Abstract
Tankyrase 1 (TNKS1; a.k.a. ARTD5) and tankyrase 2 (TNKS2; a.k.a ARTD6) are highly homologous poly(ADP‐ribose) polymerases (PARPs) that function in a wide variety of cellular processes including Wnt signaling, Src signaling, Akt signaling, Glut4 vesicle translocation, telomere length regulation, and centriole and spindle pole maturation. Tankyrase proteins include a sterile alpha motif (SAM) domain that undergoes oligomerization in vitro and in vivo. However, the SAM domains of TNKS1 and TNKS2 have not been structurally characterized and the mode of oligomerization is not yet defined. Here we model the SAM domain‐mediated oligomerization of tankyrase. The structural model, supported by mutagenesis and NMR analysis, demonstrates a helical, homotypic head‐to‐tail polymer that facilitates TNKS self‐association. Furthermore, we show that TNKS1 and TNKS2 can form (TNKS1 SAM‐TNKS2 SAM) hetero‐oligomeric structures mediated by their SAM domains. Though wild‐type tankyrase proteins have very low solubility, model‐based mutations of the SAM oligomerization interface residues allowed us to obtain soluble TNKS proteins. These structural insights will be invaluable for the functional and biophysical characterization of TNKS1/2, including the role of TNKS oligomerization in protein poly(ADP‐ribosyl)ation (PARylation) and PARylation‐dependent ubiquitylation.
Keywords: protein poly(ADP‐ribosyl)ation, PARylation, tankyrase, molecular model, sterile alpha motif, SAM, oligomerization, PARP, PARP5
Abbreviations
- ARTD
diphtheria toxin‐like ADP‐ribosyltransferase
- GLUT4
glucose transporter 4
- HSQC
heteronuclear single quantum correlation spectroscopy;
- ITC
isothermal titration calorimetry
- PAR
poly(ADP‐ribose)
- PARP
poly(ADP‐ribose) polymerase
- PARylation
poly(ADP‐ribosyl)ation
- PDB
Protein Data Bank
- PTEN
phosphatase and tensin homolog
- SAM
sterile alpha motif
- TNKS
tankyrase
Introduction
Tankyrase‐1 (TNKS1) and tankyrase‐2 (TNKS2) constitute two of the six members of the bona fide human poly(ADP‐ribose) polymerase (PARP) enzymes. TNKS1/2 poly(ADP‐ribosyl)ate (PARylate) proteins in a myriad of cellular functions including Wnt signaling,1 Src signaling,2 Hippo signaling,3 telomere length regulation,4 Glut‐4 vesicle translocation,5, 6 and mitosis.7, 8, 9, 10 Not only is the PARylation of substrate required for all observed functions of TNKS, but PAR‐dependent ubiquitylation mediated by RNF146 is also necessary for many TNKS1/2‐mediated regulatory events, including Axin,11, 12, 13 Angiomotin,3 3BP2,2 and PTEN turnover.14 These diverse regulatory functions have led to an intense focus on the development of TNKS1/2 PARP inhibitors for cancer therapies.15, 16, 17 Characterization of the many protein‐protein interactions required for TNKS1/2 function, such as substrate and RNF146 binding,13 could facilitate more targeted small molecule or mutagenic pathway manipulation. Though the mechanism is currently unclear, TNKS1/2 oligomer formation has been suggested to affect the PARylation of substrates,1, 18 implying a potential regulatory function.
Of the six PARPs, the tankyrase proteins (TNKS) have unique domain compositions and TNKS1 and TNKS2 share ∼83% identity [Fig. 1(A)]. The sterile alpha motif (SAM) domain in TNKS1/2 is of particular interest because it is adjacent to the PARP domain—a position held by the PARP regulatory domain in other human poly‐ADP‐ribosylating enzymes.19 SAM domains are composed of a ∼70 amino acid helical structure in which three or four helices cradle a c‐terminal helix and act as interaction motifs with diverse functions in signal transduction and transcriptional regulation.20, 21 SAM domains have been shown to mediate protein–protein,21 protein–RNA,22, 23, 24 and protein–lipid interactions.25 Furthermore, SAM domains can bind themselves symmetrically, as in the example of Eph4A which forms a homodimer,26 can bind other SAM domains (e.g., odin binding to EphA2, and Ste11 binding to Ste50),27, 28, 29 and can form long‐range homo‐oligomeric/polymeric structures.30, 31, 32, 33, 34, 35, 36, 37 While it is generally accepted that the SAM domain is largely responsible for the oligomerization of TNKS1/2,38, 39 the domain has not yet been structurally or biochemically characterized. It is still unknown whether the domain forms oligomers using multiple interfaces as observed for the SAM domain structure of EphB2 receptor,40 homotypic oligomers with a single, distinct head‐to‐tail interface observed for other SAM polymers,41 or utilizes some heretofore unobserved oligomeric topology. Furthermore, it is unclear whether the reported co‐localization of TNKS1 and TNKS242 is due to association through SAM domain hetero‐binding or mediated through the large ankyrin repeat cluster region [Fig. 1(A)].18, 43
Figure 1.
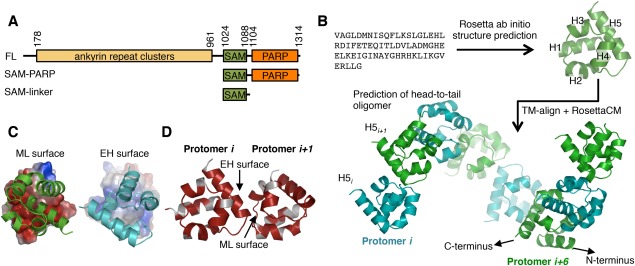
Modeling tankyrase SAM domain oligomerization. (A) The TNKS1 domain architecture and constructs used in this manuscript. TNKS2 is homologous to TNKS1, but lacks a ∼170 amino acid N‐terminal histidine serine proline rich region in Tankyrase 1. FL; full‐length. (B) Schematic of the TNKS1 SAM domain structure and oligomeric structure prediction workflow. First, the amino acid sequence was used for structure prediction using Rosetta, followed by a TM‐align search for similar folds participating in SAM‐SAM interactions used as initial docking positions for a RosettaCM TNKS1 oligomer modeling protocol with helical symmetry. H1‐5; Helix 1, 2, 3, 4, and 5. Protomers are colored either green or cyan to clarify oligomeric structure. (C) Surface charges experienced by opposing faces of the oligomer protomers. Between the two interacting surfaces, the mid‐loop (ML) surface (left) is negatively charged (red), whereas the end‐helix (EH) surface is enriched in positively (blue) charged residues (right). (D) Conservation of residues between TNKS1 and TNKS2 plotted on the surface of the TNKS1 model showing sequence identity (red) at the interface of the two protomers. White; non‐identical. See also Supporting Information Figure S1.
Here we model the tankyrase SAM oligomer structure revealing a head‐to‐tail oligomerization mode, with conserved interfaces for TNKS1 and TNKS2. Model‐based mutations of residues in the interface generate monomeric TNKS1 and TNKS2 mutants, providing confirmation of the model. Because of their head‐to‐tail association, mixtures of two different interface mutants readily form homo‐ and heteromeric dimers with high affinity, consistent with the notion that TNKS1 and TNKS2 can form homo‐ and hetero‐oligomer structures through their SAM domains. We have identified the residues involved in the SAM–SAM interaction using NMR spectroscopy as further experimental confirmation of the model. The data and the ability to generate well‐defined, well‐behaved tractable TNKS species will aid in future biophysical and functional characterization of TNKS.
Results
It is known that the tankyrase proteins are very insoluble and likely oligomerize through their SAM domains.38, 39, 44 We therefore sought to determine the mode of SAM‐domain mediated tankyrase oligomerization with the hope that controlling oligomerization might yield more soluble and therefore structurally and biochemically tractable proteins. We performed a fold prediction of the isolated TNKS1 SAM domain (residues 1,026–1,088) using Rosetta ab initio,45 which generated models that match a canonical SAM domain fold [Fig. 1(B)]. The top 10 models produced by Rosetta were very similar, with an average pairwise root mean square deviation (RMSD) of 1.99 Å.
A TNKS1 oligomer model was generated from the top‐scoring model, using clues from known SAM‐SAM interfaces. Using TM‐align,46 the top‐scoring Rosetta ab initio model was used to search the PDB for structurally similar domains, independent of sequence homology. From the top hits, all with TM‐align score ≥ 0.6, we screened for entries containing multiple SAM‐like folds. The TM‐align search returned five SAM domains with significant structural similarity to TNKS1 (PDB codes: 3BQ7,34 3SEI,47 3TAD, 3TAC,48 1PK132; 20–31% sequence identity) that contained SAM–SAM interfaces. Though the angles between SAM domains differ substantially in these structures, all contained asymmetric SAM‐SAM interfaces centered on helix 5 (H5; termed the end‐helix or EH surface) and a patch composed of helices 2, 3, and 4 (termed mid‐loop or ML surface). By performing structural alignments between our TNKS1 SAM domain model and these SAM‐SAM complex structures, we obtained initial docking positions. The RosettaCM49 protocol was used to refine and generate homotypic TNKS1 SAM oligomer models. Despite different initial SAM‐SAM orientations, the models converged on a single relative SAM‐SAM angle with a maximum pairwise RMSD for the (dimeric) models of 1.16 Å. The resulting model shows a head‐to‐tail helical oligomer [Fig. 1(B)], similar to the oligomeric structures of several other SAM domains30, 31, 32, 33, 34, 35, 36, 37 in which oligomers of SAM domains form a right‐handed helix that contains ∼6 protomers per turn. While these structures differ in their helical pitch (primarily determined by the angle between promoters), this helical form is likely adopted by many SAM oligomers.41 Importantly, in our model the N and C‐termini of the SAM domains are directed away from the core of the oligomer [Fig. 1(B)], consistent with the multidomain architecture of TNKS1/2. It is unclear how the close proximity of the PARP domain might affect long‐range polymer structure.
Inspection of the interface between protomers in our oligomer model revealed that the interaction is a combination of charge–charge and hydrophobic contacts. Notably, amino acids on opposing faces of the TNKS1/2 SAM domain are highly biased to generate electrostatic interactions [Fig. 1(C)]. Furthermore, the residues that are predicted to participate in the interaction are nearly 100 percent conserved between TNKS1 and TNKS2 [Fig. 1(D)] and tankyrase orthologs (Supporting Information Fig. S1). Hence, this model predicts that the TNKS1 and TNKS2 may form homo and hetero‐oligomers through their conserved SAM‐SAM interface in a head‐to‐tail manner.
We attempted to express and purify proteins from Escherichia coli for biochemical characterization, but were unable to purify detectable amounts of wild‐type TNKS1 or TNKS2 or fragments containing the SAM domain by conventional means. This is consistent with the reported insolubility of the protein.38 We therefore used our model to design missense mutations in TNKS1 and TNKS2 that are predicted to disrupt oligomerization. Because of the head‐to‐tail polymerization mode, we were able to identify potential positions at the interface of protomers in the oligomer, for example Y1073, D1055, and V1056 [Fig. 2(A)]. The head‐to‐tail oligomerization model predicts that mutations on one surface should generate monomeric SAM domains [see Fig. 3(A)]. Two mutants were generated in the SAM domains of both TNKS1 and TNKS2: Y1073E and D1055A/V1056K (referred to as YE, and DAVK, respectively; tankyrase 1 numbering). The resulting mutant proteins are highly soluble and NMR 1H15N‐HSQC analysis reveals that both mutant SAM‐Linker [Fig. 1(A)] fragments of TNKS are folded as indicated by well‐dispersed peaks in the 1H dimension [Fig. 2(B)]. The HSQC spectrum for TNKS1 SAM‐Linker(DAVK) was assigned using conventional three‐dimensional heteronuclear protocols, and the 13Cα/13Cβ chemical shifts were analyzed for secondary structure relative to predicted random coiled values [Fig. 2(C)]. The NMR chemical shifts strongly predict five α−helices in locations consistent with our model, though the length of Helix‐1 (H1) may be ∼1 turn shorter than the Rosetta‐predicted structure. The three N‐terminal residues of H5 likely experience conformational heterogeneity as indicated by a drop in the chemical shift differences for residue H1075, which neighbors a glycine near the end of H5 (G1074), and by the apparent lack of an NH resonance from Y1073 in the 1H 15N‐HSQC spectrum. Notably, conformational flexibility appears to be reduced in the oligomer (see below). Altogether, the NMR spectra are consistent with the predicted SAM fold for the protomers.
Figure 2.
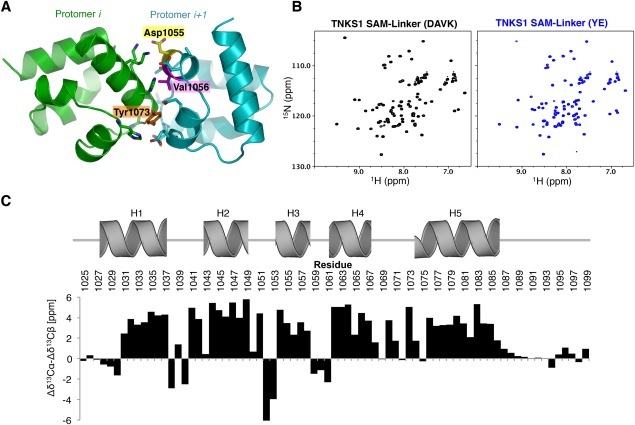
SAM mutants retain a folded structure. (A) Residues at the interface of two protomers in the predicted oligomer interface of TNKS1. Residues that were mutated to disrupt oligomerization are highlighted. (B) 1H15N‐HSQC NMR analysis of the TNKS1 SAM‐Linker(DAVK) mutant (left), and the TNKS1 SAM‐Linker(YE) mutant (right). DAVK, D1055A/V1056K; YE, Y1073E. Both spectra are well dispersed and highly similar, consistent with folded, monomeric domains. (C) Chemical shift differences between random coil and experimentally determined Cα and Cβ atoms (Δδ13Cα‐Δδ13Cβ [ppm]). Positions of Rosetta‐predicted helices are shown above histogram. High values are predictive of helical structure.
Figure 3.
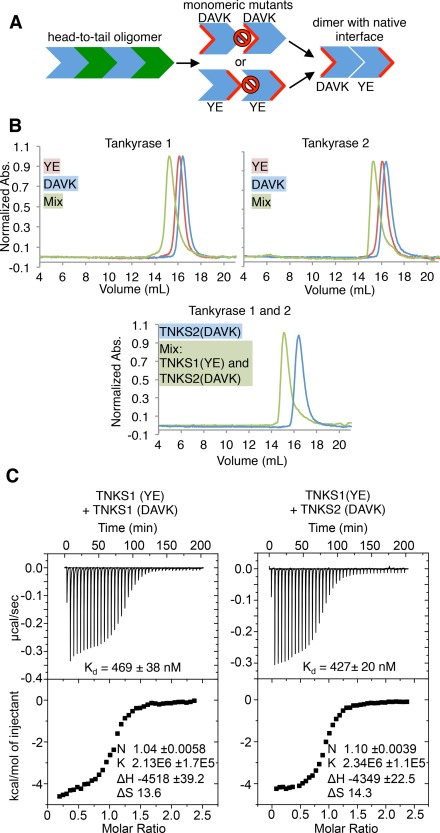
Tankyrase 1/2 SAM form strong homo and hetero‐oligomers. (A) Conceptual schematic of experiments in Figure 3. Residues are mutated on one face of the oligomeric interface to generate monomers (center). Complementary monomers can be mixed to generate dimers (right). Red indicates mutated surface. Prohibition signs indicate “no binding”. YE, TNKS1 Y1073E or TNKS2 Y920E; DAVK, TNKS1 D1055A/V1056K, or TNKS2 D902A/V903K. (B) Size exclusion chromatography (SEC) chromatograms of tankyrase 1 (top left) and tankyrase 2 (top right) SAM‐PARP YE and DAVK mutants. (Bottom) SEC trace of a 1:1 mixture of SAM‐PARP TNKS1 YE and TNKS2 DAVK, with TNKS2 DAVK elution profile shown as a reference. The maximum absorbance at 280 nm (y‐axis) is normalized to 1 for each trace. (C) Isothermal titration calorimetry (ITC) of TNKS1 SAM‐Linker YE with DAVK mutants (left; K d of 469 ± 38 nM), and TNKS1 SAM‐Linker(YE) with TNKS2 SAM‐Linker(DAVK) (right; K d of 427 ± 20 nM). K d, dissociation constants.
SEC‐MALS analysis was performed on mutant constructs that include both the SAM and PARP domains [SAM‐PARP; Fig. 1(A)]. Each of the constructs migrate at the expected retention volume on a Superdex 200 column and exhibit monomeric molecular weights: TNKS1‐YE 33.0 kDa ± 1.73%, TNKS1‐DAVK 33.7 kDa ± 0.986%, TNKS2‐YE 33.5 kDa ± 0.461%, and TNKS2‐DAVK 33.9 kDa ± 0.469% [Fig. 3(A) and Fig. 3(B) top]. When YE and DAVK mutants are mixed, the resulting species co‐migrate on a SEC column [Fig. 3(A,B) top], forming a complex near the expected molecular weight of a SAM‐PARP dimer (57.8 kDa ± 1.69% for TNKS1 and 63.0 kDa ± 1.16% for TNKS2). Consistent with the long‐lived interaction detected by SEC, the binding affinity (K d) between TNKS1 SAM‐Linker DAVK and YE mutants measured by isothermal titration calorimetry (ITC) is 469(±38) nM [Fig. 3(C) left]. As the dimers formed between the two mutants likely do so through their unmutated (i.e., native) interface, the data imply a strong propensity of TNKS to form homo‐oligomers. As predicted by the strong conservation in the oligomeric interface between TNKS1 and TNKS2, mixtures of TNKS1‐YE and TNKS2‐DAVK co‐elute as a heterodimer, with a molecular weight of 64.1 kDa ± 0.549% [Fig. 3(B) bottom]. The strength of this interaction was determined to be 427(±20) nM by ITC [Fig. 3(C) right]. Thus, our data reveal a roughly equal preference for TNKS hetero‐oligomer formation and TNKS1 homo‐oligomerization, confirming that the two proteins can form hetero‐oligomers through their SAM domains42 and are likely to do so in vivo. Furthermore, placing SAM mutations in the context of full‐length TNKS2 can generate soluble protein (Supporting Information Fig. S2), suggesting that the SAM domain makes a large contribution to the overall oligomeric assembly of TNKS.
To investigate the residues involved in the interaction interface, we performed NMR chemical shift perturbation analysis. Titration of TNKS1 SAM‐Linker(YE) into 15N‐labeled TNKS1 SAM‐Linker(DAVK) produced profound shifts in residues at the beginning of and throughout H5 [Fig. 4(A,B)]. Furthermore, while a peak is not observed for Y1073 in the initial spectrum of 15N‐labeled TNKS1 SAM‐Linker (DAVK), backbone assignments of the bound domain in a fully saturated sample confirmed the appearance of an amide peak for Y1073 [Fig. 4(A)]. This suggests that Y1073 undergoes a conformational exchange process in the monomeric SAM domain and is stabilized upon dimer formation. Mapping of the most perturbed residues [Fig. 4(B)] on the surface of the dimer model confirms the predicted surface to be directly involved [Fig. 4(C)]. Importantly, the NMR data indicate that binding only occurs at one interface, and does not appear to involve multiple surfaces between TNKS1 SAM domains in solution. These results not only provide key support for our model but also indicate that it can be used to identify other candidate residues for monomerizing mutations in the future.
Figure 4.
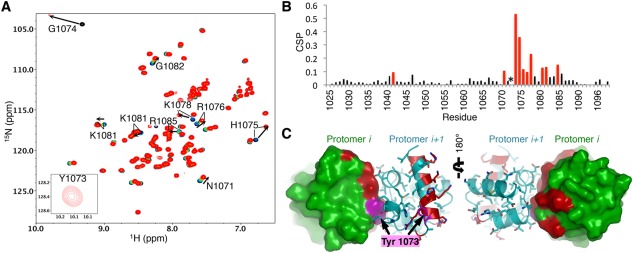
NMR mapping of the oligomeric interface supports model. (A) 1H15N‐HSQC of 150 µM TNKS1 SAM‐Linker(D1055A/V1056K) (black) with increasing quantities of TNKS1 SAM‐Linker(Y1073E): 0.25 molar equivalence (mol. eq.) (blue), 0.5 mol. eq. (green), 0.75 mol. eq. (orange), and 1.1 mol. eq. (red). A weak peak is seen for Y1073 at 1.1 mol. eq. when a spectrum was obtained at 250 µM TNKS1 SAM‐Linker (D1055A/V1056K), but not in the unbound spectrum in matching conditions (inset). (B) Chemical shift perturbations (CSPs) determined between the 0 mol. eq. (black) and 1.1 mol. eq. (red) spectra shown in (A) plotted against residue number. Red bars indicate the top 15% most perturbed residues. Asterisk (*) indicates Y1073, which only has a detectable peak in the dimeric form. (C) Residues shown in (B) are plotted (red) on the surface/cartoon representation of two neighboring protomers (cyan and green) of the oligomer. The residue highlight in magenta is Y1073. The binding surface recapitulates the predicted interface between protomers.
Discussion
We have shown that TNKS polymerizes through its SAM domain in a head‐to‐tail fashion to form a predicted helical oligomer. Mutations in the SAM domain can generate folded, monomeric proteins that can subsequently dimerize when mixed with complementary surface mutations. Furthermore, we have shown that the native SAM‐SAM interactions are relatively strong, with K d values below 500 nM for TNKS1 homodimerization and for TNKS1/TNKS2 heterodimerization. These values suggest that oligomers formed in vivo may contain both homomeric and heteromeric interfaces. Though this work indicates a major role for the SAM domain in TNKS oligomerization, the estimates of TNKS solubility (<40 nM)38 may imply that other regions, such as the third ankyrin‐repeat cluster,43 may also contribute to oligomeric assembly.
Our results and their structural implications provide a basis to investigate (1) the role of oligomerization for substrate PARylation in vivo; and (2) the potential effects of the SAM domain on inhibitor binding.15, 50, 51 How TNKS oligomerization affects substrate binding to the ankyrin repeat regions, PARylation activity, and subsequent ubiquitination of key cellular proteins such as Axin and Angiomotin remain to be investigated. It is likely that the multivalent nature of both TNKS oligomerization and its substrate or RNF146 binding will be affected by mutations in the SAM domain. This study provides a path forward towards a fine‐grained approach to studying the importance of TNKS oligomerization in vivo.
Materials and Methods
Generation of the SAM oligomer model
The TNKS1 oligomer model was generated using a combination of structural homology and computational efforts. Rosetta ab initio 45 design was performed on TNKS1 SAM domain (residues 1,018–1,092) producing 62,200 models. Models were trimmed to include only the core SAM domain (residues 1,026–1,088) for further modeling. The top model was used to search the PDB using TM‐align46 for structurally similar SAM domains that appear in deposited structures with multiple SAM folds. The top structural homologs identified by TM‐align with SAM‐SAM interfaces were used to generate starting positions for the oligomer model; these deposited structures were structurally aligned with our isolated SAM model, then used to model and refine the TNKS1 SAM oligomer in helical symmetry with RosettaCM.49 During sampling, the side‐chains, backbone, and symmetric definition were allowed to change and fragment insertion was allowed at all positions. No restraints were used, allowing for full degrees of motion. The final oligomer models converged on the same mode of interaction (SAM–SAM angle) most resembling the interface present in the PDB entry 1PK1. Therefore, while the SAM domain fold was predicted, this initial model was compared to homologous SAM structures to find clues about potential oligomeric interfaces. Hence the final model is a combination of fold prediction and homology assisted modeling.
Protein expression and purification
Mouse TNKS1 and human TNKS2 fragments or full‐length protein were cloned into a pET‐28a expression plasmid (Novagen, Madison, Wisconsin) with an N‐terminal His6 tag and tobacco etch virus (TEV) cleavage site and/or a pAL‐SUMO plasmid (Zheng Lab, University of Washington) with N‐terminal His6, SUMO tag, and TEV protease site upstream of the TNKS sequences. Site directed mutagenesis in the SAM domain was used to generate soluble TNKS fragments. All tags were removed before protein use unless otherwise noted. All regions in mouse TNKS1 used have 100 percent sequence identity to the human sequence at the protein level.
All proteins were expressed in Escherichia coli (BL‐21) by induction with 0.10–0.20 mM isopropyl β‐d−1‐thiogalactopyranoside (IPTG; Research Products International, Mt. Prospect, IL) at an attenuance (D) of 0.6–1.2 at 600 nm in either LB media or 15N/13C minimal mops media and grown over night at 16°C. TNKS1 SAM‐PARP (residues 1,024–1,314) and TNKS1 SAM‐Linker (residues 1,024–1,102) construct mutants (Y1073E and D1055A/V1056K) and TNKS2 SAM‐Linker (residues 871–952) mutants (D902A/V903K and Y920E) were purified by Ni‐NTA resin (Qiagen, Hilden, Germany), followed by dialysis in the presence of His‐tagged TEV protease to remove imidazole. TEV was captured on Ni‐NTA column and the eluates containing TNKS proteins were concentrated and further purified by SEC. TNKS2 SAM‐PARP (residues 871‐1,166) mutants (D902A/V903K and Y920E) were purified by Talon resin (Clonetech, Mountain view, CA), followed by a TEV cleavage to remove tags, and dialyzed overnight at 4°C. After capturing TEV on a Talon resin, protein was then diluted in 30 mM MES pH 6.0 buffer to a salt concentration of <50 mM and purified using an SP column (GE Healthcare, Pittsburgh, PA). Protein eluted from the SP column near 350 mM NaCl, was concentrated, and further purified by SEC into appropriate buffers.
Size exclusion chromatography and SEC‐MALS
Size exclusion chromatography (SEC) and SEC‐multi‐angle light scattering (SEC‐MALS) was performed at room temperature on a Superdex 200 10/300 GL (GE Healthcare) and a Superdex 200 Increase 3.2/300 (GE Healthcare), respectively, in running buffer (20 mM Tris‐HCl pH 7.5, 150 mM NaCl). For SEC, ∼80–100 µg of samples was protein was injected in a 100 µL volume and elution profiles were monitored at 280 nm on an AtkaPurifier (GE Healthcare). Fifteen microliters of SEC‐MALS samples were injected at 2 mg mL−1 onto a AktaPure purification system (GE Healthcare) equipped with a MiniDawn TREOS and a Optilab T‐rEX detectors (Wyatt, Santa Barbara, CA). SEC‐MALS data was processed in the Astra (Wyatt) software. Reported molecular weights are average values over the length of the peak.
ITC
ITC was performed on a VP‐ITC MicroCal colorimeter (Malvern Instruments, Worcestershire, United Kingdom) at 20°C. Proteins were dialyzed into 25 mM sodium phosphate pH 7.0, 150 mM NaCl buffer overnight before use. For TNKS1‐TNKS1 SAM binding, TNKS1 SAM‐Linker (D1055A/V1056K) was concentrated to 400 μM (titrant) and TNKS1 SAM‐Linker (Y1073E) was used at 20 μM (titrand). For TNKS1‐TNKS2 binding, TNKS1 SAM‐Linker (Y1073E) protein was concentrated to 400 µM (titrant) and the TNKS2 SAM‐Linker (D902A/V903K) mutant (residues 871–952) was concentrated to 20 µM (titrand). Proteins were degassed before use. Titrant was added in 5 µL injections with a delay between each addition of 300 s. A total of 40 injections were performed for each titration. Data was analyzed with the Origin 7.0 software (OriginLab Corp, Wellesley Hills, MA); curves were fit to a 1‐site model.
NMR spectroscopy
NMR data was collected on a Bruker Avance 600 mHz spectrometer fitted with a TCI CryoProbe (Bruker, Billerica, MA). All NMR data sets were obtained at 25°C in 25 mM sodium phosphate pH 7.0, 150 mM NaCl. A standard set of triple resonance NMR experiments (CBCACONH, HNCACB, HNCOCA, HNCA)52, 53 were used for assignments of the TNKS1 SAM‐Linker(D1055A/V1056K) protein (assigned residues: 1,025–1,099) were acquired with 250 µM 15N‐13C‐labeled TNKS1 SAM‐Linker (D1055A/V1056K), in the absence and presence of 322 µM unlabeled TNKS1 SAM‐Linker (Y1073E) (bound spectrum). For the bound spectrum, assignments were guided by the unbound spectrum and were confirmed by triple resonance experiments. Assignments can be found in the Supporting Information. 15N‐HSQC experiments were performed with 150 µM 15N‐labeled TNKS1 SAM‐Linker (D1055A/V1056K) in the absence or presence of 38, 75, 94, and 165 µM unlabeled TNKS1 SAM‐Linker (Y1073E). All NMR data was processed with NMRPipe.54 Peak intensity, chemical shift analysis, and NMR assignments were determined using NMRViewJ.55 Predicted chemical shifts for the disordered sequence of TNKS1 SAM‐Linker(D1055A/V1056K) were generated using a SBiNLab java script coded by Alex Maltsev using published calculations.56, 57, 58 Deviations from predicted intrinsically disordered chemical shifts were calculated by the formula Δδ13Cα‐Δδ13Cβ [ppm], where Δδ13Cα and Δδ13Cβ are the difference between the disordered values and experimental values for Cα chemical shifts and Cβ chemical shifts, respectively. Chemical shift perturbations were determined using the formula Δδj = [(15NΔδj/5)2 + (1HΔδj)2]1/2 where 15NΔδj and 1HΔδj are the difference in chemical shift between the bound and unbound states of TNKS1 SAM‐Linker(D1055A/V1056K), respectively. The top 15% most perturbed residues were mapped onto the surface/cartoon of two SAM domain protomers within the oligomeric model of TNKS1.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting Information
Acknowledgments
The authors thank David Baker for computational resources used to generate our model.
References
- 1. Huang S‐MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620. [DOI] [PubMed] [Google Scholar]
- 2. Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, Cong F, Simoncic PD, Ueki Y, La Rose J, Rottapel R (2011) Loss of Tankyrase‐mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147:1324–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang W, Li N, Li X, Tran MK, Han X, Chen J (2015) Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep 13:524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith S, De Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10:1299–1302. [DOI] [PubMed] [Google Scholar]
- 5. Yeh T‐YJ, Sbodio JI, Tsun ZY, Luo B, Chi NW (2007) Insulin‐stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem J 402:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo HL, Zhang C, Liu Q, Li Q, Lian G, Wu D, Li X, Zhang W, Shen Y, Ye Z, Lin S‐Y, Lin S‐C (2012) The Axin/TNKS complex interacts with KIF3A and is required for insulin‐stimulated GLUT4 translocation. Cell Res 22:1246–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dynek JN, Smith S (2004) Resolution of sister telomere association is required for progression through mitosis. Science 304:97–100. [DOI] [PubMed] [Google Scholar]
- 8. Chang P, Coughlin M, Mitchison TJ (2005) Tankyrase‐1 polymerization of poly(ADP‐ribose) is required for spindle structure and function. Nat Cell Biol 7:1133–1139. [DOI] [PubMed] [Google Scholar]
- 9. Kim MK, Dudognon C, Smith S (2012) Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep 13:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozaki Y, Matsui H, Asou H, Nagamachi A, Aki D, Honda H, Yasunaga S, Takihara Y, Yamamoto T, Izumi S, Ohsugi M, Inaba T (2012) Poly‐ADP ribosylation of Miki by tankyrase‐1 promotes centrosome maturation. Mol Cell 47:694–706. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud G. a, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang S‐MA, Cong F (2011) RNF146 is a poly(ADP‐ribose)‐directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13:623–629. [DOI] [PubMed] [Google Scholar]
- 12. Callow MG, Tran H, Phu L, Lau T, Lee J, Sandoval WN, Liu PS, Bheddah S, Tao J, Lill JR, Hongo J‐A, David D, Kirkpatrick DS, Polakis P, Costa M (2011) Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One 6:e22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W (2014) Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP‐ribosyl)ation signal. Nature 517:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, Wang W, Songyang Z, Lin C, Yang L, Yu Y, Chen J (2015) Poly‐ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev 29:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haikarainen T, Krauss S, Lehtio L (2014) Tankyrases: structure, function, and therapeutic implications in cancer. Curr Pharm Des 20:6472–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehtio L, Chi NW, Krauss S (2013) Tankyrases as drug targets. FEBS J 280:3576–3593. [DOI] [PubMed] [Google Scholar]
- 17. Riffell JL, Lord CJ, Ashworth A (2012) Tankyrase‐targeted therapeutics: expanding opportunities in the PARP family. Nat Rev Drug Discov 11:923–936. [DOI] [PubMed] [Google Scholar]
- 18. Hatsugai K, Ohishi T, Sugimoto Y, Seimiya H (2010) Tankyrase‐1 assembly to large protein complexes blocks its telomeric function. FEBS Lett 584:3885–3890. [DOI] [PubMed] [Google Scholar]
- 19. Langelier MF, Planck JL, Roy S, Pascal JM (2012) Structural basis for DNA damage‐dependent poly(ADP‐ribosyl)ation by human PARP‐1. Science 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiao F, Bowie JU (2005) The many faces of SAM. Sci Signal 2005:re7–re7. [DOI] [PubMed] [Google Scholar]
- 21. Kim CA, Bowie JU (2003) SAM domains: uniform structure, diversity of function. Trends Biochem Sci 28:625–628. [DOI] [PubMed] [Google Scholar]
- 22. Johnson PE, Donaldson LW (2006) RNA recognition by the Vts1p SAM domain. Nat Struct Mol Biol 13:177–178. [DOI] [PubMed] [Google Scholar]
- 23. Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FHT (2006) Shape‐specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol 13:160–167. [DOI] [PubMed] [Google Scholar]
- 24. Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA (2003) The RNA‐binding SAM domain of Smaug defines a new family of post‐transcriptional regulators. Nat Struct Mol Biol 10:614–621. [DOI] [PubMed] [Google Scholar]
- 25. Bhunia A, Domadia PN, Mohanram H, Bhattacharjya S (2009) NMR structural studies of the Ste11 SAM domain in the dodecyl phosphocholine micelle. Proteins 74:328–343. [DOI] [PubMed] [Google Scholar]
- 26. Stapleton D, Balan I, Pawson T, Sicheri F (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Mol Biol 6:44–49. [DOI] [PubMed] [Google Scholar]
- 27. Mercurio FA, Marasco D, Pirone L, Pedone EM, Pellecchia M, Leone M (2012) Solution structure of the first Sam domain of Odin and binding studies with the EphA2 receptor. Biochemistry 51:2136–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leone M, Cellitti J, Pellecchia M (2008) NMR studies of a geterotypic Sam‐Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor. Biochemistry 47:12721–12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwan JJ, Warner N, Pawson T, Donaldson LW (2004) The solution structure of the S. cerevisiae Ste11 MAPKKK SAM domain and its partnership with Ste50. J Mol Biol 342:681–693. [DOI] [PubMed] [Google Scholar]
- 30. Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU (2001) Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J 20:4173–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim CA, Gingery M, Pilpa RM, Bowie JU (2002) The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol 9:453–457. [DOI] [PubMed] [Google Scholar]
- 32. Kim CA, Sawaya MR, Cascio D, Kim W, Bowie JU (2005) Structural organization of a sex‐comb‐on‐midleg/polyhomeotic copolymer. J Biol Chem 280:27769–27775. [DOI] [PubMed] [Google Scholar]
- 33. Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU (2006) An architectural framework that may lie at the core of the postsynaptic density. Science 311:531–535. [DOI] [PubMed] [Google Scholar]
- 34. Harada BT, Knight MJ, Imai S, Qiao F, Ramachander R, Sawaya MR, Gingery M, Sakane F, Bowie JU (2008) Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase delta1. Structure 16:380–387. [DOI] [PubMed] [Google Scholar]
- 35. Leettola CN, Knight MJ, Cascio D, Hoffman S, Bowie JU (2014) Characterization of the SAM domain of the PKD‐related protein ANKS6 and its interaction with ANKS3. BMC Struct Biol 14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sayou C, Nanao MH, Jamin M, Posé D, Thévenon E, Grégoire L, Tichtinsky G, Denay G, Ott F, Peirats Llobet M, Schmid M, Dumas R, Parcy F (2016) A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun 7:11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nanyes DR, Junco SE, Taylor AB, Robinson AK, Patterson NL, Shivarajpur A, Halloran J, Hale SM, Kaur Y, Hart PJ (2014) Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif. Proteins 82:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Rycker M, Price CM (2004) Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP‐ribose) polymerase domains. Mol Cell Biol 24:9802–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Rycker M, Venkatesan RN, Wei C, Price CM (2003) Vertebrate tankyrase domain structure and sterile alpha motif (SAM)‐mediated multimerization. Biochem J 372:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thanos CD, Goodwill KE, Bowie JU (1999) Oligomeric structure of the human EphB2 receptor SAM domain. Science 283:833–836. [DOI] [PubMed] [Google Scholar]
- 41. Meruelo AD, Bowie JU (2009) Identifying polymer‐forming SAM domains. Proteins 74:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sbodio JI, Lodish HF, Chi NW (2002) Tankyrase‐2 oligomerizes with tankyrase‐1 and binds to both TRF1 (telomere‐repeat‐binding factor 1) and IRAP (insulin‐responsive aminopeptidase). Biochem J 361:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morrone S, Cheng Z, Moon RT, Cong F, Xu W (2012) Crystal structure of a Tankyrase‐Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA 109:1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knight MJ, Leettola C, Gingery M, Li H, Bowie JU (2011) A human sterile alpha motif domain polymerizome. Protein Sci 20:1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D (1999) Improved recognition of native‐like protein structures using a combination of sequence‐dependent and sequence‐independent features of proteins. Proteins 34:82–95. [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y, Skolnick J (2005) TM‐align: a protein structure alignment algorithm based on the TM‐score. Nucleic Acids Res 33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stafford RL, Hinde E, Knight MJ, Pennella MA, Ear J, Digman MA, Gratton E, Bowie JU (2011) Tandem SAM domain structure of human Caskin1: a presynaptic, self‐assembling scaffold for CASK. Structure 19:1826–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei Z, Zheng S, Spangler SA, Yu C, Hoogenraad CC, Zhang M (2011) Liprin‐mediated large signaling complex organization revealed by the liprin‐α/CASK and liprin‐α/Liprin‐β complex structures. Mol Cell 43:586–598. [DOI] [PubMed] [Google Scholar]
- 49. Song Y, DiMaio F, Wang RY‐R, Kim D, Miles C, Brunette TJ, Thompson J, Baker D (2013) High‐resolution comparative modeling with RosettaCM. Structure 21:1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Narwal M, Koivunen J, Haikarainen T, Obaji E, Legala OE, Venkannagari H, Joensuu P, Pihlajaniemi T, Lehtiö L (2013) Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity. J Med Chem 56:7880–7889. [DOI] [PubMed] [Google Scholar]
- 51. Narwal M, Haikarainen T, Fallarero A, Vuorela PM, Lehtiö L (2013) Screening and structural analysis of flavones inhibiting tankyrases. J Med Chem 56:3507–3517. [DOI] [PubMed] [Google Scholar]
- 52. Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution. Prog Nucl Magn Reson Spectrosc 34:93–158. [Google Scholar]
- 53. Muhandiram DR, Kay LE (1994) Gradient‐enhanced triple‐resonance three‐dimensional NMR experiments with improved sensitivity. J Magn Reson Ser B 103:203–216. [Google Scholar]
- 54. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 55. Johnson BA, Blevins RA (1994) NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614. [DOI] [PubMed] [Google Scholar]
- 56. Kjaergaard M, Poulsen FM (2011) Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR 50:157–165. [DOI] [PubMed] [Google Scholar]
- 57. Kjaergaard M, Brander S, Poulsen FM (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149. [DOI] [PubMed] [Google Scholar]
- 58. Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ (2001) Sequence‐dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


