Abstract
Based on the ability of nitrate reductase synthesis, Corynebacterium pseudotuberculosis is classified into two biovars: Ovis and Equi. Due to the presence of nitrate reductase, the Equi biovar can survive in absence of oxygen. On the other hand, Ovis biovar that does not have nitrate reductase is able to adapt to various ecological niches and can grow on certain carbon sources. Apart from these two biovars, some other strains are also able to carry out the reduction of nitrate. The enzymes that are involved in electron transport chain are also identified by in silico methods. Findings about pathogen metabolism can contribute to the identification of relationship between nitrate reductase and the C. pseudotuberculosis pathogenicity, virulence factors, and discovery of drug targets.
1. Introduction
Corynebacterium pseudotuberculosis is a Gram-positive facultative intracellular pathogen [1–11]. C. pseudotuberculosis can be classified into two biovars, based on their ability to convert nitrate to nitrite. The nitrate-positive biovar is Equi, which causes ulcerative lymphangitis in equines, while the nitrate-negative biovar is known as Ovis, which is the etiologic agent of caseous lymphadenitis in small ruminants [9]. Both diseases are globally distributed and cause large economic losses to goat, sheep, horse, and cattle farmers.
The nitrate reduction is associated with the bacterium's ability to breathe in the absence of oxygen and having two different metabolic pathways, (1) respiratory nitrate reductase and (2) dissimilatory nitrate reduction. In the first pathway, the denitrification process takes place where the nitrate is sequentially reduced to nitrite, nitric oxide, nitrous oxide, and finally to dinitrogen [12, 13]. In the second pathway nitrate is directly converted into ammonia, which is secreted from the cell; this process can be performed by organisms with the nrf gene. This is a less common method of nitrate reduction than denitrification in most ecosystems [12–14]. There are four classes of nitrate reductases, one in eukaryotes and three are in prokaryotes. Prokaryotic nitrate reductases include a class of assimilatory enzymes and two classes of respiratory enzymes; all contain a guanylate molybdenum cofactor but differ in their substructures, cellular location, and requirement for cofactor. Variability among enzyme is also found into the classes [12, 13].
Among the Actinobacteria, respiratory nitrate reductase (Nar) is observed [15–18]. This class of enzymes is mostly membrane-bound and is having three different subunits: respiratory nitrate reductase from Pseudomonas stutzeri [19, 20], nitrate reductase A, and nitrate reductase Z from Escherichia coli K-12 [21–23]. In Corynebacterium glutamicum and Mycobacterium tuberculosis the nitrate reduction reaction is catalyzed by the operons narKGHJI and narGHJI, respectively [15, 16]. In these organisms, the first step in nitrate assimilation is nitrate reduction (NO3−) to nitrite (NO2−) and the second step is nitrite reduced (N3−) to ammonia (NH4+) [15, 16].
Nitrate respiration is associated with virulence and adoption of pathogenic organisms. In Brucella suis it helps in intramacrophagic multiplication, in M. bovis it allows survival inside the host, and in M. tuberculosis it confers resistance to stress [17, 24, 25]. However, the role of nitrate respiration is not known in C. pseudotuberculosis. We presume that Nar may be associated with pathogenicity in C. pseudotuberculosis biovar Equi, providing a generate energy pathway when it grows under oxygen deprived condition allowing the survival of C. pseudotuberculosis inside the host.
With the increase in genome sequencing projects, understanding of the biological capabilities of the organisms has also increased. The availability of the annotated genomes allows the information of metabolic pathways to be interpreted and used for computational reconstruction at genomic scale. Thus the reconstruction of metabolic pathways through comparative analysis can help identify potential targets and find the mechanisms that cause a disease.
In the present study, the genomes of C. pseudotuberculosis strains deposited in GenBank were used to identify the genes involved in the nitrate reduction pathway; a comparative genomic study was performed on 15 C. pseudotuberculosis strains of the biovars Ovis and Equi to identify the molecular bases of nitrate reduction in each C. pseudotuberculosis biovar Equi. Thus, we hope that understanding of Nar's role in C. pseudotuberculosis can help in identifying potent targets both for the development of more effective diagnostics and therapeutics and for the treatment and prevention of disease.
2. Material and Methods
2.1. Identification of Nitrate Reductase Genes, Metabolic Pathways, and Nitrate Reduction Enzymes
C. pseudotuberculosis genomes Table 1 were obtained from the NCBI GenBank. The sequence and arrangement analysis of nitrate reductase genes in narKGHJI operon was performed using the Artemis software (http://www.sanger.ac.uk/Software/Artemis) and manual curation based annotations are performed. Pathway Tools software developed by SRI International [45] and MetaCyc database [46] were used for computational prediction of metabolic pathways in C. pseudotuberculosis.
Table 1.
Strains used in phenotypic and genotypic analysis regarding the nitrate reductase activity.
| Strain | Biovar | Genome size (MB) | Nitrate testa | NCBI access | Ref |
|---|---|---|---|---|---|
| 1002 | Ovis | 2.33511 | Yes | NC_017300.1 | [26] |
| C231 | Ovis | 2.32821 | Yes | NC_017301.1 | [26] |
| FRC41 | Ovis | 2.33791 | Yes | NC_014329.1 | [27] |
| I19 | Ovis | 2.33773 | Yes | NC_017303.1 | [28] |
| PAT10 | Ovis | 2.33532 | Yes | NC_017305.1 | [29] |
| 42/02-A | Ovis | 2.33761 | Yes | NC_017306.1 | [30] |
| 3/99-5 | Ovis | 2.33794 | Yes | NC_016781.1 | [30] |
| 267 | Ovis | 2.33763 | Yes | NC_017462.1 | [31] |
| P54B96 | Ovis | 2.33794 | Yes | NC_017031.1 | [32] |
| CIP5297 | Equi | 2.32059 | Yes | NC_017307.1 | [33] |
| 1/06 -A | Equi | 2.27912 | Yes | NC_017308.1 | [34] |
| 316 | Equi | 2.31041 | Yes | NC_016932.1 | [35, 36] |
| 258 | Equi | 2.36982 | Yes | NC_017945.1 | [37] |
| 162 | Equi | 2.29346 | Yes | NC_018019.1 | [38] |
| 31 | Equi | 2.38969 | Yes | NC_017730.1 | [39] |
| 262 | Equi | 2.32575 | Yes | NZ_CP012022.1 | — |
| MB20 | Equi | 2.36309 | Yes | JPUV01 | [40] |
| E19 | Equi | 2.36796 | Unknown | NZ_CP012136.1 | — |
| CCUG27541 | Equi | 2.37942 | Unknown | JPJB01 | [41] |
aBiochemical test.
2.2. narKGHJI Operon Analysis
The families of proteins as well as the conserved protein domains present in the operon narKGHJI and adjacent genes were analyzed using InterProScan database (https://www.ebi.ac.uk/interpro). Prediction of transmembrane helices in protein was performed using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The similarity analysis of genes involved in the nitrate reduction pathway was performed using the UniProtKB database in category BLASTp. Clustal Omega was used to multiple sequence alignment between operon narKGHJI of C. pseudotuberculosis biovar Equi and homologous sequences of multiple organisms (http://www.ebi.ac.uk/Tools/msa/clustalo/), and Jalview was used to analyze similarity (http://www.jalview.org/) among these sequences.
3. Results and Discussion
3.1. Correlation between Nitrate Reduction Phenotype and Genotype
Nineteen C. pseudotuberculosis strains that had genome information in GenBank are presented Table 1. To perform the phenotyping analysis, 18 strains that were sequenced in our lab were used. All these strains were biochemically tested in order to verify their ability of nitrate reduction; see Table 1. Among the C. pseudotuberculosis strains, 31, 258, 262, and MB20 strains showed concordance between phenotypic and genotypic results. Strains 1/06-A, 316, 162, and CIP52.97 are found positive phenotype for reduction of nitrate; however they do not have genes involved in nitrate reduction in their genome.
3.2. Genes Involved in Respiratory Nitrate Reductase Metabolic Pathway
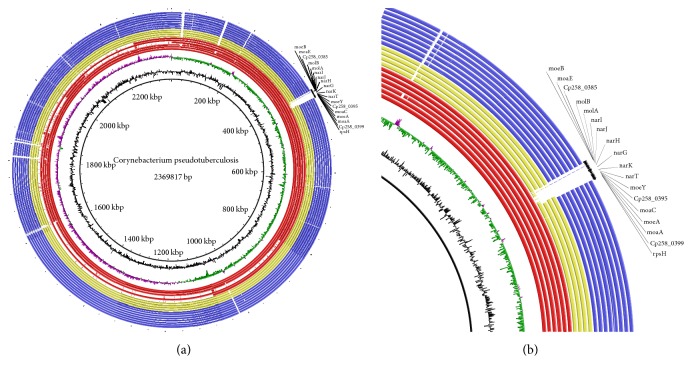
We first compared the autoassigned pathways by the Pathologic software with the pathways in databases such as KEGG (http://www.genome.jp/kegg/) and resolved possible discrepancies using manual curation. Comparative genome analysis revealed that C. pseudotuberculosis biovar Equi possess narKGHJI gene clusters that participate via the respiratory anaerobic process of the nitrate reduction similarly to E. coli (Figure S8) [47]. The C. pseudotuberculosis narKGHJI gene cluster showed great similarity with the protein sequences found in other Actinomycetes, such as C. diphtheriae, C. glutamicum, and M. tuberculosis (Table 2). All biovar Ovis strains do not present any gene of the narKGHJI operon in their genomes (Figure 1).
Table 2.
Similarity sequence analysis of narKGHJI gene clusters of C. pseudotuberculosis.
| C. pseudotuberculosis protein | Species | Length (aa) | ID (%) | Similarity (%) | Access number/UniProtKB |
|---|---|---|---|---|---|
| Nitrate reductase alpha subunit (NarG)—1240aa | Corynebacterium diphtheriae | 1,240 | 97 | 99 | Q6NJA9 |
| Corynebacterium glutamicum | 1,248 | 77 | 87 | Q8NR68 | |
| Mycobacterium tuberculosis | 1,232 | 56 | 70 | P9WJQ3 | |
| Escherichia coli | 1,247 | 46 | 64 | P09152 | |
|
| |||||
| Nitrate reductase beta subunit (NarH)—533aa | Corynebacterium diphtheriae | 533 | 95 | 98 | H2G670 |
| Corynebacterium glutamicum | 531 | 79 | 89 | S5Y1U5 | |
| Mycobacterium tuberculosis | 538 | 64 | 75 | A0A049E025 | |
| Escherichia coli | 512 | 54 | 72 | P11349 | |
|
| |||||
| Nitrate reductase gamma subunit (NarI)—259aa | Corynebacterium diphtheriae | 259 | 96 | 98 | H2HVN2 |
| Corynebacterium glutamicum | 259 | 75 | 89 | S5YHX1 | |
| Mycobacterium tuberculosis | 241 | 48 | 68 | Q7D8Q6 | |
| Escherichia coli | 225 | 28 | 51 | P11350 | |
|
| |||||
| Nitrate reductase chaperone (NarJ)—209aa | Corynebacterium diphtheriae | 209 | 89 | 96 | H2H309 |
| Corynebacterium glutamicum | 228 | 63 | 80 | Q6M5Z0 | |
| Mycobacterium tuberculosis | 206 | 37 | 49 | Q7D8Q7 | |
| Escherichia coli | 236 | 30 | 46 | P0AF26 | |
|
| |||||
| Nitrate/nitrite transporter (NarK)—443aa | C. diphtheriae | 443 | 93 | 97 | H2HGI1 |
| C. glutamicum | 445 | 66 | 78 | I0LIN6 | |
| M. tuberculosis | 395 | 22 | 35 | P9WJY7 | |
| E. coli | 463 | 33 | 51 | P10903 | |
Figure 1.
Graphical circular map showing BLAST between nineteen C. pseudotuberculosis strains. (a) From center to the outside: GC content in black and GC skew in pink and green; in red and yellow Equi strains and in blue Ovis strains. From the inner to outer circle on (a) and (b): the biovar Equi strains are the following: 258, 31, 262, MB20, E19, CCUG27541, CIP52.97, 1/06-A, 316, and 162, and the biovar Ovis strains are the following: 1002, C231, FRC41, I19, PAT10, 42/02-A, 3/99-5, 267, and P54B96. (b) Zoom to the nitrate locus, absent in the biovar Equi strains CIP52.97, 1/06 –A, 316, and 162 and all biovar Ovis strains.
The nitrate locus that contains narKGHJI operon in C. pseudotuberculosis is composed of gene cluster encoding the molybdopterins moeB, moaE, molB, molA, moeY, moaC, moeA, and moa (Figure 2). Molybdopterin is a cofactor that is indispensable for activity of nitrate reductase [48]. The organization and conservation of the narKGHJI gene cluster was assessed among C. pseudotuberculosis and C. diphtheriae, C. glutamicum, M. tuberculosis and, E. coli. The analysis showed that narKGHJI gene cluster is conserved among these species; however gene orientation of the five genes is only conserved among C. pseudotuberculosis, C. diphtheriae, and C. glutamicum. In addition, in C. pseudotuberculosis we found narT gene is located at the upstream of the narKGHJI cluster. The narT gene encodes a nitrate transporter and is not present in E. coli and C. glutamicum.
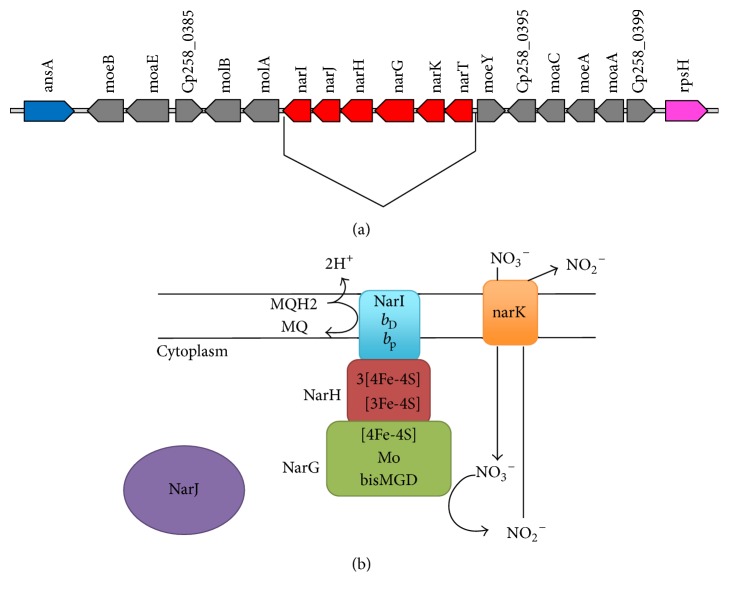
Figure 2.
Nitrate locus from C. pseudotuberculosis biovar Equi. (a) This locus contains the following: genes encoding the nitrate reductase narK, narG, narH, narJ, and narI, and the genes encoding the molybdopterins moeB, moaE, molB, molA, moeY, moaC, moeA, and moaA. Insertion between ansA and rpsH: genes encoding the nitrate reductase are lacking in nitrate-negative C. pseudotuberculosis biovar Ovis strains. Arrows represent open reading frames and their orientations. (b) Bacterial nitrate respiration. The NarG and NarH bind to the membrane via the interaction between a hydrophobic patch of NarH and NarI which is buried within the membrane. The NarJis a chaperone that is involved in folding, maturation, and molybdenum cofactor insertion of nitrate reductase.
3.3. Enzymes Involved in Electron Transport Chain
We found conserved functional domains of the genes present in the narGHI complex (involved in electron transport chain) in C. pseudotuberculosis biovar Equi (Table 3). To infer the functionality of narGHI complex, we used tridimensional structure from E. coli (PDB accession 1Q16) as a model for nar genes, as the structure of these enzymes is resolved in these bacteria.
Table 3.
Residues analyzed regarding the conservation between the narGHI protein sequences of E. coli and C. pseudotuberculosis.
| E. coli | C. pseudotuberculosis | Conservation (%) | Function | REF |
|---|---|---|---|---|
| Residues NarG ∗ | ||||
| His49(H) | His55 | 100 | Mo ion coordination | [42] |
| Cys53(C) | Cys 59 | 100 | Mo ion coordination | [42] |
| Cys57(C) | Cys 63 | 100 | Mo ion coordination | [42] |
| Cys92(C) | Cys 98 | 100 | Mo ion coordination | [42] |
| Asp222 (D) | Asp 228 | 100 | Mo-bisMGD ligand | [42] |
| Val 578 (V) | Val 584 | 100 | Left relatively hydrophobic environment to allow binding within the active site, with one face of the side chain carboxylate exposed of Asp222. Optimal interaction with the Mo of the cofactor | [43] |
| Tyr 220 (Y) | Tyr 226 | 100 | Left relatively hydrophobic environment to allow binding to within the active site, with one face of the side chain carboxylate exposed of Asp222. Allowing optimal interaction with the Mo of the cofactor | [43] |
| His1092(H) | His 1099 | 100 | Form a hydrogen bond network that links the solvent interface with AsnA52 and could be important for structural integrity and/or proton delivery to the active site | [42] |
| His1098(H) | His 1105 | 100 | Form a hydrogen bond network that links the solvent interface with AsnA52 and could be important for structural integrity and/or proton delivery to the active site | [42] |
| His1163(H) | His 1170 | 100 | Form a hydrogen bond network that links the solvent interface with AsnA52 and could be important for structural integrity and/or proton delivery to the active site | [42] |
| Arg 94(R) | Arg 100 | 100 | Forms a hydrogen bond with the Cys92 ligand of FS0 | [43] |
| Residues NarH ∗ | ||||
| Cys196 (C) | Cys 196 | 100 | Binding of iron atoms of FS4 cluster [3Fe-4S] | [44] |
| Cys217(C) | Cys 217 | 100 | Binding of iron atoms of FS4 cluster [3Fe-4S] | [44] |
| Cys223(C) | Cys 223 | 100 | Binding of iron atoms of FS4 cluster [3Fe-4S] | [44] |
| Cys184(C) | Cys 184 | 100 | Binding of iron atoms of FS3 cluster [4Fe-4S] | [44] |
| Cys187(C) | Cys 187 | 100 | Binding of iron atoms of FS3 cluster [4Fe-4S] | [44] |
| Cys192(C) | Cys 192 | 100 | Binding of iron atoms of FS3 cluster [4Fe-4S] | [44] |
| Cys 227(C) | Cys 227 | 100 | Binding of iron atoms of FS3 cluster [4Fe-4S] | [44] |
| Cys26(C) | Cys 26 | 100 | Binding of iron atoms of FS2 cluster [4Fe-4S] | [44] |
| Cys244(C) | Cys 244 | 100 | Binding of iron atoms of FS2 cluster [4Fe-4S] | [44] |
| Cys247(C) | Cys 247 | 100 | Binding of iron atoms of FS2 cluster [4Fe-4S] | [44] |
| Cys259(C) | Cys 259 | 100 | Binding of iron atoms of FS2 cluster [4Fe-4S] | [44] |
| Cys16(C) | Cys 16 | 100 | Binding of iron atoms of FS1 cluster [4Fe-4S] | [44] |
| Cys19(C) | Cys 19 | 100 | Binding of iron atoms of FS1 cluster [4Fe-4S] | [44] |
| Cys22(C) | Cys 22 | 100 | Binding of iron atoms of FS1 cluster [4Fe-4S] | [44] |
| Cys263 (C) | Cys 263 | 100 | Binding of iron atoms of FS1 cluster [4Fe-4S] | [44] |
| Residues NarI ∗ | ||||
| His66 (H) | His66 | 100 | Iron atoms coordination of heme bD | [43] |
| His187 (H) | His190 | 100 | Iron atoms coordination of heme bD | [43] |
| His56 (H) | His56 | 100 | Iron atoms coordination of heme bP | [43] |
| His205 (H) | His208 | 100 | Iron atoms coordination of heme bP | [43] |
| Arg112 | Arg113 | 100 | Hydrogen bond network electron transfer between the redox center bP, FS4, and NarG | [43] |
| Arg202 | Arg205 | 100 | Hydrogen bond network electron transfer between the redox center bP, FS4, and NarG. | [43] |
| Ser39 | Ser39 | 100 | Hydrogen bond network electron transfer between the redox center bP, FS4, and NarG. | [43] |
| Ser40 | Ser40 | 100 | Hydrogen bond network electron transfer between the redox center bP, FS4, and NarG. | [43] |
| Tyr213(Y) | Tyr216 | 100 | Involved in electrostatic interactions and hydrogen bond, are involved in the formation of NarGHI heterotrimer | [43] |
| Arg216(R) | Arg219 | 100 | Involved in electrostatic interactions and hydrogen bond, are involved in the formation of NarGHI heterotrimer | [43] |
| Arg222(R) | Arg225 | 100 | Involved in electrostatic interactions and hydrogen bond, are involved in the formation of NarGHI heterotrimer | [43] |
| Ser201(S) | Ser204 | 100 | A strong link at Ser201 of NarI allows distinctly shorter distances between the hemes in NarI than the distances observed between the hemes in the cytochrome bc1 complex | [43] |
∗Shown is the position of the residues at E. coli and C. pseudotuberculosis sequences and as well as the function of each residue in the E. colinarGHI structure.
In the narGHI complex in Figure 2, the narG gene is a member of a superfamily of enzymes that use a Mo-bisMGD cofactor (bisMGD) for their catalytic activity. Residues present in the active site of the molybdenum atom (Mo) are highly conserved in NarG subunits of Gram-negative and Gram-positive bacterial species [21]. The NarH belongs to the superfamily of electron transfer subunit (ferredoxins) of bacterial oxidoreductases. NarH has three clusters [4Fe-4S] (FS1, FS2, and FS3) and a [3Fe-4S] cluster (FS4) [21, 42, 44]. The general structure of NarH in E. coli is composed of a central region of a linker region that is inserted between the two subdomains in coordination of the iron-sulfur clusters and forms an extended connection between the NarG and NarH subunits and an extended C-terminus. In C. pseudotuberculosis it was observed that the nine cysteine residues responsible for coordinating three clusters [4Fe-4S] and one cluster [3Fe-4S] are conserved in all analyzed sequences. NarI is the transmembrane subunit that anchors NarGH to the cytoplasmic side of the membrane and provides the connection between the quinone and the site of oxidation, referred to as “Site Q.” This subunit coordinates two hemes, one of which is located towards the side of cytoplasmic end of NarI and is referred to as the proximal heme (heme bP), while the other is located towards the periplasmic side of the same and is referred to as the distal heme (heme bD). Such hemes act as mediators in electron transfer from Site Q to the clusters [Fe-S] in NarH. NarI of C. pseudotuberculosis presented five transmembrane helices S1 Figure, which is in agreement with the literature when compared to the E. coli crystal structure [49, 50]. This C-terminal segment presents highly conserved residues in the family NarI proteins and also is present in C. pseudotuberculosis. It is interesting to note that NarI is considerably preserved in relation to its primary sequence between orthologous proteins found in species of the Corynebacterium genus, particularly in relation to C. diphtheriae, which has 96% identity and 98% similarity. These results suggest that the enzymatic complex narGHI is probably very similar with respect to its three dimensional structure among the species of C. pseudotuberculosis biovar Equi and C. diphtheriae in view of the high identity found among NarG, NaH, and NarI subunits of these species. The C. pseudotuberculosis biovar Equi presents 48% and 43% identity in relation to the orthologous proteins from M. tuberculosis and B. subtilis, respectively.
3.4. NarJ (Molybdenum-Cofactor-Assembly Chaperone)
The NarJ is a chaperone that is involved in folding, maturation, and molybdenum cofactor insertion of nitrate reductase in E. coli [51, 52]. The similarity values found between C. pseudotuberculosis biovar Equi and other sequences of orthologous proteins by BLAST, including the genus Corynebacterium, were slightly lower than those found in other proteins (NarG, NarH, and NarI) that form the three subunits of the enzyme nitrate reductase (Table 2). However, narJ remained conserved between C. pseudotuberculosis biovar Equi strains, and results of biochemical tests (Table 1) indicate that its function has remained preserved, since NarJ is important for nitrate reductase expression in others organisms [53].
3.5. Nitrate and Nitrite Transporters
The nitrate and nitrite export is mediated by nitrate and nitrite transporters that are members of the major facilitator superfamily (MFS) [54]. In bacteria such carriers are the narK family represented by NarK and NarU proteins in E. coli that transport both nitrate and nitrite [47]. However, NarK is expressed at higher levels than NarU and it remains unclear till date if NarK is a nitrate/nitrite antiporter or symporter [55]. In C. pseudotuberculosis biovar Equi, we found that the two genes narK and narT are responsible for coding two probable nitrate and nitrite carriers and in C. pseudotuberculosis biovar Equi is present at the upstream of the narGHJI operon (Figure 2). Both proteins are found to be composed of 12 transmembrane helices similar to other members of the superfamily of bacterial transporters; see S2 and S3 Figures. In C. glutamicum the narK is in close proximity to narG, narH, narJ, and narI, which together form the narKGHJI operon [56], but in E. coli, narGHJI operon is transcribed individually and narT is upstream of the operon narKGHJI [57]. Since we did only the in silico analysis, we cannot state the precise transcription and regulation of these genes in vivo. We presume that this narKGHJI cluster in C. pseudotuberculosis biovar Equi may be regulated together as found in C. glutamicum. However, experimental confirmation is required.
3.6. Nitrite Reductase in C. pseudotuberculosis
Genes in C. pseudotuberculosis are found to encode two different types of nitrite reductase enzymes. One is the nitric oxide-forming enzyme that is encoded by nirK gene and shown to be a copper dependent enzyme. The second groups of enzymes are ammonium nitrite reductase that are encoded by nrfA gene that encodes catalytic subunit and nrfH that encodes for a membrane associated electron transfer subunit. In all C. pseudotuberculosis strains we found NrfA and NrfH subunits of nitrite reductase NrfAH complex that catalyze the reduction of nitrite to ammonium. Beside C. pseudotuberculosis, such genes are found only in C. ulcerans and are absent in all other species from this genus. In C. pseudotuberculosis, the two genes nrfA and nrfH are located in a “cluster” of genes, S4 Figure, responsible for coding biogenesis system I proteins cytochrome c that appears to be a common feature among bacterial genomes. C. pseudotuberculosis also has a nitrite reductase enzyme likely dependent on copper (CuNIR). The gene for CuNIR is nirK that encodes a protein composed of 882 residues and is conserved in all strains of C. pseudotuberculosis; see Figure S5 of the Supplementary Material available online at https://doi.org/10.1155/2017/9481756. The sequence similarity analysis of nirK was performed using the BLAST and it was observed that such protein has an N-terminal region of about 500 amino acids, S6 Figure, compared to the most homologous proteins that have been experimentally well characterized [58–61]. Within the genus Corynebacterium, the CuNIR proteins are found to have similar sizes to that of C. pseudotuberculosis, such as Corynebacterium durum (WP_006063127.1) and Corynebacterium vitaeruminis (WP_051483598.1), with 40% identity between the sequences; see Table S1. Furthermore, it was found that the N-terminal region of about 500 residues is probably composed of 13 transmembrane helices in C. pseudotuberculosis, S7 Figure, which indicates that it can be a membrane associated protein. To verify the residues conservation, we used G. kaustophilus CuNIR proteins as a reference sequence because its crystal structure has recently been determined (pdb∣3WIA∣A). All residues responsible for binding to the copper atoms and the catalytic site were analyzed. It is observed that the C-terminal region is 100% conserved in C. pseudotuberculosis; see S6 Figure.
3.7. Reconstruction of C. pseudotuberculosis Biovar Equi Metabolic Network
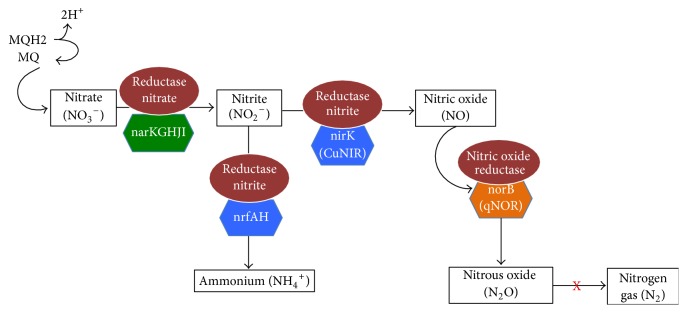
A metabolic network reconstruction is assembled piece-by-piece by compiling data on known enzymes, and annotated genome together with data from the literature is used to assemble a network reconstruction [62]. In C. pseudotuberculosis after the similarity analysis we observed that genomic sequences participating in the nitrate reduction pathway are conserved, and the residues were analyzed, indicating a possible conserved function. Thus we can infer that C. pseudotuberculosis biovar Equi is different from that occurring in C. glutamicum, where nitrate is excreted out of the cells as the major end product under the conditions of anaerobic respiration, and nitrate and nitrite are not used as sources of nitrogen for aerobic or anaerobic growth [18] so C. pseudotuberculosis biovar Equi has a likely route of denitrification composed of the nitrate reductase Nar, nitrite reductase dependent copper, and nitric oxide reductase quinol-dependent, and this is a way of incomplete denitrification, in which nitrous oxide is the final product, and, therefore, there is no presence of nitrous oxide reductase enzyme and subsequent reduction to nitrogen gas; see Figure 3. Furthermore C. pseudotuberculosis biovar Equi also shows the pathway of ammonification in dissimilatory nitrate reduction to ammonium, what has been proposed as catalyzed by the enzyme nitrate reductase Nar and nitrite reductase complex NrfAH, a membrane-associated cytochrome; see Figure 3. E. coli not only reduce nitrite to ammonia but also catalyze the reduction of nitrite to NO, which can be used as an electron acceptor for anaerobic respiration [63]. The presence of enzymes and pathways of nitrate reduction proposals was also compared between biovar Ovis and biovar Equi strains and with respect to enzymes, the main difference was the presence of the nitrate locus that contains the genes encoding the molybdopterin and the genes encoding the nitrate reductase and a region situated upstream of the ansA gene and this locus has a 19,606 pb length.
Figure 3.
Model building of nitrate reductase from C. pseudotuberculosis biovar Equi. Showed here is respiratory nitrate reduction to nitrite; incomplete denitrification of nitrite in which nitrous oxide is the final product; and Nrf-dependent ammonification.
4. Conclusions
In this study, we found that C. pseudotuberculosis strains 1/06-A, 316, 162, and CIP52.97 are able to do nitrate reduction but in their genomes they do not have genes associated with nitrate reduction, warning us about resequencing of these genomes. The C. pseudotuberculosis nitrate locus is composed of narKGHJI gene cluster and by genes encoding the molybdopterins moeB, moaE, molB, molA, moeY, moaC, moeA, and moa genes are similar to the other bacteria under Actinomycetes and all the biovar Ovis strains that lack the nitrate locus in their genomes. The narGHI complex in C. pseudotuberculosis biovar Equi shows conserved functional domains. narK and narT genes in C. pseudotuberculosis biovar Equi encode two putative nitrate and nitrite carriers, respectively. Finally, the nirK, nrfA, and nrfH encoding nitrite reductase enzymes are conserved in C. pseudotuberculosis. Therefore, the study of pathogen metabolism can contribute to the identification of pathogen virulence, discovery of drug targets, host response, and generating hypotheses for experimental investigation.
Supplementary Material
Supplementary Figures 1 to 3 and Figure 7: Transmembrane helices prediction to NarI, NarK, NarT and NirK proteins, respectively, from C. pseudotuberculosis biovar Equi. Supplementary Figure 4: Transcription unit for the nrfHA genes into C. pseudotuberculosis biovar Equi. Supplementary Figure 5: Multiple sequence alignment of C. pseudotuberculosis biovar Equi nirK gene. Supplementary Figure 6: Amino acid sequence alignment of bacterial NirKs. Supplementary Figure 8: Comparative nitrogen metabolism between E.coli and C. pseudotuberculosis biovar Equi. In red pathway to E. coli, in green to C. pseudotuberculosis. Supplementary Table 1: Similarity sequence analysis of nirK gene of C. pseudotuberculosis and bacterial CuNiRs
Acknowledgments
The authors would like to acknowledge the help of all the team members and the financing agencies. CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil), and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil). Andrey P. Lage was also supported by the Programa Pesquisador Mineiro (PPM) (00923-15) from FAPEMIG.
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Ayers J. L. Caseous lymphadenitis in goats and sheep: a review of diagnosis, pathogenesis, and immunity. Journal of the American Veterinary Medical Association. 1977;171(12):1251–1254. [PubMed] [Google Scholar]
- 2.Brown C. C., Olander H. J., Zometa C., Alves S. F. Serodiagnosis of inapparent caseous lymphadenitis in goats and sheep, using the synergistic hemolysis-inhibition test. American Journal of Veterinary Research. 1986;47(7):1461–1463. [PubMed] [Google Scholar]
- 3.Doherr M. G., Carpenter T. E., Hanson K. M. P., Wilson W. D., Gardner I. A. Risk factors associated with Corynebacterium pseudotuberculosis infection in California horses. Preventive Veterinary Medicine. 1998;35(4):229–239. doi: 10.1016/S0167-5877(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 4.Piontkowski M. D., Shivvers D. W. Evaluation of a commercially available vaccine against Corynebacterium pseudotuberculosis for use in sheep. Journal of the American Veterinary Medical Association. 1998;212(11):1765–1768. [PubMed] [Google Scholar]
- 5.Baird G., Synge B., Dercksen D. Erratum: survey of caseous lymphadenitis seroprevalence in British terminal sire sheep breeds (The Veterinary Record (505-506)) Veterinary Record. 2004;154(17):p. 530. doi: 10.1136/vr.154.17.530. [DOI] [PubMed] [Google Scholar]
- 6.Baird G. J., Fontaine M. C. Corynebacterium pseudotuberculosis and its Role in Ovine Caseous Lymphadenitis. Journal of Comparative Pathology. 2007;137(4):179–210. doi: 10.1016/j.jcpa.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Peel M. M., Palmer G. G., Stacpoole A. M., Kerr T. G. Human lymphadenitis due to Corynebacterium pseudotuberculosis: report of ten cases from Australia and review. Clinical Infectious Diseases. 1997;24(2):185–191. doi: 10.1093/clinids/24.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Selim S. A. Oedematous skin disease of buffalo in Egypt. Journal of Veterinary Medicine, Series B. 2001;48(4):241–258. doi: 10.1046/j.1439-0450.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 9.Songer J. G., Beckenbach K., Marshall M. M., Olson G. B., Kelley L. Biochemical and genetic characterization of Corynebacterium pseudotuberculosis. American Journal of Veterinary Research. 1988;49(2):223–226. [PubMed] [Google Scholar]
- 10.Shpigel N. Y., Elad D., Yeruham I., Winkler M., Saran A. An outbreak of Corynebacterium pseudotuberculosis infection in an Israeli dairy herd. Veterinary Record. 1993;133(4):89–94. doi: 10.1136/vr.133.4.89. [DOI] [PubMed] [Google Scholar]
- 11.Pratt S. M., Spier S. J., Carroll S. P., Vaughan B., Whitcomb M. B., Wilson W. D. Evaluation of clinical characteristics, diagnostic test results, and outcome in horses with internal infection caused by Corynebacterium pseudotuberculosis: 30 Cases (1995–2003) Journal of the American Veterinary Medical Association. 2005;227(3):441–448. doi: 10.2460/javma.2005.227.441. [DOI] [PubMed] [Google Scholar]
- 12.Zumft W. G. Cell biology and molecular basis of denitrification? Microbiology and Molecular Biology Reviews. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno-Vivián C., Cabello P., Martínez-Luque M., Blasco R., Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. Journal of Bacteriology. 1999;181(21):6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaspyrou S., Smith C. J., Dong L. F., Whitby C., Dumbrell A. J., Nedwell D. B. Nitrate reduction functional genes and nitrate reduction potentials persist in deeper estuarine sediments. Why? PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0094111.e94111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malm S., Tiffert Y., Micklinghoff J., et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009;155(4):1332–1339. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura T., Teramoto H., Inui M., Yukawa H. Gene expression profiling of Corynebacterium glutamicum during anaerobic nitrate respiration: induction of the SOS response for cell survival. Journal of Bacteriology. 2011;193(6):1327–1333. doi: 10.1128/jb.01453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan M. P., Sequeira P., Lin W. W., et al. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013356.e13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura T., Vertès A. A., Shinoda Y., Inui M., Yukawa H. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Applied Microbiology and Biotechnology. 2007;75(4):889–897. doi: 10.1007/s00253-007-0879-y. [DOI] [PubMed] [Google Scholar]
- 19.Blümle S., Zumft W. G. Respiratory nitrate reductase from denitrifying Pseudomonas stutzeri, purification, properties and target of proteolysis. Biochimica et Biophysica Acta (BBA)—Bioenergetics. 1991;1057(1):102–108. doi: 10.1016/s0005-2728(05)80089-1. [DOI] [Google Scholar]
- 20.Härtig E., Schiek U., Vollack K.-U., Zumft W. G. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. Journal of Bacteriology. 1999;181(12):3658–3665. doi: 10.1128/jb.181.12.3658-3665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertero M. G., Rothery R. A., Palak M., et al. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nature Structural Biology. 2003;10(9):681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 22.Avazéri C., Turner R. J., Pommier J., Weiner J. H., Giordano G., Verméglio A. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology. 1997;143(4):1181–1189. doi: 10.1099/00221287-143-4-1181. [DOI] [PubMed] [Google Scholar]
- 23.Stewart V., Lu Y., Darwin A. J. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. Journal of Bacteriology. 2002;184(5):1314–1323. doi: 10.1128/jb.184.5.1314-1323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köhler S., Foulongne V., Ouahrani-Bettache S., et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohaskey C. D. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. Journal of Bacteriology. 2008;190(8):2981–2986. doi: 10.1128/jb.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz J. C., D'Afonseca V., Silva A., et al. Evidence for reductive genome evolution and lateral acquisition of virulence functions in two Corynebacterium pseudotuberculosis strains. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0018551.e18551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trost E., Ott L., Schneider J., et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics. 2010;11(1, article no. 728) doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva A., Schneider M. P. C., Cerdeira L., et al. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with bovine mastitis. Journal of Bacteriology. 2011;193(1):323–324. doi: 10.1128/jb.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerdeira L. T., Pinto A. C., Schneider M. P. C., et al. Whole-Genome Sequence of Corynebacterium pseudotuberculosis PAT10 Strain Isolated from Sheep in Patagonia, Argentina. Journal of Bacteriology. 2011;193(22):6420–6421. doi: 10.1128/jb.06044-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pethick F. E., Lainson A. F., Yaga R., et al. Complete genome sequences of Corynebacterium pseudotuberculosis strains 3/99-5 and 42/02-A, isolated from Sheep in scotland and Australia, respectively. Journal of Bacteriology. 2012;194(17):4736–4737. doi: 10.1128/jb.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes T., Silva A., Thiago R., et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain cp267, isolated from a llama. Journal of Bacteriology. 2012;194(13):3567–3568. doi: 10.1128/jb.00461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan S. S., Guimarães L. C., Pereira U. D. P., et al. Complete genome sequence of Corynebacterium pseudotuberculosis biovar ovis strain P54B96 isolated from antelope in South Africa obtained by rapid next generation sequencing technology. Standards in Genomic Sciences. 2012;7(2):189–199. doi: 10.4056/sigs.3066455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerdeira L. T., Schneider M. P. C., Pinto A. C., et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain CIP 52.97, isolated from a horse in Kenya. Journal of Bacteriology. 2011;193(24):7025–7026. doi: 10.1128/jb.06293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pethick F., Lainson A., Yaga R., et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain 1/06-A, isolated from a Horse in North America. Journal of Bacteriology. 2012;194(16):4476–4476. doi: 10.1128/jb.00922-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos R. T. J., Silva A., Carneiro A. R., et al. Genome sequence of the Corynebacterium pseudotuberculosis Cp316 strain, isolated from the abscess of a Californian horse. Journal of Bacteriology. 2012;194(23):6620–6621. doi: 10.1128/jb.01616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos R. T. J., Carneiro A. R., Soares S. D. C., et al. Tips and tricks for the assembly of a Corynebacterium pseudotuberculosis genome using a semiconductor sequencer. Microbial Biotechnology. 2013;6(2):150–156. doi: 10.1111/1751-7915.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soares S. C., Trost E., Ramos R. T. J., et al. Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. Journal of Biotechnology. 2013;167(2):135–141. doi: 10.1016/j.jbiotec.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Hassan S. S., Schneider M. P., Ramos R. T., et al. Whole-genome sequence of Corynebacterium pseudotuberculosis strain Cp162, isolated from camel. Journal of Bacteriology. 2012;194(20):5718–5719. doi: 10.1128/jb.01373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva A., Ramos R. T. J., Carneiro A. R., et al. Complete genome sequence of Corynebacterium pseudotuberculosis Cp31, isolated from an Egyptian buffalo. Journal of Bacteriology. 2012;194(23):6663–6664. doi: 10.1128/jb.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barauna R. A., Guimaraes L. C., Veras A. A., et al. Genome sequence of Corynebacterium pseudotuberculosis MB20 bv. equi isolated from a pectoral abscess of an oldenburg horse in California. Genome Announcements. 2014;2(6) doi: 10.1128/genomea.00977-14.e00977-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Havelsrud O. E., Sorum H., Gaustad P. Genome sequences of Corynebacterium pseudotuberculosis strains 48252 (human, pneumonia), CS_10 (lab strain), Ft_2193/67 (goat, pus), and CCUG 27541. Genome Announcements. 2014;2(5) doi: 10.1128/genomea.00869-14.e00869-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jormakka M., Richardson D., Byrne B., Iwata S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure. 2004;12(1):95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Bertero M. G., Rothery R. A., Palak M., et al. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nature Structural Biology. 2003;10(9):681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 44.Fedor J. G., Rothery R. A., Weiner J. H. A new paradigm for electron transfer through Escherichia coli nitrate reductase A. Biochemistry. 2014;53(28):4549–4556. doi: 10.1021/bi500394m. [DOI] [PubMed] [Google Scholar]
- 45.Karp P. D., Paley S. M., Krummenacker M., et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Briefings in Bioinformatics. 2009;11(1):40–79. doi: 10.1093/bib/bbp043.bbp043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caspi R., Foerster H., Fulcher C. A., et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research. 2008;36(1):D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clegg S., Yu F., Griffiths L., Cole J. A. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Molecular Microbiology. 2002;44(1):143–155. doi: 10.1046/j.1365-2958.2002.02858.x. [DOI] [PubMed] [Google Scholar]
- 48.Loux V., Mariadassou M., Almeida S., et al. Mutations and genomic islands can explain the strain dependency of sugar utilization in 21 strains of Propionibacterium freudenreichii. BMC Genomics. 2015;16(1, article 296) doi: 10.1186/s12864-015-1467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magalon A., Rothery R. A., Lemesle-Meunier D., Frixon C., Weiner J. H., Blasco F. Inhibitor binding within the NarI subunit (cytochrome b(nr)) of Escherichia coli nitrate reductase A. Journal of Biological Chemistry. 1998;273(18):10851–10856. doi: 10.1074/jbc.273.18.10851. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Espinosa R. M., Dridge E. J., Bonete M. J., et al. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiology Letters. 2007;276(2):129–139. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 51.Vergnes A., Pommier J., Toci R., Blasco F., Giordano G., Magalon A. NarJ chaperone binds on two distinct sites of the aponitrate reductase of Escherichia coli to coordinate molybdenum cofactor insertion and assembly. Journal of Biological Chemistry. 2006;281(4):2170–2176. doi: 10.1074/jbc.m505902200. [DOI] [PubMed] [Google Scholar]
- 52.Zakian S., Lafitte D., Vergnes A., et al. Basis of recognition between the NarJ chaperone and the N-terminus of the NarG subunit from Escherichia coli nitrate reductase. FEBS Journal. 2010;277(8):1886–1895. doi: 10.1111/j.1742-4658.2010.07611.x. [DOI] [PubMed] [Google Scholar]
- 53.Palmer T., Santini C.-L., Iobbi-Nivol C., Eaves D. J., Boxer D. H., Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Molecular Microbiology. 1996;20(4):875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 54.Reddy V. S., Shlykov M. A., Castillo R., Sun E. I., Saier M. H., Jr. The major facilitator superfamily (MFS) revisited. FEBS Journal. 2012;279(11):2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng H., Wisedchaisri G., Gonen T. Corrigendum: crystal structure of a nitrate/nitrite exchanger. Nature. 2014;507(7491):647–651. doi: 10.1038/nature13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimura T., Teramoto H., Inui M., Yukawa H. Corynebacterium glutamicum ArnR controls expression of nitrate reductase operon narKGHJI and nitric oxide (NO)-detoxifying enzyme gene hmp in an NO-responsive manner. Journal of Bacteriology. 2014;196(1):60–69. doi: 10.1128/jb.01004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonnefoy V., Demoss J. A. Nitrate reductases in Escherichia coli. Antonie van Leeuwenhoek. 1994;66(1–3):47–56. doi: 10.1007/bf00871632. [DOI] [PubMed] [Google Scholar]
- 58.Barth K. R., Isabella V. M., Clark V. L. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology. 2009;155(12):4093–4103. doi: 10.1099/mic.0.032961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S.-W., Fushinobu S., Zhou S., Wakagi T., Shoun H. Eukaryotic nirK genes encoding copper-containing nitrite reductase: originating from the protomitochondrion? Applied and Environmental Microbiology. 2009;75(9):2652–2658. doi: 10.1128/aem.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moir J. W. B., editor. Nitrogen Cycling in Bacteria: Molecular Analysis. Norfolk, UK: Caister Academic Press; 2011. [Google Scholar]
- 61.Theerachat M., Virunanon C., Chulalaksananukul S., Sinbuathong N., Chulalaksananukul W. NirK and nirS Nitrite reductase genes from non-agricultural forest soil bacteria in Thailand. World Journal of Microbiology and Biotechnology. 2011;27(4):999–1003. doi: 10.1007/s11274-010-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chavali A. K., D'Auria K. M., Hewlett E. L., Pearson R. D., Papin J. A. A metabolic network approach for the identification and prioritization of antimicrobial drug targets. Trends in Microbiology. 2012;20(3):113–123. doi: 10.1016/j.tim.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon J., Klotz M. G. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochimica et Biophysica Acta—Bioenergetics. 2013;1827(2):114–135. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 to 3 and Figure 7: Transmembrane helices prediction to NarI, NarK, NarT and NirK proteins, respectively, from C. pseudotuberculosis biovar Equi. Supplementary Figure 4: Transcription unit for the nrfHA genes into C. pseudotuberculosis biovar Equi. Supplementary Figure 5: Multiple sequence alignment of C. pseudotuberculosis biovar Equi nirK gene. Supplementary Figure 6: Amino acid sequence alignment of bacterial NirKs. Supplementary Figure 8: Comparative nitrogen metabolism between E.coli and C. pseudotuberculosis biovar Equi. In red pathway to E. coli, in green to C. pseudotuberculosis. Supplementary Table 1: Similarity sequence analysis of nirK gene of C. pseudotuberculosis and bacterial CuNiRs