively. An Ab specific for DNA polymerase II was used as a positive control. Owing to the proximity of these binding sites, primers were designed to test multiple sites as a single unit for some of these E-box binding motifs. The locations of the putative E-box binding motifs and the primers used for the PCR amplification specifically for each site are listed in Table I. The PCR products were analyzed using agarose electrophoresis.
Table I. Putative E-box motifs of CXCL12 and RelB gene.
| Gene | Location | E-box Motif | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | PCR Product Size (bp) |
|---|---|---|---|---|---|
| RelB | −1718 to −1713 | E1 | CACATTTGTATATAGCACCCACATG | GAGAAAACTGAAACCCGGCA | 123 |
| −1701 to −1696 | E2 | ||||
| −1699 to −1694 | E3 | ||||
| −1598 to −1593 | E4 | GCCGGGTTTCAGTTTTCTCA | GTGACAGAGTGAGACCTTGTC | 128 | |
| −1558 to −1553 | E5 | ||||
| −1389 to −1384 | E6 | CCTCGGCCTCCCAAAGTATT | TCTCAGACCACAGATGCAGG | 114 | |
| −1318 to −1313 | E7 | ||||
| −1222 to −1217 | E8 | TCAGGAATATGAGAGGCTTAGGA | CCTCCGTATACATCCTGAGCT | 155 | |
| −1173 to −1168 | E9 | ||||
| −472 to −467 | E10 | ATGGATGGCAGGTGTAGAGC | TCTGGACGAGACAACTGAGG | 177 | |
| −320 to −315 | E11 | ||||
| −227 to −222 | E12 | TGCCCAACCCCTCCTGAG | GACGTCACGCCTGGGGAA | 101 | |
| CXCL12 | −1894 to −1889 | E1 | CTTTCAGGCTTCTGGGACAG | GTCCGCGGGAAATCTACAC | 201 |
| −1886 to −1881 | E2 | ||||
| −1700 to −1795 | E3 | CCTCCCGGGTTTCATCAG | ACTGAAGGCAGTGAGGTCCA | 194 | |
| −1046 to −1041 | E4 | GCTGGGAAGGACACAGAGAG | TGGGTCCAGAAAACTCTTCC | 208 | |
| −517 to −512 | E5 | ACTGCAAAGACGGGTCTCAT | GATGGCGGGAACTGAATG | 197 |
Microarray analysis
The RNA isolation and microarray procedures have been previously described (28, 29). These data were obtained from the Lung Genomics Research Consortium. One hundred thirty-four IPF patients and 108 normal controls were selected. Table II describes the LGRC cohort. These data and methods are available on the Gene Expression Omnibus database under reference number GSE47460. The data were normalized using a cyclic loess algorithm from the Bioconductor suite of R tools (30). Analyses were performed using BRB-ArrayTools 4.3.0 stable release developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. Additional gene expression data were obtained from Gene Expression Omnibus (GSE10667, https://www.ncbi.nlm.nih.gov/geo/) (31).
Table II. LGRC cohort.
| Control |
IPF |
p Value |
|
|---|---|---|---|
| N | 108 | 134 | |
| Men (%) | 49 (45) | 94 (70) | <0.0001 |
| Women (%) | 59 (55) | 40 (30) | |
| Age, y (SD) | 63.6 (11.4) | 56.4 (11.4) | <0.0001 |
| Ethnicity (%) | |||
| White | 100 (92) | 122 (91) | 0.95 |
| African American | 3 (2.8) | 5 (3.7) | |
| Hispanic | 1 (0.9) | 1 (0.7) | |
| Asian/Pacific Islander | 3 (2.8) | 2 (1.4) | |
| Other | 1 (0.9) | 1 (0.7) | |
| Smoking status (%) | |||
| Never | 32 (30) | 47 (35) | 0.3 |
| Current | 2 (1.9) | 2 (1.5) | |
| Ever | 63 (58) | 81 (60) | |
| Pulmonary function | |||
| FEV1, % predicted (SD) | 95 (12.6) | 70.5 (18.2) | <0.0001 |
| FVC, % predicted (SD) | 94.4 (13.1) | 63.4 (16.9) | <0.0001 |
| DLCO, % predicted (SD) | 84.1 (16.7) | 46.7 (19.2) | <0.0001 |
FEV1, forced expiratory volume in 1 s.
Statistical analysis
Data in these studies were analyzed by a t test, ANOVA followed by a Fisher least significant difference test or the Newman–Keuls post hoc test, or χ2 testing using Prism 6.0 (GraphPad Software).
Results
Loss of Twist1 in COL1A2+ cells leads to increased bleomycin-induced pulmonary fibrosis
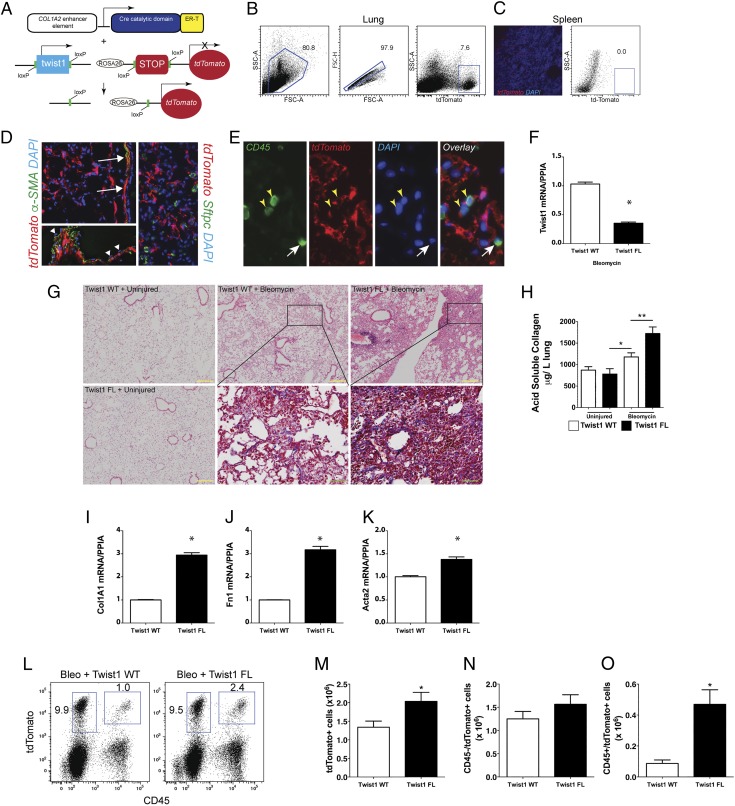
Based on our previous data showing that loss of Twist1 leads to apoptosis in lung fibroblasts (3), we hypothesized that loss of Twist1 in experimental pulmonary fibrosis would be characterized by enhanced fibroblast apoptosis and protection from pulmonary fibrosis. To test this hypothesis, we bred mice expressing tamoxifen-inducible Cre recombinase in collagen-expressing (COL1A2+) cells (17) and Twist1 FL (18) or wild-type control Twist1 (Twist1 WT). Additionally, we bred the ROSA-26 tdTomato reporter (32) to identify the cells undergoing Cre-mediated recombination. The strategy is shown in Fig. 1A–C. Following injury with bleomycin, immunofluorescent staining showed tdTomato expression in cells limited to the mesenchymal compartment (Fig. 1D), including cells that morphologically appeared to be fibroblasts as well as airway and vascular smooth muscle cells. Endothelial cells did not demonstrate tdTomato expression. α-SMA staining was essentially limited to airway and vascular smooth muscle cells. No tdTomato+ cells coexpressed the type 2 alveolar epithelial cell marker surfactant protein C (Fig. 1D). Cells coexpressing the common leukocyte Ag CD45 (33, 34) and tdTomato were also identified in situ (Fig. 1E). Expression of tdTomato was then used by flow cytometry to isolate all the cells in the lung that have undergone Cre-mediated recombination. Following administration of tamoxifen, cells were flow-sorted from Twist1 WT and Twist1 FL mice and processed for quantitative RT-PCR (Fig. 1F). Cells from Twist1 FL mice expressed significantly less Twist1 at the mRNA level. At 14 d following administration of saline by inhalation as a control for bleomycin injury, no differences were observed between Twist1 WT and Twist1 FL animals (Fig. 1G). Following injury with bleomycin, much to our surprise, the histology in the Twist1 FL animals, as compared with Twist1 WT controls, was characterized by more confluent areas of injury and matrix deposition as well as lymphoid aggregates. To quantify both injury and fibrosis, we performed the following analyses: first, we quantified acid-soluble collagen as we have done previously (Fig. 1H) (25). No difference was observed between uninjured WT and FL animals. In contrast, significantly more collagen was detected in Twist1 FL animals compared with Twist1 WT animals, suggesting that, in fact, loss of Twist1 in COL1A2+ cells was associated with more severe injury and fibrosis. Next, we analyzed the flow-sorted cells as described above for expression of several fibrotic markers by quantitative RT-PCR: COL1A1, fibronectin (Fn1), and α-SMA (Acta2) (Fig. 1I–K). Significantly increased expression of all three genes was observed in the Twist1 FL animals, further supporting the observation of increased bleomycin-induced pulmonary fibrosis in these animals. Finally, we quantified the total number of tdTomato+ cells as a marker of COL1A2+ cell accumulation in the lung (Fig. 1L, 1M). After digestion of and dispersion of lungs from bleomycin-injured Twist1 WT and Twist1 FL animals into single-cell suspensions, we found significantly more tdTomato+ cells from the Twist1 FL animals compared with the WT controls. To determine whether this population of tdTomato+ cells arose from a local population or from a bone marrow–derived population, we also quantified the number of tdTomato+CD45+ cells (Fig. 1L–O). No significant difference was observed between CD45−/tdTomato+ cells. However, CD45+tdTomato+ cells were more frequently observed in Twist1 FL animals. These data suggest that loss of Twist1 in COL1A2+ cells led to increased bleomycin-induced pulmonary fibrosis by several mechanisms, including the accumulation of a bone marrow–derived matrix-producing population of cells and increased matrix synthetic activity in these cells.
FIGURE 1.
Loss of Twist1 in COL1A2+ cells leads to increased bleomycin-induced pulmonary fibrosis. (A) Schematic representation of the triple transgenic COL1A2 Cre-ER(T) Twist1 fl/fl ROSA26-tdTomato mouse (Twist1 FL). (B) Gating strategy for tdTomato+ cells in the lung. (C) Negative control fluorescent images of spleen showing rare tdTomato+ cells (left) and dot plots of splenocytes showing absence of tdTomato+ cells (right). (D) Fluorescent images of lungs from bleomycin-injured animals showing tdTomato+ (red) cells and staining α-SMA (left, green) or surfactant protein C (SFTPC; right, green). Original magnification, ×200. Arrows indicate α-SMA+tdTomato+ airway or vascular smooth muscle cells. Arrowheads indicate tdTomato− endothelial cells overlying vascular smooth muscle. Nuclei are counterstained with DAPI. (E) Immunofluorescent images of CD45 expression (green). Yellow arrowheads identify CD45+tdTomato+ cells and the white arrow identifies a CD45+tdTomato− cell. Original magnification, ×400. (F) At 14 d after injury, tdTomato+ cells from Twist1 WT or Twist1 FL injured with bleomycin were flow sorted and processed immediately for quantitative RT-PCR of Twist1 (*p < 0.0001, n = 3). (G) H&E staining of lungs at 14 d after bleomycin injury in Twist1 WT or Twist1 FL animals (yellow inset scale bar, 200 μm; original magnification, ×100). Masson trichrome images from bleomycin-injured are magnified (green inset scale bars, 50 μm; original magnification. ×400). (H) Left lungs were processed for detection of acid-soluble collagen. Bleomycin-induced deposition of collagen was increased in Twist1 FL animals compared with WT controls (*p = 0.03 saline plus Twist1 FL versus bleomycin plus Twist1 WT, and **p < 0.003, bleomycin plus Twist1 WT versus bleomycin plus Twist1 FL, by ANOVA, n = 10–14 per group). Quantitative RT-PCR of flow-sorted cells from bleomycin-injured Twist1 WT or FL animals for (I) COL1A1 (*p < 0.0001, n = 3, by t test), (J) FN1 (*p = 0.0001, n = 3, by t test), and (K) Acta2 (α-SMA, *p = 0.033, n = 3, by t test). (L and M) Flow cytometry was performed to quantify the number of CD45+ and tdTomato+ cells. Total tdTomato+ cells were significantly higher in the bleomycin-injured Twist1 FL mice than in their WT counterparts (*p < 0.04, n = 8–9). No significant difference was observed between tdTomato+CD45− cells in (N). (O) Significantly more CD45+tdTomato+ cells were detected in the Twist1 FL animals than in the WT (*p = 0.002, n = 8–9).
Loss of Twist1 in COL1A2+ cells was associated with the increased accumulation of T cells in the lung
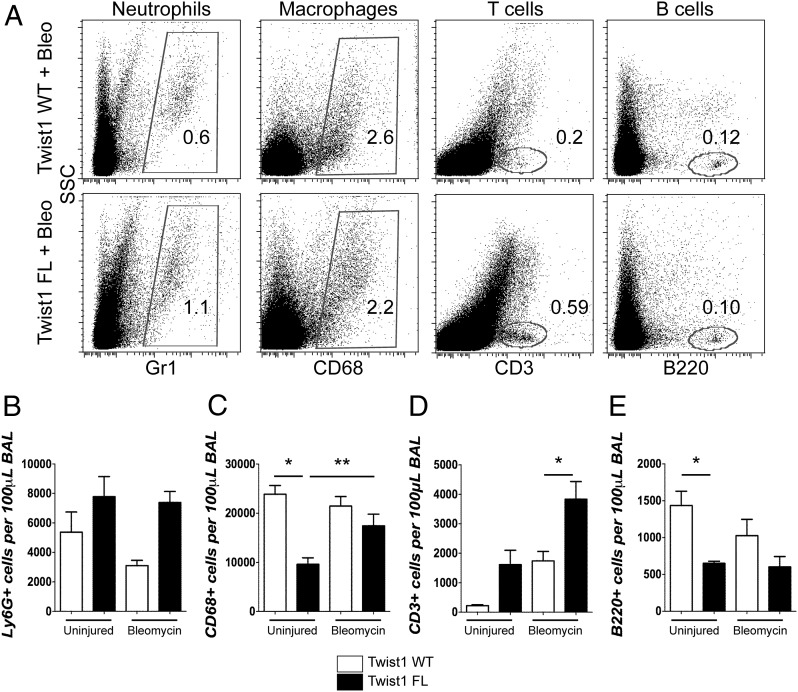
Histologically, the Twist1 FL mice injured with bleomycin appeared to have a more extensive inflammatory infiltrate (Fig. 1). To further characterize the inflammatory infiltrate of bleomycin and uninjured controls, we analyzed the cells recovered from BAL by flow cytometry (Fig. 2). BAL cells were stained for Ly6G to mark neutrophils, CD68 to mark macrophages, CD3 to mark T cells, and B220 to mark B cells. Although not statistically significant, Twist1 FL mice were characterized by a trend toward more neutrophils between the uninjured and the bleomycin-injured mice (Fig. 2A, 2B). This is consistent with increased injury in the Twist1 FL mice. Uninjured Twist1 FL mice had significantly fewer CD68+ cells compared with WT. There was a significant accumulation of CD68+ cells in Twist1 FL mice after bleomycin injury compared with the uninjured (Fig. 2A, 2C). Following bleomycin injury, the most significant observation was the large accumulation of CD3+ cells in bleomycin-injured Twist1 FL mice compared with bleomycin-injured Twist1 WT controls (Fig. 2A, 2D). Although it did not reach statistical significance, more CD3+ cells were observed in Twist1 FL mice in the absence of injury compared with Twist1 WT mice. There were significantly fewer B220+ cells in uninjured Twist1 FL mice compared with uninjured WT controls, but the same difference was not observed following bleomycin injury (Fig. 2A, 2E). These data show significant shifts in the inflammatory infiltrate between Twist1 WT and FL animals.
FIGURE 2.
Loss of Twist1 in COL1A2+ cells is associated with enhanced accumulation of T cells following bleomycin injury. (A–E) BAL was processed for flow cytometry for markers of neutrophils, macrophages, and T and B cells. For these experiments, n = 5 per uninjured condition and n = 11–12 for injured conditions. BAL was collected at 14 d after injury. (A) Dot plots of uninjured and bleomycin-injured animals for neutrophils, macrophages, T cells, and B cells. Quantification of (B) Ly6G, (C) CD68 (*p = 0.006, uninjured Twist1 WT versus uninjured Twist1 FL and **p < 0.025 by ANOVA, uninjured plus Twist1 FL versus bleomycin plus Twist1 FL), (D) CD3 (*p = 0.0021, bleomycin plus Twist1 WT versus bleomycin plus Twist1 FL), and (E) B220 (*p = 0.03, uninjured plus Twist1 WT versus uninjured plus Twist1 FL) is shown.
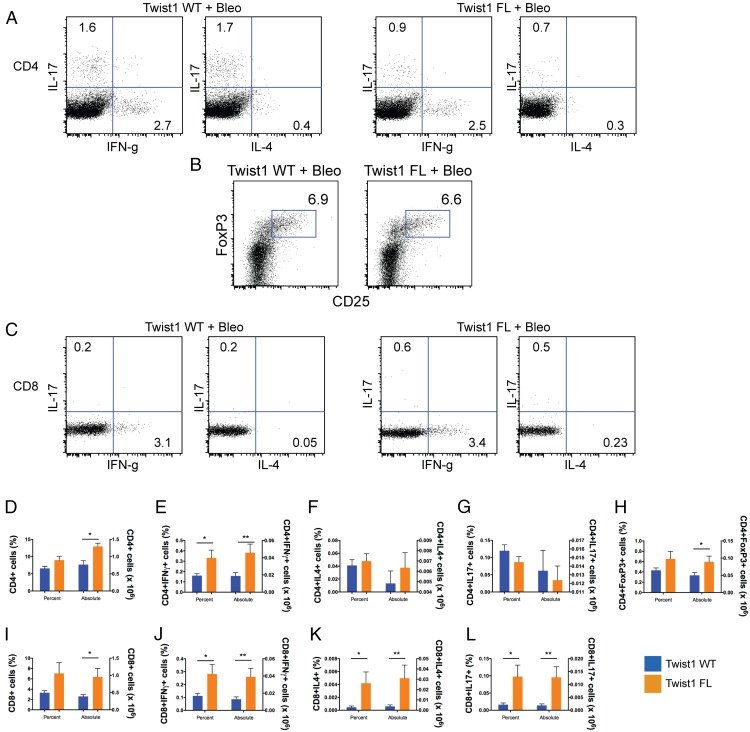
To further subphenotype the infiltrating T cells based on their functionality, we generated single-cell suspensions of bleomycin-injured lungs from Twist1 WT and FL mice, and the cells were stimulated for intracellular cytokine staining (Fig. 3). Consistent with previous results, the average number of cells isolated from the single-cell suspension of bleomycin-injured Twist1 FL mice was higher than the WT controls (data not shown). This result largely drives the differences between the genotypes for absolute counts of cells. There was no significant difference in terms of the percentage of CD4+ cells. However, more CD4+ cells were stained from the bleomycin-injured Twist1 FL mice (Fig. 3A, 3D). Of the cells isolated, more cells in the Twist1 FL mice were CD4+IFN-γ+ (Fig. 3A, 3E), indicating a Th1 phenotype of Th cells population. However, no significant differences were observed between genotypes for CD4+IL-4+ (Th2) or CD4+IL-17+ cells (Th17) (Fig. 3A, 3F, 3G). The percentage of CD4+Foxp3+ cells did not differ between genotypes (Fig. 3B, 3H). However, there were absolutely more CD4+Foxp3+ cells in the Twist1 FL mice compared with WT controls. By absolute numbers, CD4+Foxp3+ cells appeared to be the most enriched subphenotype of T cell isolated. As with CD4, there was no significant difference between the percentage of CD8+ cells between genotypes, but more CD8+ cells were detected in the Twist1 FL mice (Fig. 3C, 3I). For all subphenotypes of CD8+ cells, more IFN-γ+, IL-4+, and IL-17+ cells were present in the Twist1 FL mice compared with WT controls (Fig. 3C, 3J–L). Overall, these data show that the loss of Twist1 FL mice in COL1A2+ cells leads to derangements in the inflammatory infiltrate, including increased accumulation of CD68+ and CD3+ cells in bleomycin-injured lungs. The T cells, which include both Th cells (CD4+ T cells) as well as cytotoxic T cells (CD8+) that express IFN-γ, IL-4 and IL-17, likely contribute to the enhanced tissue injury in Twist1 FL mice.
FIGURE 3.
Subphenotyping of T cells from bleomycin-injured Twist1 FL and WT mice. Flow cytometry of single-cell suspensions of bleomycin-injured Twist1 WT or Twist1 FL mouse lungs to describe the T cell subphenotypes following stimulation and intracellular cytokine staining is as described in Materials and Methods. (A) Dot plots for CD4+ IFN-γ, IL-4, and IL-17 from bleomycin-injured Twist1 WT and Twist1 FL animals. (B) Dot plot for CD4+Foxp3+ cells. (C) Dot plots for CD8+ IFN-γ, IL-4, and IL-17. Percentage of cells (left y-axis) and the absolute numbers of cells (right y-axis) are reported, n = 8 per condition, and data were analyzed by an unpaired t test. (D) CD4 (*p = 0.003). (E) CD4 plus IFN-γ (*p = 0.04, **p = 0.02). (F) CD4 plus IL-4. (G) CD4 plus IL-17. (H) CD4 plus Foxp3 (*p < 0.05). (I) CD8 (*p = 0.04). (J) CD8 plus IFN-γ (*p = 0.04, **p = 0.03). (K) CD8 plus IL-4 (*p < 0.05, **p = 0.05). (L) CD8 plus IL-17 (*p < 0.03, **p = 0.02).
Loss of Twist1 in COL1A2+ cells is associated with increased expression of the T cell chemoattractant CXCL12
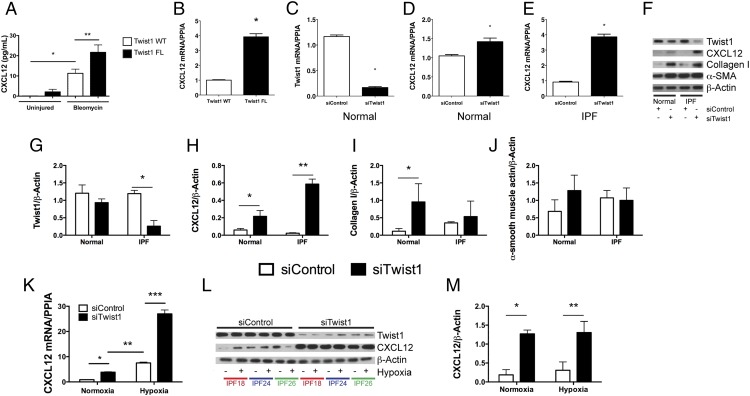
If T cells and tdTomato+ cells accumulate in bleomycin-injured lungs following loss of Twist1 in COL1A2+ cells, we reasoned that COL1A2+ Twist1-null cells must secrete a soluble signal to call these cells into the lung. To investigate potential soluble signals that might be secreted by Twist1-null cells, we analyzed the BAL for multiple cytokines and chemokines by the Luminex platform (Supplemental Fig. 1). Consistent with the derangement of T cells reported in Fig. 3, we found multiple inflammatory mediators expressed at higher levels in Twist1-null mice. To determine whether any of these factors are expressed in COL1A2+Cre-ER(T) Twist1 FL cells, we subjected the RNA from the flow-sorted cells to the Nanostring platform (Supplemental Table I). We found no concordant results between the Luminex and the Nanostring data. We also performed an ELISA for active TGF-β and found no differences between bleomycin-injured Twist1 WT and Twist1 FL animals (Supplemental Fig. 1P). One potentially important chemokine candidate that we considered is CXCL12. CXCL12 is a potent chemoattractant (33) for T cells and so-called fibrocytes. First, we analyzed the concentration of CXCL12 in the BAL ELISA (Fig. 4A). We found significantly more CXCL12 in the BAL of Twist1 FL mice compared with Twist1 WT controls following bleomycin injury. To confirm that the increase in CXCL12 may be in part due to increased expression in tdTomato+ Twist1 FL cells, we analyzed CXCL12 by quantitative RT-PCR from the flow-sorted cells (Fig. 4B). We found a nearly 4-fold increase in CXCL12 mRNA in tdTomato+ Twist1 FL cells compared with tdTomato plus Twist1 WT controls. We also tested other chemokine candidates with T cell chemoattractant activity, including CXCL9, CXCL10, CXCL11, CCL19, and CCL21. However, we found no differences between Twist1 FL and WT mice (data not shown). To explore a possible link between Twist1 and CXCL12, we then moved to human fibroblasts isolated from normal and IPF lungs. In these experiments, we determined whether silencing of Twist1 expression by siRNA was associated with increased expression of CXCL12. In lung fibroblasts from normal lungs, silencing of Twist1 led to a significant reduction in Twist1 mRNA (Fig. 4C). However, a small but significant increase in CXCL12 mRNA was observed (Fig. 4D). In IPF lung fibroblasts, however, silencing of Twist1 by siRNA was associated with a 4-fold increase in CXCL12 mRNA (Fig. 4E). We repeated these experiments and processed the cells for immunoblotting for Twist1 and CXCL12 in addition to the fibrosis markers collagen I and α-SMA (Fig. 4F–J). Silencing of Twist1 protein was more pronounced in IPF fibroblasts as shown in the representative immunoblot and by quantitative densitometry (Fig. 4F, 4G). Loss of Twist1 was associated with increased CXCL12 in normal lung fibroblasts and to an even greater extent in IPF lung fibroblasts (Fig. 4F, 4H). Loss of Twist1 in normal lung fibroblasts led to a very significant increase in expression of collagen I. This increase was much less pronounced in IPF lung fibroblasts (Fig. 4F, 4I). Loss of Twist1 in normal lung fibroblasts was associated, possibly, with a small increase in expression of α-SMA. However, no difference in α-SMA expression was observed in IPF lung fibroblasts (Fig. 4F, 4J). Hypoxia is a known and potent inducer of CXCL12 expression (35, 36). We silenced Twist1 expression in IPF fibroblasts in the presence or absence of hypoxia (Fig. 4K–M). Silencing of Twist1 led to a significant increase in CXCL12 mRNA in the presence of normoxia by quantitative PCR, and this difference was exaggerated in the presence of 5% O2 atmosphere. We observed a nearly 25-fold increase in CXCL12 mRNA. However, by immunoblotting, no difference was observed in intracellular CXCL12. Taken together, these data indicate that in both COL1A2+ mouse cells and human lung fibroblasts, loss of Twist1 is associated with increased expression of CXCL12.
FIGURE 4.
Loss of Twist1 in COL1A2+ cells and in human lung fibroblasts leads to increased expression of CXCL12. (A) BAL from Twist1 WT and Twist1 FL animals at 14 d following bleomycin injury was analyzed by ELISA for CXCL12 as described in Materials and Methods (*p < 0.002, n = 5–8, Twist1 WT plus uninjured versus Twist1 WT plus bleomycin, **p < 0.004, Twist1 WT plus bleomycin versus Twist1 FL plus bleomycin, n = 4–5 per group). (B) Quantitative RT-PCR for CXCL12 was performed on tdTomato+ cells flow sorted from Twist1 WT or Twist1 FL animals following bleomycin injury (*p < 0.0002, by t test, n = 3). Normal human lung fibroblasts were cultured in the presence of Twist1 siRNA (siTwist1) or nontargeting controls (siControl) and processed for quantitative RT-PCR for (C) Twist1 (*p < 0.0001, by t test, n = 3) and for (D) CXCL12 (*p < 0.022, by t test, n = 3). (E) IPF-derived lung fibroblasts were also treated with siTwist1 or siControl, and quantitative PCR was performed for CXCL12 (*p < 0.0001, by t test, n = 3). (F) Normal and IPF lung fibroblasts were incubated with siTwist1 or siControl and subjected to immunoblotting for Twist1, CXCL12, collagen I, and α-SMA. All experiments reflect fibroblasts from three independent normal and three IPF lungs. Data were analyzed by two-way ANOVA followed by a Newman–Keuls post hoc test. Band intensity was quantified for (G) Twist1 (*p < 0.0001, IPF siTwist1 versus siControl, n = 3), (H) CXCL12 (*p < 0.03, normal siTwist1 versus siControl, n = 3 and **p < 0.0001, IPF siTwist1 versus siControl, n = 3), (I) collagen I (*p < 0.02, normal siTwist1 versus siControl, n = 3), and (J) α-SMA. (K) Quantitative RT-PCR for CXCL12 was performed on IPF lung fibroblasts in the presence of siControl or siTwist1 with and without hypoxia (*p < 0.03, IPF siTwist1 versus siControl under normoxic conditions, **p < 0.0003, IPF normoxia and hypoxia, and ***p < 0.0001, siControl versus siTwist1 under hypoxic conditions, n = 3). (L) Immunoblot for Twist1 and CXCL12 with siControl or siTwist1 in the presence or absence of hypoxia. (M) ImageJ quantification of the blots in (L) for CXCl12 (*p < 0.0002, IPF siControl versus siTwist1 in normoxia and **p < 0.0001, siControl versus siTwist1 under hypoxic conditions, n = 3).
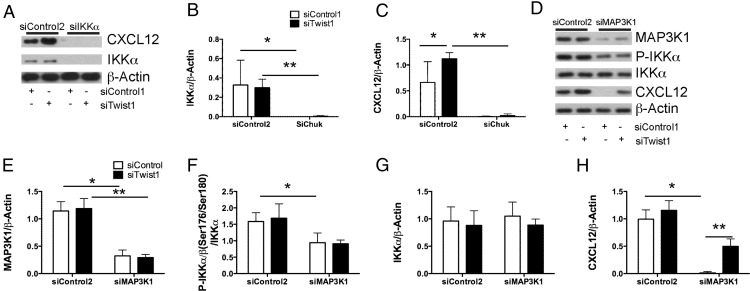
Expression of CXCL12 mediated by the loss of Twist1 is associated with events that occur downstream of activation of the noncanonical NF-κB complex by IKKα
CXCL12 has been shown to be regulated preferentially in response to noncanonical NF-κB signaling (p52/RelB) (37). Signaling specific for NF-κB2 activates several upstream kinases that phosphorylate IKKα homodimers, leading to phosphorylation of the transcriptionally inactive NF-κB2/p100/RelB complex and proteasomal processing to the transcriptionally active NF-κB2/p52/RelB heterodimer (38), nuclear translocation, and transcription of downstream targets, including CXCL12 (37). We have shown that loss of Twist1 is associated with increased expression of CXCL12 at the level of both mRNA and protein. However, how loss of Twist1 regulates CXCL12 is unknown. In this experiment, we sought to understand the effect of Twist1 silencing on activation of IKKα and one of its upstream kinases MEK kinase 1 (Map3k1) (39). Silencing of Twist1 in IPF fibroblasts did not affect IKKα protein levels (Fig. 5A). Simultaneous silencing of Twist1 and IKKα completely blocked CXCL12 expression (Fig. 5A–C), indicating that IKKα is central to the expression of CXCL12. IKKα expression appears to be independent of Twist1. Twist1 silencing did not affect Map3k1 protein expression (Fig. 5D, 5E). Silencing of Twist1 alone had no effect on IKKα phosphorylation (Fig. 5D, 5F). Silencing of Map3k1 does partially block phosphorylation of IKKα without an effect on total IKKα (Fig. 5G). Silencing of Map3k1 completely blocks expression of CXCL12 whereas simultaneous silencing of Twist1 partially overcomes this inhibition (Fig. 5H). These data indicate that Map3k1 is a principal kinase regulating IKKα phosphorylation in IPF fibroblasts. Following silencing of Map3k1, IKKα remains phosphorylated and appears to be adequate for expression of CXCL12 following the silencing of Twist1. Taken together, these data suggest that the actions of Twist1 are distal to IKKα and may be related to the function of Twist1 as a transcription factor.
FIGURE 5.
Twist1-mediated regulation of CXCL12 expression is downstream of IKKα. (A) Immunoblotting (IB) of CXCL12 and IKKα (encoded by Chuk) from IPF fibroblasts incubated with siTwist1, siChuk, or both. Data were analyzed by two-way ANOVA followed by a Newman–Keuls post hoc test unless otherwise indicated. (B) Band densitometry for IKKα (Chuk) (*p < 0.017, siControl1 plus siControl2 versus siControl1 plus siChuk and **p < 0.03, siTwist1 plus siControl2 versus siTwist1 plus siChuk, n = 3) and (C) CXCL12 (*p < 0.029, siControl1 plus siControl2 versus siTwist1 plus siControl2 and **p = 0.0002, siTwist1 plus siControl2 versus siTwist1 plus siChuk, n = 3). (D) IB for MAP3K1, phospho-IKKα/β, total IKKα, and CXCL12 following silencing of Twist1, MAP3K1, or both. (E–H) Band densitometry was performed for (D). (E) MAP3K1 (*p < 0.0001, siControl1 plus siControl2 versus siControl1 plus siMAP3K1 and **p < 0.0001, siTwist1 plus siControl2 versus siTwist1 plus siMAP3K1, n = 3). (F) Phospho-IKKα/β (*p < 0.031, siControl1 plus siControl2 versus siControl1 plus siMAP3K1, n = 3). (G) Total IKKα. (H) CXCL12 (*p < 0.0001, siControl1 plus siControl2 versus siControl1 plus siMAP3K1 and **p = 0.003, siControl1 plus siMAP3K1 versus siTwist1 plus siMAP3K1).
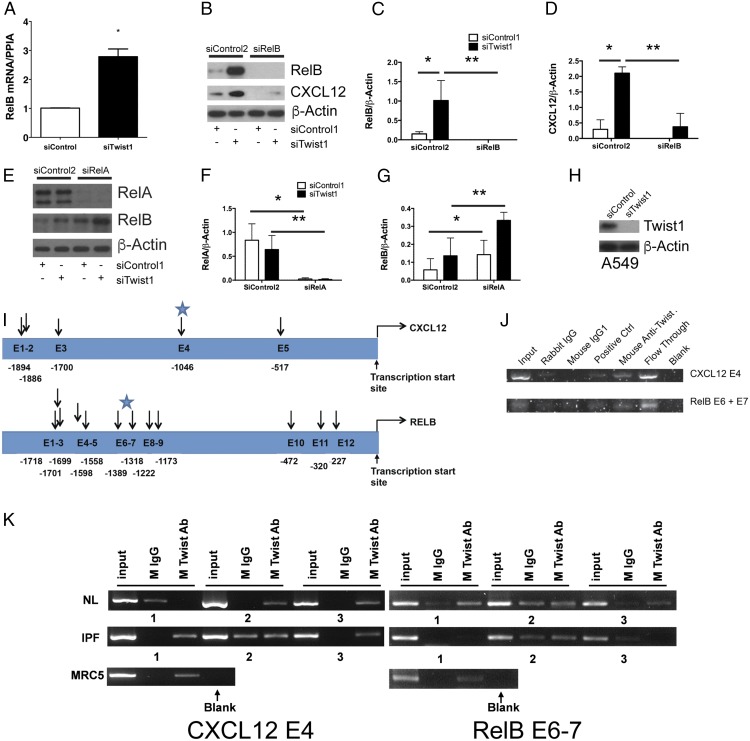
Loss of Twist1 is associated with increased expression of the noncanonical NF-κB transcription factor RelB
In these next experiments we focused on the role of Twist1 as a transcription factor assaying targets downstream of IKKα (Fig. 6). First, we determined the effect of Twist1 on the noncanonical NF-κB family member RelB. Silencing of Twist1 in IPF lung fibroblasts led to a nearly 3-fold increase in RelB mRNA (Fig. 6A). Similarly, at the level of protein there was a dramatic increase in RelB protein upon silencing of Twist1 (Fig. 6B–D). Silencing of RelB in the presence or absence of Twist1 expression completely blocks both RelB expression and CXCL12 (Fig. 6B, 6D). This effect led us to suspect that Twist1 may affect expression of RelA, a principal canonical NF-κB transcription factor regulating RelB expression in immune cells (40). Silencing of Twist1 had no effect on RelA expression (Fig. 6E–G). Simultaneous silencing of RelA completely blocked RelA whereas expression of RelB actually increased in the absence of RelA. Silencing of both RelA and Twist1 were additive, leading to the highest expression of RelB. These data indicate that RelA expression in IPF fibroblasts is unrelated to Twist1 and may even act as an inhibitor of RelB expression.
FIGURE 6.
Loss of Twist1 in human lung fibroblasts leads to increased expression of RelB. (A) IPF fibroblasts were incubated with siTwist1 or siControl. Cells were processed for quantitative RT-PCR for the noncanonical NF-κB transcription factor RelB (*p < 0.003, n = 3, by t test). (B) Immunoblot of IPF fibroblasts following silencing of Twist1, RelB, or both. Data were analyzed by two-way ANOVA followed by a Newman–Keuls post hoc test. (C) ImageJ quantification of RelB (*p < 0.004, siTwist1 plus siControl2 versus siControl1 plus siControl2 and **p = 0.0014, siTwist1 plus siControl2 versus siTwist1 plus siRelB, n = 3) and (D) CXCL12 (*p < 0.0001, siTwist1 plus siControl2 versus siControl1 plus siControl2 and **p < 0.0001, siTwist1 plus siControl2 versus siTwist1 plus siRelB, n = 3). (E) Immunoblot of IPF fibroblasts following silencing of Twist1, the RelB regulator, RelA, or both. (F) ImageJ quantification of RelA (*p = 0.002, siRelA plus siControl2 versus siControl1 plus siControl2 and **p < 0.01, siTwist1 plus siControl2 versus siTwist1 plus siRelA, by two-way ANOVA, n = 3) and (G) RelB (*p < 0.002, siControl1 plus siRelA versus siControl1 plus siControl2 and **p = 0.01, siTwist1 plus siControl2 versus siTwist1 plus siRelA, by two-way ANOVA, n = 3. (H) Immunoblot of A549 cells with siTwist1 or siControl. (I) Locations of potential E-box motifs identified in the CXCL12 (top) and RelB (bottom) gene. The starting nucleotide position relative to the transcription start site was listed for each of the potential E-box motifs identified in the upstream sequences of each gene. Blue stars identify regions of DNA that were amplified by PCR of ChIP products. (J and K) PCR amplification of CXCL12 and RelB upstream sequences using ChIP products from (J) A549 cells and (K) normal, IPF, and MRC5 fibroblasts. For normal and IPF lung fibroblasts, n = 3. ChIP assay of the CXCL12 and RelB upstream sequences were performed using mouse Ab specific for Twist1. Ab specific for DNA polymerase II was used as a positive control and IgG1 purified from rabbit and mouse serum, respectively, were used as negative controls.
In these next experiments, we sought to determine whether Twist1 has a direct interaction with the promoter regions of CXCL12 and RelB. Twist1 binds to E-box motifs, CANNTG, and is most typically associated with transcriptional inhibition (41). Functional E-box motifs have been characterized upstream of the transcriptional start site for CXCL12 (35). To this end, we first employed A549 cells to obtain an adequate number of cells for ChIP. We show that A549 cells express Twist1 (Fig. 6H). For these experiments we scanned the regions directly upstream of the transcriptional start site for CXCL12 and RelB. E-box motifs were identified at the locations noted in Fig. 6I. We hypothesized that we would detect possible interactions between Twist1 and the E-boxes in these regions. To determine whether the potential E-box motifs identified in the upstream sequences of CXCL12 and RelB gene bind to Twist1, we conducted Twist1 protein-specific ChIP assays and amplified the regions spanning these potential E-box motif sites using multiple sets of primers (Table I). PCR amplification (Fig. 6J) was observed in both the input DNA as well as the flow-through fraction of the ChIP assay. As a control, no PCR amplification was observed after ChIP with nonimmune mouse IgG1 or rabbit IgG. Specific PCR products were detected only in the Twist Ab ChIP assay for the CXCL12 E4 and RelB E6 plus E7 sites. We then probed for these same sites in ChIP products from normal, IPF, and MRC5 lung fibroblasts (Fig. 6K). We detected amplification of the CXCL12 E4 region in two of three normal lung fibroblasts sampled, all three IPF fibroblasts, and in the MRC5 cells. Amplification of the RelB E6–7 site was detected in all three normal lung fibroblasts and the MRC5 cells but in only one of three IPF fibroblasts. Therefore, these data suggest that Twist1 interacts with E-box motifs within the promoter regions of CXCL12 and RelB.
Inhibition of CXCR4, the receptor for CXCL12, with AMD3100 blocks pulmonary fibrosis and accumulation of T cells in Twist1 FL mice
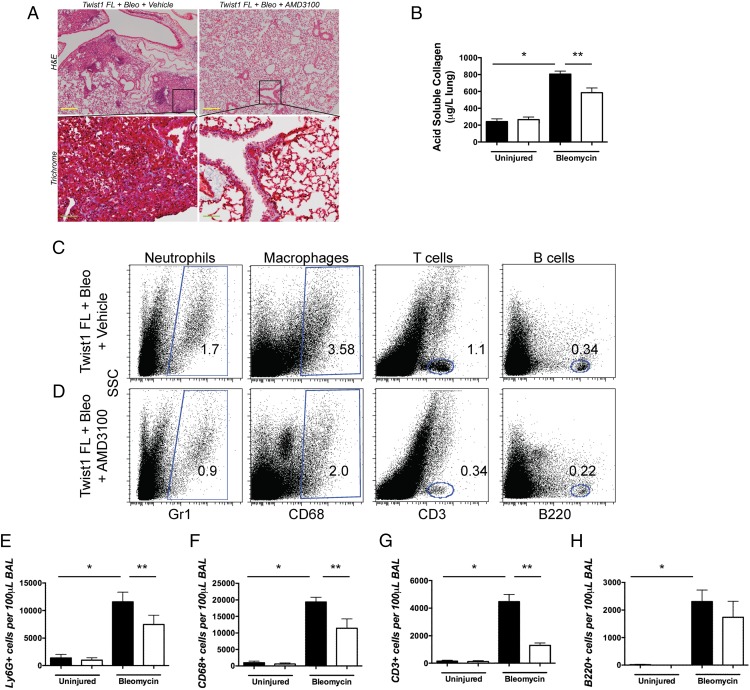
We have shown that the loss of Twist1 leads to increased bleomycin-induced pulmonary fibrosis, possibly through a mechanism that involves expression of CXCL12 and the downstream recruitment of T cells and bone marrow–derived matrix-producing cells to the lung. We next hypothesized that if the loss of Twist1 drives increased expression of CXCL12 and pulmonary fibrosis in Twist1 FL mice, then blockade of CXCL12 would protect from the exaggerated fibrosis. To test this hypothesis, we determined the effect of Twist1 silencing on expression of CXCR4, the receptor for CXCL12. First, we analyzed whether bone marrow–derived COL1A2+ cells expressed CXCR4, a G protein–coupled receptor for CXCL12 (42) (Supplemental Fig. 2A–E). Bone marrows from Twist1 WT and Twist1 FL mice were obtained at 14 d after bleomycin injury or saline control. A small percentage of CD45+ bone marrow cells were tdTomato+ (Supplemental Fig. 2A–C). No significant differences were observed with injury or genotype. Of the CD45+tdTomato+ cells, >95% of the cells were CXCR4+ (Supplemental Fig. 2A, 2B, 2D, 2E), and 40–50% of these cells were Ki67+, indicating that half of the CXCR4+ cells in the bone marrow are proliferating. No differences were observed between genotypes and injury. Next we determined CXCR4 expression in normal and IPF lung fibroblasts after silencing of Twist1 by siRNA (Supplemental Fig. 2F, 2G). Following silencing of Twist1, we found no effect on CXCR4 expression. These data show that CXCR4 is expressed on bone marrow–derived collagen-producing cells in mice and in human lung fibroblasts. Expression of CXCR4 is unaffected by Twist1 expression.
In addition to CXCR4, CXCL12 also binds to CXCR7, but CXCR4 may mediate the more profibrotic effects of CXCL12 (43). Neutralization of CXCL12 signaling (44) and CXCR4 blockade by AMD3100 have been shown to decrease bleomycin-induced pulmonary fibrosis (45). In this experiment, we treated bleomycin-injured or uninjured Twist1 FL mice only with AMD3100 or a vehicle control (Fig. 7). No effect of AMD3100 was observed in the absence of bleomycin injury. After bleomycin, significant injury and fibrosis were observed in the vehicle-treated mice (Fig. 7A). This injury was largely absent from the AMD3100-treated bleomycin-injured Twist1 FL mice. We determined the acid-soluble collagen content following injury (Fig. 7B). Bleomycin injury was associated with a nearly 4-fold increase in collagen content in the Twist1 FL mice. Treatment with AMD3100 significantly blocked but did not completely reverse bleomycin-induced fibrosis in these mice. Next, we reasoned that if CXCL12 is responsible for the homing of inflammatory cells to the lung in the Twist1 FL mice, then blockade of CXCR4 would block this accumulation. We then subjected the cells isolated from the BAL for flow cytometry to detect Ly6G, CD68, CD3, and B220 (Fig. 7C–H) as described above for Fig. 2. Following bleomycin injury, we found that AMD3100 significantly decreased the accumulation of CD3+ cells in the lung. These data suggest that blockade of CXCR4, the CXCL12 receptor, blocks the accumulation of T cells in Twist1 FL mice and partially blocks fibrosis. Additional reductions in Ly6G+ and CD68+ cells with AMD3100 treatment were also observed. AMD3100 may also block binding of MIF to CXCR4 (46). However, by quantitative RT-PCR, we found no significant difference in MIF expression from flow-sorted tdTomato+ cells from Twist1 WT and Twist1 FL animals (data not shown).
FIGURE 7.
Treatment of bleomycin-injured Twist1 FL animals with the CXCR4 blocker AMD3100 decreased the enhanced pulmonary fibrosis associated with the loss of Twist1 and significantly reduced the accumulation of T cells to the lung. Twist1 FL mice were injured with bleomycin or saline control and then treated with or without the CXCR4 blocker AMD3100. Animals were sacrificed on day 14. (A) H&E staining and Masson trichrome staining of lung sections (inset scale bars, 200 μm; original magnification, ×200). (B) The left lungs were processed for collagen determination by the Sircol assay (*p < 0.0001, uninjured versus bleomycin, n = 8–9 and **p = 0.0004 by ANOVA, bleomycin plus vehicle versus bleomycin plus AMD3100, n = 8–9). BAL was processed for flow cytometry for (C) and (D). Dot plots for Ly6G+ neutrophils, CD68+ macrophages, CD3+ T cells, and B220+ B cells from bleomycin-injured Twist1 FL mice treated with or without AMD3100. (E) Ly6G (*p < 0.0001, uninjured plus vehicle versus bleomycin plus vehicle, n = 8–9 and **p < 0.04, bleomycin plus vehicle versus bleomycin plus AMD3100, n = 8–9), (F) CD68 (*p < 0.0001, uninjured plus vehicle versus bleomycin plus vehicle, n = 8–9 and **p < 0.002, bleomycin plus vehicle versus bleomycin plus AMD3100, n = 8–9), (G) CD3 (*p < 0.0001, uninjured plus vehicle versus bleomycin plus vehicle, n = 8–9 and **p < 0.0001, bleomycin plus vehicle versus bleomycin plus AMD3100, n = 8–9), and (H) B220 (*p < 0.0001, uninjured plus vehicle versus bleomycin plus vehicle, n = 8–9). All data were analyzed by ANOVA.
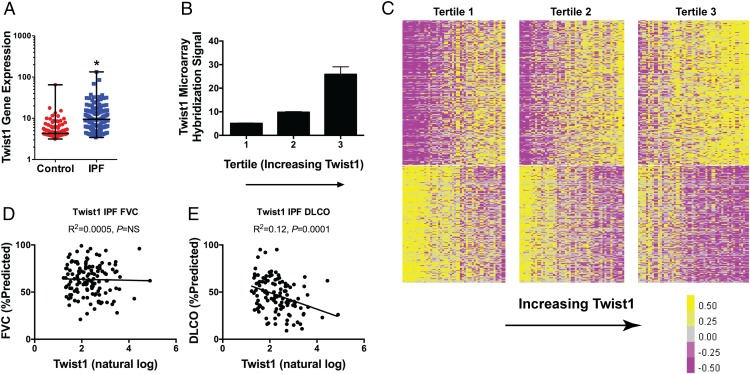
Low Twist1 expression in IPF is associated with enrichment of T cell pathways
We have shown that the loss of Twist1 in the mesenchymal compartment in mice is associated with the accumulation of T cells and enhanced bleomycin-induced pulmonary fibrosis. We next asked whether the loss of Twist1 in bleomycin injury can be modeled in actual IPF clinical data. To explore this question, we analyzed data from the LGRC as described in Materials and Methods. The LGRC cohort (47) is described in Table II. These data combine gene expression analysis from whole-lung homogenates with cross-sectional clinical data. We determined the relative expression for Twist1 between IPF and controls (Fig. 8A). Verifying our original data (3) in a new cohort, by microarray, from 134 patients with IPF compared with 108 normal controls, we found that Twist1 was 2.3-fold increased in IPF. Additionally, we have found that Twist1 expression is increased in IPF compared with controls in an additional cohort (Supplemental Fig. 3A). These gene expression studies have employed whole-lung homogenates. We subjected IPF and normal lung fibroblasts to immunoblotting for baseline Twist1 expression in full media. We found that Twist1 expression was higher in IPF fibroblasts compared with normal lung fibroblasts (Supplemental Fig. 3B). Because of the association of Twist1 with CXCL12 and RelB, we queried the LGRC data for gene expression for these genes (Supplemental Fig. 3C, 3D). From whole lung, we found that CXCL12 goes up in IPF 1.5-fold (p < 0.001) and that RelB goes down 30% (p < 0.05). Next, we arranged the IPF patients into three equal tertiles based on increasing Twist1 gene expression. In Fig. 8B, Twist1 expression increased from quartile 1 to 3 (p = 0.003 for trend by one-way ANOVA, n = 44–45 per tertile). Seeing a broad range of Twist1 gene expression in this dataset, we hypothesized that IPF patients would be transcriptomically distinct based on Twist1 gene expression. To test this hypothesis, we generated a heat map of the IPF patients and arranged the patients in columns based on increasing Twist1 expression (Fig. 8C). Each of the tertiles is marked by a break. Patients in tertile 1 were defined as Twist1-low, and patients in tertile 3 were defined as Twist1-high. With this analysis, we found that 387 genes (at a false discovery rate of <0.05) were differentially expressed between the Twist1-high and Twist1-low patients. The 30 most upregulated and most downregulated genes are presented in Tables III and IV, respectively. We next subjected the differentially expressed gene list to Ingenuity Pathway Analysis. Table V is an analysis of the differentially expressed genes and represents a calculation and a prediction whether certain pathways are activated or inhibited. These data suggest that patients in the Twist1-low tertile are transcriptomically distinct from patients in the Twist1-high tertile and are characterized by activation of T cell pathways downstream of IFN-γ and STAT1. Taken together, Twist1-low IPF patients and bleomycin-injured Twist1 FL mice are characterized by enrichment of T cell pathways.
FIGURE 8.
Twist1 gene expression is increased in IPF and, among IPF patients, negatively correlates with DLCO in IPF and is associated with a distinct gene expression profile. (A) Twist1 gene expression was measured in IPF patients (n = 134) and control patients (n = 107). Gene expression was 2.3-fold higher in IPF (*p < 0.0001, by unpaired t test). (B) IPF patients were organized into three tertiles based on increasing expression of Twist1 (p = 0.003 for trend by one way ANOVA, n = 44–45 per tertile). (C) Heat map of differentially expressed genes in IPF patients based on Twist1 expression. Each column represents a patient, and each row represents a gene. Between the lowest and the highest tertiles, 387 genes are differentially expressed. (D) FVC (percent predicted) and (E) DLCO (percent predicted) as a function of natural log-transformed Twist1 gene expression.
Table III. Top 30 upregulated genes in the Twist1-low tertile (tertile 1).
| Gene Symbol | Gene Name | Entrez ID | Fold Change | −Log (p Value) |
|---|---|---|---|---|
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter), member 4 | 6532 | 4.54 | 3.68 |
| CA4 | Carbonic anhydrase IV | 762 | 2.99 | 3.37 |
| HTR3C | 5-Hydroxytryptamine (serotonin) receptor 3C, ionotropic | 170572 | 2.94 | 3.27 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 6373 | 2.78 | 3.67 |
| SOSTDC1 | Sclerostin domain containing 1 | 25928 | 2.60 | 3.25 |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 4283 | 2.51 | 4.21 |
| RTKN2 | Rhotekin 2 | 219790 | 2.50 | 3.18 |
| FGFBP2 | Fibroblast growth factor binding protein 2 | 83888 | 2.37 | 4.08 |
| GRM8 | Glutamate receptor, metabotropic 8 | 2918 | 2.20 | 4.12 |
| LRRTM4 | Leucine-rich repeat transmembrane neuronal 4 | 80059 | 2.04 | 4.94 |
| KLRF1 | Killer cell lectin-like receptor subfamily F, member 1 | 51348 | 2.04 | 4.38 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 3620 | 2.00 | 3.29 |
| CD160 | CD160 molecule | 11126 | 1.98 | 4.24 |
| SH2D1B | SH2 domain containing 1B | 117157 | 1.89 | 3.35 |
| GBP4 | Guanylate binding protein 4 | 115361 | 1.85 | 6.40 |
| GCOM1 | GRINL1A complex locus 1 | 145781 | 1.84 | 3.22 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 1524 | 1.84 | 4.33 |
| IZUMO1 | Izumo sperm-egg fusion 1 | 284359 | 1.83 | 3.50 |
| COLEC10 | Collectin sub-family member 10 (C-type lectin) | 10584 | 1.82 | 3.94 |
| STXBP6 | Syntaxin binding protein 6 (amisyn) | 29091 | 1.81 | 3.25 |
| fam70a | Transmembrane protein 255A | 55026 | 1.80 | 4.26 |
| FIGF | c-Fos induced growth factor (vascular endothelial growth factor D) | 2277 | 1.78 | 3.11 |
| LRRN3 | Leucine-rich repeat neuronal 3 | 54674 | 1.76 | 3.95 |
| KCNH6 | Potassium channel, voltage gated eag-related subfamily H, member 6 | 81033 | 1.74 | 3.54 |
| CLIC5 | Chloride intracellular channel 5 | 53405 | 1.73 | 3.77 |
| USHBP1 | Usher syndrome 1C binding protein 1 | 83878 | 1.72 | 3.10 |
| SAMD3 | Sterile alpha motif domain containing 3 | 154075 | 1.70 | 4.18 |
| KLRD1 | Killer cell lectin-like receptor subfamily D, member 1 | 3824 | 1.70 | 3.12 |
| TNFSF10 | TNF (ligand) superfamily, member 10 | 8743 | 1.67 | >7 |
| P2RY13 | Purinergic receptor P2Y, G-protein coupled, 13 | 53829 | 1.67 | 4.22 |
Table IV. Top 30 upregulated genes in the Twist1-high tertile (tertile 3).
| Gene Symbol | Gene Name | Entrez ID | Fold Change | −Log (p Value) |
|---|---|---|---|---|
| GREM1 | Gremlin 1, DAN family BMP antagonist | 26585 | 2.86 | 4.62 |
| PLA2G2A | Phospholipase A2, group IIA (platelets, synovial fluid) | 5320 | 2.84 | 3.95 |
| COMP | Cartilage oligomeric matrix protein | 1311 | 2.48 | >7 |
| PCSK1 | Proprotein convertase subtilisin/kexin type 1 | 5122 | 2.46 | 3.23 |
| SLN | Sarcolipin | 6588 | 2.43 | 3.30 |
| c13orf33 | Mesenteric estrogen-dependent adipogenesis | 84935 | 2.34 | 4.94 |
| MMP3 | Matrix metallopeptidase 3 | 4314 | 2.32 | 3.36 |
| ECEL1 | Endothelin converting enzyme-like 1 | 9427 | 2.31 | 3.99 |
| RIMS2 | Regulating synaptic membrane exocytosis 2 | 9699 | 2.27 | 3.93 |
| SNX31 | Sorting nexin 31 | 169166 | 2.26 | 3.15 |
| BDKRB1 | Bradykinin receptor B1 | 623 | 2.26 | 4.08 |
| SCRG1 | Stimulator of chondrogenesis 1 | 11341 | 2.26 | 4.44 |
| CHRDL2 | Chordin-like 2 | 25884 | 2.26 | 3.18 |
| SFRP2 | Secreted frizzled-related protein 2 | 6423 | 2.25 | 5.47 |
| DIO2 | Deiodinase, iodothyronine, type II | 1734 | 2.22 | 6.52 |
| SLC38A11 | Solute carrier family 38, member 11 | 151258 | 2.19 | 5.26 |
| MMP11 | Matrix metallopeptidase 11 | 4320 | 2.12 | 4.08 |
| AVPR1A | Arginine vasopressin receptor 1A | 552 | 2.07 | 5.05 |
| CLEC4G | CCR4-NOT transcription complex, subunit 8 | 339390 | 2.06 | 3.50 |
| ASTN1 | Astrotactin 1 | 460 | 2.06 | 3.80 |
| GJB2 | Gap junction protein, beta 2, 26 kDa | 2706 | 2.00 | 4.03 |
| AGT | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 183 | 1.95 | 5.03 |
| TUBB3 | Tubulin, beta 3 class III | 10381 | 1.95 | 4.48 |
| CTHRC1 | Collagen triple helix repeat containing 1 | 115908 | 1.94 | 5.70 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | 3479 | 1.93 | 4.88 |
| FAM55D | Neurexophilin and PC-esterase domain family, member 4 | 54827 | 1.90 | 3.47 |
| C6orf142 | Muscular LMNA-interacting protein | 90523 | 1.87 | 3.78 |
| BDKRB2 | Bradykinin receptor B2 | 624 | 1.85 | 4.51 |
| ATP10B | ATPase, class V, type 10B | 23120 | 1.84 | 3.29 |
| KCTD8 | Potassium channel tetramerization domain containing 8 | 386617 | 1.83 | 3.60 |
Table V. Predicted upstream regulators of Twist1-low (tertile 1) versus Twist1-high (tertile 3).
| Upstream Regulator | Molecule Type | Predicted Activation State | Activation z-Score | −Log (p Value of Overlap) | Target Molecules in Dataset |
|---|---|---|---|---|---|
| IFNG | Cytokine | Activated | 2.431 | 12.66 | ACTB, ADGRG2, AGT, ALDH1A3, AVPR1A, B2M, BATF2, BST2, BTN3A1, C1R, CASP1, CCL11, CD2, CHST7, CHST8, CLIC5, CX3CR1, CXCL11, CXCL9, FHL2, GBP1, GBP4, GCH1, GJB2, GLIS1, GRM8, HIF1A, HMGCR, IDO1, IFI44L, IFIH1, IGF1, IGFBP4, IL23R, ITGAL, KLRC4-KLRK1/KLRK1, LOX, MMP11, MMP3, OAS2, OAS3, PCSK1, PLA2G2A, PRDX2, PSMB9, SCG5, SMAD1, TACR2, TBX21, TIMP1, TLR3, TLR7, TMEM158, TNFAIP6, TNFSF11, TRPC4, WARS |
| TNF | Cytokine | 0.767 | 10.34 | ACTB, ADRB2, AGT, ALDH1A3, APC, AVPR1A, B2M, BDKRB1, BDKRB2, BST2, CASP1, CCL11, COL15A1, COL3A1, CX3CR1, CXCL11, CXCL9, FERMT1, FIGF, FST, FZD5, GADD45A, GBP1, GCH1, HIF1A, HMGCR, IDO1, IFIH1, IGF1, IGFBP4, IGFBP5, IGFBP6, ITGAL, LOX, MMP3, NGFR, NME1, NPPA, NUMB, OAS2, OAS3, OSMR, PLA2G2A, PPAP2A, PSMB9, RBPMS, SERPINF1, SLC2A4, SMAD1, TBX21, TDRD7, TGFB3, TIMP1, TLR3, TLR7, TNC, TNFAIP6, TNFSF11, TPST1, VIP | |
| LPS | Chemical drug | −0.073 | 9.05 | ADRB2, AGT, AK4, AVPR1A, BDKRB1, CASP1, CCL11, CD160, CHST7, COL3A1, COX4I1, CX3CR1, CXCL11, CXCL9, DIO2, DRD5, EHHADH, FBLN2, FST, GADD45A, GBP1, GBP4, GCH1, GJB2, HIF1A, HMGCR, IDO1, IFI44L, IFIH1, IGF1, IGFBP4, IGFBP5, IL23R, ITGAL, LGALS1, MMP3, NGFR, NPPA, OAS2, OAS3, OSMR, PLA2G2A, PSMB9, RNF19A, RXRG, SERPINF1, SLC39A14, TBX21, TIMP1, TLR3, TLR7, TNC, TNFAIP6, TNFSF11, TP53BP2, TPBG,VCAN, VIP, WARS | |
| IL1B | Cytokine | 0.178 | 9.04 | ADRB2, AVPR1A, BDKRB1, C1R, CCL11, CX3CR1, CXCL11, CXCL9, FST, GADD45A, GBP1, GCH1, GYS1, HIF1A, IDO1,IGF1, IGFBP4, IGFBP5, IGFBP6, IL23R, LOX, MMP11, MMP3, NPPA, OAS2, OSMR, PCSK1, PLA2G2A, PSMB9, SLC2A4, SLC6A4, TBX21, TGFB3, TIMP1, TLR3, TLR7, TNFAIP6, TNFSF11, VCAN | |
| STAT3 | Transcription regulator | Inhibited | −2.600 | 8.85 | AGT, BOC, BST2, CASP1, CCL11, COL3A1, CXCL9, FST, GADD45A, HIF1A, IFIH1, IGFBP5, IL23R, KLRC4-KLRK1/KLRK1, MMP3, NPPA, OAS2, OAS3, PCSK1, PLA2G2A, PSMB9, TBX21, TIMP1, TLR3, TNFSF11, VCAN, VIP, WARS |
| TGFB1 | Growth factor | Inhibited | −3.305 | 8.77 | ACKR1, ADGRG2, B2M, CASP1, CCL11, CHST8, COL3A1, CPXM1, CTNND1, CX3CR1, CYP21A2, ECM1, FBLN2, FERMT1, GADD45A, GBP1, GDF6, GJB2, GREM1, HIF1A, IFIH1, IGF1, IGFBP4, IGFBP5, IGFBP6, IL23R, ITGA7, ITGAL, KDELR2, KLRC4-KLRK1/KLRK1, KLRD1, KRT10, LOX, LTBP1, MMP11, MMP3, MYO1C, NME1, NPPA, OSR2, PPP2R5A, RBPMS, RHOD, SCG5, SLC39A14, SLN, TBX21, TGFB3, TIMP1, TNC, TNFAIP6, TNFSF11, TUBB3, UCK2, USP25, VCAN, VIP, XPNPEP2 |
| SB203580 | Chemical, kinase inhibitor | 0.741 | 8.27 | ADAMTS9, ADRB2, AGT, BDKRB1, BST2, CCL11, COL3A1, CX3CR1, CXCL11, CXCL9, GADD45A, GPI, HIF1A, IDO1, LMO2, MMP3, NPPA, TGFB3, TIMP1, TLR3, TNFAIP6, TNFSF11, ZFYVE26 | |
| IL6 | Cytokine | −1.288 | 7.97 | AGT, BST2, CASP1, CCL11, COL3A1, COX4I1, CX3CR1, GADD45A, GREM1, HIF1A, IDO1, IGF1, IGFBP4, IGFBP5, IGFBP6, IL23R, KLRD1, LGALS1, MMP3, PLA2G2A, PSMB9, PTP4A3, SLC2A4, SLC39A14, SMOX, TIMP1, TLR3, TLR7, TNC, TNFSF11, VIP, WARS, XPNPEP2 | |
| Estrogen | Chemical drug | −0.763 | 7.96 | AGT, COL14A1, COL15A1, COL3A1, GJB2, HIF1A, HMGCR, IDO1, IGF1, IGFBP4, IGFBP5, IGFBP6, LGALS1, MTA3, NGFR, NME1, SLC6A4, TBX21, TGFB3, TNFSF11 | |
| IFNL1 | Cytokine | Activated | 3.293 | 7.76 | BST2, CXCL11, CXCL9, DDX60, GBP1, IFI44L, IFIH1, OAS2, OAS3, PSMB9, TDRD7 |
| Tretinoin | Chemical, endogenous mammalian | 0.159 | 7.51 | AGT, ALDH1A3, CASP1, COL3A1, CRABP2, CX3CR1, CYP21A2, DAPK2, DDX60, DIO2, FERMT1, FST, GBP4, GIMAP2, GREM1, IFI44L, IFIH1, IGF1, IGFBP4, IGFBP5, IGFBP6, IL23R, ITGAL, LGALS1, MMP11, MMP3, MYO1C, NPPA, NUMB, OAS2, OAS3, PRDX2, PRKCH, PSMB9, PTP4A3, RTKN2, RXRG, SLC2A4, SMOX, TGFB3, TIMP1, TLR3, TNC, TNFAIP6, TNFSF11, TRPC4, TUBB3, UBA7, VIP, YPEL1 | |
| Rosiglitazone | Chemical drug | 0.836 | 7.48 | AGT, APOL3, CCL11, COX4I1, CXCL11, CXCL9, DIO2, GBP1, GPR65, IGF1, IGFBP5, IGFBP6, ITGAL, KLF11, LOX, NPPA, OAS2, OAS3, SERPINF1, SLC2A4, TBX21, TGFB3, TIMP1, TNC | |
| IL1RN | Cytokine | Inhibited | −2.514 | 7.32 | BDKRB1, BTN3A1, CXCL11, GBP1, IFI44L, IFIH1, IGF1, IGFBP4, IGFBP5, OAS2, OAS3, SLC6A4, TIMP1 |
| STAT1 | Transcription regulator | Activated | 2.169 | 7.31 | AGT, ALDH1A3, B2M, BATF2, CASP1, CLIC5, CXCL11, CXCL9, GBP1, HIF1A, IDO1, IGF1, NPPA, OAS2, PSMB9, TBX21, TLR3, TP53BP2, WARS |
| IL1 | Group | −0.269 | 7.29 | ADRB2, CASP1, CXCL11, CXCL9, FST, FZD5, HIF1A, HMGCR, IDO1, IGF1, LOX, MMP3, NGFR, PLA2G2A, TGFB3, TIMP1, TLR7, TNC, TNFAIP6, TNFSF11, VCAN |
Although Twist1-high and Twist1-low patients are transcriptomically distinct, are they clinically distinct? We returned to the LGRC data and plotted forced vital capacity (FVC) and diffusing capacity for carbon monoxide (DLCO) as a function of Twist1 expression (Fig. 8D, 8E). We found that Twist1 expression was not associated with FVC but was negatively associated with DLCO. Taken together, these data show that the transcriptomic differences between Twist1-high and Twist1-low patients are associated with a clinically relevant measure of gas exchange in IPF patients.
Discussion
In this study, we tested the hypothesis that the loss of Twist1 in the mesenchymal compartment would promote a fibroblast that is susceptible to apoptosis and thus protect these mice from pulmonary fibrosis. To our surprise, we found quite the opposite. Bleomycin-induced pulmonary fibrosis was exaggerated and characterized by T cell–dominant inflammation in the lung. To date, the association of Twist1 with canonical NF-κB activity and chronic systemic inflammation in Twist1 and Twist2 heterozygous mice has been known (12). However, our data show a very clear role for Twist1 as a regulator of noncanonical NF-κB signaling and, through CXCL12, leads to several potentially profibrotic effects downstream. Increased CXCL12 in IPF fibroblasts is the result of de-repression of both CXCL12 and RelB transcription.
A relationship between Twist1 and CXCL12 has recently been identified in cancer-associated fibroblasts (CAF) (48). The authors show that high expression of Twist1 is associated with increased expression of CXCL12. These observations stand in contrast to our data where Twist1 acts as a repressor of CXCL12 expression. The authors do show a decrease in CXCL12 luciferase activity with induction of Twist1 in 293T cells, which would go along with our observation of Twist1 as a transcriptional inhibitor. Increased CXCL12 in CAF may be related to other mechanisms not present in IPF lung fibroblasts. The increase in CXCL12 that we observed in IPF fibroblasts may be related principally to increased RelB. To our knowledge, this association between RelB and Twist1 has not been described previously. In our system, we actually found that RelA, known to be a transcriptional regulator of RelB expression (49), appears to inhibit expression of RelB. Further study is needed to determine whether this relationship is present in other inflammatory cells and to explore the extent to which this mechanism may operate in other disease processes, where fibroblast-like cells are known to express CXCL12 such as rheumatoid arthritis and CAF (35, 50). These data support the concept that mesenchymal cells in the lung are immunologically active and can act as potent regulators of the local inflammatory infiltrate with critical effects on phenotypes in the lung (36, 45, 51).
If the Twist1-null mesenchymal cells enhance the accumulation of T cells into the injured lung, to what extent is fibrosis driven by these T cells? It is possible that the T cell infiltrate may drive fibrosis, as has been suggested previously (52). We recovered more CD4+ and CD8+ cells from bleomycin-injured Twist1 FL animals compared with the Twist1 WT controls. Th1 and regulatory T cells appeared to be most highly represented group among the CD4+ cells. All CD8 subphenotypes were more highly represented in the Twist1 FL animals. However, the implications of increased numbers of CD8+ cells in bleomycin injury and IPF in general are unclear. The data on the role of T cells in animal models of fibrosis do not clearly identify whether they are pathologic. CXCL10 signaling appears to be protective against pulmonary fibrosis (53, 54). Note that we did not detect altered expression of CXCL9, CXCL10, or CXCL11 in tdTomato+ cells isolated from bleomycin-injured mice (data not shown). Conversely, deficiency of Th1 cells may be deleterious. T-bet–null mice have increased susceptibility to bleomycin injury (55), suggesting a positive role for Th1 cells in this model. In contrast, loss of T regulatory CD4+CD25+ cells protected against bleomycin injury (56). This suggests that the balance of CD4+ cell subphenotypes is crucial to regulating pulmonary fibrosis. Future studies to deplete T cells in Twist1 FL mice or simultaneous conditional knockout of CXCL12 may determine to what extent T cells are necessary for the increased fibrosis observed in bleomycin-injured Twist1 FL mice.
Finally, we have found that a cohort of IPF patients with low expression of Twist1 is characterized by activation of T cell pathways. This is entirely consistent with our experimental model where loss of Twist1 in the mesenchymal compartment is associated with accumulation of T cells. However, we are left to reconcile what appears to be a paradox between the animal model and IPF: the animal is worse, but is the IPF patient better? We can only speculate whether the Twist1-low IPF patient is indeed better for several reasons. First, the LGRC data are cross-sectional, so the outcomes for these patients are unknown. It would seem that enhancement of inflammation as exemplified in the Twist1-low patient would be a negative prognostic factor in IPF and might contribute to reduced survival. Second, although the difference in DLCO between the tertiles suggests that the transcriptomic distinctiveness of Twist1-high and Twist1-low patients is associated with an actual clinical phenotype, the lack of association of Twist1 with FVC (57)—the most reproducible measure of prognosis in IPF—may argue against the idea that Twist1-low patients are healthier. Third, we have modeled loss of Twist1 in the mesenchymal compartment. However, because the LGRC data are from whole-lung gene expression, the cell or cells that express Twist1 at a low level are unknown. Future studies, perhaps with single-cell RNA sequencing from suspensions of whole IPF lungs may provide the granularity to correlate Twist1 with CXCL12 and RelB in vivo. Although epithelial cells are potentially an important cell type to study in the context of Twist1 expression, it has recently been found that epithelial cell–specific loss of Twist1 does not have a fibrosis phenotype (58). The association of Twist1 with DLCO may also argue for a pulmonary vascular phenotype, as may be suggested by experimental data with adventitial fibroblasts (36, 51). Further evidence in support of the association of Twist1 with a pulmonary vascular phenotype comes from recent data that show that knockout of Twist1 in endothelial cells is protective from bleomycin injury (59). Inflammation may be reduced in with loss of Twist1 in endothelial cells, which may, in part, explain the protection from bleomycin injury that the authors observed. Finally, this apparent paradox between our observations in mice and in patients highlights the contrasts between bleomycin injury in mice and IPF. We suggest that the critical observation from our experimental data is the effect of T cell activation on the phenotype of bleomycin-injured mice and IPF patients. Further study is needed to understand the role of Twist1 in other compartments in the lung in other experimental models of pulmonary fibrosis in addition to the role that inflammation plays in IPF.
IPF is a disease classically defined as independent of inflammation (60). However, in studies of both experimental pulmonary fibrosis and IPF (29, 52, 61–64), there is now increased recognition of the role that inflammation can play (65). We have found that the loss of Twist1 in the mesenchymal compartment in mice leads to increased expression of CXCL12, which promotes crosstalk with and accumulation of T cells in the lung and increased pulmonary fibrosis. Twist1 may be one of the factors that shape the fibrotic phenotype in IPF and in experimental lung injury.
Supplementary Material
This work was supported by the Dorothy P. and Richard P. Simmons Center for Interstitial Lung Disease as funded by National Institutes of Health Grants R01HL095397 and RC2HL101715 (to N.K.), DK104680-01A1 (to P.S.B.), and R01HL126990 (to D.J.K.). The Human Airway Cell and Tissue Core (supported in part by National Institutes of Health Grant P30 DK072506 and by a Cystic Fibrosis Foundation research development program grant) provided human lung fibroblasts.
The online version of this article contains supplemental material.
- BAL
- bronchoalveolar lavage
- CAF
- cancer-associated fibroblast
- ChIP
- chromatin immunoprecipitation
- DLCO
- diffusing capacity for carbon monoxide
- FVC
- forced vital capacity
- IPF
- idiopathic pulmonary fibrosis
- LGRC
- Lung Genomics Research Consortium
- MIF
- macrophage migration inhibitory factor
- siRNA
- short interfering RNA
- α-SMA
- α-smooth muscle actin
- Twist1 FL
- Twist1 locus flanked by loxP sites
- Twist1 WT
- wild-type control Twist1.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Thannickal V. J., Horowitz J. C. 2006. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc. Am. Thorac. Soc. 3: 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawson W. E., Crossno P. F., Polosukhin V. V., Roldan J., Cheng D. S., Lane K. B., Blackwell T. R., Xu C., Markin C., Ware L. B., et al. 2008. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: association with altered surfactant protein processing and herpesvirus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 294: L1119–L1126. [DOI] [PubMed] [Google Scholar]
- 3.Bridges R. S., Kass D., Loh K., Glackin C., Borczuk A. C., Greenberg S. 2009. Gene expression profiling of pulmonary fibrosis identifies Twist1 as an antiapoptotic molecular “rectifier” of growth factor signaling. Am. J. Pathol. 175: 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajayi I. O., Sisson T. H., Higgins P. D., Booth A. J., Sagana R. L., Huang S. K., White E. S., King J. E., Moore B. B., Horowitz J. C. 2013. X-linked inhibitor of apoptosis regulates lung fibroblast resistance to Fas-mediated apoptosis. Am. J. Respir. Cell Mol. Biol. 49: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker M. W., Rossi D., Peterson M., Smith K., Sikström K., White E. S., Connett J. E., Henke C. A., Larsson O., Bitterman P. B. 2014. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 124: 1622–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Z., Sasaoka T., Fujimori T., Oya T., Ishii Y., Sabit H., Kawaguchi M., Kurotaki Y., Naito M., Wada T., et al. 2005. Deletion of the PDGFR-β gene affects key fibroblast functions important for wound healing. J. Biol. Chem. 280: 9375–9389. [DOI] [PubMed] [Google Scholar]
- 7.Xia H., Khalil W., Kahm J., Jessurun J., Kleidon J., Henke C. A. 2010. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am. J. Pathol. 176: 2626–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia H., Diebold D., Nho R., Perlman D., Kleidon J., Kahm J., Avdulov S., Peterson M., Nerva J., Bitterman P., Henke C. 2008. Pathological integrin signaling enhances proliferation of primary lung fibroblasts from patients with idiopathic pulmonary fibrosis. J. Exp. Med. 205: 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thisse B., el Messal M., Perrin-Schmitt F. 1987. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 15: 3439–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard T. D., Paznekas W. A., Green E. D., Chiang L. C., Ma N., Ortiz de Luna R. I., Garcia Delgado C., Gonzalez-Ramos M., Kline A. D., Jabs E. W. 1997. Mutations in TWIST, a basic helix–loop–helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 15: 36–41. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z. F., Behringer R. R. 1995. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9: 686–699. [DOI] [PubMed] [Google Scholar]
- 12.Šošić D., Richardson J. A., Yu K., Ornitz D. M., Olson E. N. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell 112: 169–180. [DOI] [PubMed] [Google Scholar]
- 13.Okada T., Sugie I., Aisaka K. 1993. Effects of gamma-interferon on collagen and histamine content in bleomycin-induced lung fibrosis in rats. Lymphokine Cytokine Res. 12: 87–91. [PubMed] [Google Scholar]
- 14.Cheng G. Z., Chan J., Wang Q., Zhang W., Sun C. D., Wang L. H. 2007. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 67: 1979–1987. [DOI] [PubMed] [Google Scholar]
- 15.Karreth F., Tuveson D. A. 2004. Twist induces an epithelial-mesenchymal transition to facilitate tumor metastasis. Cancer Biol. Ther. 3: 1058–1059. [DOI] [PubMed] [Google Scholar]
- 16.Pozharskaya V., Torres-González E., Rojas M., Gal A., Amin M., Dollard S., Roman J., Stecenko A. A., Mora A. L. 2009. Twist: a regulator of epithelial-mesenchymal transition in lung fibrosis. PLoS One 4: e7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng B., Zhang Z., Black C. M., de Crombrugghe B., Denton C. P. 2002. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160: 1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D. M., Olson E. N., et al. 2004. A twist code determines the onset of osteoblast differentiation. Dev. Cell 6: 423–435. [DOI] [PubMed] [Google Scholar]
- 19.Devor D. C., Bridges R. J., Pilewski J. M. 2000. Pharmacological modulation of ion transport across wild-type and ΔF508 CFTR-expressing human bronchial epithelia. Am. J. Physiol. Cell Physiol. 279: C461–C479. [DOI] [PubMed] [Google Scholar]
- 20.Pilewski J. M., Liu L., Henry A. C., Knauer A. V., Feghali-Bostwick C. A. 2005. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am. J. Pathol. 166: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kass D., Bridges R. S., Borczuk A., Greenberg S. 2007. Methionine aminopeptidase-2 as a selective target of myofibroblasts in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 37: 193–201. [DOI] [PubMed] [Google Scholar]
- 22.Shu H. K., Yoon Y., Hong S., Xu K., Gao H., Hao C., Torres-Gonzalez E., Nayra C., Rojas M., Shim H. 2013. Inhibition of the CXCL12/CXCR4-axis as preventive therapy for radiation-induced pulmonary fibrosis. PLoS One 8: e79768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramani K., Garg A. V., Jawale C. V., Conti H. R., Whibley N., Jackson E. K., Shiva S. S., Horne W., Kolls J. K., Gaffen S. L., Biswas P. S. 2016. The Kallikrein-Kinin system: a novel mediator of IL-17-driven anti-Candida immunity in the kidney. PLoS Pathog. 12: e1005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramani K., Pawaria S., Maers K., Huppler A. R., Gaffen S. L., Biswas P. S. 2014. An essential role of interleukin-17 receptor signaling in the development of autoimmune glomerulonephritis. J. Leukoc. Biol. 96: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kass D. J., Yu G., Loh K. S., Savir A., Borczuk A., Kahloon R., Juan-Guardela B., Deiuliis G., Tedrow J., Choi J., et al. 2012. Cytokine-like factor 1 gene expression is enriched in idiopathic pulmonary fibrosis and drives the accumulation of CD4+ T cells in murine lungs: evidence for an antifibrotic role in bleomycin injury. Am. J. Pathol. 180: 1963–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S., Yerges-Armstrong L. M., Chu Y., Zmuda J. M., Zhang Y. 2015. AP2 suppresses osteoblast differentiation and mineralization through down-regulation of Frizzled-1. Biochem. J. 465: 395–404. [DOI] [PubMed] [Google Scholar]
- 27.Yu S., Yerges-Armstrong L. M., Chu Y., Zmuda J. M., Zhang Y. 2013. E2F1 effects on osteoblast differentiation and mineralization are mediated through up-regulation of frizzled-1. Bone 56: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer Y., Tedrow J., de Bernard S., Birker-Robaczewska M., Gibson K. F., Guardela B. J., Hess P., Klenk A., Lindell K. O., Poirey S., et al. 2015. A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 52: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herazo-Maya J. D., Noth I., Duncan S. R., Kim S., Ma S. F., Tseng G. C., Feingold E., Juan-Guardela B. M., Richards T. J., Lussier Y., et al. 2013. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci. Transl. Med. 5: 205ra136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu W., Dave N., Tseng G. C., Richards T., Xing E. P., Kaminski N. 2005. Comparison of normalization methods for CodeLink Bioarray data. BMC Bioinformatics 6: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konishi K., Gibson K. F., Lindell K. O., Richards T. J., Zhang Y., Dhir R., Bisceglia M., Gilbert S., Yousem S. A., Song J. W., et al. 2009. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 180: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock J. R., Barkauskas C. E., Cronce M. J., Xue Y., Harris J. R., Liang J., Noble P. W., Hogan B. L. 2011. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA 108: E1475–E1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips R. J., Burdick M. D., Hong K., Lutz M. A., Murray L. A., Xue Y. Y., Belperio J. A., Keane M. P., Strieter R. M. 2004. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. J. Clin. Invest. 114: 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto N., Jin H., Liu T., Chensue S. W., Phan S. H. 2004. Bone marrow-derived progenitor cells in pulmonary fibrosis. J. Clin. Invest. 113: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago B., Calonge E., Del Rey M. J., Gutierrez-Cañas I., Izquierdo E., Usategui A., Galindo M., Alcamí J., Pablos J. L. 2011. CXCL12 gene expression is upregulated by hypoxia and growth arrest but not by inflammatory cytokines in rheumatoid synovial fibroblasts. Cytokine 53: 184–190. [DOI] [PubMed] [Google Scholar]
- 36.Li M., Riddle S. R., Frid M. G., El Kasmi K. C., McKinsey T. A., Sokol R. J., Strassheim D., Meyrick B., Yeager M. E., Flockton A. R., et al. 2011. Emergence of fibroblasts with a proinflammatory epigenetically altered phenotype in severe hypoxic pulmonary hypertension. J. Immunol. 187: 2711–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madge L. A., May M. J. 2010. Classical NF-κB activation negatively regulates noncanonical NF-κB-dependent CXCL12 expression. J. Biol. Chem. 285: 38069–38077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun S. C. 2012. The noncanonical NF-κB pathway. Immunol. Rev. 246: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee F. S., Peters R. T., Dang L. C., Maniatis T. 1998. MEKK1 activates both IκB kinase α and IκB kinase β. Proc. Natl. Acad. Sci. USA 95: 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millet P., McCall C., Yoza B. 2013. RelB: an outlier in leukocyte biology. J. Leukoc. Biol. 94: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laursen K. B., Mielke E., Iannaccone P., Füchtbauer E. M. 2007. Mechanism of transcriptional activation by the proto-oncogene Twist1. J. Biol. Chem. 282: 34623–34633. [DOI] [PubMed] [Google Scholar]
- 42.Balabanian K., Lagane B., Infantino S., Chow K. Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. 2005. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280: 35760–35766. [DOI] [PubMed] [Google Scholar]
- 43.Ding B. S., Cao Z., Lis R., Nolan D. J., Guo P., Simons M., Penfold M. E., Shido K., Rabbany S. Y., Rafii S. 2014. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 505: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J., Mora A., Shim H., Stecenko A., Brigham K. L., Rojas M. 2007. Role of the SDF-1/CXCR4 axis in the pathogenesis of lung injury and fibrosis. Am. J. Respir. Cell Mol. Biol. 37: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song J. S., Kang C. M., Kang H. H., Yoon H. K., Kim Y. K., Kim K. H., Moon H. S., Park S. H. 2010. Inhibitory effect of CXC chemokine receptor 4 antagonist AMD3100 on bleomycin induced murine pulmonary fibrosis. Exp. Mol. Med. 42: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz V., Krüttgen A., Weis J., Weber C., Ostendorf T., Lue H., Bernhagen J. 2012. Role for CD74 and CXCR4 in clathrin-dependent endocytosis of the cytokine MIF. Eur. J. Cell Biol. 91: 435–449. [DOI] [PubMed] [Google Scholar]
- 47.Tan J., Tedrow J. R., Dutta J. A., Juan-Guardela B., Nouraie M., Chu Y., Trejo Bittar H., Ramani K., Biswas P. S., Veraldi K. L., et al. 2016. Expression of RXFP1 is decreased in idiopathic pulmonary fibrosis. Implications for relaxin-based therapies. Am. J. Respir. Crit. Care Med. 194: 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee K. W., Yeo S. Y., Sung C. O., Kim S. H. 2015. Twist1 is a key regulator of cancer-associated fibroblasts. Cancer Res. 75: 73–85. [DOI] [PubMed] [Google Scholar]
- 49.Bren G. D., Solan N. J., Miyoshi H., Pennington K. N., Pobst L. J., Paya C. V. 2001. Transcription of the RelB gene is regulated by NF-κB. Oncogene 20: 7722–7733. [DOI] [PubMed] [Google Scholar]
- 50.Kojima Y., Acar A., Eaton E. N., Mellody K. T., Scheel C., Ben-Porath I., Onder T. T., Wang Z. C., Richardson A. L., Weinberg R. A., Orimo A. 2010. Autocrine TGF-β and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. USA 107: 20009–20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frid M. G., Li M., Gnanasekharan M., Burke D. L., Fragoso M., Strassheim D., Sylman J. L., Stenmark K. R. 2009. Sustained hypoxia leads to the emergence of cells with enhanced growth, migratory, and promitogenic potentials within the distal pulmonary artery wall. Am. J. Physiol. Lung Cell. Mol. Physiol. 297: L1059–L1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reilkoff R. A., Peng H., Murray L. A., Peng X., Russell T., Montgomery R., Feghali-Bostwick C., Shaw A., Homer R. J., Gulati M., et al. 2013. Semaphorin 7a+ regulatory T cells are associated with progressive idiopathic pulmonary fibrosis and are implicated in transforming growth factor-β1–induced pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 187: 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang D., Liang J., Hodge J., Lu B., Zhu Z., Yu S., Fan J., Gao Y., Yin Z., Homer R., et al. 2004. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J. Clin. Invest. 114: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang D., Liang J., Campanella G. S., Guo R., Yu S., Xie T., Liu N., Jung Y., Homer R., Meltzer E. B., et al. 2010. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J. Clin. Invest. 120: 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J., Mora A. L., LaVoy J., Brigham K. L., Rojas M. 2006. Increased bleomycin-induced lung injury in mice deficient in the transcription factor T-bet. Am. J. Physiol. Lung Cell. Mol. Physiol. 291: L658–L667. [DOI] [PubMed] [Google Scholar]
- 56.Boveda-Ruiz D., D’Alessandro-Gabazza C. N., Toda M., Takagi T., Naito M., Matsushima Y., Matsumoto T., Kobayashi T., Gil-Bernabe P., Chelakkot-Govindalayathil A. L., et al. 2013. Differential role of regulatory T cells in early and late stages of pulmonary fibrosis. Immunobiology 218: 245–254. [DOI] [PubMed] [Google Scholar]
- 57.du Bois R. M., Weycker D., Albera C., Bradford W. Z., Costabel U., Kartashov A., King T. E., Jr., Lancaster L., Noble P. W., Sahn S. A., et al. 2011. Forced vital capacity in patients with idiopathic pulmonary fibrosis: test properties and minimal clinically important difference. Am. J. Respir. Crit. Care Med. 184: 1382–1389. [DOI] [PubMed] [Google Scholar]
- 58.Yang J., Velikoff M., Agarwal M., Disayabutr S., Wolters P. J., Kim K. K. 2015. Overexpression of inhibitor of DNA-binding 2 attenuates pulmonary fibrosis through regulation of c-Abl and Twist. Am. J. Pathol. 185: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mammoto T., Jiang A., Jiang E., Mammoto A. 2016. Role of twist1 phosphorylation in angiogenesis and pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 55: 633–644. [DOI] [PubMed] [Google Scholar]
- 60.Selman M., King T. E., Pardo A., American Thoracic S., European Respiratory S., American Thoracic Society. European Respiratory Society. American College of Chest Physicians 2001. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann. Intern. Med. 134: 136–151. [DOI] [PubMed] [Google Scholar]
- 61.Xue J., Kass D. J., Bon J., Vuga L., Tan J., Csizmadia E., Otterbein L., Soejima M., Levesque M. C., Gibson K. F., et al. 2013. Plasma B lymphocyte stimulator and B cell differentiation in idiopathic pulmonary fibrosis patients. J. Immunol. 191: 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prasse A., Probst C., Bargagli E., Zissel G., Toews G. B., Flaherty K. R., Olschewski M., Rottoli P., Müller-Quernheim J. 2009. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 179: 717–723. [DOI] [PubMed] [Google Scholar]
- 63.Luzina I. G., Kopach P., Lockatell V., Kang P. H., Nagarsekar A., Burke A. P., Hasday J. D., Todd N. W., Atamas S. P. 2013. Interleukin-33 potentiates bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 49: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donahoe M., Valentine V. G., Chien N., Gibson K. F., Raval J. S., Saul M., Xue J., Zhang Y., Duncan S. R. 2015. Autoantibody-targeted treatments for acute exacerbations of idiopathic pulmonary fibrosis. [Published erratum appears in 2015 PLos One 10: e0133684.] PLoS One 10: e0127771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King T. E., Jr., Pardo A., Selman M. 2011. Idiopathic pulmonary fibrosis. Lancet 378: 1949–1961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.