Abstract
The highly pathogenic avian influenza (HPAI) H5N1 virus poses a significant potential threat to human society due to its wide spread and rapid evolution. In this study, we present a comprehensive antigenic map for HPAI H5N1 viruses including 218 newly sequenced isolates from diverse regions of mainland China, by computationally separating almost all HPAI H5N1 viruses into 15 major antigenic clusters (ACs) based on their hemagglutinin sequences. Phylogenetic analysis showed that 12 of these 15 ACs originated in China in a divergent pattern. Further analysis of the dissemination of HPAI H5N1 virus in China identified that the virus’s geographic expansion was co-incident with a significant divergence in antigenicity. Moreover, this antigenic diversification leads to global antigenic complexity, as typified by the recent HPAI H5N1 spread, showing extensive co-circulation and local persistence. This analysis has highlighted the challenge in H5N1 prevention and control that requires different planning strategies even inside China.
The highly pathogenic avian influenza (HPAI) H5N1 virus has become of global concern since the isolation and identification of the strain A/Goose/Guangdong/1/1996 (GsGD) in Guangdong province of China in 19961,2,3. Since then, the GsGD lineage of HPAI H5N1 virus has spread into many countries and regions in Asia, Europe, Africa and North America, causing epizootic and panzootic infections in birds of many species, killing tens of millions of birds and spurring the culling of hundreds of millions of poultry to halt its spread1,3. Moreover, as of 21 November 2016, sporadic infections of the HPAI H5N1 virus have been responsible for 452 known fatalities among 856 confirmed human infections4.
Given the wide spread of the virus among animals of many species and a relatively high fatality rate in humans following zoonotic infection, the concern of a host jump that would allow human-to-human spread has led to global efforts to prepare for a potential devastating threat5,6. However, due to its propagation in multiple hosts, diverse H5N1 viral populations exist that comprise of genetic variants, shaped by collected mutations and frequent re-assortments of genes from different strains7,8,9,10,11. This diversity is higher than that observed for seasonal influenza viruses like human H1N1 and H3N212,13. Given this diversity, H5N1 antigenic variants can rapidly evolve to escape host immune surveillance. Moreover, the dissemination of the virus is complicated. Previous studies have shown that the global persistence of the HPAI H5N1 virus results from the interplay between a high capacity to persist in domestic poultry in localized areas, combined with sporadic long-distance introduction events involving migratory birds1,14,15. This makes the battle against the HPAI H5N1 virus quite a challenge.
Because vaccination is currently the most effective way to prevent and control infections by influenza viruses, several variants of the HPAI H5N1 virus have been recommended as vaccine strains for protection of poultry3,16, and it has been proposed that such vaccines should be stockpiled to be prepared for future outbreaks3,17. However, due to the rapid evolution of the virus and its unknown evolutionary patterns, in many cases vaccines for poultry are not well matched to the strains in circulation, and such vaccines could actually drive the evolution of the virus18,19,20,21. Therefore, understanding the evolution of HPAI H5N1, especially the evolution of its antigenicity in a temporal-spatial manner, is critical for efficient prevention and control of the virus. Despite multiple global efforts, the antigenic evolution of HPAI H5N1 is not adequately understood22,23.
For ease of tracking the evolution of the virus, the H5N1 Evolution Working Group (HEWG), a joint effort of the World Health Organization (WHO), World Organisation for Animal Health (OIE) and Food and Agriculture Organization (FAO), has designed a nomenclature to classify the GsGD lineage of Eurasian HPAI H5N1 viruses24,25, based on the phylogeny of the antigen hemagglutinin (HA). According to this nomenclature, all viruses of the GsGD lineage are classified into 10 clades (numbered 0 to 9), which are further subdivided into second-order, third-order and even fourth-order subclades. Although this system is very comprehensive, it is more reflective of the genetic than of the antigenic properties of the virus. For example, based on the cross-reactivity to a panel of 17 monoclonal antibodies raised against HPAI H5N1 strains, Wu and colleagues found that the seven recognized genetic clades of HPAI H5N1 (isolated between 2002 and 2007 in Asia) could actually be grouped into four distinct antigenic groups26. Antigenic grouping of virus strains would facilitate the recognition of emerging antigenic variants, thus aiding to the selection of vaccine strains27. Moreover, due to the rapid evolution of the virus, the classification of HPAI virus based on phylogenic analysis will become very complicated and hard to interpret over time.
China, especially southern China, is often quoted as the source for the HPAI H5N1 virus, owing to its complex ecology and diverse geographical features15,28,29. However, influenza virus antigenic evolution in China and its impact on global influenza dynamics is not adequately understood, in part due to a lack of sufficient viral data originating from China. In this study, we carried out the large-scale sequencing of the HA genes of 218 HPAI H5N1 viruses, isolated from representative regions of mainland China. Through accurate modeling of these newly sequenced virus and viruses with HA gene sequences available from public databases, we developed a comprehensive picture of the antigenic evolution of HPAI H5N1 viruses across the globe.
Results
Sequencing of HPAI H5N1 viruses from mainland China
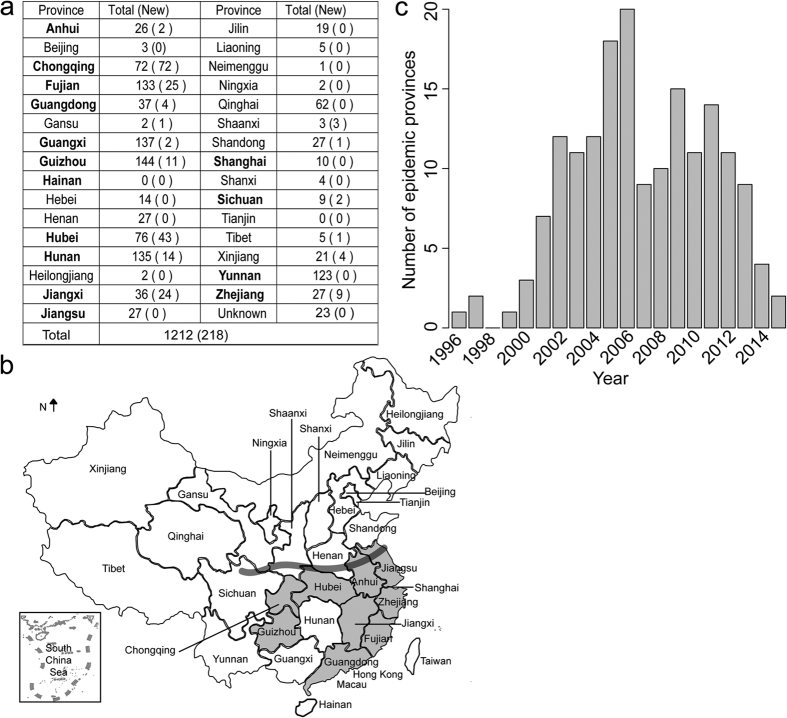
To obtain a better understanding of the evolution of HPAI H5N1, we sequenced HA of 218 HPAI H5N1 viruses isolated from mainland China (Fig. 1a and Supplementary Table S1). These newly derived sequences constituted 18% of the sequences from mainland China available until now (Fig. 1a). Most of the sequences represent isolates from 2006 onwards, due to the difficulty to derive virus samples from earlier years. From a geographical point of view, the areas we sampled the viruses covered 16 provinces (Fig. 1a), which represent nearly half of the provinces of the country. Important to note is that, except for Hainan, Jiangsu, Shanghai and Yunnan, the sampling covered all provinces of southern China (area south of the heavy gray line in Fig. 1b), a region supposed to be the epicenter of the HPAI H5N1 virus15,28,29. In addition, a number of samples were derived from provinces in northern China (north of the heavy gray line in Fig. 1b) which experienced human infections or frequent H5N1 epidemics, such as Xinjiang, Shaanxi, Shandong provinces. For provinces Chongqing, Hubei, Jiangxi and Shaanxi, the newly derived sequences outnumber those available in the public database. Complementing the newly derived sequences with sequences available in the public databases allowed for an in-depth investigation on the evolutionary dynamics of HPAI H5N1 virus in China.
Figure 1. The sequencing of 218 HPAI H5N1 viruses sampled from mainland China.
(a) Number of HA sequences for HPAI H5N1 virus sampled per province of mainland China. The number in the brackets refers to the number of newly sequenced viruses. Provinces in southern China are in bold text. (b) A map of China showing the all provinces, and, in grey, provinces in which epidemics caused by HPAI H5N1 virus were identified in the year 2013. The heavy gray line indicates the position of the Qin Mountains and Huai River that divide northern and southern China. The map was reconstructed using OpenStreetMap (http://www.openstreetmap.org/), licensed on terms of the Open Database License, “ODbL” 1.0 (http://opendatacommons.org/licenses/odbl/), and is for illustrative purposes only. (c) The number of provinces with epidemics caused by HPAI H5N1 virus inferred from HA sequences in mainland China from 1996 to 2015.
The time distribution of the sequenced isolates roughly reflected the epidemic dynamics of HPAI H5N1 in mainland China (Fig. 1c). In earlier years from 1996–1999, HPAI H5N1 virus only caused sporadic epidemics in a few provinces in southern China, such as Guangdong and Guangxi15. The virus became more prevalent in poultry and wild birds in mainland China after the year 2000, as shown by the increasing number of provinces experiencing epidemics. In 2005 and 2006, widespread outbreaks occurred in mainland China. In these two years, more than 15 provinces reported HPAI H5N1 epidemics, inferred from the sequences available. Although the number of provinces suffering from H5N1 epidemics decreased strongly after 2006, the virus was not eliminated: between 2007 and 2015, there were on average nine provinces with H5N1 outbreaks each year (Fig. 1c). The situation in 2013, during which nine provinces experienced H5N1 epidemics, exemplified by the grey color in Fig. 1b, showing all were located in southern China.
High-confidence modeling of antigenic clusters of HPAI H5N1 viruses
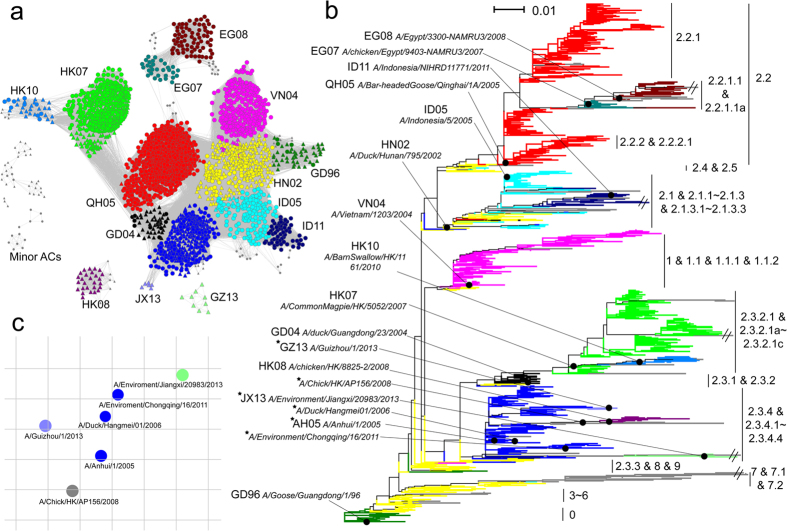
We developed an antigenic modeling tool, which we called PREDAC-H5-C, to divide viruses into antigenic clusters (AC), based on the immunogenic part of HA protein sequences (HA1, see Methods). The PREDAC-H5-C tool facilitated a systematic investigation of the antigenic evolution of all investigated HPAI H5N1 viruses. To determine which clustering most accurately reflected the actual antigenic evolution, an antigenic dataset including 798 pairs of HPAI H5N1 viruses with known antigenic relationship (see Methods) was constructed. It should be noted that none of the viruses in the antigenic dataset had been used in training the PREDAC-H5-C model. The rationale behind this was that if the antigenic clustering accurately captured the actual antigenic relationship of HPAI H5N1 viruses, the viruses within predicted ACs should be antigenically similar, while viruses grouped in different ACs should differ more in antigenicity (Supplementary Methods). Thus, with the help the antigenic dataset, the best antigenic clustering by PREDAC-H5-C was obtained, which achieved an agreement of 0.81 with the antigenic dataset (Supplementary Fig. S1). It separated a total of 5605 viruses, including 218 newly sequenced isolates and 5387 sequences collected from public databases, into 36 ACs (Fig. 2a and Supplementary Table S2). Among them, 15 major ACs were defined which covered 97% of all viruses analyzed. They were named after their representative viruses (in most cases the WHO-recommended vaccine strain), by two letters that refer to the country/region of isolation followed by two digits that refer to the year of isolation. For example, the AC GD96 was named after the strain A/Goose/Guangdong/1/1996 (Fig. 2). Besides for the major ACs, the other ACs were defined as minor ACs (see Methods).
Figure 2. High-confidence modeling of ACs of HPAI H5N1 viruses.
(a) Predicted antigenic correlation network (ACnet) of the ACs defined for 2441 HPAI H5N1 viruses with unique HA1 protein sequences. All pairs of viruses which were predicted to be antigenically similar were connected in ACnet. Triangles in the network refer to the viruses from China. The names for the major ACs (in color) are indicated, while minor ACs are shown in gray. (b) Phylogenetic tree of the 2441 HA1 sequences, colored according to the predicted ACs. The sub-clades to which the viruses belong (H5N1 Evolution Working Group nomenclature) are shown to the right. The branch length was scaled according to the legend in the top left. The strains listed to the left of the tree refer to the strains used for naming the predicted major ACs. The stars indicate strains used in the HI assay. (c) The antigenic cartography for six representative viruses of four antigenic clusters which were mainly composed of viruses of clade 2.3.4 and its sub-clades. The viruses were colored according to the antigenic clusters they belong to.
When mapping the predicted ACs to the phylogenetic tree of these viruses (comparing Fig. 2a and b), we found that a predicted AC generally covered a side lineage or comprised several closely related side lineages. We further tried to compare the predicted ACs with clade IDs as per the nomenclature system of the HEWG, but since the designated clades for H5N1 are hierarchically organized, this was not straightforward. Table S2 summarizes the correspondence between our predicted ACs and the designated clades or sub-clades. Overall, a good correspondence was observed: a pair of viruses had a probability of 0.94 to be found in the same predicted AC if they were from the same phylogenetic clade/sub-clade, and that probability was 0.90 for strains to be divided in different ACs if they belonged to different phylogentic clades/sub-clades. We further assessed how well the predicted ACs and designated HEWG clades matched with the antigenic data. The ratio of antigenically similar pairs within a predicted AC was similar to that based on HEWG clades (0.78), while the ratio of antigenically different pairs between the predicted ACs was much larger than that based on HEWG clades (0.85 versus 0.73). This suggests the predicted ACs are more accurate in describing antigenic relationships than the HEWG nomenclature system. This can be exemplified with HEWG clade 2.2, as shown in Fig. 2b. The clade is divided into tertiary sub-clades 2.2.1 and 2.2.2 and further into quaternary clade 2.2.1.1, but most of these viruses fell into our predicted AC QH05 (marked in red) except for members of the quaternary sub-clade 2.2.1.1. Experimental data confirmed that indeed only the quaternary 2.2.1.1 was antigenically different from the other viruses in clade 2.2, while the clades 2.2.1 and 2.2.2 contained viruses that are antigenically similar to clade 2.2 (Supplementary Table S3). Interestingly, the quaternary clade 2.2.1.1 comprises two predicted ACs, EG07 and EG08, which were also reported to be antigenically different (Supplementary Table S3).
To further demonstrate the accuracy of the predicted ACs, the antigenic relationship between viruses of clade 2.3.4 and its sub-clades 2.3.4.1 to 2.3.4.4, which circulated most extensively in mainland China in recent years, were determined with the HA-inhibition (HI) assay. The viruses belonged to 8 ACs (Supplementary Table S2); of these, four (AH05, GZ13, JX13 and Minor-28) were included in the assay, as shown in Fig. 2c (also see Supplementary Table S4). Although some antigenic heterogeneity was observed in AH05, these four ACs were antigenically all distinguishable with each other by the HI assay, confirming that by and large the predicted ACs reflect true antigenic relationships.
The origins of ACs of HPAI H5N1 viruses
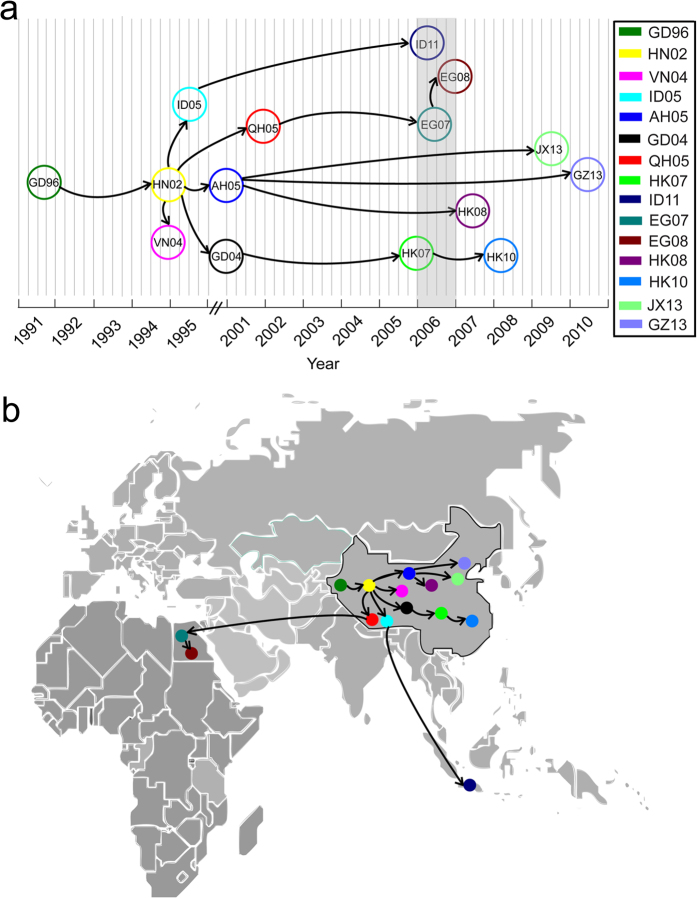
To find out how the predicted ACs had originated, we dated the timing of their most recent common ancestors (tMRCA) and inferred the most probable source countries for the major ACs (Supplementary Table S5). As summarized in Fig. 3a, although the first strain of GsGD lineage was isolated in 1996 in the Guangdong province of China, the emergence of the first AC GD96 could be dated back to the end of 1991. Then GD96 gave rise to HN02 in the beginning of 1995. The HN02 had been hidden for years until its discovery in 2000. Remarkably, HN02 had five propagations. Two (VN04 and ID05) were generated right after HN02 appeared, while the other three (QH05, AH05, and GD04) were generated around 2001–2002. Interestingly, there was a burst of generation of four new ACs around 2006, when widespread outbreaks of HPAI H5N1 viruses were observed in China and Southeast Asia. The latest two ACs (JX13 and GZ13) were generated around 2010. We further mapped the source countries of these predicted major ACs (Fig. 3b). Overall, 12 of the 15 major ACs were generated in China, while ID11 originated in Indonesia and EG07 and EG08 evolved in Egypt.
Figure 3. The origins and evolutionary pathways of predicted major ACs of HPAI H5N1 virus.
(a) The tMRCA for major ACs (encircled) and their phylogenetic relationship, as inferred from Fig. 2b. (b) The most probable source country for each major AC. The map was reconstructed using OpenStreetMap (http://www.openstreetmap.org/), licensed on terms of the Open Database License, “ODbL” 1.0 (http://opendatacommons.org/licenses/odbl/), and is for illustrative purposes only.
Antigenic diversification with the spread of the virus in China
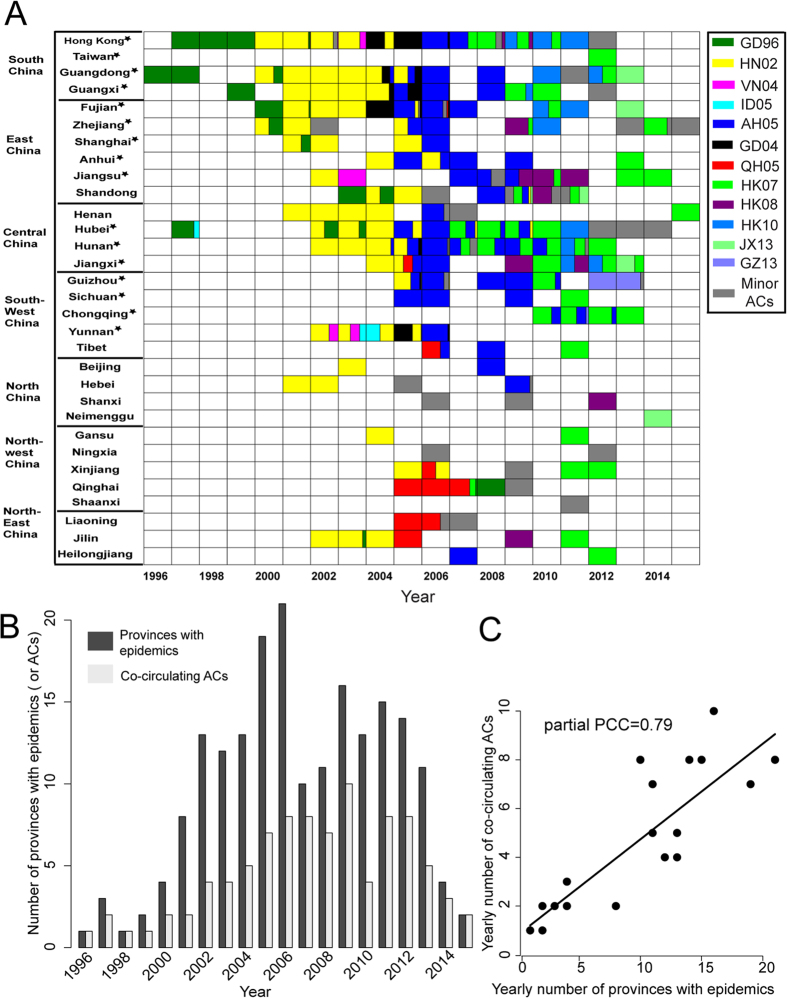
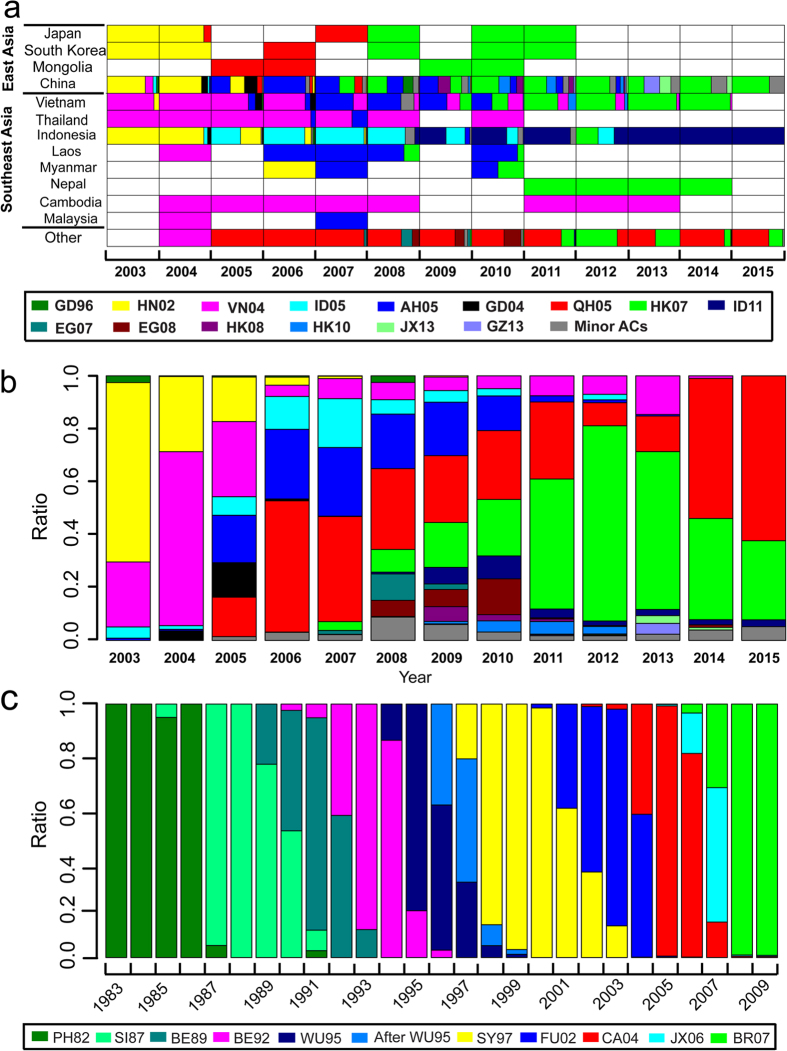
We sought to investigate in detail the antigenic evolution of the virus in China. Figure 4A shows the overall evolutionary dynamics of the predicted ACs in China (here combining mainland China with Hong Kong) from 1996 to 2015. In the early years during 1996–1999, HPAI H5N1 virus consistently belonged to AC GD96 and mainly circulated in South China (including Hong Kong). In 2000, GD96 was replaced by the predicted AC HN02 which spread rapidly across the country. HN02 continued to dominate until 2004. During 2005–2009, AH05 became predominant and in 2010 HK07 became predominant. Also note that besides the dominant ACs, quite a few major and minor ACs co-circulated which added up to a significant portion in each year since 2002; they could actually dominate in some provinces. For example, in 2006, although the dominant AC AH05 occupied almost the whole southern part of China, the major AC QH05 and a few minor ACs became dominant in northern China. This exemplifies the antigenic complexity of HPAI H5N1 virus populations in the country. Panels b and c of Fig. 4 clearly show that the antigenic diversification of the virus was co-incident with its geographic expansion in China. We observed a significant correlation between the number of predicted ACs in circulation and the number of provinces in epidemic inferred from the HA-sequenced viruses, with a partial Pearson Correlation Coefficient (PCC) of 0.79 (p-value 6.9e-8). On a global scale, however, there was no significant correlation between the number of predicted ACs in circulation and the number of countries with epidemics inferred from the HA-sequenced viruses (data not shown).
Figure 4. Translocation of antigenic types of HPAI H5N1 virus in China.
(A) Tempo-spatial dynamics of ACs within China, showing dynamic changes in the fraction of ACs (colored as in Fig. 2) recorded on a yearly basis for each province. White indicates no virus is isolated in that province in that year. (B) The number of provinces with epidemics (dark gray) and co-circulating ACs (light gray) between 1996 and 2015. (C) The partial Pearson Correlation Coefficient (PCC) indicating the correlation between the number of province with epidemics and co-ciruclating ACs after controlling for the influence of the number of HA sequences per year.
Global co-circulation of ACs of recent H5N1 viruses
As demonstrated above, since the detection/discovery of the first HPAI H5N1 virus in South China in 1996, its antigenic types have altered dramatically, not only in China but also across the globe. Therefore, we further investigated the antigenic evolution of recent H5N1 viruses across the globe. As shown in Fig. 5a, the antigenic diversity in East and Southeast Asia is much larger than that in other regions, including Europe, Africa, North America and the other regions of Asia (more details see Supplementary Fig. S2). In addition to China in East Asia, some countries in Southeast Asia, mainly Vietnam and Indonesia, also maintained a great antigenic diversity since 2003 based on the HA-sequenced viruses. However, the composition and dominance of the antigenic types varies between countries. Taking 2006 as an example, AH05 dominated in China, while VN04 was dominant in Vietnam and Thailand, and ID05 in Indonesia; other countries reported yet other dominant ACs. Thus, antigenic differences between countries/regions further contributed to the overall antigenic diversity in East and Southeast Asia. Co-circulation of multiple ACs coincided with local persistence of some ACs. For example, ID11 was only detectable in HA-sequenced viruses in Indonesia, where it was detected over multiple years (Fig. 5a and Supplementary Fig. S2).
Figure 5. Global co-circulation of multiple ACs of recent H5N1 viruses compared to the seasonal human influenza H3N2 virus.
(a) Global tempo-spatial dynamics of HPAI H5N1 ACs from 2003 to 2015 (for details see Supplementary Fig. S2). Coloring is similar as in Fig. 4. (b) The yearly global proportion of ACs of HPAI H5N1 virus from 2003 to 2015. (c) The yearly global composition of ACs of human influenza H3N2 virus, covering the years 1983 to 2009 (adapted from Du’s work27).
At a global scale, the co-circulation of various ACs is even more evident. Figure 5b summarizes the global composition of ACs in each year between 2003 and 2015. As a word of warning, the relative ratios displayed here are based on the HA-sequenced isolates, which may not reflect their true ratios due to sampling bias. Nevertheless, co-circulation of up to 11 ACs throughout the globe per year was observed since 2003 (detailed information regarding circulation of these ACs in different countries/regions over time is shown in Supplementary Figs S2 and S3).
Finally, for comparison we analysed antigenic clusters of human influenza H3N2 viruses (Fig. 5c), which does not show the large antigenic complexity of avian H5N1. For the human influenza H3N2 virus, there were typically one or two ACs circulating across the globe per year. The replacement of dominant ACs in the global population of human influenza H3N2 viruses in successive periods of approximately 3 to 4 years is also obvious.
Discussion
In this study antigenic modeling was performed, based on existing HA1 sequences of HPAI H5N1 viruses and supplemented with 218 newly sequenced strains isolated during 2004–2013 from mainland China. This analysis provides a comprehensive picture of the antigenic evolution of HPAI H5N1 virus across the globe. We not only tracked the origins of different antigenic ACs, but also found that generation of antigenic diversity came with the spread of the virus in China. The continuous antigenic divergence of the virus has lead to extensive co-circulation and local persistence of ACs in recent years.
The antigenic evolution of human influenza H3N2 and H1N1 viruses is reported to be in a cluster-wise and trunk-like pattern, i.e., it can be viewed as the serial replacement of one AC by another13,30. In contrast, the HPAI H5N1 virus seems to evolve according to a divergent pattern, whereby ACs can evolve in multiple directions, as visualized in Figs 2b and 3. New ACs of this strain seem to emerge at high frequencies, suggested by the emergence of 15 major ACs and 21 minor ACs from a single lineage since 1996, while for human H3N2 viruses only 7 ACs circulated during 1996–2009 (Fig. 5c). This makes a sharp contrast to the antigenic evolution of other avian and swine influenza viruses, such as avian H9N220, H731,32 and swine H3N2 viruses33, for which only a few antigenic clusters were observed and the rate of antigenic evolution is much lower than that of HPAI H5N1 viruses. The rapid generation of new ACs of the HPAI H5N1 virus imposes a larger challenge for HPAI H5N1 virus surveillance and vaccination strategies.
It has been suggested that the source of HPAI H5N1 virus was southern China28,29, and the first HPAI H5N1 virus of the GsGD lineage was indeed detected in Guangdong province of southern China in 19962. Then, the ‘Qinghai’ virus34 and ‘Fujian’ virus35 also emerged in China. In this study, we produce a more detailed picture of the antigenic evolution of HPAI H5N1 virus in China (Fig. 4a). The result showed that the development of its antigenic diversity was co-incident with its geographic expansion in China. This may be caused in part by the continuing pressure imposed by vaccination across the country, since mass vaccination campaigns have been conducted as a routine measure for the control of avian influenza viruses in China since 20043,17. Despite these efforts, the virus has caused epidemics nearly throughout the complete country.
Besides for China, Southeast Asia and Egypt are two major host-spot regions for epidemics caused by HPAI H5N1 viruses (Fig. 5a and Supplementary Fig. S2). Large amounts of poultry were raised in these regions36,37. Although large antigenic diversity was observed in these regions, most of the ACs circulating there were introduced from China28,38. Few or minor antigenic drifts were observed in most countries except in Vietnam39,40, Indonesia41 and Egypt42,43 where significant antigenic drift happened. It is noteworthy that, three novel ACs ID11, EG07 and EG08 were generated in Indonesia and Egypt, respectively. It is of great concern that these new ACs can cause a new round of epidemics if they move out of these countries. Therefore, it is important to strengthen the surveillance for HPAI H5N1 virus in these countries. In addition, new ACs may also be generated in the countries with dense human and poultry populations like Bangladesh and India44,45.
To control the HPAI H5N1 virus, vaccinations have been conducted worldwide, especially in China, Indonesia, Vietnam and Egypt, where more than 99% of avian influenza vaccines were used. As is reported in previous studies19,20,21, vaccination programs could induce faster rates of antigenic drift in avian influenza viruses, especially when there are antigenic differences between the vaccine strain and the epidemic viruses. The large antigenic diversity and frequent drifts in the above four countries suggested that vaccination may drive the antigenic evolution of this virus in these countries. To promote the efficiency of vaccination, better vaccination strategies should be adapted to match the vaccines with circulating strains. Systematic antigenic grouping of this virus could facilitate such a vaccination strategy. As is shown above, local persistence and co-circulation of ACs are widely observed for the virus, which suggests that the application of vaccines should be based on the epidemic ACs in a country/region. For countries or regions with multiple ACs co-circulating, such as China and Southeast Asia, multiple vaccines or the universal vaccine should be provided; while for most countries in Europe, Africa and Middle East where QH05 mainly circulated, the vaccine against QH05 should be enough for protection of infections by HPAI H5N1 virus.
Our study could be biased towards the sequence distribution by region. As shown in our study, most of the sequence data (70%) came from China (including Hong Kong), Vietnam, Indonesia and Egypt (Supplementary Fig. S4). This could reflect their hot spot roles in HPAI H5N1 dissemination, as is exemplified by the observation that all the major ACs originated in these four countries, but it could also be a reflection of less thorough surveillance and sequencing efforts in other countries. Anyway, the viruses in these four countries captured almost all the antigenic diversity (Supplementary Table S6), suggesting the importance of strengthening the surveillance in these countries.
Overall, the coupling of large-scale HA sequencing and high-accuracy antigenic modeling will be a valuable tool not only for systematic understanding of the antigenic evolution of influenza viruses, but also for timely surveillance of new ACs, which could help for vaccine recommendations for HPAI H5N1 prevention and control.
Methods
HA sequence data
HA sequencing of 218 HPAI H5N1 viruses sampled from diverse regions of mainland China between 2004 and 2013 (Supplementary Table S1) were carried out according to the methods described in Supplemental Information. Other HA sequences of HPAI H5N1 virus with length greater than 900 were collected from the EpiFlu database of Global Initiative on Sharing All Influenza Data (GISAID)46 on January 27, 2016. The acknowledgement table for these viruses was available at http://www.computationalbiology.cn/material/. Only the sequences of HPAI H5N1 viruses in GsGD lineage were kept. All the remaining sequences were aligned using the software MAFFT47 with an additional manual check. The non-coding regions and the region coding for signal peptide were removed for each sequence. After removing the re-assortment sequences synthesized in laboratory, the sequences with gap content greater than 10% and the sequences stopped abnormally when translated into protein, we obtained in total 5387 DNA sequences with each 960 nucleotides long. They were then translated into protein sequences. The software cd-hit48 was used to remove the redundant sequences of 100% similarity. Finally, 2441 unique protein sequences were obtained. The information of all the viruses used in this study was available at http://www.computationalbiology.cn/material/.
Antigenic clustering based on RPEDAC-H5-C
The ACs were predicted with the computational method PREDAC-H5-C which was adapted from PREDAC, a computational method for prediction of ACs for the human influenza H3N2 virus27. It included four steps: firstly, the antigenic relationship between any pairs of viruses used in this study were predicted based on HA1 protein sequences with the computational method PREDAC-H5 developed in our previous study49,50; secondly, any pair of viruses which was predicted to be antigenically similar was connected by an edge, which resulted in the antigenic correlation network (ACnet); thirdly, the ACnet would be separated into clusters using the software MCL51; finally, an antigenic dataset composed of 798 pairs of HPAI H5N1 viruses with known antigenic relationship were used to help determine the best antigenic clustering (details see Supplementary Methods).
Based on PREDAC-H5-C, all the HPAI H5N1 viruses used in this study were grouped into 36 ACs in total (Supplementary Table S2). Among them, 15 ACs were considered as major ACs, which circulated in no less than three years and caused epidemics (ratio greater than 20%) in its dominant country (the country covering most viruses of the cluster) in at least one year. The major ACs included over 97% of the viruses. The remaining ACs were defined as minor ACs.
ACnet visualization
The ACnet was visualized using the yFiles Organic layout in Cytoscape52. To display the network clearly in Fig. 2a, only the unique protein sequences described above and the connections between them were used (Fig. 2a). To better display predicted ACs, the positions of some nodes were manually adjusted so that each predicted major AC had a clear boundary from the others.
Clade determination
The clade of each virus belongs to in the nomenclature system was determined with Highly Pathogenic H5N1 Clade Classification Tool on Influenza Research Database53, which is available at http://www.fludb.org/brc/h5n1Classifier.spg?method=ShowCleanInputPage&decorator=influenza.
Phylogenetic reconstruction and bayesian coalescent analysis
To infer the phylogenetic relationship between the HPAI H5N1 viruses used in the ACnet, the DNA sequences of the viruses used in the network were used to determine the phylogenetic tree with the help of software MEGA 5.254. The maximum likelihood method was used with the general reversible GTR+I+c4 model. The tree was rooted with the virus GsGD. The coloring and visualization of the tree were done with script colorTree.pl55 and Dendroscope56, respectively. The detailed phylogenetic tree file is available at http://www.computationalbiology.cn/material/.
The time and country for the MRCAs of major ACs were inferred by Bayesian MCMC sampling using the software BEAST v1.7557 with the SRD06 codon position model and the uncorrelated exponential clock model. To reduce the computational cost, the earliest 20 viruses in each AC were chosen for analysis after removing the redundancy by country and month. Bayesian MCMC sampling was run for up to 100,000,000 times to achieve convergence. The summary tree file is available at http://www.computationalbiology.cn/material/.
HI experiment
The antigenic characterization of six representative viruses of four antigenic clusters which were mainly composed of viruses of clade 2.3.4 and its subclades (2.3.4.1~2.3.4.4), including A/Anhui/1/2005 (AH05), A/Chicken/Hong Kong/AP156/2008 (Minor-28), A/Guizhou/1/2013 (GZ13), A/duck/Hubei/Hangmei01/2006 (AH05), A/Environment/Chongqing/16/2011 (AH05) and A/Environment/Jiangxi/20983/2013 (JX13), were determined by HI assays according to standard protocols proposed by WHO58 with ferret antisera and 0.5% turkey red blood cells. The antigenic cartography was generated with Smith’s method30.
Additional Information
How to cite this article: Peng, Y. et al. Continual Antigenic Diversification in China Leads to Global Antigenic Complexity of Avian Influenza H5N1 Viruses. Sci. Rep. 7, 43566; doi: 10.1038/srep43566 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31500126), the Chinese Academy of Medical Sciences (2016-I2M-1–005), the National Key Plan for Scientific Research and Development of China (2016YFD0500300 and 2016YFC1200200). The authors would like to thank the members of the Jiang and Shu’s labs for their help and deliberations.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.J., Y.S., and M.J. conceived and designed the study. Y.P. and A.W. did the computational analysis. X.D.L., H.Z., L.D., Y.Z., R.G., H.B., L.Y., D.W. and X.L. did the HPAI H5N1 surveillance and sequencing. L.D. and Y.Z. did the H.I. experiment. Y.P. and T.J. wrote the paper. Y.S. and M.J. reviewed and edited the manuscript. All authors read and approved the manuscript.
References
- Li X. et al. Global and local persistence of influenza A(H5N1) virus. Emerg Infect Dis 20, 1287–1295, doi: 10.3201/eid2008.130910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Subbarao, Cox N. J. & Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261, 15–19 (1999). [DOI] [PubMed] [Google Scholar]
- Li C., Bu Z. & Chen H. Avian influenza vaccines against H5N1 ‘bird flu’. Trends in biotechnology 32, 147–156, doi: 10.1016/j.tibtech.2014.01.001 (2014). [DOI] [PubMed] [Google Scholar]
- World Health Organization. Cumulative number of confirmed human cases of avian influenza A (H5N1) reported to WHO as of Nov 21st, 2016, http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/ (2016).
- Ungchusak K. et al. Probable person-to-person transmission of avian influenza A (H5N1). The New England journal of medicine 352, 333–340, doi: 10.1056/NEJMoa044021 (2005). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet 371, 1427–1434, doi: 10.1016/S0140-6736(08)60493-6 (2008). [DOI] [PubMed] [Google Scholar]
- Duan L. et al. The development and genetic diversity of H5N1 influenza virus in China, 1996–2006. Virology 380, 243–254, doi: 10.1016/j.virol.2008.07.038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan V. G. et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS pathogens 4, e1000076, doi: 10.1371/journal.ppat.1000076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D. et al. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS pathogens 4, e1000161, doi: 10.1371/journal.ppat.1000161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y. et al. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proceedings of the National Academy of Sciences of the United States of America 99, 8950–8955, doi: 10.1073/pnas.132268999 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proceedings of the National Academy of Sciences of the United States of America 103, 2845–2850, doi: 10.1073/pnas.0511120103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. I. & Holmes E. C. The evolution of epidemic influenza. Nature reviews. Genetics 8, 196–205, doi: 10.1038/nrg2053 (2007). [DOI] [PubMed] [Google Scholar]
- Liu M. et al. Antigenic Patterns and Evolution of the Human Influenza A (H1N1) Virus. Scientific reports 5, doi: 10.1038/Srep14171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. et al. Flying over an infected landscape: distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. EcoHealth 7, 448–458, doi: 10.1007/s10393-010-0672-8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. S. et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213, doi: 10.1038/nature02746 (2004). [DOI] [PubMed] [Google Scholar]
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness, February 2015. (2015). [PubMed] [Google Scholar]
- Swayne D. E., Pavade G., Hamilton K., Vallat B. & Miyagishima K. Assessment of national strategies for control of high-pathogenicity avian influenza and low-pathogenicity notifiable avian influenza in poultry, with emphasis on vaccines and vaccination. Rev Sci Tech 30, 839–870 (2011). [DOI] [PubMed] [Google Scholar]
- Kim J. K. et al. Puzzling inefficiency of H5N1 influenza vaccines in Egyptian poultry. Proceedings of the National Academy of Sciences of the United States of America 107, 11044–11049, doi: 10.1073/pnas.1006419107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. W., Senne D. A. & Suarez D. L. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. Journal of virology 78, 8372–8381, doi: 10.1128/jvi.78.15.8372-8381.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. P. et al. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Veterinary microbiology 146, 215–225, doi: 10.1016/j.vetmic.2010.05.010 (2010). [DOI] [PubMed] [Google Scholar]
- Kayali G. et al. Avian Influenza A(H5N1) Virus in Egypt. Emerg Infect Dis 22, 379–388, doi: 10.3201/eid2203.150593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish A. L. et al. Antigenic and genetic diversity of highly pathogenic avian influenza A (H5N1) viruses isolated in Egypt. Avian Dis 54, 329–334 (2010). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. Journal of virology 84, 8389–8397, doi: 10.1128/JVI.00413-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/OIE/FAO H5N1 Evolution Working Group. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 14, e1, doi: 10.3201/eid1407.071681 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/OIE/FAO H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza and other respiratory viruses 8, 384–388, doi: 10.1111/irv.12230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. L. et al. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. Journal of virology 82, 1798–1807, doi: 10.1128/JVI.02256-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X. et al. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nature communications 3, 709, doi: 10.1038/ncomms1710 (2012). [DOI] [PubMed] [Google Scholar]
- Wallace R. G., Hodac H., Lathrop R. H. & Fitch W. M. A statistical phylogeography of influenza A H5N1. Proceedings of the National Academy of Sciences of the United States of America 104, 4473–4478, doi: 10.1073/pnas.0700435104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. et al. New evidence suggests Southern China as a common source of multiple clusters of highly pathogenic H5N1 avian influenza virus. The Journal of infectious diseases 202, 452–458, doi: 10.1086/653709 (2010). [DOI] [PubMed] [Google Scholar]
- Smith D. J. et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376, doi: 10.1126/science.1097211 (2004). [DOI] [PubMed] [Google Scholar]
- Xu Y. F. et al. Limited Antigenic Diversity in Contemporary H7 Avian-Origin Influenza A Viruses from North America. Scientific reports 6, doi: 10.1038/Srep20688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M. S., Xu Y. F., Long L. P., Capua I. & Wan X. F. Antigenic and Genetic Evolution of Low-Pathogenicity Avian Influenza Viruses of Subtype H7N3 following Heterologous Vaccination. Clinical and Vaccine Immunology 21, 603–612, doi: 10.1128/Cvi.00647-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J. C. et al. Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. Journal of virology 81, 4315–4322, doi: 10.1128/Jvi.02458-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309, 1206, doi: 10.1126/science.1115273 (2005). [DOI] [PubMed] [Google Scholar]
- Smith G. J. et al. Emergence and predominance of an H5N1 influenza variant in China. Proceedings of the National Academy of Sciences of the United States of America 103, 16936–16941, doi: 10.1073/pnas.0608157103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. et al. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proceedings of the National Academy of Sciences of the United States of America 105, 4769–4774, doi: 10.1073/pnas.0710581105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M. et al. Antigenic analysis of H5N1 highly pathogenic avian influenza viruses circulating in Egypt (2006–2012). Veterinary microbiology 167, 651–661, doi: 10.1016/j.vetmic.2013.09.022 (2013). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Identification of the progenitors of Indonesian and Vietnamese avian influenza A (H5N1) viruses from southern China. Journal of virology 82, 3405–3414, doi: 10.1128/Jvi.02468-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamatsu M. et al. The genetic and antigenic diversity of avian influenza viruses isolated from domestic ducks, muscovy ducks, and chickens in northern and southern Vietnam, 2010–2012. Virus genes 47, 317–329, doi: 10.1007/s11262-013-0954-7 (2013). [DOI] [PubMed] [Google Scholar]
- Wan X. F. et al. Evolution of Highly Pathogenic H5N1 Avian Influenza Viruses in Vietnam between 2001 and 2007. PloS one 3, doi: 10.1371/journal.pone.0003462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel B. F. et al. Antigenic Variation of Clade 2.1 H5N1 Virus Is Determined by a Few Amino Acid Substitutions Immediately Adjacent to the Receptor Binding Site. mBio 5, doi: 10.1128/mBio. 01070–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shesheny R. et al. Antigenic diversity and cross-reactivity of avian influenza H5N1 viruses in Egypt between 2006 and 2011. Journal of General Virology 93, 2564–2574, doi: 10.1099/vir.0.043299-0 (2012). [DOI] [PubMed] [Google Scholar]
- Watanabe Y. et al. Antigenic analysis of highly pathogenic avian influenza virus H5N1 sublineages co-circulating in Egypt. Journal of General Virology 93, 2215–2226, doi: 10.1099/vir.0.044032-0 (2012). [DOI] [PubMed] [Google Scholar]
- Hogerwerf L. et al. Persistence of highly pathogenic avian influenza H5N1 virus defined by agro-ecological niche. EcoHealth 7, 213–225, doi: 10.1007/s10393-010-0324-z (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. R. et al. New introduction of clade 2.3.2.1 avian influenza virus (H5N1) into Bangladesh. Transboundary and emerging diseases 59, 460–463, doi: 10.1111/j.1865-1682.2011.01297.x (2012). [DOI] [PubMed] [Google Scholar]
- Peter Bogner I. C., David J. L. & Nancy J. C. A global initiative on sharing avian flu data. Nature 442 (2006). [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. & Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659, doi: 10.1093/bioinformatics/btl158 (2006). [DOI] [PubMed] [Google Scholar]
- Li H. et al. PREDAC-H5: a user-friendly tool for the automated surveillance of antigenic variants for the HPAI H5N1 virus. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 28, 62–63, doi: 10.1016/j.meegid.2014.08.030 (2014). [DOI] [PubMed] [Google Scholar]
- Peng Y., Zou Y., Li H., Li K. & Jiang T. Inferring the antigenic epitopes for highly pathogenic avian influenza H5N1 viruses. Vaccine 32, 671–676 (2014). [DOI] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S. & Ouzounis C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires R. B. et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza and other respiratory viruses 6, 404–416, doi: 10.1111/j.1750-2659.2011.00331.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. H. & Lercher M. J. ColorTree: a batch customization tool for phylogenic trees. BMC research notes 2, 155, doi: 10.1186/1756-0500-2-155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H. et al. Dendroscope: An interactive viewer for large phylogenetic trees. Bmc Bioinformatics 8, 460, doi: 10.1186/1471-2105-8-460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology 7, 214, doi: 10.1186/1471-2148-7-214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.