Abstract
Cysteine residues ubiquitously stabilize tertiary and quaternary protein structure by formation of disulfide bridges. Here we investigate another linking interaction that involves sulfhydryl groups of cysteines, namely intra‐ and intermolecular methylene‐bridges between cysteine and lysine residues. A number of crystal structures possessing such a linkage were identified in the Protein Data Bank. Inspection of the electron density maps and re‐refinement of the nominated structures unequivocally confirmed the presence of Lys‐CH2‐Cys bonds in several cases.
Keywords: cysteine, lysine, crosslink, methylene bridge, intermolecular, intramolecular
Due to the presence of nucleophilic sulfhydryl group, cysteine is a very reactive amino acid. Formation of disulfide bridges, between two cysteine residues, has been long known as the post‐translational, fold‐stabilizing modification of proteins. But are there any other proteinogenic amino acids that can react with cysteines to create intra‐ or intermolecular crosslinks? So far, there have been no reports describing other cysteine‐involving crosslinks in protein structures, whereas, theoretically, a functional group with at least a partial, local positive charge should be prone to nucleophilic attack by the sulfhydryl group.
Recently, we have discovered methylene bridging between cysteine and lysine residues in Medicago truncatula histidinol‐phosphate phosphatase, MtHPP.1 In that case, two intermolecular methylene bridges formed between Cys245 and Lys158 residues of two monomers within a symmetric dimer, apparently stabilizing the quaternary structure. Formation of such crosslinks was observed in the presence of either carbon dioxide or formaldehyde. It is also of note that the bond was present (at ∼60% occupancy) in the crystal structures even if neither of these two agents was intentionally added to the sample, except for the carbon dioxide naturally present in the atmosphere (below 0.4%).
We have searched the Protein Data Bank, PDB,2 released on April 21, 2015, to find further examples of such Lys‐CH2‐Cys bonds. The data‐mining was performed with the program CONTACT, from the CCP4 package.3 The following input settings were applied: resolution limit 2.0 Å, maximal Sγ‐Nζ distance 3.0 Å, and search between symmetry mates was allowed. Among the 96062 crystal structures in total (including non‐protein), 46,260 entries satisfied the resolution limit. Unique Sγ‐Nζ pairs within 3.0 Å distance, excluding multiple instances in different protein subunits, were found in 99 PDB entries (132, including different chains in the same entries). For 15 of these, the structure factors were unavailable, and in such cases we were unable to verify the existence of crosslinks. The electron density maps of remaining 84 candidate structures were examined for the presence of positive peaks suggesting methylene groups. The promising hits were re‐refined in REFMAC4 with and without the bridging methylene. To model the methylene bridges for refinement, the original Lys residues were replaced with N‐monomethylated lysines (ligand ID: MLZ) to add the CM atoms. The LINK records in .pdb files were added to disable anti‐bumping for Sγ…CM distances, which were additionally restrained to 1.82 Å. Refinements (10 cycles) of the methylene‐bridges containing structures was carried out as for the original structures.
The visual inspection of the electron density maps (Fig. 1) from refined structures showed methylene bridges in 14 unique PDB entries (Table 1), with twenty Lys‐CH2‐Cys crosslinks in total, in addition to MtHPP (PDB ID: 5eqa ). In most cases, the bonds have intramolecular character, and only in 4v3l and MtHPP, the crosslink bridges connect different protein subunits. Interestingly, even though the electron density maps clearly indicate the presence of —CH2— group, it was neglected in all cases, except for MtHPP. It is also of note that an analogous, unintended, carbon‐mediated junction of Cys Sγ with a primary amine was observed in case of N‐carbamoylputrescine amidohydrolase from M. truncatula.5 In this example, the amine group was not a part of a lysine residue but of 1,4‐diaminobutane (putrescine) which apparently reacted with carbon dioxide, absorbed by the crystallization solution from air. The process was however stopped at the geminal diol intermediate, stabilized by the protein oxyanion hole, observed in the structure of PDB ID: 5h8i.
Figure 1.
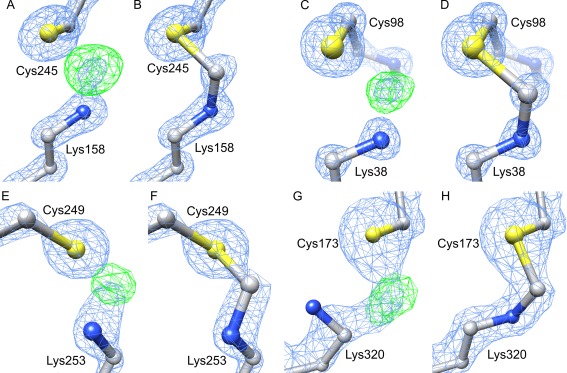
Four clear examples of the presence of Lys‐CH2‐Cys crosslinks in the PDB. A, B: MtHPP structure (PDB ID: 5eqa) original and with the bridging methylene group, respectively. The remaining panels are organized in the same manner and present: (C,D) 1es5 structure; (E, F), 2xhi; (G, H), 3czg. In (A–F) the 2F o‐F c maps (blue) are contoured at 3σ, F o‐F c maps (green) at 7σ, whereas the respective levels in panels (G,H) are 2σ and 6σ.
Table 1.
Clear Examples of Lys‐CH2‐Cys Bonds From the PDB Sorted by Difference Map Peak heights
| PDB ID | Data resolution (Å) | Nζ (chain number) | Sγ (chain number) | Nζ… Sγ distance in PDB (Å) | Peak height(s) [σ]; (other chains) |
|---|---|---|---|---|---|
| 5eqa | 1.32 | A 158 | B 245 | 2.8 | 16.9 |
| 1es5 | 1.12a | A 38 | A 98 | 2.9 | 11.2 |
| 3u7z | 1.3 | B 128 | B 100 | 2.7 | 9.5; (A) 7.1 |
| 2xhi | 1.55 | A 253 | A 249 | 2.6 | 9.4 |
| 2y8k | 1.47 | A 106 | A 95 | 2.8 | 9.4 |
| 3czg | 1.8 | A 320 | A 173 | 2.9 | 9.1 |
| 1es2 | 1.50b | A 38 | A 98 | 2.9 | 7.8 |
| 2wpg | 1.9 | A 321 | A 174 | 2.8 | 7.3 |
| 1uxb | 1.75 | C 295 | C 333 | 2.8 | 6.2; (B) 5.5; (A) 3.4 |
| 4ndb | 2.0 | A 41 | A 24 | 2.8 | 5.8; (B) 5.7 |
| 1m3q | 1.9 | A 249 | A 253 | 2.8 | 5.4 |
| 3k9d | 2.0 | C 392 | C 359 | 2.9 | 5.3; (A) 5; (B) 3.6 |
| 4v3l | 1.53 | A 8 | C 418 | 2.9 | 5.1 |
| 2py5 | 1.6 | A 114 | A 106 | 2.6 | 5.1 |
| 3cze | 1.9 | A 320 | A 173 | 2.9 | 5.1 |
Underlined PDB IDs indicate that the structure is shown in the figure, whereas those in bold represent intermolecular crosslinks.
Reported data resolution is 1.4 Å.
Reported data resolution is 1.55 Å.
The exact mechanism of Lys‐CH2‐Cys crosslinking is quite perplexing, although its initial steps are relatively simple to deduce: the Lys Nζ atoms may react with carbon dioxide6 to form carbamic acids, with a partial positive charge on carbamic C atom. The latter is susceptible to a nucleophilic attack by Cys Sγ. Further dehydration should result in S‐alkyl thiocarbamate bridge, and it is unknown why only a methylene is observed between the Lys Nζ and Cys Sγ atoms in the crystal structures. We may only assume the role of reducing agents, commonly used during protein crystallization and/or the reducing power of synchrotron radiation.
The situation is much clearer when formaldehyde, a different, biologically relevant crosslinking agent is considered. Lys, coupled with HCHO in the presence of H+, can be dehydrated to form iminium cation, as in the initial steps of Mannich reaction.7 Iminium cations are very prone to react with nucleophilic Cys residues and form methylene bridges.8
We hope that this article will inspire researchers to investigate the presence of Lys‐CH2‐Cys links in nature. At this point it is difficult to predict the putative biological significance of Lys‐CH2‐Cys crosslinks, although at least a few conceivable scenarios come to mind. First, such bonds may stabilize protein fold, as do disulfide bridges. Second, they may be linked with formaldehyde scavenging and aid or supplement other natural ways used to neutralize this toxic compound. Third, Lys‐CH2‐Cys linking might impact protein half‐life or be related to modulation of enzyme activity if it involved active site residues. Unquestionably, the presence of methylene bridges in the crystal structures may simply reflect artifacts linked to the nature of long‐term crystallization experiments. Nonetheless, we believe that if Lys‐CH2‐Cys crosslinks are as clear as in the presented examples they should be included in protein model building and refinement.
Acknowledgement
Authors are grateful for fruitful discussions with Dr. Jacek Kolanowski and Dr. Maura Malinska.
References
- 1. Ruszkowski M, Dauter Z (2016) Structural studies of Medicago truncatula histidinol‐phosphate phosphatase from inositol monophosphatase superfamily reveal details of penultimate step of histidine biosynthesis in plants. J Biol Chem 291:9960–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Cryst D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst D 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sekula B, Ruszkowski M, Malinska M, Dauter Z (2016) Structural investigations of N‐carbamoylputrescine amidohydrolase from Medicago truncatula: Insights into the ultimate step of putrescine biosynthesis in plants. Front Plant Sci 7:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Penny DE, Ritter TJ (1983) Kinetic‐study of the reaction between carbon‐dioxide and primary amines. JCS‐Faraday Trans I 79:2103–2109. [Google Scholar]
- 7. Mannich C, Krösche W (1912) Ueber ein Kondensationsprodukt aus Formaldehyd, Ammoniak und Antipyrin. Archiv Pharmaz 250:647–667. [Google Scholar]
- 8. Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W (2004) Identification of formaldehyde‐induced modifications in proteins: reactions with model peptides. J Biol Chem 279:6235–6243. [DOI] [PubMed] [Google Scholar]


