Abstract
Here we investigated the relationship between local bacterial colonization and anti-bacterial immune responses in pre-school asthmatic and control children within the EU-wide study PreDicta. In this cohort of pre-school asthmatic children, nasopharyngeal colonization with Gram-negative bacteria such as Haemophilus influenzae and Moraxella catarrhalis was found to be associated with the highest interferon beta (IFNβ) and IL-33 levels in the nasal pharyngeal fluids (NPF). IL33R-ST2 was found induced in the blood of asthmatic children with additional Gram + bacteria in the nasopharynx (Gr+/−). Furthermore, asthmatic children had more episodes of infection that required antibiotic therapy than the control group. Treatment with antibiotics associated with reduced ST2 in blood cells of both asthmatic and control children and reduced IL-33 levels in the airways of asthmatic children. In the absence of Staphylococcus (S.) aureus in NPF, antibiotic therapy associated with decreased IL-33 levels in the NPF and lower ST2 values in the blood of control children but not of asthmatic children. These data suggest that, in asthmatic children, Gram- bacteria, which persist after antibiotic therapy, contributes to IL-33 locally and associated with Gr + bacteria colonization in the airways, inhibited IFN-β and in the absence of Staphylococcus (S.) aureus, induced ST2 bearing cells in their blood.
Bacterial infections are known to trigger asthma exacerbations1,2,3. It is also known that recurrent viral and bacterial infections during childhood can promote the susceptibility to allergy and asthma4. The microbial milieu has been shown to influence the priming of the immune system already in utero as well as in early childhood5,6,7,8. However, we do not know yet which immune responses are present in combination with distinct bacteria colonization in pediatric asthma.
Interferon beta (IFNβ) is a protein that is induced by both viral and non-viral pathogens and best known for its strong antiviral, antibacterial and immunoregulatory effects9,10,11. IL-33 is a cytokine known to be released by damaged epithelial cells where infectious agents and/or allergens can spread through blood and into tissues. It is a cytokine of the innate immune response and activates innate lymphoid cells type 2 (ILC2) and Th2 cells to produce IL-5 and IL-13 after binding to its receptor suppression of tumorigenicity (ST2), also termed IL-33 receptor (IL-33R). These Th2 cytokines were found to be increased in the nasopharynx of asthmatic patients12. Moreover, recently IFN-β has been shown to inhibit ILC2 cells which carry the type I IFN-R13.
The relationship between the bacterial colonization and the pathogenesis of allergic asthma has become a field of intense research2,14,15. Recently, it has been found that the lower respiratory tract, which previously was assumed to be sterile, is colonized by several microorganisms16. The microbiota in the airways of asthmatic subjects showed a higher bacterial diversity compared with the one of healthy subjects16,17.
At the moment there is increasing evidence that the colonization of the airways with microorganisms can trigger the onset and the development of asthma, but can also have protective effects. In this study we analyzed two cohorts of pre-school children one with and the second without asthma recruited within the Europe-wide study PreDicta (Post-infectious immune reprogramming and its association with persistence and chronicity of respiratory allergic diseases) and addressed the question whether the presence of distinct bacteria in their nasal pharyngeal fluids (NPF) as well as previous antibiotic therapy were associated with different antibacterial immune responses such as IL-33 and IFNβ production in their NPF and IL-33R/ST2 in the blood.
Results
Differences in bacterial nasopharyngeal colonization of pre-school children with asthma compared to healthy children
The demographic and clinical data of the pre-school children analyzed in this study are reported in Table 1. To investigate the relationship between bacterial nasopharyngeal colonization and the immune responses, we first looked for the common bacterial colonization of the nasopharynx, as well as for Gram positive and Gram negative bacteria, which might cause airway infections, especially in pre-school age in two cohorts of pre-school children with and without asthma.
Table 1. Demographic and clinical data of the cohorts of WP1-UK-ER*.
| Control Children | Asthma Children | |
|---|---|---|
| Number of subjects | 21 | 24 |
| Gender | 12 (M) = 57% 9 (F) = 42,8% | 15 (M) = 62,5% 9 (F) = 37,5% |
| Age [years] | 4,7 ± 0,18 | 4,9 ± 0,1 |
| Skin Prick Test positive | 25% (2/8) | 77% (17/22)*** |
| Average age at onset of symptoms [years] | — | 2,2 |
| **FEV1 < 100% | 23,8% (5/21) | 54% (13/24) |
| Allergic Rhinitis | 0 | 33% (8/24) |
| Atopic eczema | 4,76% (1/21) | 4% (1/24) |
| Allergic rhinitis and atopic eczema | 0 | 38% (9/24) |
| without allergic co-morbidity | 95% (20/21) | 25% (6/24) |
| family predisposition to asthma | 14% (3/21 maternal) | 38% (9/24) |
| family predisposition to atopy | 76% (16/21) | 83% (20/24) |
| both parents with atopy | 23,8% (5/21) | 46% (11/24) |
| without family predisposition | 23% (5/21) | 17% (4/24) |
*WP1-UK-ER = Work Package 1 within the PreDicta Study at the Universitätsklinikum Erlangen and the University of Erlangen-Nürnberg; **FEV1 = Forced Expiratory Volume in 1 second; ***2 children: skin prick test not done.
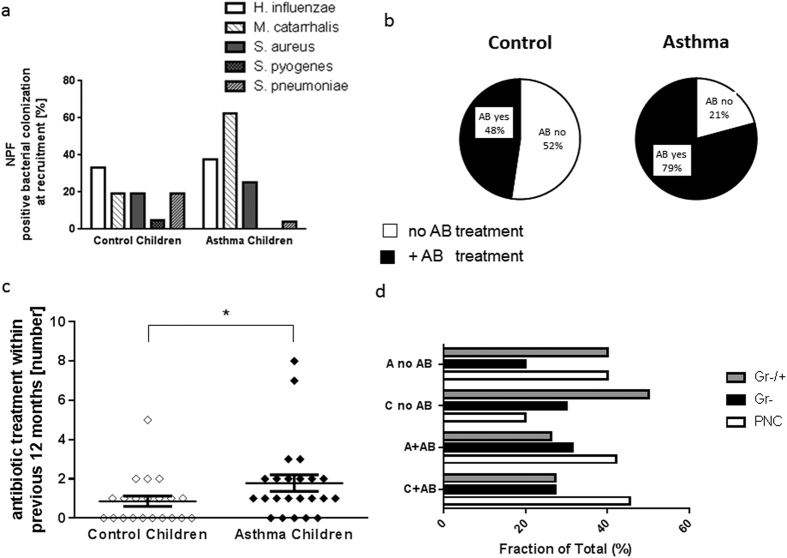
Here we found a prevalence of five facultative pathogen bacteria that are known to be associated with asthma development (Fig. 1a).
Figure 1. More frequent antibiotic treatment in children with asthma (A) within 12 months before recruitment.
(a) Percentage of colonization of the five main bacteria Haemophilus influenza (H. influenzae: Gram-), Moraxella catarrhalis (M. catarrhalis: Gram-), Staphylococcus aureus (S. aureus: Gram+), Streptococcus pyogenes (S. pyogenes: Gram+) and pneumoniae (S. pneumonia: Gram+). NPF: Nasalpharyngeal fluid. (b,c). More frequent antibiotic treatment in children with asthma (A) within 12 months before recruitment (d) Higher percentage of colonization with diverse Gram positive and Gram negative bacteria in children without antibiotic therapy. (a: Control (C): n = 21; Asthma (A): n = 24 ; b: C: n = 10,11; A: n = 19, 5 ; c: n = 21, 24, p = 0,037; d: beginning from the top A no AB: n = 2,1,2,8,3,2,5,6,8,3,3,5).
In order to start analysing the influence of different bacterial colonization on asthma, we subdivided both cohorts in accordance to their bacterial nasopharyngeal colonization: Children that had saprophytic germs and bacteria that are physiological in the nasopharyngeal microbiome (PNC), or children that had additional or exclusively Gram negative bacteria in their nasopharyngeal fluid (Gram−). The Gram negative respiratory bacteria for which the nasopharyngeal fluid was analysed are Haemophilus influenzae and Moraxella catarrhalis with or without physiological flora. The third subgroup consists of children who have additional to physiological and/or Gram negative bacterial colonization also special Gram positive bacteria in their nasopharynx (Gram−/+). These Gram positive bacteria are Staphylococcus aureus and Streptococcus species.
As shown in Fig. 1a, the percentage of special bacterial colonization was increased in the asthma group as compared to the control group at the recruitment (B0). Moraxella catarrhalis was detected more frequently in asthmatics as compared to control children.
Increased frequency of antibiotic therapy in asthmatic children as compared to healthy controls
As antibiotic treatment has a profound influence on the bacterial flora, we first established for every child the number of antibiotic courses received during the past twelve months before recruitment (Table 2 and Fig. 1b).
Table 2. Antibiotic treatment and bacterial nasopharyngeal colonization of the cohorts of WP1-UK-ER.
| Control Children | Asthma Children | |
|---|---|---|
| >1 antibiotic treatment within last 12 months before recruitment | 19% (4/21) | 42% (10/24) |
| ≥3 antibiotic treatments within last 12 months before recruitment* | 9,5% (2/21) | 17% (4/24) |
| antibiotic treatment in children with S. aureus | 0% (0/3) | 83% (5/6) |
| antibiotic treatment in children with only H. influenza and/or M. catarrhalis | 67% (2/3) | 100% (5/5) |
*Most frequently used antibiotics by pediatricians: Control Children: cefaclor, amoxicillin; Asthma Children: clarithromycin, erythromycin, cefpodoxime (justified by the diagnosis: pneumonia).
While half of the control children did not receive antibiotic treatment during infections in the previous twelve months, only 21% of children with asthma were not treated with an antibiotic therapy (Fig. 1b). As a result, asthmatic children received more often antibiotic therapy than control children (Fig. 1c). The more striking increase in antibiotic treatment was observed in asthmatic children with S. aureus detected in their NPF (3.3 fold induction) as compared to the control group (Table 2). Moreover, 84% of the asthmatic children with antibiotic treatment were also under steroid therapy (Table 3). Finally, we analyzed the fraction of children with different bacteria flora without antibiotic treatment (no AB) and with antibiotic treatment (+AB) (Fig. 1d). Here we observed a reducing effect of antibiotic on the frequency of both asthmatic and control children with a mixed bacterial Gram positive and Gram negative colonization in NPF. Moreover, antibiotics did not decrease the percentage of children with Gram negative bacterial colonization in the NPF. Table 4 shows antibiotic treatment of control children at recruitment and Table 5 contains the relevant bacterial and medication data of the study children at the end of the study course (F4: 24 month follow-up visit).
Table 3. Treatment of asthmatic children at the time of recruitment of the cohorts of WP1-UK-ER.
| Asthma children | Gram + bacteria: S. aureus | Gram- bacteria: H. influenzae and M. catarrhalis | Number of antibiotic courses within previous 12 months before recruitment | Antibiotic treatment [days] | Antibiotic treatment [component] | Allergic rhinitis | Inhalative treatment | Oral and nasal treatment | Steroid treatment |
|---|---|---|---|---|---|---|---|---|---|
| 201 | Yes | Yes | 3 | 10 | No data | Yes | Fluticasone, Salmeterol 1–0–1 | Montelukast 5 mg/d | Yes |
| 202 | No | No | 1 | 10 | Ciprofloxacin | Yes | Fluticason 1–0–1 | Mometason (nasal) | Yes |
| 203 | Yes | Yes | 1 | No data | No data | Yes | Budesonid 0,5 mg 1–0–1 | Cetirizin | Yes |
| 204 | Yes | No | 0 | 0 | — | Yes | Fluticasone, Salmeterol 1–0–1 | — | Yes |
| 205 | No | Yes | 0 | 0 | — | No | Fluticasone, Salmeterol 1–0–1 | — | Yes |
| 206 | No | No | 1 | 7–10 | No data | No | Beclomethasone | Yes | |
| 207 | No | No | 0 | 0 | — | No | Fluticasone, Salmeterol 1–0–1 | — | Yes |
| 209 | No | Yes | 1 | 7 | No data | yes | Budesonide | — | Yes |
| 210 | Yes | Yes | 2 | 7 | Clarithromycin, Cefpodoxime | yes | Salbutamol | — | No |
| 212 | No | No | 8 | No data | No data | No | Fluticasone, Salmeterol 1–0–1 | — | Yes |
| 213 | No | No | 1 | 10 | No data | No | Fluticasone, Salmeterol 1–0–1 | — | Yes |
| 216 | No | No | 2 | No data | No data | Yes | Fluticasone, Salmeterol | — | Yes |
| 217 | Yes | Yes | 1 | 7 | No data | Yes | Betamethasone 0,1 mg, 1–0–1 | — | Yes |
| 223 | No | No | 2 | 10 | No data | Yes | Budesonide, 2 × 0,2 mg | Montelukast 5 mg | Yes |
| 224 | No | No | 1 | 5 | No data | Yes | Fluticasone 1–0–1 | — | Yes |
| 225 | No | Yes | 2 | 10 | No data | No | Fluticasone 1–0–1 | — | Yes |
| 228 | No | No | 0 | 0 | — | Yes | Rescue treatment: Salbutamol | — | No |
| 229 | No | Yes | 1 | 7–10 | Erythromycin, Amoxicillin | Yes | Salbutamol 1x/d | — | No |
| 230 | No | No | 2 | 14 | No data | Yes | Rescue treatment: Salbutamol | — | No |
| 231 | No | Yes | 1 | 4 | No data | Yes | Budesonide 0,5 mg, Salbutamol | — | Yes |
| 238 | No | Yes | 7 | 7 | No data | Yes | Salmeterol, Fluticasone | — | Yes |
| 239 | No | Yes | 0 | 0 | — | Yes | Rescue treatment | Cetirizin | No |
| 242 | Yes | Yes | 3 | 7–10 | No data | Yes | Budesonide 0,5 mg | Cetirizin | Yes |
| 243 | No | Yes | 2 | 10 | Erythromycin | No | Salbutamol, Budesonid 0,5 mg | — | Yes |
Table 4. Treatment of control children at the time of recruitment of the cohorts of WP1-UK-ER.
| Control Children | Gram + bacteria: S. aureus | Gram- bacteria: H. influenzae and M. catarrhalis | Number of antibiotic courses within previous 12 months before B0 | Antibiotic treatment [days] | Antibiotic treatment: Component |
|---|---|---|---|---|---|
| 208 | No | No | 2 | 7 | No data |
| 211 | No | Yes | 1 | 28 | amoxicillin + clavulanic acid |
| 214 | No | No | 0 | 0 | — |
| 215 | No | No | 0 | 0 | — |
| 218 | No | Yes | 1 | 5 | No data |
| 219 | No | Yes | 0 | 0 | — |
| 220 | No | No | 0 | 0 | — |
| 221 | No | Yes | 1 | 7 | Cefaclor |
| 222 | No | No | 0 | 0 | Cefaclor |
| 226 | No | No | 1 | 10 | — |
| 227 | Yes | No | 0 | 0 | — |
| 232 | Yes | Yes | 0 | 0 | — |
| 233 | No | No | 5 | 10 | phenoxymethylpenicillin, erythromycin, cefaclor |
| 234 | No | Yes | 0 | 0 | — |
| 235 | No | Yes | 0 | 0 | — |
| 236 | No | Yes | 1 | 10 | amoxicillin |
| 237 | No | No | 4 | 7 | amoxicillin |
| 240 | No | Yes | 2 | 7 | Cefaclor |
| 241 | No | No | 1 | 10 | No data |
| 245 | Yes | No | 0 | 0 | — |
Table 5. Bacterial nasopharyngeal colonization, antibiotic and asthma medication at the 24 months follow-up visit of the cohorts WP1-UK-ER.
| Child number | Asthma (A)/Control (C) | Gram + bacteria: S. aureus | Gram- bacteria: H. influenzae and or M. catarrhalis | subgrouped in: | Days of antibiotic treatment in the previous 12 months | Component of antibiotic treatment | Asthma medication |
|---|---|---|---|---|---|---|---|
| 214 | C | No | Yes | Gr- | 7 | no data | no medication |
| 215 | C | Yes | Yes | Gr−/+ | 7 | no data | no medication |
| 220 | C | Yes | No | Gr−/+ | 0 | no medication | |
| 221 | C | Yes | No | Gr−/+ | 0 | no medication | |
| 222 | C | Yes | No | Gr−/+ | 0 | no medication | |
| 226 | C | No | No | PNC | 0 | no medication | |
| 227 | C | No | Yes | Gr- | 0 | no medication | |
| 201 | A | Yes | Yes | Gr−/+ | 0 | steroid | |
| 202 | A | No | Yes | Gr−/+ | 6 | no data | steroid |
| 204 | A | No | No | PNC | 6 | Amoxicillin, Cefaclor | no medication |
| 205 | A | No | Yes | Gr- | 0 | no medication | |
| 210 | A | No | No | PNC | 0 | no medication | |
| 212 | A | No | Yes | Gr- | 8 | no data | steroid |
| 216 | A | No | No | PNC | 3 | no data | steroid |
| 217 | A | Yes | No | Gr−/+ | 10 | infectocillin | non-steroid |
| 223 | A | No | No | PNC | 0 | steroid |
Higher IFNβ production in nasopharyngeal fluid of children with asthma and a Gram negative (Gr-) bacterial colonization in the nasopharynx
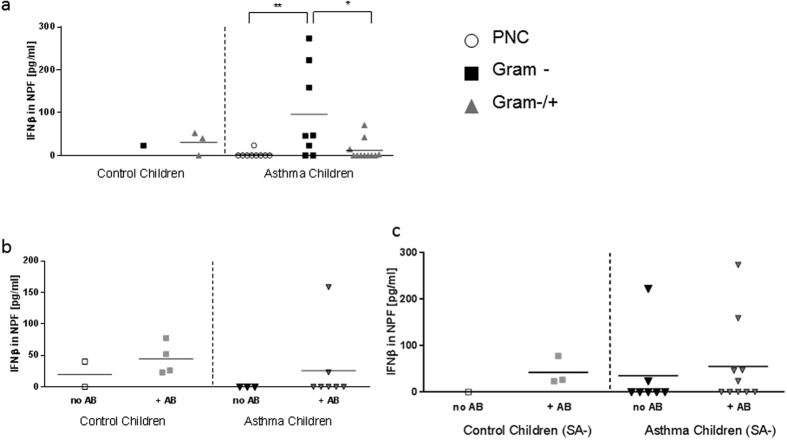
To understand the influence of local nasopharyngeal bacterial colonization on IFNβ production, we analyzed the production of IFNβ in the NPF of the study children (baseline and follow up visits). Here, we found significantly higher amounts of IFNβ in the NPF of asthmatic children with Gram negative nasopharyngeal colonization as compared to asthmatic children with physiological bacterial flora (physiological nasalpharyngeal colonization: PNC; p = 0.0095) or additional Gram-positive bacteria in the nasopharynx (Gram−/+; p = 0.0101; Fig. 2a). IFNβ levels in NPF were not affected by previous antibiotic treatment (Fig. 2b). Finally, the absence of S. aureus (SA-) in the NPF did not influence IFN-β release (Fig. 2c).
Figure 2. Gram negative germs are associated with higher levels of IFNβ in NPF of asthmatic children.
(a) Increased IFNβ production in asthmatic children with a Gram negative bacterial colonization in nasopharyngeal fluid during the study (p = 0.0095, p = 0.0101; baseline and follow up visits). (b) Treatment with antibiotics ( + AB) or (c) in absence of S. aureus (SA-) had no significant effect on IFNβ in NPF of control and asthmatic children. (a: n = 1,3,9,8,11; b: n = 2,4,3,7; c: n = 1,3,7,10). Statistic values: (a) Control children (C) Gram+/− vs Asthma children (A) Gram+/−: p = 0.12; (b) C no AB vs C + AB: p = 0.16; C no AB vs A no AB: p = 0.13; C + AB vs A + AB: p = 0.28; A no AB vs A + AB: p = 0.24; (c) C + AB-SA vs A + AB-SA: p = 0.41; A no AB-SA vs A + AB-SA: p = 0.33.
Higher IL-33 production in nasopharyngeal fluid of children with asthma and a Gram negative (Gram-) bacterial colonization in the nasopharynx
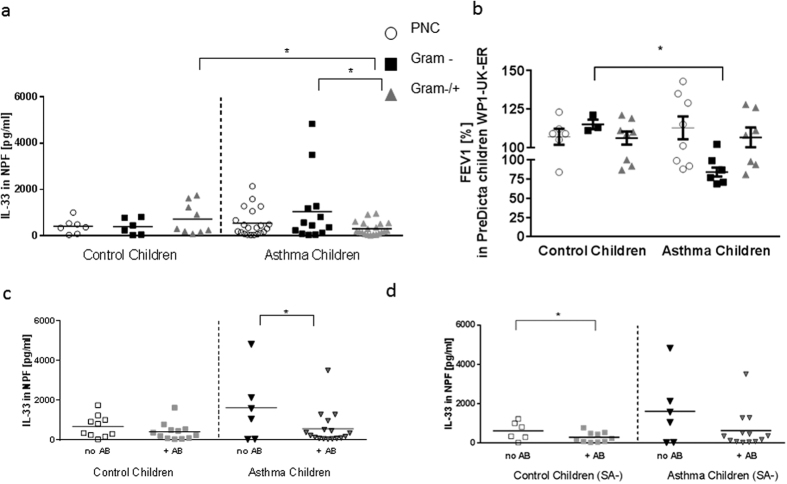
IL-33 is known to be produced by damaged epithelial cells and to shift the immune response towards the Th2 cytokines IL-5 and IL-1318. To test whether there is also a correlation between IL-33 production and bacterial colonization, we evaluated the IL-33 production in the NPF of the study children. By analyzing the NPF at different study visits (baseline and follow up visit) with regard to bacterial colonization, we could observe an increase in IL-33 production in the NPF of asthmatic children with a Gram- bacterial colonization as compared to children with additional Gram positive bacteria in the nasopharynx (Gram−/+; Fig. 3a, p = 0.0181). The asthmatic children with additional Gram + bacteria in the nasopharynx had also a significant reduction of IL-33 in their airways as compared to the correspondent control group of children (Fig. 3a, p = 0.0121).
Figure 3. Increased IL-33 production in nasopharyngeal fluid of children with Asthma and Gram negative bacterial colonization in NPF.
Decreased IL-33 production in children with Asthma after antibiotic therapy and in control children after antibiotic therapy in absence of S. aureus (a) Increased IL-33 production in the NPF of asthmatic children with Gram negative bacterial colonization at baseline and follow up visit as compared to asthmatic children with Gram negative and positive bacterial colonization (Gram−/+) (p = 0.0181). IL-33 levels in asthmatic children Gram−/+were significantly decreased as compared to the corresponding control group (p = 0.0121). (b) Lower values of FEV1 in children with asthma are associated with Gram negative nasopharyngeal bacterial colonization at time of recruitment (B0). Significantly lower percentage of FEV1 in children with asthma and Haemophilus influenzae or Moraxella catarrhalis than in healthy children (n = 4–8; p = 0,0238, Mann-Whitney U test). (c) Decreased IL-33 production measured in the NPF of asthmatic children after antibiotic therapy (p = 0.0274). (d) Decreased level of IL-33 measured in the NPF of control children in absence of S. aureus (SA-) in NPF after antibiotic treatment ( + AB) p = 0.045. (a: n = 7,6,9,21,13,20; b: n = 11,11,7,16; c: n = 6,10,6,13). Statistic values: (a) Control children (C) Gram+/− vs Asthma children (A) Gram+/−: p = 0.0121; C Gram- vs A Gram-: p = 0.15; C PNC vs A PNC: p = 0.29; C PNC vs C Gram-: p = 0.46; C PNC vs C Gram−/+: p = 0.14; C Gram- vs C Gram−/+: p = 0.14; A PNC vs A Gram-: p = 0.086; A PNC vs A Gram−/+: p = 0.058. (c) C no AB vs C + AB: p = 0.13 ; C + AB vs A + AB: p = 0.29; (d) C + AB-SA vs A + AB-SA: p = 0.14; A no AB-SA vs A + AB-SA: p = 0.067; C no AB-SA vs A no AB-SA: p = 0.10.
The presence of Gram negative bacteria in the nasopharyngeal tract is associated with lower FEV1 in children with asthma at the time of recruitment into the study
Since IL-33 is associated with asthma exacerbations19, we next looked at the forced expiratory volume of one second (FEV1). Here we found that children with asthma and Gram negative bacterial nasopharyngeal colonization had less relative FEV1 as compared to the healthy controls (Fig. 3b, p = 0.0238, n = 4–8). In conclusions, we observed that colonization with Gram negative respiratory pathogens is associated with an increased airway hyperresponsiveness in our cohort of asthmatic children (Fig. 3b) and induced IL-33 in the airways.
Therapy with antibiotics is associated with decreased IL-33 in asthmatic children and in control children in the absence of S. aureus in nasopharyngeal fluid
We next reasoned that antibiotic therapy could influence IL-33 expression and release. We thus analyzed the baseline and follow-up data with regard to the antibiotic therapy. Here we could observe that only within the asthmatic group, antibiotic treatment was associated with reduced IL-33 levels in the NPF (Fig. 3c, p = 0.0274).
S. aureus is a common human pathogen which can cause severe infections in various tissues including the respiratory tract. Especially, some of the exotoxins (superantigens) produced by S. aureus are thought to be involved in the modulation and aggravation of airway inflammation20,21. We thus analyzed antibiotic therapy along with the absence of S. aureus and IL-33 production in NPF. We found that antibiotic therapy was associated with a significant reduction of IL-33 production only in control children that did not carry S. aureus in their NPF (Fig. 3d, p = 0.045).
Increased ST2 mRNA expression in the blood cells of asthmatic children with a Gram positive and negative (Gram+/−) bacterial colonization in the nasopharynx
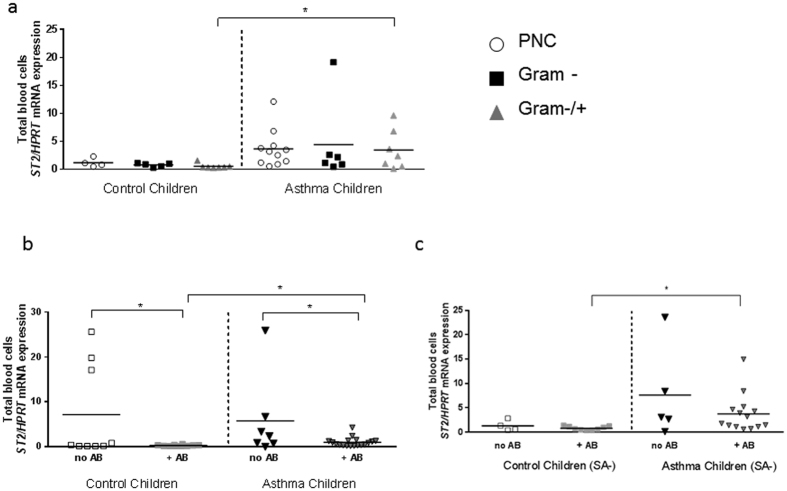
As IFN-beta was found to be upregulated in the NPF of asthmatic children with Gram-negative colonization as compared to those children with asthma and a Gram−/+ bacteria colonization and has recently been shown to directly blunt IL-33 induced proliferation of bone marrow-derived ILC2 cells13, we next analyzed the expression of IL-33 receptor (R) (ST2: suppressor of tumorigenicity 2), signature of ILC2 cells and Th2 cells, in the peripheral blood of the cohorts of pre-school children taking into consideration their bacterial colonization in the NPF. Here we found that, the group of asthmatic children with a Gram negative and positive colonization (Gram−/+) in their airways, despite of the fact that they had lower IL-33 levels as compared to other subgroups of asthmatic children with different nasal-pharyngeal bacteria colonization (Fig. 3a), had a higher expression of ST2 mRNA in the peripheral blood as compared to the corresponding subgroup of control children (Fig. 4a, p = 0.027). One possible explanation for this finding, to be further investigated in the future by expanding the number of children analyzed, could be the reduced IFNβ in the airways of these children (Fig. 2a) or other not yet identified factors regulating ST2 positive cells. Although the number of samples analyzed were limited, these children with sole Gram negative colonization of H.influenzae and/or M.catarrhalis had the highest IFNβ among the children analyzed (Fig. 2a) but also high IL-33 levels thus, as result, ST2 mRNA in this group was not significantly regulated. In the systemic circulation, ST2 mRNA expression is significantly higher in children with asthma than in their corresponding controls, so that the nasopharyngeal colonization might not predominantly influence systemic mRNA expression. Yet, antibiotic therapy might provide a target to reduce inflammation. Consequently, the influence of antibiotic therapy as it effects blood circulation was examined next to find a target to reduce inflammatory pathways.
Figure 4. Higher levels of ST2 mRNA expression in asthmatic children with Gram positive and negative bacterial colonization in their airways.
Decreased ST2 mRNA expression in the blood of control children and asthmatic children after antibiotic treatment. (a) Higher Level of ST2 mRNA expression in the blood of asthmatic children and diverse Gram positive and Gram negative bacterial nasopharyngeal colonization at baseline visit and follow-up visit (p = 0.027). (b) Decreased ST2 mRNA measured in the blood of control (p = 0.027) and asthmatic (p = 0.018) children who were treated with antibiotics (+AB) compared to not treated (no AB) children. (c) Higher ST2 mRNA expression in asthmatic children with antibiotic therapy independently of S. aureus (SA) (SA-: p = 0.018, SA included see b: p = 0.0174). (a: n = 4,5,10,12,6,7; b: n = 9,10,7,18; c: n = 4,9,6,14). Statistic values: (a) C Gram- vs A Gram-: p = 0.08; C PNC vs A PNC: p = 0.087; C PNC vs C Gram-: p = 0.17; C PNC vs C Gram−/+: p = 0.06; C Gram- vs C Gram−/+: p = 0.18; A PNC vs A Gram-: p = 0.38; A PNC vs A Gram−/+: p = 0.45; A Gram-vs A Gram−/+: p = 0.38 (b) C no AB vs C + AB: p = 0.026; C + AB vs A + AB: p = 0.0174; (c) C + AB-SA vs A + AB-SA: p = 0.0177; A no AB-SA vs A + AB-SA: p = 0.10; C no AB-SA vs A no AB-SA: p = 0.12.
Therapy with antibiotics is associated with decreased ST2 in control and asthmatic children
We next analyzed the children in accordance to antibiotics treatment and then looked at ST2 mRNA expression in the blood. Here, we found that ST2 mRNA was down-regulated in the blood of control children, as well as asthmatic children treated with antibiotic as compared to the corresponding group without antibiotic treatment (no AB). Moreover, asthmatic children treated with antibiotics had higher levels of ST2 mRNA compared to the corresponding control children treated with antibiotics (Fig. 4b, p = 0.0174).
After antibiotic treatment, in the absence of S. aureus colonization in nasopharyngeal fluid, asthmatic children had higher values of ST2 mRNA as compared to control children
To analyze the effect of S. aureus colonization on ST2 mRNA we analyzed the antibiotic treatment effect without S. aureus. Here we found ST2 mRNA upregulated in the blood of asthmatic children treated with antibiotics in the absence of S. aureus in their NPF compared to the corresponding control children subgroup (Fig. 4c, p = 0.018). Together these data suggest an inhibitory function of the antibiotic therapy on the IL-33/ST2 pathway in the absence of S. aureus in control children but not asthmatic children.
Discussion
In this study we analyzed the immune responses of pre-school children with and without asthma taking into consideration their bacteria colonization in the NPF. Because the bacteria detected in the airways of these children are affected by the use of antibiotics during infections, we first investigated the use of antibiotics 12 months before the point of blood and NPF withdrawal. Not surprisingly, we noticed an increased use of antibiotics in asthmatic children as compared to control children. The most striking increase in antibiotic treatment was observed in asthmatic children with S. aureus detected in their NPF (3.3 fold induction) as compared to the control group. It is known that S. aureus can produce exotoxins with super-antigenic properties which represent a serious risk for human health22,23. These super-antigens are able to stimulate T cells and antigen-presenting cells already at very low concentrations and thus initiate a cascade of pro-inflammatory cytokines. It has been suggested that S. aureus is able to “hide” in host cells and therefore could survive standard antibiotic treatment24,25,26. Therefore, it is possible that the asthmatic disease generates the necessary micromilieu that allows S. aureus to escape antibiotic eradication. Furthermore, we could show that antibiotic therapy in control children without S. aureus colonization in the nasopharynx resulted in lower levels of IL-33 in the NPF than in the corresponding control children who were not treated with antibiotics (Fig. 3d). Previous studies could already show that the immunosuppressant rapamycin (which is chemically related to macrolide antibiotics) could inhibit IL-33 induced airway inflammation27. Furthermore, we found that therapy with antibiotics is associated with decreased IL-33 only in control children in the absence of S. aureus in nasopharyngeal fluid. However, IL-33R/ST2 mRNA was found to be downregulated in the control group of children treated with antibiotics as compared to the antibiotic untreated paired group. These data suggest an inhibitory effect of antibiotics on ST2 mRNA pathway in blood cells and only in the absence of S. aureus on IL-33 in the airways of control but not asthmatic children that need further investigations.
IFNβ production in the airways was not increased after antibiotic treatment. Since antibiotic treatment has a huge systemic effect, regulation of mRNA gene expression in blood cells might be better seen than effects of local bacterial colonization. These results suggest a difference in local immune response to antibiotic treatment in children with an atopic airway as compared to non-atopic children.
The micromilieu of the airways in children with asthma might differ significantly from the airway milieu in non-asthmatic children. This provides opportunities in form of biological niches for commensal germs to persist. As it is shown in Fig. 1d, antibiotic treatment in Asthma children was not able to eliminate Gram negative facultative pathogen bacteria of the airways. In contrast to control children, colonization with H. influenzae or M. catarrhalis was higher after antibiotic treatment in asthmatic children. This might be a hint that bacteria of asthmatic children are either often resistant to common antibiotic treatment with macrolide or cephalosporine antibiotic due to frequent treatment in the individuals, or that an asthmatic airway provides further options for Gram negative bacteria to survive and modify immune response. Further studies in this direction should be performed in a larger population.
It is already known that lipopolysaccharide (LPS), a component of the cell wall of Gram-negative bacteria, exerts a huge variety of immune modulating effects2,28,29. The main pattern recognition receptor (PRR) for LPS is Toll-like receptor 4 (TLR4). High doses of LPS induce a shift towards T helper cell type 1 immune response30. By contrast, low doses of LPS induce a shift towards a T helper cell type 2 (Th2) immune response31. In our studies, we could show that Gram negative bacteria preferentially induce the release of IFNβ in the NPF of asthmatic children. These children had also induced levels of IL-33 in their airways (Fig. 3a), thus indicating a possible activating effect of IL-33 on circulating ILC2 blunting the inhibiting effect of IFNβ in this group.
ST2, the receptor for IL-33 on T and ILC2 cells, is an important effector molecule of Th2 responses. It negatively regulates type I interleukin 1 receptor (IL-1RI) and TLR4 but not TLR3 signaling by sequestrating the adaptor molecules like MyD88. Inhibition of TLRs by IL-33/ST2 promotes a Th2 response, and also identifies IL-33/ST2 as a key regulator of endotoxin tolerance32. TLR3 can therefore activate interferon regulating factor 3 (IRF3) and thus induces IFNβ. The mechanism by which Gram negative respiratory pathogens (H. influenzae, M. catarrhalis) induce the elevated IFNβ levels seen in the respective children, is currently unknown.
The contact of bacterial endotoxins like LPS with epithelial cells creates a cellular damage and necrosis33. At that point IL-33 is produced by these cells. IL-33 is associated with recurrent wheezing in children with asthma and currently, a polymorphism in IL-33/ST2 has been connected to asthma development in childhood34. IL-33 together with thymic stromal lymphopoietin (TSLP) induces inflammation in a Th2 immune response35. Thus, IL-33 production induces IL-5 and IL-13 release in asthma, cytokines that are produced by Th2 and ILC2 cells. We could show that asthmatic children were sensitive to antibiotic therapy in terms of down-regulation of IL-33, whereas they do not so in the absence of S. aureus. Consistently, IL-33R remained upregulated in asthmatic children in the presence of antibiotic treatment and absence of S. aureus. The mechanism of action of antibiotics inhibiting IL-33/IL-33R-ST2 in control, but not in asthmatic children is unknown and needs further investigation. Moreover, we describe here that children with a Gram negative bacterial colonization have increased IFNβ in their NPF. Both these findings could be explained by upregulation of IRF336.
By contrast, when Gram negative bacterial colonization was associated with S. aureus and Streptococcus species (Gram positive), IFNβ was suppressed thus resulting in elevated ST2 values inspite of the fact that IL-33 was found reduced in this group. This could be explained by immune evasion mechanisms of S. aureus and certain Streptococcus species24,37,38.
In summary, we have found that IFNβ and IL-33 are associated with Gram negative colonization in the airways of asthmatic children. Moreover, ST2 mRNA expression in blood was found induced in asthmatic children associated with a mixed Gram- and Gram + bacteria flora in the airways. Furthermore, treatment with antibiotics was found associated with reduced ST2 mRNA in the blood and IL-33 levels in the airways of control children without S. aureus in NPF but not significantly in asthmatic children, although the latter were more often treated with antibiotics.
Material and Methods
The methods described in this manuscript were carried out in accordance with the approved local and European guidelines.
Study subjects
Children with or without asthma at the age of four to six years were recruited in the Department of Allergy and Pediatric Pneumology of the Children’s Hospital of Erlangen as part of the Europe-wide study PreDicta (Post-infectious immune reprogramming and its association with persistence and chronicity of respiratory allergic diseases). One aim of PreDicta is to identify altered host-pathogen interactions, molecules and pathways that mediate the establishment and persistence of chronic inflammation in allergic diseases, thus to help to develop new preventive, diagnostic and therapeutic strategies. Therefore, we and the other study centers established and followed up a cohort of preschool children with or without asthma for two years.
Inclusion criteria for cases and controls were (a) the need of a written informed consent from the child’s parents or guardians (the legal custodian must have the verbal, writing and mental ability to understand the intent and character of the study); (b) an age of four to six years at the baseline visit (beginning at the day of the 4th birthday and ending at the day of the 6th birthday); (c) a gestational age of 36 weeks or above; and (d) the diagnosis of asthma within the last two years, confirmed by a pediatrician of the Children’s Hospital in Erlangen. The asthma should be of mild to moderate persistent severity according to the GINA guidelines (2005). There must be three asthma episodes during the preceding 12 months and one of them within the last six months. In addition, the child had to be able to perform at least one peak expiratory flow (PEF) manoeuvre. The controls should not have a history of asthma or wheezing and atopic illness.
Exclusion criteria for the cases of this study were (a) severe or brittle asthma; (b) present immunotherapy or more than six courses of oral steroids during the previous 12 months; (c) other underlying chronic respiratory diseases (e.g., cystic fibrosis, bronchopulmonary dysplasia, immunodeficiencies) except for allergic rhinitis and (d) other chronic diseases except for atopic eczema or chronic medication use.
We recruited 24 asthmatic patients and 21 healthy subjects at our study center in Erlangen. Atopy was proven by at least one positive skin prick test, while asthma was defined in accordance to a physician’s diagnosis of mucus production, bronchial hyperresponsiveness and dyspnoea. The healthy control subjects did not have a history of atopy or asthma. This study was approved by the ethics committee of the Friedrich-Alexander University Erlangen-Nürnberg, Germany (Re-No 4435) and is also registered in the German Clinical Trial Register (Registration number: DRKS00004914). Informed consent was obtained from the parents of all children of the PreDicta study.
Blinding
Every single participant of the study was assigned a specific number. Only the clinical investigators and study nurses of the Children’s Hospital had access to the full name.
Nasopharyngeal samples
A nasopharyngeal specimen from control and asthmatic children was collected using a per-nasal applicator swab, which has a tip with flocked soft nylon fiber (E-Swab 482CE, Copan, Italy). Swabs were passed through the nostrils until resistance was felt and they were slowly rotated for five seconds to allow for mucus absorption. In addition, swabs were also rotated against the mucosa of the anterior nares before exiting the nose. The nylon tip was eluted by placing into the E-Swab’s medium.
The nasopharyngeal fluid was divided into aliquots under sterile conditions. A 100 μl aliquot was used for bacterial culture, while the other aliquots were stored at −80 °C until further analysis by ELISA. Within four hours from collection the nasopharyngeal fluid was brought to the Microbiology Institute and used to inoculate two microbiological plates. 50 μl each of the nasopharyngeal fluid were used to inoculate a sheep blood agar and a chocolate agar plate and streaked out using a Drigalski spatula and applying the “triple streak technique”. The plates were incubated at 36 °C in an atmosphere supplemented with 5% CO2 for 48 hours. After 24 hours and 48 hours the plates were evaluated and the bacterial growth was semiquantified (growth in the 1st streak only = few; 1st and 2nd streak = moderate; 1st, 2nd and 3rd streak = numerous). In addition, the following bacterial species were identified: Streptococcus (S.) pneumoniae, Moraxella (M.) catarrhalis, Haemophilus (H.) influenzae, Staphylococcus (S.) aureus and Streptococcus (S.) pyogenes.
Antibiotic therapy
With the help of a questionnaire, filled out by the parents after receiving written information from the childrens’ doctors, the number of antibiotic therapies within twelve months prior to the baseline visit and the maximal duration (days) of a particular antibiotic treatment was recorded.
ELISA
Nasopharyngeal fluids were analyzed for proteins by using ELISA. Human IFNβ was detected by using human ELISA Set Verikine (25–2000 pg/ml, PBL Assay Science, New Jersey, USA). Human IL-33 (23.44–1500 pg/ml) was detected by using a DuoSet sandwich ELISA kit (R&D Systems, Wiesbaden, Germany).
Isolation of RNA from total blood
Blood samples were collected in Tempus Blood RNA Tubes (Life Technologies, GmbH, Darmstadt, Germany) containing a stabilizing reagent, starting lysis almost immediately after mixing with venous blood, inactivating cellular RNases and precipitating RNA. Isolation of RNA was performed with the MagMAX Kit according to the manufacturer’s protocol (life technologies™, GmbH, Darmstadt, Germany). After, with 1x PBS, the total volume was brought to the desired final volume and probes were centrifuged (15 min, 4 °C, 5000 × g). Crude RNA pellets were washed with Tempus Pre-Digestion Wash and centrifuged again (10 min, 4 °C, 5000 × g). Afterwards RNA was diluted in a mixture consisting of Tempus Resuspension Solution and Tempus Proteinase. Subsequent purification of RNA was performed by using a magnetic bead-based technology. Briefly, binding solution-concentrate, RNA binding beads and isopropanol were added and the samples brought in a magnetic field. Separation of RNA from the solution was automatically performed by the magnetic RNA binding beads. Beads were washed away twice with washing solution 1 and 2 in the magnetic field. After adding elution buffer and using the magnetic field again, supernatant contained the purified RNA.
After isolation, RNA from total blood was reverse transcribed using the first strand cDNA synthesis kit (Thermo Scientific, Darmstadt, Germany).
Quantitative real-time PCR
Quantitative PCR was used for analyzing mRNA as described for expression of ST2 by using the following set of primers: hST-2 fwd:5′CACGGTCAAGGATGAGCAAG3′ hST-2 rev:5′GCAGAGCAAGTTAGGTTTGCG3′. Hypoxanthine Guanine Phosphoribosyl Transferase (HPRT) served as reference gene and was amplified by using forward (5′-TGA CAC TGG CAA AAC AAT GCA-3′) and reverse (5′-GGT CCT TTT CAC CAG CAA GCT-3′) primers. Reactions contained SYBR® Green (Bio-Rad Laboratories, Munich, Germany), forward and reverse primers, DEPC water and cDNA. Analysis was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Munich, Germany) with incubation for 2 minutes at 98 °C followed by 50 cycles of 5 seconds at 95 °C and 10 seconds at 60 °C. Afterwards the reaction was stopped with 5 seconds at 65 °C and 5 seconds at 95 °C. The data were analyzed using the 2(−∆∆Ct) method. Primers were purchased from Eurofins-MWG-Operon (Ebersberg, Germany).
Statistical analysis
The data were evaluated for significant differences by using the one-tailed T test for unpaired data (Graphpad Prism 6, Graphpad Software, Inc., La Jolla, CA, USA). Data are shown as means values ± SEMs. *p < 0.05; **p < 0.01, ***p < 0.001.
Additional Information
How to cite this article: Hentschke, I. et al. IL-33/ST2 immune responses to respiratory bacteria in pediatric asthma. Sci. Rep. 7, 43426; doi: 10.1038/srep43426 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
The authors thank all children and their parents/guardians who took part in our Europe-wide study PreDicta. Moreover, the authors are grateful to S. Mittler, C. Holzinger, S. Mousset at the Division of Molecular Pneumology and to E. Muschiol, Jawa and L. Schramm at the Paediatric Pneumology-Allergology, Department of aediatrics and Adolescent Medicine, Universitätsklinikum Erlangen, Erlangen Children Hospital in Erlangen for their technical support. Moreover, the authors thank Drs. M. Wölfel, C. Reinhardt, A. Neubert and Prof. Dr. W. Rascher from the Department of Pediatrics and Adolescent Medicine, Universitätsklinikum Erlangen, Erlangen for their assistance in the PreDicta UKER wp1 study. Furthermore, we thank R. Stergiou and S. Taka from the Allergy and Clinical Immunology Unit of the National and Kapodistrian University of Athens and the team at the Institute of Clinical Microbiology, Immunology and Hygiene in Erlangen. This work was supported by the European Grant PreDicta (Post-infectious immune reprogramming and its association with persistence and chronicity of respiratory allergic diseases) in Erlangen and in the other European Centers, by the Department of Molecular Pneumology in Erlangen, and by a DFG grant (SFB 643/TP 12) in Erlangen.
Footnotes
The authors declare no competing financial interests.
Author Contributions I.H. took care of the microbiology data and organized information of the clinical questionnaire. She divided the study cohorts in subgroups according to their bacterial colonization and referred them to different cytokine and clinical parameters. She did analysis and part of the IFNβ measurement and wrote the versions of the manuscript. A.G. took care of the sample material collection. She analyzed the data, contributed to the generation of the figures and tables, revised the manuscript and approved the final version of the manuscript. S.F. conceptualized and designed the study, coordinated and supervised the collection of data, contributed to the data interpretation and writing of the manuscript. She revised the manuscript and approved the final version of the manuscript. C.B. conceived the microbiological analyses and contributed to the data interpretation and writing of the manuscript. N.P. designed the PreDicta-Study and is the coordinator of PreDicta P.X. contributed to the design of the Predicta-Study WP1. F.A., generated the microbiological analyses. P.H. took care of the antibiotic data and analyzed ST2. B.K. took care of all PreDicta’s probes, isolated RNA and performed IFN-beta ELISA in the NFP. V.O.M., K.A. and T.Z. conducted and are responsible of the clinical part of the study. V.O.M. also contributed to the data interpretation and writing of the manuscript.
References
- Huang Y. J. et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology 127, 372–381, e371–373, doi: 10.1016/j.jaci.2010.10.048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M. R., Bartlett N. W., Hussell T., Openshaw P. & Johnston S. L. The microbiology of asthma. Nat Rev Microbiol 10, 459–471, doi: 10.1038/nrmicro2801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P. G. & Sly P. D. Viral infections and atopy in asthma pathogenesis: new rationales for asthma prevention and treatment. Nature medicine 18, 726–735, doi: 10.1038/nm.2768 (2012). [DOI] [PubMed] [Google Scholar]
- Gavala M. L., Bertics P. J. & Gern J. E. Rhinoviruses, allergic inflammation, and asthma. Immunological reviews 242, 69–90, doi: 10.1111/j.1600-065X.2011.01031.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H. et al. Childhood asthma after bacterial colonization of the airway in neonates. The New England journal of medicine 357, 1487–1495, doi: 10.1056/NEJMoa052632 (2007). [DOI] [PubMed] [Google Scholar]
- von Mutius E. & Vercelli D. Farm living: effects on childhood asthma and allergy. Nature reviews. Immunology 10, 861–868, doi: 10.1038/nri2871 (2010). [DOI] [PubMed] [Google Scholar]
- Ege M. J. et al. Exposure to environmental microorganisms and childhood asthma. The New England journal of medicine 364, 701–709, doi: 10.1056/NEJMoa1007302 (2011). [DOI] [PubMed] [Google Scholar]
- Heederik D. & von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? The Journal of allergy and clinical immunology 130, 44–50, doi: 10.1016/j.jaci.2012.01.067 (2012). [DOI] [PubMed] [Google Scholar]
- Bogdan C., Mattner J. & Schleicher U. The role of type I interferons in non-viral infections. Immunological reviews 202, 33–48, doi: 10.1111/j.0105-2896.2004.00207.x (2004). [DOI] [PubMed] [Google Scholar]
- Stetson D. B. & Medzhitov R. Type I interferons in host defense. Immunity 25, 373–381, doi: 10.1016/j.immuni.2006.08.007 (2006). [DOI] [PubMed] [Google Scholar]
- Monroe K. M., McWhirter S. M. & Vance R. E. Induction of type I interferons by bacteria. Cellular microbiology 12, 881–890, doi: 10.1111/j.1462-5822.2010.01478.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrinioti H., Toussaint M., Jackson D. J., Walton R. P. & Johnston S. L. Role of interleukin 33 in respiratory allergy and asthma. The Lancet. Respiratory medicine 2, 226–237, doi: 10.1016/S2213-2600(13)70261-3 (2014). [DOI] [PubMed] [Google Scholar]
- Duerr C. U. et al. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nature immunology 17, 65−+, doi: 10.1038/ni.3308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mutius E. Gene-environment interactions in asthma. The Journal of allergy and clinical immunology 123, 3–11; quiz 12–13, doi: 10.1016/j.jaci.2008.10.046 (2009). [DOI] [PubMed] [Google Scholar]
- Beigelman A., Weinstock G. M. & Bacharier L. B. The relationships between environmental bacterial exposure, airway bacterial colonization, and asthma. Current opinion in allergy and clinical immunology 14, 137–142, doi: 10.1097/ACI.0000000000000036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty M. et al. Disordered microbial communities in asthmatic airways. PloS one 5, e8578, doi: 10.1371/journal.pone.0008578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri P. R., Stern D. A., Wright A. L., Billheimer D. & Martinez F. D. Asthma-associated differences in microbial composition of induced sputum. The Journal of allergy and clinical immunology 131, 346–352, e341–343, doi: 10.1016/j.jaci.2012.11.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboki K., Nakae S., Matsumoto K. & Saito H. IL-33 and Airway Inflammation. Allergy, asthma & immunology research 3, 81–88, doi: 10.4168/aair.2011.3.2.81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. K. & Herbert C. IL-33-dependent type 2 inflammation in asthma exacerbations. American journal of respiratory and critical care medicine 191, 237–238, doi: 10.1164/rccm.201411-2042LE (2015). [DOI] [PubMed] [Google Scholar]
- Davis M. F., Peng R. D., McCormack M. C. & Matsui E. C. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. The Journal of allergy and clinical immunology 135, 811–813, e815, doi: 10.1016/j.jaci.2014.10.052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert C. et al. IgE to Staphylococcus aureus enterotoxins in serum is related to severity of asthma. The Journal of allergy and clinical immunology 111, 1131–1132 (2003). [PubMed] [Google Scholar]
- Bachert C., Gevaert P., Zhang N., van Zele T. & Perez-Novo C. Role of staphylococcal superantigens in airway disease. Chemical immunology and allergy 93, 214–236, doi: 10.1159/0000100897 (2007). [DOI] [PubMed] [Google Scholar]
- White J. et al. The V beta-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell 56, 27–35 (1989). [DOI] [PubMed] [Google Scholar]
- Hardt W. D. Antibiotics: Homed to the hideout. Nature 527, 309–310, doi: 10.1038/nature15647 (2015). [DOI] [PubMed] [Google Scholar]
- Organization W. H. Antimicrobial Resistance: Global Report on Surveillance (2014). [Google Scholar]
- Lehar S. M. et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328, doi: 10.1038/nature16057 (2015). [DOI] [PubMed] [Google Scholar]
- Ananda Mirchandani R. S., Foo Liew. Rapamycin inhibits IL-33-induced airway inflammation. European Respiratory Journal (2011). [Google Scholar]
- Saraiva M. & O’Garra A. The regulation of IL-10 production by immune cells. Nature reviews. Immunology 10, 170–181, doi: 10.1038/nri2711 (2010). [DOI] [PubMed] [Google Scholar]
- Takeuchi O. & Akira S. Toll-like receptors; their physiological role and signal transduction system. International immunopharmacology 1, 625–635 (2001). [DOI] [PubMed] [Google Scholar]
- Kim Y. K. et al. Airway exposure levels of lipopolysaccharide determine type 1 versus type 2 experimental asthma. J Immunol 178, 5375–5382 (2007). [DOI] [PubMed] [Google Scholar]
- Dabbagh K., Dahl M. E., Stepick-Biek P. & Lewis D. B. Toll-like receptor 4 is required for optimal development of Th2 immune responses: role of dendritic cells. J Immunol 168, 4524–4530 (2002). [DOI] [PubMed] [Google Scholar]
- Brint E. K. et al. ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nature immunology 5, 373–379, doi: 10.1038/ni1050 (2004). [DOI] [PubMed] [Google Scholar]
- Weber J. R. & Tuomanen E. I. Cellular damage in bacterial meningitis: an interplay of bacterial and host driven toxicity. Journal of neuroimmunology 184, 45–52, doi: 10.1016/j.jneuroim.2006.11.016 (2007). [DOI] [PubMed] [Google Scholar]
- Savenije O. E. et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. The Journal of allergy and clinical immunology 134, 170–177, doi: 10.1016/j.jaci.2013.12.1080 (2014). [DOI] [PubMed] [Google Scholar]
- Tsicopoulos A., de Nadai P. & Glineur C. Environmental and genetic contribution in airway epithelial barrier in asthma pathogenesis. Current opinion in allergy and clinical immunology 13, 495–499, doi: 10.1097/ACI.0b013e328364e9fe (2013). [DOI] [PubMed] [Google Scholar]
- Polumuri S. K. et al. Transcriptional Regulation of Murine IL-33 by TLR and Non-TLR Agonists. J Immunol 189, 50–60, doi: 10.4049/jimmunol.1003554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcina M. et al. Pathogen-triggered activation of plasmacytoid dendritic cells induces IL-10-producing B cells in response to Staphylococcus aureus. J Immunol 190, 1591–1602, doi: 10.4049/jimmunol.1201222 (2013). [DOI] [PubMed] [Google Scholar]
- Thorburn A. N. et al. Pneumococcal components induce regulatory T cells that attenuate the development of allergic airways disease by deviating and suppressing the immune response to allergen. J Immunol 191, 4112–4120, doi: 10.4049/jimmunol.1201232 (2013). [DOI] [PubMed] [Google Scholar]