Abstract
Gastric cancer is the sixth leading cause of cancer‐related death in Taiwan, and the identification of related factors is essential to increase patient survival. ADP‐ribosylation factor 1 (ARF1) was initially identified using 2‐D electrophoresis combined with MALDI–time‐of‐flight mass spectrometry. ADP‐ribosylation factor 1 belongs to the Ras superfamily or GTP‐binding protein family and has been shown to enhance cell proliferation. In the current study, we evaluated the potential of ARF1 as a biomarker for gastric cancer detection. ADP‐ribosylation factor 1 mRNA was upregulated in tumor tissues (compared with adjacent non‐tumor tissues, n = 55) in approximately 67.2% of gastric cancer patients. Expression of ARF1 protein was additionally observed using Western blot and immunohistochemistry (IHC) analyses. The clinicopathological correlations of ARF1 were further evaluated. Elevated ARF1 expression was strongly correlated with lymph node metastasis (P = 0.008), serosal invasion (P = 0.046), lymphatic invasion (P = 0.035), and pathological staging (P = 0.010). Moreover, the 5‐year survival rate for the lower ARF1 expression group (n = 50; IHC score < 90) was higher than that of the higher expression group (n = 60; IHC score ≥ 90) (P = 0.0228, log–rank test). To establish the specific function of ARF1 in human gastric cancer, isogenic ARF1‐overexpressing cell lines were prepared. Our results showed that ARF1‐overexpressing clones display enhanced cell proliferation, migration, and invasion. Furthermore, ARF1‐overexpression might contribute to poor prognosis of patients. These findings collectively support the utility of ARF1 as a novel prognostic marker for gastric cancer and its role in cell invasion. Cancer Sci 2012; 103: 1136–1144)
Gastric cancer is the second most common cancer worldwide, and the sixth leading cause of cancer‐related death in Taiwan. Surgery remains the only effective cure for this disease. In a recent study, more than 30% of surgical patients presented with the disease to an extent that was too advanced to receive curative resection.1 To improve poor survival outcomes and permit earlier diagnosis, new prognostic indicators or tumor markers are essential.2
Gastric cancer is divided into two histomorphologic types, intestinal‐differentiated and diffuse‐undifferentiated.3, 4 Regardless of the similar histomorphologic lesions, the tumor cells may still differ in their aggressiveness or response to chemotherapy.5 The molecular events involved in the development and progression of gastric cancer are complex, involving multiple genes and steps that operate sequentially or in concert.4 Several risk factors, including genetic alterations, chromosomal instability, and Helicobacter pylori infections, have been determined for gastric cancer.6, 7, 8 Moreover, the identification of numerous biomarkers has contributed to our knowledge of the molecular and cellular mechanisms of gastric carcinogenesis and progression.8 The majority of biomarkers are effective prognostic factors used to identify groups of patients at risk of relapse or metastasis.9 However, the useful biomarkers to elucidate the molecular mechanism of gastric cancer or to monitor the disease progression are still needed.
Protein expression profiling is another relatively recent approach for cancer marker detection and facilitates elucidation of the mechanisms underlying gastric cancer.10, 11, 12 To achieve these goals, relevant subsets of differentially expressed proteins must be identified, cloned, and investigated in detail. Proteomics is a highly effective and sensitive procedure that allows the identification of novel diagnostic, prognostic, or therapeutic biomarkers.
In this study, we have attempted to identify novel putative diagnostic or prognostic markers using 2‐D gel electrophoresis followed by MALDI–time‐of‐flight mass spectrometry (MALDI‐TOF/TOF MS) analysis. The protein ADP ribosylation factor 1 (ARF1) displaying high expression in tumor specimens was selected for further study. Expression of ARF1 mRNA was significantly upregulated in 67.2% of gastric cancer patients. Accordingly, our study focused largely on ARF1 expression, with a view to establishing its role in gastric cancer.
ADP‐ribosylation reactions play important roles in a wide range of physiological and pathophysiological processes, including cell differentiation, proliferation, necrosis, apoptosis, inter‐ and intracellular signaling.13 However, its role in carcinogenesis is still unknown. ADP‐ribosylation factor represents a branch of the small GTPase family that regulates vesicular traffic and organelle structure, consisting of six isoforms. Among these, ARF1 and ARF6 have been the most widely characterized. ADP‐ribosylation factor 1 is associated with the Golgi apparatus to regulate vesicle trafficking, whereas ARF6 is located in the plasma membrane and is involved in receptor endocytosis and actin remodeling.14 Moreover, ARF6 is overexpressed in highly invasive breast cancer cells, and plays an essential role during invasion15 by ERK signaling, which leads to Rac1 activation in melanoma cells16 or glioma.17
Materials and Methods
Subjects
In total, 110 patients (69 males, 41 females; median age, 66 years; range, 28–86 years) diagnosed with gastric cancer at Chang‐Gung Memorial Hospital (Chiayi, Taiwan) from 2000 to 2006 were enrolled in this study. All patients provided informed consent. Individual patients were subjected to gastric resection (32 had total gastrectomy and 78 had partial gastrectomy). No preoperative chemotherapy was used in our patients. Postoperatively, the patients with stages II/III disease received adjuvant chemotherapy after curative resection, whereas those with stage IV received therapeutic chemotherapy. The study protocol was approved by the Medical Ethics and Human Clinical Trial Committee at Chang‐Gung Memorial Hospital.
Clinicopathology
Resected specimens were examined pathologically using the criteria from the Japanese General Rules for Gastric Cancer Study18 and the International Union Against Cancer pTNM classification system.19 Data included patient age, gender, tumor location, size, gross (Borrmann) type, wall invasion, resection margin, histological type, lymph node metastasis, vascular invasion, lymphatic invasion, and perineural invasion. After discharge, all patients were scheduled for periodic follow‐up visits at the outpatient department at Chang‐Gung Memorial Hospital until death or the beginning of preparation of this article.
Tumor samples
Fresh samples of tumor tissue and adjacent non‐cancerous mucosa were harvested immediately after gastric resection. Samples dissected from resected specimens were immediately snap‐frozen in individual vials using liquid nitrogen. Frozen specimens were stored at −70°C in a tumor bank until use.
Two‐dimensional gel electrophoresis analysis
Tumor and non‐cancerous tissue protein samples (150 μg each) were separated by 13 cm Immobiline DryStrip 3–10 linear on the IPGphor Isoelectric Focusing System (Amersham Bioscience, Uppsala, Sweden) in the first dimension. Following equilibration, the IPG gel strips were transferred onto vertical gels (10% SDS‐PAGE, Hoefer SE600; Amersham Bioscience) for the second dimension as described.20
Mass spectrometric analysis of differentially expressed proteins
The silver‐stained spots were excised and in‐gel digested with trypsin according to procedures described previously.21 Following trypsin digestion, the tryptic peptides were acidified with 0.5% trichloroacetic acid (TCA) and loaded onto an MTP AnchorChip 600/384 TF (Bruker‐Daltonik, Bremen, Germany). Analysis by MALDI‐TOF MS was carried out using an Ultraflex MALDI‐TOF mass spectrometer (Bruker‐Daltonik). Monoisotopic peptide masses were identified and applied for database searches using the MASCOT search engine (Matrix Science, London, UK).
Real‐time quantitative RT‐PCR (qRT‐PCR)
Total RNA was extracted from cells using Trizol Reagent (Life Technologies, Rockville, MD, USA), and qRT‐PCR carried out as described earlier.22
Immunoblot analysis
Total cell lysates from tumors and adjacent non‐cancerous mucosa were prepared and the protein concentrations determined with the method described by Bradford.23 Equal amounts of protein per lane were fractioned with SDS‐PAGE on a 10% or 15% gel. Separated proteins were transferred to a nitrocellulose membrane. The membrane was blocked for 2 h at room temperature in 5% (w/v) non‐fat dried milk in Tris‐buffered saline (TBS). Subsequently, the membrane was washed three times with TBS, and incubated for 18 h with rabbit mAbs to ARF1 (1:2000 dilution in TBS; Epitomics, Burlingame, CA, USA). After further washing, the membrane was incubated for 1 h with HRP‐conjugated, affinity‐purified antibodies to mouse (1:6000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Immune complexes were visualized by chemiluminescence with an ECL detection kit (Amersham Bioscience). The following antibodies were used: N‐cadherin (1:1000 dilution; Invitrogen, Carlsbad, CA, USA); vimentin (1:6000 dilution; Santa Cruz Biotechnology); snail (1:3000 dilution; Cell Signaling Technology, Beverly, MA, USA); slug (1:3000 dilution; Cell Signaling Technology); twist (1:3000 dilution; Abcam, Cambridge, MA, USA), active β‐catenin (1:5000 dilution; Millipore, Billerica, MA, USA), GAPDH (1:8000 dilution; Chemicon, Lansing, NC, USA); lamin A/C (1:8000 dilution; Santa Cruz Biotechnology); and β‐actin (1:8000 dilution; Chemicon).
Immunohistochemistry (IHC)
Formalin‐fixed and paraffin‐embedded tissues were examined with IHC using a mAb to human ARF1 (dilution 1:150; Epitomics) following the avidin–biotin complex method, as described previously.24 Staining intensities of carcinoma cells and benign superficial epithelium on the same slide were compared. The negative group consisted of cancer cells with no detectable (−) or trace ARF1 immunoreactivity (+1); the positive group consisted of cancer cells with moderate (+2) or high levels (+3) of ARF1 immunoreactivity. Numerical scoring (Q) was confirmed by a second independent examiner who was blinded to the initial score. Results were scored by multiplying the percentage of positive cells (P) by intensity (I), that is, Q = P × I. For example, for a tissue section in which 10%, 60%, and 30% of cells showed intensities of +1, +2, and +3, respectively, Q is calculated as 10 × 1 + 60 × 2 + 30 × 3 = 220.
Establishing stable ARF1‐overexpressing AGS cell lines
ARF1 cDNA was amplified by RT‐PCR and cloned into the pcDNA3 vector. Transfection of the pcDNA3‐ARF1 gene was carried out using Lipofectamine reagent (Invitrogen). After 24 h of incubation, cells were transferred to G418 medium for selection. Overexpression of the ARF1 gene was confirmed by detection from culture medium using Western blot analysis. The pcDNA3 vector served as the control.
In vitro assay of invasive activity
The effect of ARF1 overexpression on the invasive activity of the AGS cell line was assessed using a rapid in vitro assay (Transwell technique), as described previously.25 Briefly, cell density was adjusted to 7 × 104/mL, and 250 μL of this suspension added to each well coated with Matrigel (Becton Dickinson, Franklin Lakes, NJ, USA) in duplicate. The upper chamber contained serum‐free RPMI 1640, whereas the lower chamber contained RPMI supplemented with 20% FBS. After incubation for 20 h at 37°C, the number of viable cells that had traversed the filter to the lower chamber was determined. Colony formation ability or proliferation assays using cell counting were carried out as described previously.25
Statistical analysis
Where appropriate, the Mann–Whitney U‐test or Fisher's exact test was used for comparisons between two independent groups, and the Kruskal–Wallis test or Pearson's chi‐square‐test was used for comparing more than two independent groups. The correlation between the results of two different examinations was analyzed with the Spearman's correlation test. Patients were monitored until the time of manuscript preparation or death. Cancer‐specific survival outcomes were expressed by applying the Kaplan–Meier method for all patients, except those who died from surgical complications. The log–rank test was used to compare the prognostic significance of individual variables on survival. Cox's hazards model was used in a multivariate analysis to identify the independent predictors of survival. A P‐value of <0.05 was considered statistically significant.
Results
Proteomic results for gastric patients
Earlier 2‐D gel electrophoresis and MALDI TOF/TOF MS analysis of gastric cancer tissues, compared with normal stomach tissues, revealed more than 200 upregulated proteins (Fig. 1A).20 ADP‐ribosylation factor 1 was selected for study due to its high expression in tumor cells and unknown function in gastric carcinogenesis. The tryptic peptide mass fingerprint and matched peptide are underlined in Figure 1(B). Two‐dimensional gel analysis was carried out on at least three specimens. Figure 1(C) shows cropped images from three paired 2‐D gels indicating differential expression of ARF1 proteins. Overexpression of ARF1 in tumor versus normal tissue in the three gels was 3.1, 1.8, and 4.7‐fold.
Figure 1.
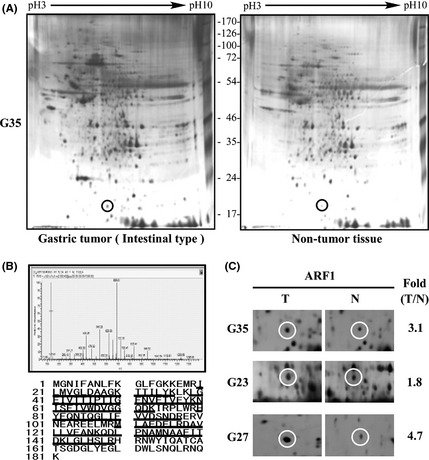
Representative images of 2‐D gel electrophoresis. (A) Total protein (150 μg) extracted from intestinal‐type carcinoma (T) and adjacent non‐tumor (N) tissues were separated on linear strips with a pH range of 3.0–10.0, followed by protein separation using 10% SDS‐PAGE. The ADP‐ribosylation factor 1 (ARF1) protein is marked with circles. Molecular weight standards are indicated. (B) Analysis of ARF1 protein with MALDI–time‐of‐flight mass spectrometry. Matched peptides from mass spectrometric analysis of the amino acid sequence of human ARF1 are underlined. (C) Expression of ARF1 was cropped from three pairs of gastric carcinoma tissues and non‐cancerous counterparts (the patient number is indicated on the left). Elevated density of ARF1 protein (circles) is shown on the right.
Clinical characteristics of patients
Table 1 lists the characteristics of study patients (n = 110). The average tumor size (maximum diameter) was 5.2 cm (median, 4.0 cm; range, 0.5–18 cm). Tumors were located in the proximal third of the stomach in 24 cases (21.8%), middle third in 28 (25.5%), distal third in 55 (50%), and the whole stomach in 3 (2.7%) cases. Histologically, tumor types were classified as intestinal (n = 41, 37.3%) or diffuse (n = 69, 62.7%). Early gastric cancer (T1; mucosa and submucosa), defined by the depth of wall invasion, was diagnosed in 24 (21.8%) cases, advanced cancers (T2; muscle proper and subserosa) in 17 (15.5%) cases, serosa (T3) in 56 (50.9%), and invasion into adjacent organs (T4) in 13 (11.8%) cases. Lymph node metastasis was identified in 78 cases (70.9%). During surgery, peritoneal seeding was detected in 19 (17.3%) patients and liver metastasis in 2 (1.8%) patients. Pathological staging was distributed as follows: stage I in 30 cases (27.3%); stage II in nine cases (8.2%); stage III in 42 cases (38.2%); and stage IV in 29 cases (26.4%).
Table 1.
Clinicopathological correlations of ADP‐ribosylation factor 1 (ARF1) expressions detected by immunohistochemistry (IHC) in 110 gastric cancer patients
| Parameter | No. | Mean ± SEa | P ‡ | Parameter | No. | Mean ± SEa | P ‡ |
|---|---|---|---|---|---|---|---|
| Age (years) | Lymph node metastasis | ||||||
| <65 | 53 | 94.5 ± 6.6 | NS | No (N0) | 32 | 76.9 ± 5.1 | 0.008 |
| ≥65 | 57 | 99.7 ± 6.1 | Yes (N1, N2, N3) | 78 | 105.5 ± 5.7 | ||
| Gender | Distant metastasis(pM) | ||||||
| Male | 69 | 101.6 ± 6.2 | NS | No | 85 | 96.0 ± 5.0 | NS |
| Female | 41 | 89.7 ± 5.9 | Yes | 25 | 101.2 ± 10.3 | ||
| Location | Pathological stage (pStage) | ||||||
| Upper third | 24 | 95.0 ± 8.5 | NS | Stage I | 30 | 78.3 ± 6.1 | 0.039 |
| Middle third | 28 | 91.1 ± 8.9 | Stage II | 9 | 86.7 ± 13.2 | ||
| Lower third | 55 | 99.6 ± 6.3 | Stage III | 42 | 112.0 ± 7.9 | ||
| Whole | 3 | 126.7 ± 57.8 | Stage IV | 29 | 93.5 ± 9.0 | ||
| Gross type | Pathological stage | ||||||
| Localized | 43 | 85.4 ± 6.0 | NS | Stages I, II | 39 | 80.3 ± 5.5 | 0.010 |
| Infiltrative | 67 | 104.8 ± 6.1 | Stages III, IV | 71 | 106.5 ± 6.0 | ||
| Size (cm) | Liver metastasis | ||||||
| <5 | 60 | 99.2 ± 6.4 | NS | No | 108 | 96.5 ± 4.5 | NS |
| ≥5 | 50 | 94.8 ± 6.2 | Yes | 2 | 135.0 ± 45.0 | ||
| Histological type | Peritoneal seeding | ||||||
| Intestinal | 41 | 100.2 ± 7.5 | NS | No | 91 | 97.8 ± 4.8 | NS |
| Diffuse | 69 | 95.4 ± 5.6 | Yes | 19 | 94.2 ± 12.5 | ||
| Depth of invasion (pT) | Vascular invasion | ||||||
| T1 | 24 | 78.3 ± 7.5 | NS | No | 91 | 97.2 ± 5.0 | NS |
| T2 | 17 | 98.8 ± 12.5 | Yes | 19 | 96.8 ± 10.7 | ||
| T3 | 56 | 103.8 ± 6.3 | Lymphatic invasion | ||||
| T4 | 13 | 101.5 ± 14.6 | No | 45 | 84.0 ± 5.3 | 0.035 | |
| Serosal invasion | Yes | 65 | 106.3 ± 6.4 | ||||
| No (T1, T2) | 43 | 88.8 ± 6.8 | 0.046 | Perineural invasion | |||
| Yes (T3, T4) | 72 | 100.7 ± 5.9 | No | 71 | 94.5 ± 5.0 | NS | |
| Lymph node status (pN) | Yes | 39 | 102.0 ± 8.7 | ||||
| N0 | 32 | 76.9 ± 5.1 | 0.043 | ARF1 (IHC score) | |||
| N1 | 36 | 106.9 ± 8.6 | <90 (median) | 50 | – | – | |
| N2 | 22 | 108.2 ± 9.9 | ≥90 | 60 | – | ||
| N3 | 20 | 100.0 ± 12.2 | |||||
ADP‐ribosylation factor 1 scores detected by IHC method in mean ± standard error (SE). ‡Mann–Whitney U‐test (for two groups) and Kruskal–Wallis test (for >2 groups). P < 0.05 was considered significant. NS, not significant.
Expression of ARF1 in gastric cancer tissues
To validate the expression of ARF1 mRNA, real‐time qRT‐PCR was carried out in 55 frozen tumor samples and adjacent non‐tumorous mucosa. The median of ARF1 mRNA expression estimated using qRT‐PCR was 2.3‐fold higher in tumor samples (range, 0.04–49.87). Based on the definition of overexpression as ≥1.5‐fold higher than the normal level in qRT‐PCR analyses, ARF1 was found to be overexpressed in 67.2% of patients (Fig. 2A). Thus, ARF1 mRNA expression at stages III/IV was significantly (P < 0.001, Mann–Whitney U‐test) higher than that in stages I/II (Fig. 2B). Moreover, we further confirmed ARF1 protein expression with Western blot analysis. Expression of ARF1 in eight representative patients is presented in Figure 2(C,D). The 20 kDa ARF1 protein was detected in all cancer tissues. Cancer tissues from gastric cancer stage I (G86, G95) or stage II (G29, G43) displayed no changes of ARF1 expression. However, stage III (G99, G44) and stage IV (G40, G02) tissues showed increased ARF1 expression (1.46‐ to 6.56‐fold), compared with matched non‐cancerous adjacent mucosa (Fig. 2C).
Figure 2.
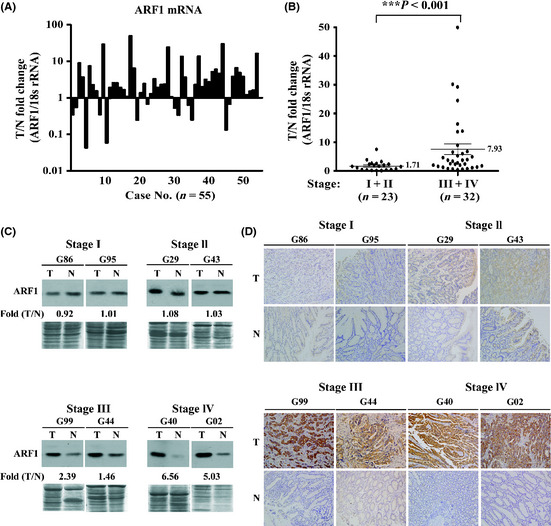
Overexpression of ADP‐ribosylation factor 1 (ARF1) mRNA in gastric carcinoma. The ARF1 gene is overexpressed in gastric carcinoma, as determined with quantitative RT‐PCR. (A) Quantitative RT‐PCR results. Levels of ARF1 mRNA were examined in 55 paired tumor (T) and adjacent normal (N) tissues. (B) Comparison of ARF1 mRNA expression in the advanced stages (III/IV) and early stages (I/II) of gastric carcinoma. (C) Western blot analysis of the 20‐kDa ARF1 protein. An equal amount (60 μg) of protein was loaded for each specimen and Coomassie blue stain was used as a loading control. The blots were exposed to film for ~3 min for stages I/II, and ~30 s for stages III/IV. (D) Immunohistochemical staining of ARF1 in two types of gastric carcinomas including intestinal (G02, G29) and diffuse type (G86, G95, G43, G99, G44, G40), stages I–IV. Positive staining of ARF1 is indicated by a dark‐brown color. ADP‐ribosylation factor 1 was observed mainly in the gastric cancer cells and barely in stromal cells.
Immunostaining reveals overexpression of ARF1 protein in gastric cancerous tissues
To further validate the expression and localization of ARF1 protein in surgical specimens, IHC was carried out in 110 patients. Figure 2(D) shows the ARF1 IHC staining of the same specimens with their Western blots to validate the staining intensity in eight representative patients. Dark‐brown immunostaining was most prevalent in cancer cells, and low levels were observed in stromal cells or fibroblasts in gastric cancer tissues. Notably, no or weak staining for ARF1 was observed in tumor or normal gastric epithelial cells in stages I/II (Fig. 2D, G86, G95, G29, and G43). Staining was more intensive in the advanced stages (III/IV) of the tumors. Stages III/IV are depicted in tumor tissues of Figure 2(D) (G99, G44, G40, and G02) compared to adjacent normal tissues (Fig. 2D). The mean IHC score of gastric cancer tissues was 97.2 (median, 90; range, 0–240), whereas that of the matching adjacent non‐cancerous mucosa was 43.1 (median, 50; range, 0–150), which was significantly different (P < 0.001, Wilcoxon signed rank test) (Fig. 3A). Paired comparison of immunoreactivity for ARF1 (n = 68) revealed that the IHC scores of cancerous tissues were higher than those of their non‐tumorous counterparts in 52 (76.5%), equivalent in 7 (10.3%), and lower in 9 (13.2%) patients. However, ARF1 expression levels in surgical specimens determined by IHC show no significant difference between intestinal and diffuse types (Fig. 3B, Table 1). Expression of ARF1 was significantly higher (P = 0.01 for IHC) in patients with more advanced pathologic stage (stages III/IV) than for those in an earlier pathologic stage (stages I/II) (Fig. 3C and Table 1).
Figure 3.
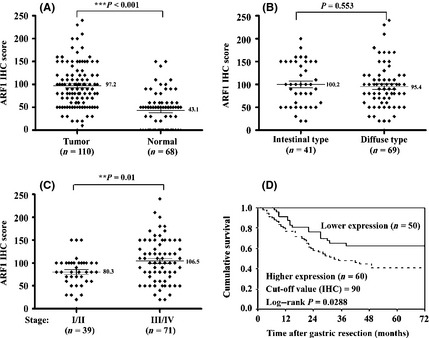
Immunohistochemical (IHC) detection of ADP‐ribosylation factor 1 (ARF1) expression in human gastric cancer tissues (T) and matching non‐cancerous mucosa (N). (A) Comparison of the ARF1 IHC scores in T/N. The differences were analyzed with the Wilcoxon signed rank test. Comparison of the ARF1 expression in two types (B) or various stages (C) of cancer by the Mann–Whitney U‐test. (D) Kaplan–Meier survival curves of two groups of gastric cancer patients defined by the ARF1 expression level cut‐off value of 90, as determined with IHC scoring. The 5‐year survival rate of the lower expression group (n = 50) was significantly better than that of the higher expression group (n = 60; P = 0.0288, log–rank test).
Expression of ARF1 and clinicopathological correlations
Expression of ARF1 in tumor tissue was not significantly associated with age, gender, tumor location, tumor size, or histological type (Table 1). However, elevated ARF1 expression was strongly correlated with lymph node metastasis (P = 0.008), serosal invasion (P = 0.046), lymphatic invasion (P = 0.035), and pathological staging (P = 0.010) (Table 1 and Fig. S1). Increased ARF1 expression was not associated with vascular invasion or distant metastasis, including peritoneal seeding or liver metastasis.
Survival outcomes
The mean duration of the follow‐up period for 50 survivors was 65.5 months (range, 26–126 months). Two patients died of postoperative complications, and six died of other diseases. In total, death occurred in 52 cases as a result of gastric cancer progression. The overall cumulative 5‐year survival rate of 110 patients with gastric resection was 50.3%. Figure 3(D) show the cumulative survival curves of patients in the lower and higher ARF1 expression groups. The lower expression group included patients with scores below median (IHC score = 90) ARF1 expression, whereas the higher expression group comprised patients expressing levels above the median value. The 5‐year survival rate of the lower expression group was significantly improved relative to that of the higher expression group (62.9% vs 40.9%; P = 0.0288, log–rank test).
Univariate analysis disclosed a number of significant prognostic factors, including status of lymph node metastasis, serosal invasion, distant metastasis, peritoneal seeding, vascular invasion, lymphatic invasion, and perineural invasion, in addition to ARF1 expression. Other significant parameters were histological type, tumor size, and gross type. Further, in multivariate analysis, the independent prognostic factors influencing patient survival were lymph node metastasis (P = 0.006), distant metastasis (P = 0.034), and higher ARF1 expression (IHC score ≥ 90) (P = 0.036) (Table 2). Although the pTNM factor is the most useful indicator to predict outcome for survival, the current results indicate that higher ARF1 expression can be used as a novel prognostic factor influencing gastric cancer patient survival.
Table 2.
Analysis of risk factors for 5‐year survival rate in 110 gastric cancer patients
| Risk factors | No. | Univariate analysisa | Multivariate analysis‡ | ||
|---|---|---|---|---|---|
| P | HR | 95% CI | P | ||
| ARF1 (IHC ≥ 90 score) | |||||
|
Low expression |
62.9 | 0.0288 | 1.987 | 1.045–3.776 | 0.036 |
|
High expression |
40.9 | ||||
| Gross type | |||||
| Localized | 78.8 | <0.0001 | 0.556 | 0.214–1.441 | NS |
| Infiltrative | 31.0 | ||||
| Size (cm) | |||||
| <5 | 69.3 | <0.0001 | 1.986 | 0.929–4.247 | NS |
| ≥5 | 25.3 | ||||
| Histological type | |||||
| Intestinal | 72.2 | 0.0001 | 2.063 | 0.924–4.607 | NS |
| Diffuse | 36.7 | ||||
| Serosal invasion | |||||
| No (T1, T2) | 82.3 | <0.0001 | 1.584 | 0.578–4.340 | NS |
| Yes (T3, T4) | 30.3 | ||||
| Lymph node metastasis | |||||
| No (N0) | 93.7 | <0.0001 | 9.985 | 1.913–52.125 | 0.006 |
| Yes (N1, N2, N3) | 32.4 | ||||
| Distant metastasis (pM) | |||||
| No | 64.0 | <0.0001 | 2.502 | 1.071–5.848 | 0.034 |
| Yes | 4.2 | ||||
| Peritoneal seeding | |||||
| No | 59.6 | <0.0001 | 1.070 | 0.441–2.597 | NS |
| Yes | 5.6 | ||||
| Vascular invasion | |||||
| No | 57.7 | <0.0001 | 1.435 | 0.726–2.836 | NS |
| Yes | 12.1 | ||||
| Lymphatic invasion | |||||
| No | 79.2 | <0.0001 | 0.769 | 0.295–2.004 | NS |
| Yes | 29.5 | ||||
| Perineural invasion | |||||
| No | 64.9 | <0.0001 | 1.138 | 0.589–2.197 | NS |
| Yes | 22.7 | ||||
Log–rank test, P < 0.05.‡Cox regression method, P < 0.05. ARF1, ADP–ribosylation factor 1; CI, confidence interval; HR, multivariate‐adjusted hazard ratio; IHC, immunohistochemistry; NS, not significant.
Influence of ectopic ARF1 overexpression on AGS cell proliferation and migration
The AGS cells expressing low levels of endogenous ARF1 were transfected with pcDNA3‐ARF1. After 2 weeks of transfection, stable expression of ARF1 protein was established. Figure 4(A) shows 2.08‐fold and 2.35‐fold higher ARF1 expression in two AGS‐ARF1 sublines (ARF‐1 and ARF‐2), respectively, compared to cells transfected with control vector (C‐1 and C‐2). To determine the effects of overexpression of ARF1 in AGS cells, cell proliferation, migration, and invasion activities were assayed. Cell proliferation was determined by cell counting and indicated as a percentage of the control for up to 5 days. Cells overexpressing ARF1 showed significantly (P < 0.01) higher proliferation rates (1.77‐ or 1.61‐fold) and colony formation ability (2.19‐ or 2.14‐fold) than those transfected with control vector on day 5 and day 10, respectively (Fig. 4B,C). Moreover, ARF1‐overexpressing cells showed significantly (P < 0.01) higher migration rates (4.53‐ or 4.8‐fold) and invasive abilities (3.53‐ or 3.8‐fold) than their control counterparts (Fig. 4D,E). Therefore, several MMPs were examined. Higher proMMP2, MMP2 activities were observed in the ARF1‐overexpressing but not in the control AGS lines (Fig. 4F). Furthermore, higher expression of epithelial–mesenchymal transition (EMT) markers including N‐cadherin, vimentin, snail, slug, twist, as well as β‐catenin, were also detected in the ARF1‐overexpressing lines (Fig. 4G). However, the AGS cell line does not express detectable endogenous E‐cadherin (data not shown). Collectively, our results indicate that ARF1‐overexpressing cells show enhanced cell proliferation, migration, and invasion.
Figure 4.
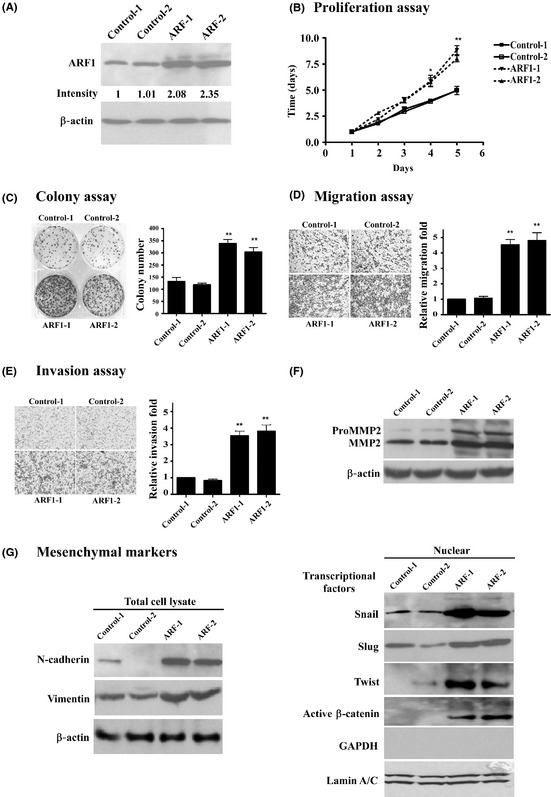
Influence of ectopic overexpression of ADP‐ribosylation factor 1 (ARF1) on AGS gastric cancer cells. Two AGS‐ARF1 sublines (ARF‐1 and ‐2) and control lines (C‐1 and C‐2) were established, as described in “Materials and Methods(A) Expression of ARF1 was determined using Western blot analysis. (B) Proliferation, (C) colony formation, (D) migration, and (E) invasion abilities were assayed as described in “Materials and MethodsData are presented as means ± SE obtained from at least three independent experiments carried out in duplicate. Values are presented as colony numbers or fold (C–E) of vector control, and differences examined with Student's t‐test, compared to the average of vector. *P < 0.05; **P < 0.01. (F) Western blot of MMP2, N‐cadherin, vimentin, snail, slug, twist, and β‐catenin proteins. Actin was used as a loading control.
Discussion
Gastric cancer, the second most frequent cause of cancer‐related death,1 remains a significant therapeutic challenge, and the molecular pathways implicated in pathogenesis require further elucidation. With the aid of proteomic technology, we identified ARF1 as a protein upregulated in gastric cancer patients. Upregulation of ARF1 expression was strongly correlated with depth of invasion, lymph node metastasis, pathological stage, and lymphatic invasion. Furthermore, increased ARF1 expression correlated strongly with poor survival rate.
Numerous biomarkers for diagnosis or prognosis, including SPARC,26 claudin‐18,27 CD54,28 hepatoma‐derived growth factor,29 vascular endothelial growth factor (VEGF)‐D and its receptor VEGFR‐3,30 melanoma inhibitory activity and MMP‐10,31 secretory leukocyte protease inhibitor,22 chloride intracellular channel 1 (CLIC1),20and chemokine (C‐X‐C motif) ligand 1 (CXCL1, also termed GRO‐1)32 have recently been identified for gastric cancer. Some of these biomarkers have been effectively applied to elucidate the molecular and cellular mechanisms of gastric carcinogenesis and progression.33
Data from the current study confirmed that ARF1 is frequently overexpressed in gastric cancer cells. Among the patients examined, higher ARF1 expression was significantly associated with tumor progression and advanced stages of gastric cancer. In addition, patients with lower ARF1 expression had better prognosis. ADP‐ribosylation factor appears to be involved in several important cellular processes, including membrane trafficking and activation of phospholipase D.34, 35 Further recent studies have shown that ARF1 controls cell proliferation, dependent on its ability to regulate pRB/E2F1 activity and gene expression for enhanced proliferation and breast cancer progression.36 Similarly, inhibition of endogenous ARF1 expression results in the suppression of breast cancer cell migration and proliferation through activation of the phosphatidylinositol 3‐kinase pathway.37
Similar to our results, Kon et al.38 reported that ARF6 isoform SMAP1 regulated E‐cadherin turnover involved in EMT. Jang et al.39 found that PMA‐stimulated ARF4 expression increases AP‐1 promoter activity, leading to induction of breast cancer cell migration. However, the underlying mechanism is still unknown. Our findings showed that ARF1 shows oncogenic potential, which is mediated at least partially by the EMT.37
Several studies have used similar techniques to identify differentially expressed proteins in gastric cancer. In a study by Ebert et al.,40 cathepsin B expression was upregulated in 60% of patients and associated with distant metastases and reduced survival rate. Upregulation of CLIC1 was strongly correlated with lymph node metastasis, lymphatic invasion, perineural invasion, advanced pathological stage, and poor survival. The expression of 14‐3‐3β was highly correlated with the number of lymph node metastases, tumor size, and reduced survival rate.41 However, to our knowledge, the significance of those proteins during carcinogenesis has not been further elucidated. The possible roles underlying the effects on carcinogenesis, development of human gastric carcinoma, or interactions with cellular ARF1 are yet to be established.42
In summary, we have confirmed the dysregulation of ARF1 in gastric carcinoma and a strong association with more advanced stages of the disease. As ARF1 is overexpressed in a variety of malignancies, it is unsuitable as a diagnostic biomarker for a specific tumor, although the protein shows promise as a prognostic marker of tumor progression and advanced cancers. Further investigation of ARF1 is warranted in view of its potential as a prognostic and therapeutic agent.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Scatter plots of the immunohistochemistry (IHC) scores for ADP‐ribosylation factor 1 (ARF1) and various clinicopathological features.
Acknowledgments
This work was supported by grants from Chang‐Gung University, Taoyuan, Taiwan (NMRPD 150311, NMRPD 170441‐42, CMRPG 640042‐43, CMRPG 670291‐93) and the National Science Council of the Republic of China (Grant Nos. NSC 95‐2314‐B‐182‐027, 97‐2314‐B‐182‐009‐MY2, 99‐2314‐B‐182‐022).
References
- 1. Wang CS, Hsieh CC, Chao TC et al Resectable gastric cancer: operative mortality and survival analysis. Chang Gung Med J 2002; 25: 216–27. [PubMed] [Google Scholar]
- 2. Marrelli D, Roviello F, De Stefano A et al Prognostic significance of CEA, CA 19‐9 and CA 72‐4 preoperative serum levels in gastric carcinoma. Oncology 1999; 57: 55–62. [DOI] [PubMed] [Google Scholar]
- 3. Lauren P. Histogenesis of intestinal and diffuse types of gastric carcinoma. Scand J Gastroenterol Suppl 1991; 180: 160–4. [PubMed] [Google Scholar]
- 4. Tahara E. Molecular mechanism of stomach carcinogenesis. J Cancer Res Clin Oncol 1993; 119: 265–72. [DOI] [PubMed] [Google Scholar]
- 5. Cervantes A, Rodriguez Braun E, Perez Fidalgo A, Chirivella Gonzalez I. Molecular biology of gastric cancer. Clin Transl Oncol 2007; 9: 208–15. [DOI] [PubMed] [Google Scholar]
- 6. Barreto‐Zuniga R, Maruyama M, Kato Y et al Significance of Helicobacter pylori infection as a risk factor in gastric cancer: serological and histological studies. J Gastroenterol 1997; 32: 289–94. [DOI] [PubMed] [Google Scholar]
- 7. Kim KM, Kwon MS, Hong SJ et al Genetic classification of intestinal‐type and diffuse‐type gastric cancers based on chromosomal loss and microsatellite instability. Virchows Arch 2003a; 443: 491–500. [DOI] [PubMed] [Google Scholar]
- 8. Yasui W, Oue N, Kuniyasu H, Ito R, Tahara E, Yokozaki H. Molecular diagnosis of gastric cancer: present and future. Gastric Cancer 2001; 4: 113–21. [DOI] [PubMed] [Google Scholar]
- 9. Allgayer H, Heiss MM, Schildberg FW. Prognostic factors in gastric cancer. Br J Surg 1997; 84: 1651–64. [PubMed] [Google Scholar]
- 10. Chen J, Kahne T, Rocken C et al Proteome analysis of gastric cancer metastasis by two‐dimensional gel electrophoresis and matrix assisted laser desorption/ionization‐mass spectrometry for identification of metastasis‐related proteins. J Proteome Res 2004; 3: 1009–16. [DOI] [PubMed] [Google Scholar]
- 11. Nishigaki R, Osaki M, Hiratsuka M et al Proteomic identification of differentially‐expressed genes in human gastric carcinomas. Proteomics 2005; 5: 3205–13. [DOI] [PubMed] [Google Scholar]
- 12. Yim EK, Park JS. Role of proteomics in translational research in cervical cancer. Expert Rev Proteomics 2006; 3: 21–36. [DOI] [PubMed] [Google Scholar]
- 13. Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP‐ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 2006; 70: 789–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D'Souza‐Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 2006; 7: 347–58. [DOI] [PubMed] [Google Scholar]
- 15. Morishige M, Hashimoto S, Ogawa E et al GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat Cell Biol 2008; 10: 85–92. [DOI] [PubMed] [Google Scholar]
- 16. Muralidharan‐Chari V, Hoover H, Clancy J et al ADP‐ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res 2009; 69: 2201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu B, Shi B, Jarzynka MJ, Yiin JJ, D'Souza‐Schorey C, Cheng SY. ADP‐ribosylation factor 6 regulates glioma cell invasion through the IQ‐domain GTPase‐activating protein 1‐Rac1‐mediated pathway. Cancer Res 2009; 69: 794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Japanese Gastric Cancer Association . Japanese classification of gastric carcinoma – 2nd English edition. Gastric Cancer 1998; 1: 10–24. [DOI] [PubMed] [Google Scholar]
- 19. Sobin LH, Fleming ID. TNM classification of malignant tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer 1997; 80: 1803–4. [DOI] [PubMed] [Google Scholar]
- 20. Chen CD, Wang CS, Huang YH et al Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics 2007; 7: 155–67. [DOI] [PubMed] [Google Scholar]
- 21. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver‐stained polyacrylamide gels. Anal Chem 1996; 68: 850–8. [DOI] [PubMed] [Google Scholar]
- 22. Cheng WL, Wang CS, Huang YH et al Overexpression of a secretory leukocyte protease inhibitor in human gastric cancer. Int J Cancer 2008; 123: 1787–96. [DOI] [PubMed] [Google Scholar]
- 23. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 1976; 72: 248–54. [DOI] [PubMed] [Google Scholar]
- 24. Hsu SM, Raine L, Fanger H. Use of avidin‐biotin‐peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem 1981; 29: 577–80. [DOI] [PubMed] [Google Scholar]
- 25. Repesh LA. A new in vitro assay for quantitating tumor cell invasion. Invasion Metastasis 1989; 9: 192–208. [PubMed] [Google Scholar]
- 26. Wang CS, Lin KH, Chen SL, Chan YF, Hsueh S. Overexpression of SPARC gene in human gastric carcinoma and its clinic‐pathologic significance. Br J Cancer 2004; 91: 1924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W. Down‐regulation of the claudin‐18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol 2006; 208: 633–42. [DOI] [PubMed] [Google Scholar]
- 28. Yashiro M, Sunami T, Hirakawa K. CD54 expression is predictive for lymphatic spread in human gastric carcinoma. Dig Dis Sci 2005; 50: 2224–30. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto S, Tomita Y, Hoshida Y et al Expression of hepatoma‐derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin Cancer Res 2006; 12: 117–22. [DOI] [PubMed] [Google Scholar]
- 30. Juttner S, Wissmann C, Jons T et al Vascular endothelial growth factor‐D and its receptor VEGFR‐3: two novel independent prognostic markers in gastric adenocarcinoma. J Clin Oncol 2006; 24: 228–40. [DOI] [PubMed] [Google Scholar]
- 31. Aung PP, Oue N, Mitani Y et al Systematic search for gastric cancer‐specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase‐10 are novel prognostic factors in patients with gastric cancer. Oncogene 2005; 25: 2546–57. [DOI] [PubMed] [Google Scholar]
- 32. Cheng WL, Wang CS, Huang YH, Tsai MM, Liang Y, Lin KH. Overexpression of CXCL1 and its receptor CXCR2 promote tumor invasion in gastric cancer. Ann Oncol 2011; 22: 2267–76. [DOI] [PubMed] [Google Scholar]
- 33. Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular‐pathological prognostic factors of gastric cancer: a review. Gastric Cancer 2005; 8: 86–94. [DOI] [PubMed] [Google Scholar]
- 34. Kim SW, Hayashi M, Lo JF, Yang Y, Yoo JS, Lee JD. ADP‐ribosylation factor 4 small GTPase mediates epidermal growth factor receptor‐dependent phospholipase D2 activation. J Biol Chem 2003b; 278: 2661–8. [DOI] [PubMed] [Google Scholar]
- 35. Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem 1995; 270: 12327–30. [DOI] [PubMed] [Google Scholar]
- 36. Boulay PL, Schlienger S, Lewis‐Saravalli S, Vitale N, Ferbeyre G, Claing A. ARF1 controls proliferation of breast cancer cells by regulating the retinoblastoma protein. Oncogene 2011; 30: 3846–61. [DOI] [PubMed] [Google Scholar]
- 37. Boulay PL, Cotton M, Melancon P, Claing A. ADP‐ribosylation factor 1 controls the activation of the phosphatidylinositol 3‐kinase pathway to regulate epidermal growth factor‐dependent growth and migration of breast cancer cells. J Biol Chem 2008; 283: 36425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kon S, Tanabe K, Watanabe T, Sabe H, Satake M. Clathrin dependent endocytosis of E‐cadherin is regulated by the Arf6GAP isoform SMAP1. Exp Cell Res 2008; 314: 1415–28. [DOI] [PubMed] [Google Scholar]
- 39. Jang SY, Jang SW, Ko J. Regulation of ADP‐ribosylation factor 4 expression by small leucine zipper protein and involvement in breast cancer cell migration. Cancer Lett 2012; 314: 185–97. [DOI] [PubMed] [Google Scholar]
- 40. Ebert MP, Kruger S, Fogeron ML et al Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics 2005; 5: 1693–704. [DOI] [PubMed] [Google Scholar]
- 41. Tseng CW, Yang JC, Chen CN et al Identification of 14‐3‐3beta in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics 2011; 11: 2423–39. [DOI] [PubMed] [Google Scholar]
- 42. Akai T, Nabeya Y, Yahiro K et al Helicobacter pylori induces mono‐(adenosine 5′‐diphosphate)‐ribosylation in human gastric adenocarcinoma. Int J Oncol 2006; 29: 965–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Scatter plots of the immunohistochemistry (IHC) scores for ADP‐ribosylation factor 1 (ARF1) and various clinicopathological features.


