Abstract
Epigenetic mechanisms control phenotypic commitment of mesenchymal stromal/stem cells (MSCs) into osteogenic, chondrogenic or adipogenic lineages. To investigate enzymes and chromatin binding proteins controlling the epigenome, we developed a hybrid expression screening strategy that combines semi-automatic real-time qPCR (RT-qPCR), next generation RNA sequencing (RNA-seq), and a novel data management application (FileMerge). This strategy was used to interrogate expression of a large cohort (n>300) of human epigenetic regulators (EpiRegs) that generate, interpret and/or edit the histone code. We find that EpiRegs with similar enzymatic functions are variably expressed and specific isoforms dominate over others in human MSCs. This principle is exemplified by analysis of key histone acetyl transferases (HATs) and deacetylases (HDACs), H3 lysine methyl transferases (e.g., EHMTs) and demethylases (KDMs), as well as bromodomain (BRDs) and chromobox (CBX) proteins. Our results show gender-specific expression of H3 lysine 9 [H3K9] demethylases (e.g., KDM5D and UTY) as expected and upregulation of distinct EpiRegs (n>30) during osteogenic differentiation of MSCs (e.g., HDAC5 and HDAC7). The functional significance of HDACs in osteogenic lineage commitment of MSCs was functionally validated using panobinostat (LBH-589). This pan-deacetylase inhibitor suppresses osteoblastic differentiation as evidenced by reductions in bone-specific mRNA markers (e.g., ALPL), alkaline phosphatase activity and calcium deposition (i.e., Alizarin Red staining). Thus, our RT-qPCR platform identifies candidate EpiRegs by expression screening, predicts biological outcomes of their corresponding inhibitors, and enables manipulation of the human epigenome using molecular or pharmacological approaches to control stem cell differentiation.
Keywords: Mesenchymal stem cell, adipose-tissue derived stromal cells, epigenetics, histone, epigenetic regulators, deacetylase, methyltransferase
1 Introduction
Epigenetic mechanisms are critical during germ-line transmission and development to ensure that cells maintain a correct spatio-temporal memory of their environmental conditions and identity through meiotic or mitotic inheritance. Heritable patterns of gene expression in progeny cells can be transferred through a number of major epigenetic mechanisms, including methylation of genomic DNA, post-translational modifications of histone tails, transcription factors that bookmark genes at mitosis [Zaidi et al., 2010], non-coding RNAs [Gibney and Nolan, 2010], and the transmission of mRNAs during mitosis [Varela et al., 2016]. The advent of next-generation genomic and proteomic technologies has allowed a greater understanding of the epigenetic states of different cell types [de Wit and de Laat, 2012; Laird, 2010; Rando and Chang, 2009]. Epigenetic mechanisms respond to environmental cues and external biological factors, and can be effectively applied in stem cell-based engineering strategies for musculoskeletal tissue regeneration [Dudakovic et al., 2015a; Dudakovic et al., 2016; Dudakovic et al., 2015b; Dudakovic et al., 2013].
Abnormal skeletal development and/or maintenance, such as non-healing fractures and osteoporosis, are debilitating biological conditions that diminish the quality of life. Epigenetic events may contribute to these bone disorders and therefore the enzymes that mediate epigenetic gene regulation represent potential therapeutic targets. Key osteogenic genes are controlled by DNA methylation [Lee et al., 2006b; Villagra et al., 2002] and critical osteogenic transcription factors that associate with chromatin modifying enzymes [Kang et al., 2005; Schroeder et al., 2004; Westendorf et al., 2002]. Several studies have shown that modifications in the epigenetic code contribute to osteogenic differentiation. For example, modulating the activity of histone deacetylases (HDACs), the WD-repeat domain protein WDR5, which is associated with a histone 3 lysine 4 (H3K4) methyltransferase complex, and the H3K9 methyl transferase SUV420H2 can stimulate osteogenic differentiation of MSCs and/or pre-osteoblasts [Di Bernardo et al., 2009; Dudakovic et al., 2013; Farzaneh et al., 2016; Gordon et al., 2015; Gori et al., 2001; Gori et al., 2006; Lee et al., 2009; Schroeder and Westendorf, 2005]. Other studies have demonstrated that suppression of histone 3 lysine 27 trimethylation (H3K27me3) or altering CpG methylation can significantly enhance osteogenic differentiation and inhibit adipogenic differentiation of MSCs [Dudakovic et al., 2016; Dudakovic et al., 2015b; Hemming et al., 2014; Jing et al., 2015; Thaler et al., 2016; Wei et al., 2011]. These studies demonstrate that the epigenetic landscape is deformable to favor selective commitment of mesenchymal stromal/stem cells to the osteogenic lineage.
Genome-wide studies have been performed to assess the epigenetic state of cells at different stages of osteogenesis. We have utilized ChIP-seq analysis in osteoblasts to demonstrate that the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) modifies the epigenomic fingerprint of histone H4 acetylation in differentially regulated genes [Dudakovic et al., 2013]. Other genomic studies have successfully mapped multiple histone modifications and chromatin conformations in pre-osteoblasts, differentiated osteoblasts, and/or osteocytes [Barutcu et al., 2014; Dudakovic et al., 2016; Meyer et al., 2014; Pike et al., 2015; St John et al., 2014; Tai et al., 2014; Wu et al., 2014]. To improve our understanding of the epigenome in supporting differentiation of mesenchymal cell types, it is necessary to characterize which EpiRegs are expressed at specific biological stages. Therefore, we assessed the expression pattern of more than 300 EpiRegs by RNA-seq and RT-qPCR analysis in MSCs. Our data reveal which EpiRegs are most highly expressed, variable among patients and/or modulated in a lineage-specific manner.
2 Materials and Methods
2.1 Culture conditions for adipose tissue-derived MSCs
Platelet-lysate expanded MSCs derived from the stromal vascular fraction of adipose-tissue are `Good Manufacturing Practice'-compliant pericyte-like immature fibroblasts that are used in clinical trials, have multi-lineage potential and express all relevant markers expected of mesenchymal stem cells [Camilleri et al., 2016; Dudakovic et al., 2014; Dudakovic et al., 2015a; Riester et al., 2016]. MSCs were harvested from lipo-aspirates obtained from consenting healthy donors as previously described [Crespo-Diaz et al., 2011; Mader et al., 2013] with approval from the Mayo Clinic Institutional Review Board. Fat tissue was enzymatically digested using 0.075% Type I collagenase (Worthington Biochemicals) for 1.5 h at 37°C. Adipocytes were separated from the stromal vascular fraction by low speed centrifugation (400 g for 5 min). The adipose supernatant was removed and the cell pellet was rinsed with PBS and passed successively through 70 μm and 40 μm cell strainers (BD Biosciences). The resulting MSC cell fraction was maintained in Advanced MEM Medium containing 5% PLTMax (a clinical grade commercial platelet lysate product [MillCreekLifeSciences]), 2 mM Glutamax (Invitrogen), 2 U/ml heparin (hospital pharmacy), 100 U/ml penicillin, and 100 μg/ml streptomycin (Cellgro) as described previously [Crespo-Diaz et al., 2011].
2.2 Osteogenic differentiation
MSCs were plated in 6-well plates in maintenance medium (4,000 cells/cm2). The following day (day 1), maintenance medium was replaced with osteogenic medium (maintenance medium supplemented with 10 μM beta-glycerol-phosphate, 50 μg/μl ascorbic acid, and 0.1 μM dexamethasone). Media were changed every three days. RNA was isolated at the indicated times. The pan-HDAC inhibitor LBH-589 (10 nM) or vehicle was added three days after induction of osteogenic differentiation (day 4) and administered for two consecutive three-day periods (until day 10). On day 7, cells were fixed in 10% neutral buffered formalin and stained with 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium to monitor the enzymatic activity of alkaline phosphatase (Promega). On day 14, cells were fixed in 10% neutral buffered formalin and stained with 2% Alizarin Red to visualize calcium deposition. Absorption of alizarin red and alkaline phosphatase stains was quantified with ImageJ software [Schneider et al., 2012].
2.3 Western blotting
MSCs (4,000 cells/cm2) were plated in 6-well plates in maintenance medium. Cells were treated with vehicle or HDAC inhibitor (10 nM LBH-589) as described. Cells were lysed in radio-immunoprecipitation buffer (150 mM NaCl, 50 mM Tris pH 7.4, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma) and phenylmethylsulphonyl fluoride (Sigma). Lysates were cleared by centrifugation. Protein concentrations were determined by the DC Protein Assay (Bio-Rad). Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. After blocking in 5% non-fat dry milk for 45 minutes at room temperature, primary antibodies were added overnight at 4°C, followed by secondary antibodies for 1 hour at room temperature. Proteins were visualized using an ECL Prime detection kit. Primary antibodies used were: Actin (1:10,000; sc-1616; Santa Cruz), H3 (1:10,000; 05-928; Millipore), and Ac-H3 (1:10,000; 06-599; Millipore).
2.4 Analysis of mRNA expression by real-time reverse transcriptase quantitative PCR (RT-qPCR)
Total cellular RNA was isolated using the miRNeasy kit (Qiagen) and subjected to reverse transcription into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Gene expression was quantified using RT-qPCR whereby each reaction was performed with 10 ng cDNA per 10 μl, QuantiTect SYBR Green PCR Kit (Qiagen), and the CFX384 Real-Time System machine (BioRad). Transcript levels were analyzed using gene specific primers (Supplementary Table 1), quantified using the 2ΔΔCt method and initially normalized to the housekeeping gene GAPDH (set at 100). None of multiple other housekeeping genes we tested (e.g., HPRT, ACTB) was truly constant across all samples, and exhibited changes in expression relative to GAPDH. Therefore, to account for biological variation in expression of GAPDH and other housekeeping genes, we standardized expression values using sample averages across a matrix of all probes and RNA samples by log2 transformation, row centering and normalization.
2.5 High throughput RNA sequencing and bioinformatic analysis
RNA was isolated from non-proliferative (confluent) MSCs derived from three different donors. High throughput next generation RNA-sequencing (RNA-seq) of polyA mRNAs and bioinformatic analyses were performed as previously reported [Camilleri et al., 2016; Dudakovic et al., 2014]. Gene expression is expressed in reads per kilobasepair per million mapped reads (RPKM) and are accessible through the NCBI Gene Expression Omnibus using series accession number GSE84322.
2.6 Database analysis using FileMerge
To facilitate comparison of RNA-seq data (n > 20,000 genes) with results from our multi-gene qPCR platform (n > 300 genes), we developed an application (FileMerge) with a user-friendly Windows-based graphical interface for rapid extraction of a large subset of genes from any larger dataset. In essence, the application merges a simple text file with a list of gene identifiers (Gene IDs) and any associated meta-data (e.g., gene ontology terms) with a text file that contains all expression and meta-data from other large data sets (e.g., micro-arrays, RNA-seq, ChIP-seq). The merged data sets are then available for import into Excel or other spreadsheet applications.
The FileMerge application was built using the Microsoft technology stack within the Microsoft Visual Studio Integrated Development Environment (IDE, Microsoft). The graphical components of the application were built using Windows Presentation Foundation (WPF, Windows), while all of the remaining code is in C# (Microsoft). Additional open source frameworks used to build the application include the MVVM Light toolkit, which was used to separate logical layers within the application, and the Metro UI toolkit of MahApps, which was used to generate a user-friendly graphical interface. The FileMerge application and instructions for use are available upon request.
2.7 Comparison of expression profiling using RT-qPCR versus high throughput RNA sequencing
FileMerge was used to compare expression data obtained by RNA-seq analysis with results from semi-automated RT-qPCR using a large panel of gene-specific primer pairs for many annotated human epigenetic regulators (Supplementary Tables 2 and 3). FileMerge acquired normalized read data (i.e., RPKM values) for all epigenetic regulators (321 genes) within our RNA-seq data set (23,398 genes) and aligned these data with the corresponding relative expression values obtained by RT-qPCR (normalized expression, GAPDH = 100). For most genes (n = 295), the arbitrary ratio of RNA-seq and RT-qPCR values represents a continuous variable (i.e., ratios ranging between 50 and 0.5) (Fig. 1). Because RNA-seq detection depends on amplification of bar-coded cDNA libraries, while RT-qPCR amplifies cDNAs using gene-specific primer pairs, differences in the RNA-seq/RT-qPCR ratio may reflect the relative efficiency by which each method detects a given gene.
Figure 1. Comparison of expression patterns of human epigenetic regulators by RNA-seq and RT-qPCR.
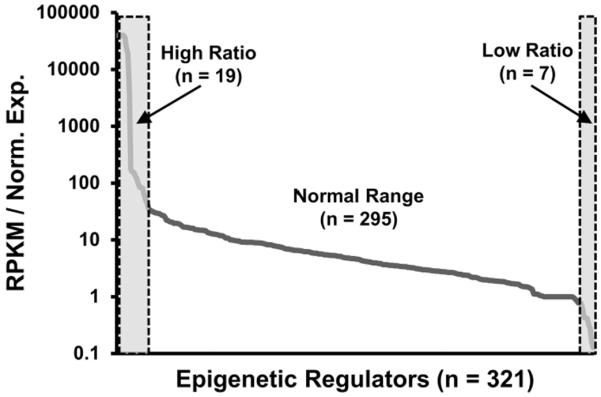
Expression data (n = 6 donors: #211, #222, # 234, #237, #258, #283) obtained using a semi-automated RT-qPCR platform that examines 321 human epigenetic regulators was compared to RNA-seq data for the same genes (n = 3 donors: #211, #258, #283) by generating a joint database using FileMerge. The figure depicts the ratio of RNA-seq expression values (in RPKMs) to arbitrarily standardized values obtained by RT-qPCR (that are normalized to GAPDH set at a value of 100). The resulting arbitrary ratio is not constant but represents a continuous variable that typically ranges from 50 to 0.5 (sloping line). The gray boxes with dashed boundaries on the left or right of the graph indicate that there are two subsets of genes with either disproportionally high (n = 19; ratio > 50) or low (n = 7; ratio < 0.5) ratios of the arbitrary values obtained by RNA-seq versus RT-qPCR.
While both methods exhibit fairly gradual differences in detection for each EpiReg, there are two subsets of genes with either disproportionally high (n = 19; ratio > 50) or low (n = 7; ratio < 0.5) ratios of the arbitrary values obtained by RNA-seq versus RT-qPCR. This observation indicates that either one of the two methods is not reliable for detection of these minor subsets of genes (e.g., highly inefficient RT-qPCR primers or technical bias in the preparation of the RNA-seq library). More importantly, combined analysis of RNA-seq and the RT-qPCR platform validates the relative expression levels of mRNAs for almost 300 EpiRegs mRNAs, thus permitting quantitative expression screens for these annotated epigenetic regulators in human cell or tissue samples.
2.8 Statistics
Statistical analysis was performed with unpaired Student's t-test. When applicable, significance is noted in the figures with a standard asterisk convention (*: p < 0.05, **: p < 0.01, and ***: p < 0.001).
3 Results
3.1 Expression pattern of epigenetic regulators in MSCs
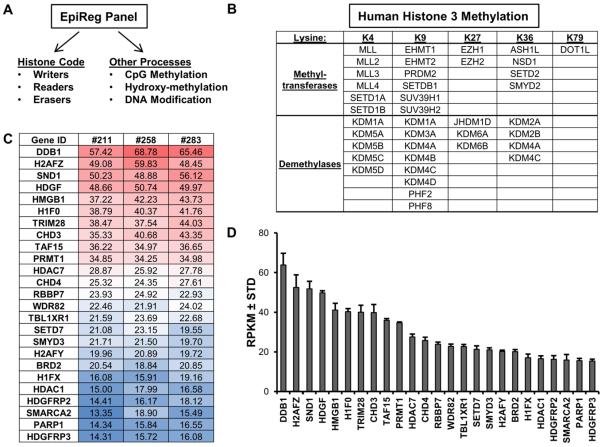
To understand which epigenetic regulators define the epigenome of a given cell type, we generated a hybrid strategy that combines semi-automated qPCR with RNA-seq and a novel data management application (FileMerge). This strategy permits expression screening for a large cohort of human epigenetic regulators [Liu et al., 2012] and includes all major proteins that affect chromatin by chemical alterations of DNA, post-translational modifications (PTMs) of histone proteins, or changes in the topology or architecture of chromatin [Kouzarides, 2007] (Fig. 2A). EpiRegs that functionally and directly regulate the transcriptional competency of chromatin are of particular importance, and this subset includes methyltransferases and demethylases that add, read and/or remove methyl-moieties on biologically important lysine (K) residues in histone H3 (i.e., K4, K9, K27, K36 and K79) (Fig. 2B).
Figure 2. Robustly expressed epigenetic regulators in MSCs.
The major classes of human epigenetic regulators (A). The histone code can be modified and interpreted by several classes of genes, including enzymes that add (writers: e.g., methyltransferases, acetyltransferases) or remove (erasers: e.g., demethylases, deacetylases) functional groups on histones. Other proteins have domains (readers: e.g., bromodomain, chromodomain) that bind to specific histone modifications and control gene expression by recruiting other transcriptional modulators. Methylation of H3 on K4, K9, K27 or K36, which represents a critical epigenetic post-translational modification that controls gene expression, is modulated by several methyltransferases and demethylases (B). The table (C) and graph (D) depict the expression patterns of the 25 most robustly expressed epigenetic regulators in non-proliferative (confluent) MSCs (n = 3). The table and graph show expression values expressed as reads per kilobasepair per million mapped reads (RPKM).
To define base-line expression values of EpiRegs, we assessed their expression in confluent (non-proliferating) MSCs. The MSCs used in this study were isolated from the stromal vascular fraction of adipose tissue and represent a cell culture model for the basal state of non-induced undifferentiated mesenchymal progenitor cells. These cells have tri-lineage potential and can differentiate into osteogenic, adipogenic and chondrogenic lineages [Dudakovic et al., 2014; Dudakovic et al., 2015a; Dudakovic et al., 2015b].
We performed RNA-seq with MSCs derived from three healthy donors [Camilleri et al., 2016; Dudakovic et al., 2014] and used a novel database application (FileMerge) to extract expression values of a large set of epigenetic regulators (>300 genes) (Supplementary Table 2) from the total RNA-seq data set (> 23,000 genes). RNA-seq values (in RPKM) for MSCs (n = 3) were rank ordered to identify epigenetic regulators with highest expression (Fig. 2C). Our analysis revealed robust expression of several epigenetic enzymes that can be potentially inhibited by small molecules, such as histone deacetylases (e.g., HDAC1 and HDAC7) and methyltransferases (e.g., PRMT1, SETD7, and SMYD3) that are expressed at higher levels than their known isoforms in MSCs.
The highest expressing epigenetic regulators are DDB1, H2AFZ, and SND1 (average expression > 50 RPKM), while many genes such as SP140, SMYD1 and the testis-specific bromodomain protein BRDT have relatively low expression (< 0.3 RPKM) (Supplementary Table 2). The most highly expressed EpiRegs that were analyzed have a similar expression pattern, as reflected by minimal standard deviations in mRNA levels observed in MSCs from three donors (Fig. 2D and Supplementary Table 2). This subset of constitutively and highly expressed genes may represent a core set of epigenetic regulators that execute cellular housekeeping functions, maintain the pericyte-like stromal fibroblast phenotype and/or multi-lineage differentiation potential of MSCs.
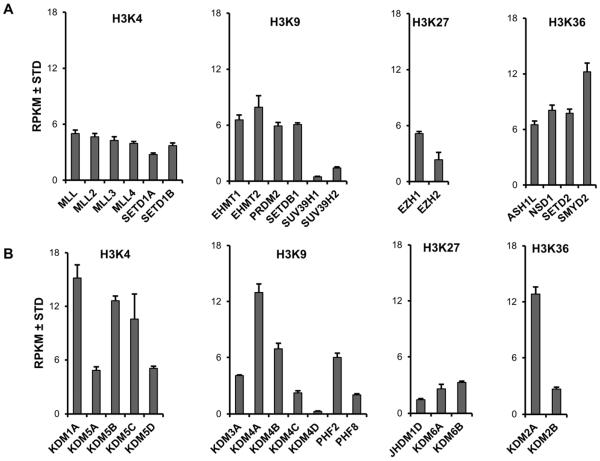
Modifications on H3, especially acetylation and methylation, are major epigenetic events that control gene expression in mesenchymal progenitor cells [Gordon et al., 2015]. To understand which epigenetic regulators contribute to the epigenetic landscape in MSCs, we assessed the expression pattern of the principal H3 methyltransferases and demethylases in post-proliferative confluent MSCs (Fig. 3). We find similar levels of expression for H3K4 methyltransferases, but the H3K9 methyltransferases SUV39H1 and SUV39H2 are expressed at less pronounced levels (Fig. 3A). EZH1 and SMYD2 are the highest expressed H3K27 and H3K36 methyltransferases, respectively, and are expressed at levels higher than their isoforms (e.g., EZH2 and SMYD1). Interestingly, unlike the H3 methyltransferases that appear to be expressed at fairly uniform levels, distinct types of H3 demethylases that target different lysine residues appear to exhibit greater variation in expression (Fig. 3B). For example, specific isoforms of the H3K4 demethylases (i.e., KDM1A, KDM5B, and KDM5C), H3K9 demethylases (i.e., KDM4A, KDM4B, and PHF2) and H3K36 demethylases (i.e., KDM2A) in each case are the highest expressed isoforms in their respective classes, while the three known H3K27 demethylases (i.e., JHDM1D, KDM6A, KDM6B) are expressed at relatively similar levels. Together, our results indicate that H3 demethylases are more variably expressed compared to H3 methyltransferases in undifferentiated non-proliferating MSCs.
Figure 3. Differential expression pattern of H3 methyltransferases and demethylases in MSCs.
Expression pattern of H3 methyltransferases (A) and demethylases (B) in non-proliferative MSCs (n = 3). The methyltransferases and demethylases are grouped by the lysine (K) residue targeted by these epigenetic regulators. The figure shows expression values as reads per kilobasepair per million mapped reads (RPKM).
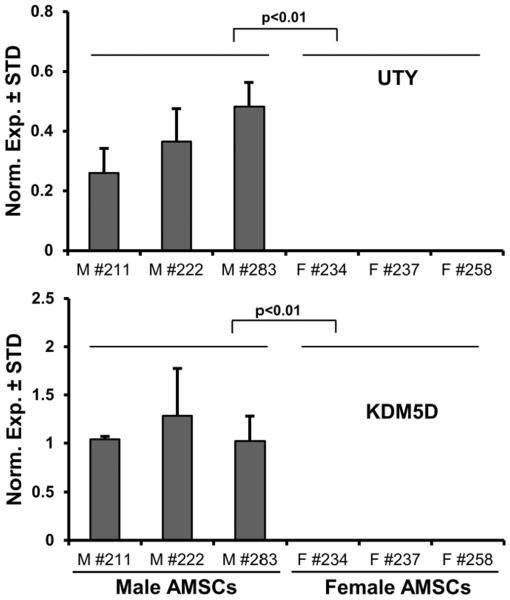
One key capability of our RT-qPCR platform is the rapid characterization of variations in EpiReg expression in cells and tissues under different biological or clinical conditions. For example, variations in EpiReg expression among different patients may provide insight into disease-related epigenomic events. As a proof-of-principle for achieving patient stratification using EpiReg data, we investigated gender-related expression differences between MSCs from six distinct donors (i.e., female: n = 3 & male: n = 3). We removed all genes exhibiting non-significant differences in expression between female and male donors (P < 0.05 using Student's t-test). Of the remaining genes (n = 9), seven EpiRegs had only marginal differences in expression (less than two-fold) between males and females. The remaining two EpiRegs exhibited robust expression in male but not female patients (i.e., KDM5D and UTY) (Fig. 4). Hence, EpiReg expression analysis reveals that KDM5D and UTY are male-specific epigenetic regulators, consistent with their known localization on the Y-chromosome. Furthermore, these results provide proof-of-concept that patient-stratification (e.g., in this case by gender) can be achieved using mRNAs for EpiRegs as biomarkers.
Figure 4. Gender-specific expression of epigenetic regulators in MSCs.
RT-qPCR expression analysis of all epigenetic regulators (n = 321) was performed on six (three male, three female) non-proliferative (confluent) MSCs. Genes were sorted for fold change differences (>2 fold) between males and females, average normalized expression values in six patients (arbitrary value > 0.1 with GAPDH set at 100), as well as statistical confidence based on p-values for differences between male and female patients (p < 0.01). Two epigenetic regulators, KDM5D and UTY, are differentially expressed MSCs derived from male and female patients.
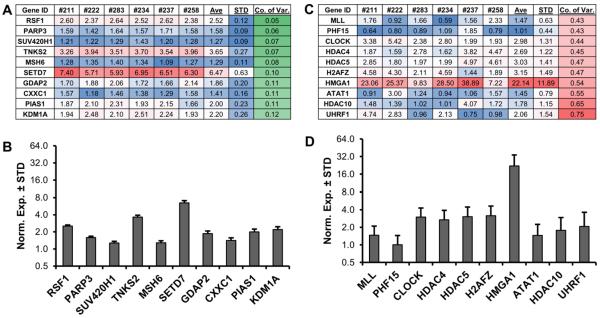
To assess which epigenetic regulators have the greatest variation in expression, we calculated the coefficient of variation (i.e., ratio of the standard deviation to the mean) for their expression in our panel of six MSCs. We selected epigenetic regulators showing clearly detectable expression (n = 134, average expression value > 1; the arbitrary expression value for GAPDH was set at 100) (Fig. 5). The expression profiles of the ten most stable genes across the six MSC samples that displayed the highest (Fig. 5A and B) or lowest (Fig. 5C and D) variability in relation to the mean. Three genes with very low standard deviations represent lysine methylome-associated genes (i.e., SUV420H1, SETD7, and KDM1A), while three representative genes with a high coefficient of variation are lysine acetylome-related genes (i.e., HDAC4, HDAC5, and HDAC10). Thus, while some epigenetic regulators show limited variation in expression among different patients, genes that exhibit highly variable expression across MSCs may be useful as epigenome-related biomarkers for patient stratification and/or different biological conditions.
Figure 5. Stable and differentially expressed epigenetic regulators in MSCs.
RT-qPCR expression analysis of all epigenetic regulators (n = 321) was performed on six (three male, three female) non-proliferative (confluent) MSCs. Genes were sorted for average normalized expression value in six patients (value >1 with GAPDH set at 100). To assess the level of variability, the standard deviation (STD) was divided by the average expression (Ave) to generate the coefficient of variation (Co. of Var.) for each epigenetic regulator in the six MSCs. The ten most stable epigenetic regulators are shown (A). To demonstrate low levels of inter-patient variability, these ten highly stable genes are graphed (B). The ten most variable (`unstable') epigenetic regulators are shown for comparison (C) and further illustrated using bar graphs (D).
3.2 Altered expression of epigenetic regulators during osteogenic differentiation of MSCs
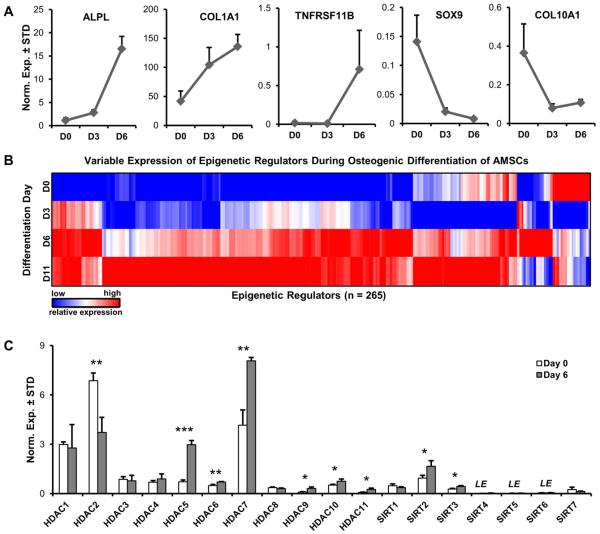
To identify epigenetic regulators that may contribute to the osteoblast phenotype, we performed RT-qPCR analysis of EpiRegs and selected bone-related mRNA markers during osteogenic differentiation of MSCs (Fig. 6). Progression of differentiation is evidenced by enhanced expression of osteoblast-related genes, including Alkaline Phosphatase (ALPL), Collagen Type I α1 chain (COL1A1), and Osteoprotegerin (OPG/TNFRSF11B) during osteogenic commitment of MSCs (Fig. 6A). For comparison, chondrogenic markers including the transcription factor `SRY (Sex Determining Region Y)-Box 9' (SOX9) and Collagen Type X α1 chain (COL10A1) are suppressed during osteogenic differentiation in MSCs. These data are generally consistent with the osteogenic potential of these multi-potent MSCs that was established in previous studies [Dudakovic et al., 2014; Dudakovic et al., 2015a; Dudakovic et al., 2015b].
Figure 6. Differentially expressed epigenetic regulators during osteogenic differentiation of MSCs.
RT-qPCR expression analysis of all epigenetic regulators and phenotypic genes was performed during an osteogenic time-course (D0, D3, D6, and D11) in MSCs (female patient #258, n = 3). Markers of osteogenic differentiation (ALPL, COL1A1, and TNFRSF11B) are up-regulated, while chondrogenic differentiation markers (SOX9 and COL10A1) are down-regulated during osteogenic differentiation of MSCs (A). Heat-map showing the expression trend of robustly expressed epigenetic regulators (n = 258) in differentiating MSCs (value >0.1 with GAPDH set at 100, out of a total of set of mRNAs of n = 321) (B). Expression pattern of HDAC in undifferentiated (D0) and differentiating (D6) MSCs (C). LE = low expression (<0.01, GAPDH = 100).
We performed RT-qPCR expression profiling using our entire RT-qPCR primer panel for detection of human epigenetic regulators (321 genes) across all four time points of the differentiation time-course to define which EpiReg isoforms control the epigenome during lineage-commitment of MSCs. To increase confidence of identifying differentially expressed genes, we sorted for genes with clearly detectable levels of expression (arbitrary average signal > 0.1 (n = 258), GAPDH was arbitrarily set at 100) (Fig. 6B and Supplementary Table 4).
Hierarchical clustering analysis and heat map presentation shows that there are at least two different groups of EpiRegs that are either down-regulated or up-regulated during osteogenic differentiation. Further analysis of our datasets using numerical filters yielded several tens of epigenetic regulators that are either up-regulated (e.g., the H3K27 demethylases JHDM1D and KDM6B) or down-regulated (e.g., H3K27me3 transferase EZH2) (Table 1). Several genes up-regulated during MSC differentiation are HDACs (i.e., HDAC5, HDAC9, and HDAC11) each are elevated by 3 to 4 fold relative to undifferentiated cells, while HDAC2 is the only member of this class that is significantly down-regulated by more than 2 fold when MSCs commit to the osteogenic lineage (Fig. 6C). Taken together, our analyses demonstrate that several epigenetic regulators, including HDACs, are differentially expressed between undifferentiated and differentiating MSCs.
Table 1.
Altered expression of epigenetic regulators during osteogenic differentiation (six days).
| Up on Day 6 | Down on Day 6 | ||
|---|---|---|---|
| Gene | Fold Change | Gene | Fold Change |
| ZCWPW1 | 7.26 | EZH2 | 19.86 |
| NCOA1 | 5.16 | H2AFX | 12.63 |
| SMARCA2 | 4.57 | SP140 | 12.49 |
| KDM4B | 4.56 | TCF19 | 9.83 |
| TRIM66 | 4.54 | SUV39H1 | 9.44 |
| MBD5 | 4.52 | CHAF1B | 8.27 |
| HDAC5 | 4.17 | HELLS | 7.88 |
| MLL3 | 3.63 | DPF1 | 6.76 |
| HDAC9 | 3.47 | UHRF1 | 6.51 |
| JHDM1D | 3.38 | H2AFZ | 6.36 |
| KDM3A | 3.38 | ATAD2 | 5.90 |
| HDAC11 | 3.34 | PHF19 | 5.83 |
| KDM6B | 3.12 | WHSC1 | 4.36 |
| HMGB1 | 3.89 | ||
| HMGB3 | 3.42 | ||
| DNMT1 | 3.27 | ||
| SUV39H2 | 3.25 | ||
3.3 Histone deacetylases inhibition suppresses osteogenic differentiation of MSCs
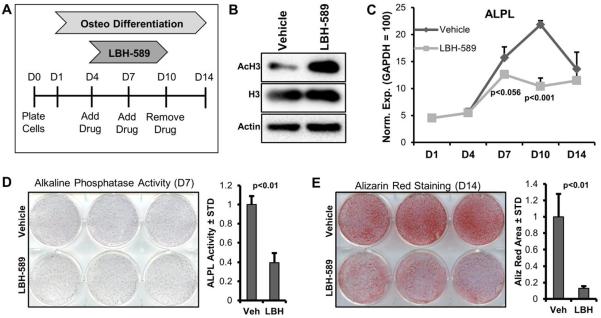
The expression pattern of HDACs is consistent with the overall idea that these enzymes control osteogenic lineage commitment of MSCs as we have shown in our previous studies [Dudakovic et al., 2015a; Dudakovic et al., 2013]. Indeed, treatment of MSCs with the pan-HDAC inhibitor LBH-589 (Panobinostat, Farydak) at low non-cytotoxic concentrations (10 nM) for six days alters osteogenic lineage commitment of MSCs (Fig. 7A). This result corroborates our previous findings for MSCs obtained with another clinically relevant HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA, Vorinostat, Zolinza) [Dudakovic et al., 2015a]. Addition of LBH-589 (10 nM) for 24 hours in differentiating MSCs enhances acetylation of H3 as expected (Fig. 7B). Furthermore, RT-qPCR analysis shows that HDAC inhibition suppresses the expression of the osteogenic marker ALPL in particular at the stage of peak mRNA expression (day 10) (Fig. 7C). The reduced levels of ALPL mRNA are supported by suppressed activity of this protein on day 7 of MSCs differentiation based on colorimetric assays (Fig. 7D). Inhibition of HDACs also reduces calcium deposition as detected by Alizarin Red staining (at day 14) in MSCs undergoing osteogenic differentiation (Fig. 7D). Taken together with previous studies (Dudakovic et al., 2015a), inhibition of HDAC activity by either SAHA or LBH-589 prevents osteogenic cell fate determination of MSCs.
Figure 7. HDAC Inhibition attenuates osteogenic differentiation of MSCs.
The experimental procedure for treatment and differentiation of MSCs involves treatment of cells at confluence for six days (d4-10) with vehicle (DMSO) or 10nM LBH-589. (A). Western blot analysis of vehicle and LBH-589 treated (24 hours) MSCs (B). RT-qPCR expression analysis of ALPL (n=3) in differentiating MSCs in the presence of vehicle or LBH-589 (C). Alkaline phosphatase staining (D) and alizarin-red staining (E) for MSCs treated with vehicle or LBH-589.
4 Discussion
In this study, we used an integrated strategy that encompasses semi-automated RT-qPCR assays, RNA-seq analysis and a data management application (FileMerge) we developed to assess expression of epigenetic regulators in different biological conditions. While this expression screening strategy is generally applicable to any biological problem, as a test case for the robustness of the methodology we applied it to uncommitted MSCs from different patients and during osteogenic differentiation. Among our key findings are results showing that uncommitted MSCs (i) robustly express a large number of EpiRegs at a broad range of expression levels (e.g., HDACs, HATs, KMTs and KDMs), (ii) differentially express distinct EpiReg isoforms with similar molecular functions (e.g., H3K9 demethylases, and (iii) show a proof-of-principle for patient-to-patient differences in EpiReg expression (i.e., expression of KDM5D and UTY is gender-related, as previously reported due to the location of these genes on the Y-chromosome [Greenfield et al., 1996; Skaletsky et al., 2003; Wang et al., 1995]. Furthermore, we show that (iv) some epigenetic regulators are stably expressed across patients and biological conditions while others have more variable expression, (v) that EpiRegs exhibit differential expression of isoforms during osteogenic differentiation, and that (vi) selective inhibition of EpiRegs impedes osteogenic differentiation. Taken together, our study establishes that expression screening of EpiRegs using our RT-qPCR platform is useful for the identification of principal regulatory factors capable of modifying the epigenetic landscape and phenotypic memory of progenitor cells during lineage commitment.
To develop novel biological strategies for manipulating the epigenome, it is necessary to understand which epigenetic regulators are expressed in different cell types and biological conditions. Beyond studies from our own group [Dudakovic et al., 2016; Dudakovic et al., 2015b; Dudakovic et al., 2013; Farzaneh et al., 2016], other studies have used various screening methods to assess expression and function of epigenetic regulators. These studies utilized technologies such RNA interference [Bajpe et al., 2015; Cellot et al., 2013; Fazzio et al., 2008], high-content cell-spot microarrays [Bjorkman et al., 2012], as well as large-scale reverse genetic screening [Huang et al., 2013] to screen for epigenetic regulators in various tissues, organism, and disease states. Our approach integrates RNA-seq and RT-qPCR technologies with data management application (FileMerge) to screen for epigenetic regulators. The advantage of RNA-seq is the absolute quantification of expression levels relative to the entire transcriptome and its apparent robustness (i.e., expression analyses are remarkably reproducible). The advantage of the RT-qPCR platform we developed is that it complements and validates RNA-seq analysis, but also circumvents some of the major challenges with RNA-seq (e.g., technically demanding, expensive and time-consuming). Our side-by-side analysis of the two methods revealed that a small fraction of EpiRegs (n=321 total analyzed) are not properly detected by RNA-seq (n=7) or RT-qPCR (n=19), presumably due to technical limitations (e.g., qPCR primer efficiency and/or RNA-seq library preparation). More importantly, our studies validated the relative expression of >90% of all epigenetic regulators examined in human MSCs using both RNA-seq and RT-qPCR, and analysis of these genes by RT-qPCR will now suffice for future studies.
Accumulating evidence indicates that epigenetic mechanisms control osteoblast differentiation [Gordon et al., 2015]. Therefore, one major goal of our laboratory is to characterize epigenetic regulators that are differentially expressed in the basal ground state of undifferentiated MSCs versus and early stages of mesenchymal lineage commitment in MSCs (e.g., during differentiation into osteoblastic cells). Using the method we described here in detail, we identified the H3K27 methyltransferase EZH2 as the most prominently down-regulated epigenetic regulator during osteogenic commitment of MSCs [Dudakovic et al., 2015b]. Our group and others collectively showed that EZH2 is a principal epigenetic regulator of osteogenic lineage commitment of MSCs and a suppressor of osteoblast maturation [Dudakovic et al., 2016; Dudakovic et al., 2015b; Hemming et al., 2014; Hui et al., 2014; Wei et al., 2011].
Our EpiReg expression screen also identified several HDACs that are differentially expressed between uncommitted and differentiated MSCs. For example, our results show that HDAC2 is down-regulated during osteogenic differentiation of clinical-grade human MSCs, similar to observations for immortalized mouse MC3T3 osteoblasts [Choo et al., 2009; Lee et al., 2006a]. We also find that several HDACs (e.g., HDAC5 and HDAC7) are up-regulated during osteogenic differentiation of human MSCs. Interestingly, previous studies have demonstrated that HDAC5 suppresses the WNT-signaling inhibitor sclerostin (SOST) in murine osteocytes, while HDAC7 controls the transcriptional activity of RUNX2 in mouse C2C12 mesenchymal cells [Jensen et al., 2008; Wein et al., 2015]. These activities of HDAC5 and HDAC7 may perhaps complement the RUNX2-dependent cell growth regulatory role of HDAC6 described in mouse MC3T3 osteoblasts [Westendorf et al., 2002]. The differentiation-related up-regulation of HDAC5 and HDAC7 we have observed in our study suggests that these HDAC isoforms may have a novel biological role during osteogenic lineage progression of human MSCs that is perhaps linked to WNT-signaling and RUNX activity.
Enhanced expression of bone-related extracellular matrix proteins (e.g., osteocalcin/BGLAP) is associated with increased histone acetylation of the corresponding genes [Shen et al., 2003; Shen et al., 2002]. Therefore, inhibition of HDAC enzymatic activity which will stimulate genome-wide acetylation of H4 proteins near transcriptional start sites [Dudakovic et al., 2013], or gene inactivation of HDACs can stimulate differentiation of pre-committed osteoblasts in vitro in different cell culture models [Di Bernardo et al., 2009; Dudakovic et al., 2013; Haberland et al., 2010; Hu et al., 2013; Iwami and Moriyama, 1993; Lee et al., 2009; Schroeder and Westendorf, 2005]. However, inhibition of HDACs with LBH-589 in human MSCs (this study) or SAHA in either human or mouse MSCs [Dudakovic et al., 2015a; McGee-Lawrence et al., 2011], interferes with lineage-progression at least in part by effects on cell proliferation. Consequently, Hdac inactivation by gene knock-out, knock-down or chemical inhibition in mouse models typically has negative effects on skeletal development and bone mineral density [Bradley et al., 2015; Gordon et al., 2015; McGee-Lawrence et al., 2011; Pratap et al., 2010; Razidlo et al., 2010; Senn et al., 2010; Zhang et al., 2008]. Similarly, HDAC inhibitors have negative effects on bone parameters in the human skeleton [Boluk et al., 2004; Elliott et al., 2007; Sheth et al., 1995; Vestergaard et al., 2004]. Because expression screening using our RT-qPCR expression platform indicates that different HDAC isoforms are maximally expressed at either uncommitted (e.g., HDAC2) or phenotype committed (e.g., HDAC5 and HDAC7) stages of human MSC differentiation, it appears that the negative biological consequences of pharmacological inhibition using pan-HDAC drugs are directly attributable to robust HDAC isoform expression at multiple stages of osteogenesis.
In conclusion, the integration of semi-automated RT-qPCR, RNA-seq and FileMerge represents a generally applicable expression screening strategy for understanding which epigenetic regulators are expressed in a given human cell type or biological condition. The RT-qPCR platform itself, which we have validated by RNA-seq, provides a rapid high-throughput assay to enables focus on those isoforms of epigenetic regulators that are most relevant to a given biological process. This understanding is also clinically relevant because it identifies specific EpiReg isoforms that could be considered as targets for drug development to mitigate or reverse human diseases that are controlled by epigenetic mechanisms.
Supplementary Material
Highlights.
Development of a hybrid expression screening platform for epigenetic regulators.
Epigenetic regulators are variably expressed in human MSCs.
Osteogenic differentiation of MSCs alters expression of epigenetic regulators.
Pan-deacetylase inhibitor LBH-589 inhibits osteogenic differentiation of AMSCs.
Acknowledgments
We thank present and former members of our laboratories, including Scott Riester, Emily Camilleri, Eric Lewallen, Janet Denbeigh, Rebekah Samsonraj, Catalina Galeano-Garces, Bashar Hasan, Oksana Pichurin, and David Razidlo for assistance with the experimentation and/or stimulating discussions. We also acknowledge the support of Asha Nair and Jared Evans from the Bioinformatics Core, as well as Bruce Eckloff of the Medical Genome Facility.
Funding: This work was supported, in whole or in part, by National Institutes of Health grants F32 AR066508 (AD), R01 AR049069 (AJvW), R01 AR068103 (JJW), R01 DE020194 (JJW), and R01 AR039588 (GSS), as well as by grants from Fondo de Finaciamiento de Centros de Investigación en Áreas Prioritarias (15090007; to MM) and Fondo Nacional de Ciencia y Tecnologia (FONDECYT 1130706; to MM). Additional support was provided by intramural grants from the Center for Regenerative Medicine at Mayo Clinic. We also appreciate the generous philanthropic support of William H. and Karen J. Eby.
ABBREVIATIONS
- SC
mesenchymal stromal/stem cell
- AMSC
adipose-tissue derived mesenchymal stromal/stem cell
- RT-qPCR
real-time reverse transcriptase quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- EpiReg
epigenetic regulator
- HAT
histone acetyl transferase
- KMT
lysine methyltransferase
- HDAC
histone deacetylase
- EHMT
euchromatic histone lysine methyltransferase
- KDM
lysine demethylase
- BRD
dromodomain
- CBX
chromobox
- KDM5D
lysine demethylase 5D
- UTY
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked
- HDAC1
histone deacetylase 1
- HDAC2
histone deacetylase 2
- HDAC4
histone deacetylase 4
- HDAC5
histone deacetylase 5
- HDAC7
histone deacetylase 7
- HDAC9
histone deacetylase 9
- HDAC10
histone deacetylase 10
- HDAC11
histone deacetylase 11
- LBH-589
panobinostat
- ALPL
alkaline phosphatase
- WDR5
WD repeat domain 5
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors have competing financial interests relevant to the content of this paper.
References
- Bajpe PK, Prahallad A, Horlings H, Nagtegaal I, Beijersbergen R, Bernards R. A chromatin modifier genetic screen identifies SIRT2 as a modulator of response to targeted therapies through the regulation of MEK kinase activity. Oncogene. 2015;34:531–6. doi: 10.1038/onc.2013.588. [DOI] [PubMed] [Google Scholar]
- Barutcu AR, Tai PW, Wu H, Gordon JA, Whitfield TW, Dobson JR, Imbalzano AN, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The bone-specific Runx2-P1 promoter displays conserved three-dimensional chromatin structure with the syntenic Supt3h promoter. Nucleic acids research. 2014;42:10360–72. doi: 10.1093/nar/gku712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman M, Ostling P, Harma V, Virtanen J, Mpindi JP, Rantala J, Mirtti T, Vesterinen T, Lundin M, Sankila A, Rannikko A, Kaivanto E, Kohonen P, Kallioniemi O, Nees M. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene. 2012;31:3444–56. doi: 10.1038/onc.2011.512. [DOI] [PubMed] [Google Scholar]
- Boluk A, Guzelipek M, Savli H, Temel I, Ozisik HI, Kaygusuz A. The effect of valproate on bone mineral density in adult epileptic patients. Pharmacological research : the official journal of the Italian Pharmacological Society. 2004;50:93–7. doi: 10.1016/j.phrs.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME, Westendorf JJ. Histone Deacetylases in Bone Development and Skeletal Disorders. Physiol Rev. 2015;95:1359–81. doi: 10.1152/physrev.00004.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri ET, Gustafson MP, Dudakovic A, Riester SM, Garces CG, Paradise CR, Takai H, Karperien M, Cool S, Sampen HJ, Larson AN, Qu W, Smith J, Dietz AB, van Wijnen AJ. Identification and validation of multiple cell surface markers of clinical-grade adipose-derived mesenchymal stromal cells as novel release criteria for good manufacturing practice-compliant production. Stem cell research & therapy. 2016;7:107. doi: 10.1186/s13287-016-0370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot S, Hope KJ, Chagraoui J, Sauvageau M, Deneault E, MacRae T, Mayotte N, Wilhelm BT, Landry JR, Ting SB, Krosl J, Humphries K, Thompson A, Sauvageau G. RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity. Blood. 2013;122:1545–55. doi: 10.1182/blood-2013-04-496281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo MK, Yeo H, Zayzafoon M. NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone. 2009;45:579–89. doi: 10.1016/j.bone.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Diaz R, Behfar A, Butler GW, Padley DJ, Sarr MG, Bartunek J, Dietz AB, Terzic A. Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell transplantation. 2011;20:797–811. doi: 10.3727/096368910X543376. [DOI] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes & development. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bernardo G, Squillaro T, Dell'Aversana C, Miceli M, Cipollaro M, Cascino A, Altucci L, Galderisi U. Histone deacetylase inhibitors promote apoptosis and senescence in human mesenchymal stem cells. Stem cells and development. 2009;18:573–81. doi: 10.1089/scd.2008.0172. [DOI] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im HJ, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. Journal of cellular biochemistry. 2014;115:1816–28. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Lewallen EA, McGee-Lawrence ME, Riester SM, Kakar S, Montecino M, Stein GS, Ryoo HM, Dietz AB, Westendorf JJ, van Wijnen AJ. Histone deacetylase inhibition destabilizes the multi-potent state of uncommitted adipose-derived mesenchymal stromal cells. Journal of cellular physiology. 2015a;230:52–62. doi: 10.1002/jcp.24680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O'Toole TM, Thaler R, Evans JM, Yan H, Subramaniam M, Hawse JR, Stein GS, Montecino MA, McGee-Lawrence ME, Westendorf JJ, van Wijnen AJ. Enhancer of Zeste Homolog 2 Inhibition Stimulates Bone Formation and Mitigates Bone Loss Caused by Ovariectomy in Skeletally Mature Mice. The Journal of biological chemistry. 2016;291:24594–24606. doi: 10.1074/jbc.M116.740571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, van Wijnen AJ. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. The Journal of biological chemistry. 2015b doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Evans JM, Li Y, Middha S, McGee-Lawrence ME, van Wijnen AJ, Westendorf JJ. Histone Deacetylase Inhibition Promotes Osteoblast Maturation by Altering the Histone H4 Epigenome and Reduces Akt Phosphorylation. The Journal of biological chemistry. 2013;288:28783–28791. doi: 10.1074/jbc.M113.489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JO, Jacobson MP, Haneef Z. Homocysteine and bone loss in epilepsy. Seizure. 2007;16:22–34. doi: 10.1016/j.seizure.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Farzaneh K, Thaler R, Paradise CR, Deyle DR, Kruijthof-de Julio M, Galindo M, Gordon JA, Stein GS, Dudakovic A, van Wijnen AJ. Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation. Journal of cellular biochemistry. 2016 doi: 10.1002/jcb.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–74. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- Gordon JA, Stein JL, Westendorf JJ, van Wijnen AJ. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone. 2015;81:739–45. doi: 10.1016/j.bone.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori F, Divieti P, Demay MB. Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. The Journal of biological chemistry. 2001;276:46515–22. doi: 10.1074/jbc.M105757200. [DOI] [PubMed] [Google Scholar]
- Gori F, Friedman LG, Demay MB. Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Developmental biology. 2006;295:498–506. doi: 10.1016/j.ydbio.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Greenfield A, Scott D, Pennisi D, Ehrmann I, Ellis P, Cooper L, Simpson E, Koopman P. An H-YDb epitope is encoded by a novel mouse Y chromosome gene. Nature genetics. 1996;14:474–8. doi: 10.1038/ng1296-474. [DOI] [PubMed] [Google Scholar]
- Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. The Journal of biological chemistry. 2010;285:14663–70. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, Gronthos S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem cells. 2014;32:802–15. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang X, Dai L, Zhu J, Jia Z, Wang W, Zhou C, Ao Y. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem cells and development. 2013;22:248–55. doi: 10.1089/scd.2012.0105. [DOI] [PubMed] [Google Scholar]
- Huang HT, Kathrein KL, Barton A, Gitlin Z, Huang YH, Ward TP, Hofmann O, Dibiase A, Song A, Tyekucheva S, Hide W, Zhou Y, Zon LI. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nature cell biology. 2013;15:1516–25. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui T, A P, Zhao Y, Wang C, Gao B, Zhang P, Wang J, Zhou X, Ye L. EZH2, a potential regulator of dental pulp inflammation and regeneration. Journal of endodontics. 2014;40:1132–8. doi: 10.1016/j.joen.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Iwami K, Moriyama T. Effects of short chain fatty acid, sodium butyrate, on osteoblastic cells and osteoclastic cells. The International journal of biochemistry. 1993;25:1631–5. doi: 10.1016/0020-711x(93)90522-g. [DOI] [PubMed] [Google Scholar]
- Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2008;23:361–72. doi: 10.1359/JBMR.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Liao L, An Y, Su X, Liu S, Shuai Y, Zhang X, Jin Y. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Molecular therapy : the journal of the American Society of Gene Therapy. 2015 doi: 10.1038/mt.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. The EMBO journal. 2005;24:2543–55. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nature reviews. Genetics. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Lee HW, Suh JH, Kim AY, Lee YS, Park SY, Kim JB. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Molecular endocrinology. 2006a;20:2432–43. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lee YM, Kim MJ, Choi JY, Park EK, Kim SY, Lee SP, Yang JS, Kim DS. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Molecules and cells. 2006b;22:182–8. [PubMed] [Google Scholar]
- Lee S, Park JR, Seo MS, Roh KH, Park SB, Hwang JW, Sun B, Seo K, Lee YS, Kang SK, Jung JW, Kang KS. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell proliferation. 2009;42:711–20. doi: 10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhen XT, Denton E, Marsden BD, Schapira M. ChromoHub: a data hub for navigators of chromatin-mediated signalling. Bioinformatics. 2012;28:2205–6. doi: 10.1093/bioinformatics/bts340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader EK, Butler G, Dowdy SC, Mariani A, Knutson KL, Federspiel MJ, Russell SJ, Galanis E, Dietz AB, Peng KW. Optimizing patient derived mesenchymal stem cells as virus carriers for a phase I clinical trial in ovarian cancer. Journal of translational medicine. 2013;11:20. doi: 10.1186/1479-5876-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee-Lawrence ME, McCleary-Wheeler AL, Secreto FJ, Razidlo DF, Zhang M, Stensgard BA, Li X, Stein GS, Lian JB, Westendorf JJ. Suberoylanilide hydroxamic acid (SAHA; vorinostat) causes bone loss by inhibiting immature osteoblasts. Bone. 2011;48:1117–26. doi: 10.1016/j.bone.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. The Journal of biological chemistry. 2014;289:16016–31. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Meyer MB, St John HC, Benkusky NA. Epigenetic histone modifications and master regulators as determinants of context dependent nuclear receptor activity in bone cells. Bone. 2015 doi: 10.1016/j.bone.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Akech J, Wixted JJ, Szabo G, Hussain S, McGee-Lawrence ME, Li X, Bedard K, Dhillon RJ, van Wijnen AJ, Stein JL, Stein GS, Westendorf JJ, Lian JB. The histone deacetylase inhibitor, vorinostat, reduces tumor growth at the metastatic bone site and associated osteolysis, but promotes normal bone loss. Molecular cancer therapeutics. 2010;9:3210–20. doi: 10.1158/1535-7163.MCT-10-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annual review of biochemistry. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo DF, Whitney TJ, Casper ME, McGee-Lawrence ME, Stensgard BA, Li X, Secreto FJ, Knutson SK, Hiebert SW, Westendorf JJ. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PloS one. 2010;5:0011492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester SM, Denbeigh JM, Lin Y, Jones DL, de Mooij T, Lewallen EA, Nie H, Paradise CR, Radel DJ, Dudakovic A, Camilleri ET, Larson DR, Qu W, Krych AJ, Frick MA, Im HJ, Dietz AB, Smith J, van Wijnen AJ. Safety Studies for Use of Adipose Tissue-Derived Mesenchymal Stromal/Stem Cells in a Rabbit Model for Osteoarthritis to Support a Phase I Clinical Trial. Stem cells translational medicine. 2016 doi: 10.5966/sctm.2016-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. The Journal of biological chemistry. 2004;279:41998–2007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2005;20:2254–63. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- Senn SM, Kantor S, Poulton IJ, Morris MJ, Sims NA, O'Brien TJ, Wark JD. Adverse effects of valproate on bone: defining a model to investigate the pathophysiology. Epilepsia. 2010;51:984–93. doi: 10.1111/j.1528-1167.2009.02516.x. [DOI] [PubMed] [Google Scholar]
- Shen J, Hovhannisyan H, Lian JB, Montecino MA, Stein GS, Stein JL, Van Wijnen AJ. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Molecular endocrinology. 2003;17:743–56. doi: 10.1210/me.2002-0122. [DOI] [PubMed] [Google Scholar]
- Shen J, Montecino M, Lian JB, Stein GS, Van Wijnen AJ, Stein JL. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. The Journal of biological chemistry. 2002;277:20284–92. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- Sheth RD, Wesolowski CA, Jacob JC, Penney S, Hobbs GR, Riggs JE, Bodensteiner JB. Effect of carbamazepine and valproate on bone mineral density. The Journal of pediatrics. 1995;127:256–62. doi: 10.1016/s0022-3476(95)70304-7. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C, Bonewald LF, Pike JW. The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin D3 hormone. Molecular endocrinology. 2014;28:1150–65. doi: 10.1210/me.2014-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai PW, Wu H, Gordon JA, Whitfield TW, Barutcu AR, van Wijnen AJ, Lian JB, Stein GS, Stein JL. Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene. 2014;550:1–9. doi: 10.1016/j.gene.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler R, Maurizi A, Roschger P, Sturmlechner I, Khani F, Spitzer S, Rumpler M, Zwerina J, Karlic H, Dudakovic A, Klaushofer K, Teti A, Rucci N, Varga F, van Wijnen AJ. Anabolic and anti-resorptive modulation of bone homeostasis by the epigenetic modulator sulforaphane, a naturally occurring isothiocyanate. The Journal of biological chemistry. 2016 doi: 10.1074/jbc.M115.678235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela N, Aranguiz A, Lizama C, Sepulveda H, Antonelli M, Thaler R, Moreno RD, Montecino M, Stein GS, van Wijnen AJ, Galindo M. Mitotic Inheritance of mRNA Facilitates Translational Activation of the Osteogenic-Lineage Commitment Factor Runx2 in Progeny of Osteoblastic Cells. Journal of cellular physiology. 2016;231:1001–14. doi: 10.1002/jcp.25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard P, Rejnmark L, Mosekilde L. Fracture risk associated with use of antiepileptic drugs. Epilepsia. 2004;45:1330–7. doi: 10.1111/j.0013-9580.2004.18804.x. [DOI] [PubMed] [Google Scholar]
- Villagra A, Gutierrez J, Paredes R, Sierra J, Puchi M, Imschenetzky M, Wijnen Av A, Lian J, Stein G, Stein J, Montecino M. Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. Journal of cellular biochemistry. 2002;85:112–22. [PubMed] [Google Scholar]
- Wang W, Meadows LR, den Haan JM, Sherman NE, Chen Y, Blokland E, Shabanowitz J, Agulnik AI, Hendrickson RC, Bishop CE, et al. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. 1995;269:1588–90. doi: 10.1126/science.7667640. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nature cell biology. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, Nagano K, Baron R, Brooks D, Bouxsein M, Pajevic PD, Kronenberg HM. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2015;30:400–11. doi: 10.1002/jbmr.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Molecular and cellular biology. 2002;22:7982–92. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome biology. 2014;15:R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nature reviews. Genetics. 2010;11:583–9. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Molecular and cellular biology. 2008;28:1688–701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.