Abstract
Angiosperm mitochondrial genomes (mtDNA) exhibit variable quantities of alien sequences. Many of these sequences are acquired by intracellular gene transfer (IGT) from the plastid. In addition, frequent events of horizontal gene transfer (HGT) between mitochondria of different species also contribute to their expanded genomes. In contrast, alien sequences are rarely found in plastid genomes. Most of the plant-to-plant HGT events involve mitochondrion-to-mitochondrion transfers. Occasionally, foreign sequences in mtDNAs are plastid-derived (MTPT), raising questions about their origin, frequency, and mechanism of transfer. The rising number of complete mtDNAs allowed us to address these questions. We identified 15 new foreign MTPTs, increasing significantly the number of those previously reported. One out of five of the angiosperm species analyzed contained at least one foreign MTPT, suggesting a remarkable frequency of HGT among plants. By analyzing the flanking regions of the foreign MTPTs, we found strong evidence for mt-to-mt transfers in 65% of the cases. We hypothesize that plastid sequences were initially acquired by the native mtDNA via IGT and then transferred to a distantly-related plant via mitochondrial HGT, rather than directly from a foreign plastid to the mitochondrial genome. Finally, we describe three novel putative cases of mitochondrial-derived sequences among angiosperm plastomes.
Since the endosymbiotic events that shaped the eukaryotic cells, cytoplasmic organelles - plastids and mitochondria - have transferred large part of their eubacterial genomes to the nucleus1. Today, DNA exchange between organelles and with the nuclear genome, known as intracellular gene transfer (IGT), continues to take place within plant cells at variable frequencies2,3. In addition, horizontal gene transfer (HGT), the genetic movement of DNA between unrelated species, is now accepted as a driving force in the evolution of land plants4. Flowering plants present exceptionally high rates of HGT, mainly involving the mitochondrial genome5,6. Plant mitochondrial genomes (mtDNA) commonly incorporate nuclear and plastid sequences acquired by IGT as well as foreign mitochondrial DNA from other plant species obtained by HGT.
Plastid-derived DNA is found in angiosperm mtDNAs (MTPTs) in variable amounts representing 0.1 to 10.3% of the mtDNAs and covering 0.5 to 87.2% of the plastid genomes7,8. Plastid-to-mitochondria transfers have been ongoing since the colonization of land plants9. Despite that most of the plastid-derived sequences result in non-functional sequences, it is now accepted that once integrated into the mitochondrial genome, MTPTs can impact mitochondrial function. For example, MTPTs can create new gene forms or promoters, or may introduce novel functional tRNA genes10,11,12,13. Interestingly, some MTPTs were acquired by HGT from distant angiosperm species8,14,15,16,17. Whether these sequences were acquired directly from the donor plastid or indirectly from the donor mitochondria is still unclear and it is the focus of the present study.
In contrast to mtDNAs, plastid genomes (ptDNAs) exhibit very low rates of alien DNA18. Lately, four mitochondrial-derived sequences located in angiosperm ptDNAs (PTMT) have been reported19,20,21,22. Here, we take advantage of the recent increase in plant organellar sequences in public databases to study the extent of MTPTs and PTMTs among flowering plants, and to weigh evidence on the genomic origin of foreign MTPTs.
Results and Discussion
MTPTs are invariably present in seed plants but are infrequent among non-seed plants
We analyzed the mitochondrial genomes of 136 diverse species of the green lineage and only identified MTPTs in gymnosperms (13 sequences) and angiosperms (1,372 sequences), and none among non-seed plants (Table S1). This is consistent with the ‘limited transfer window hypothesis’ that argues that species with a single plastid per cell, such as the majority of green algae, or species with monoplastidic meiosis, such as bryophytes and most lycophytes23, present less IGT events, if any, from the plastid to the nucleus or to the mitochondria24. Angiosperms showed the highest relative contents of MTPTs within the green lineage. Geranium maderense and Phoenix dactylifera ranked first with plastid-derived sequences covering 10.38% and 9.86% of their mtDNA, respectively (Table S1).
To evaluate the relationship between the size of the mtDNA and the MTPT content, we performed a Spearman non-parametric test (Figure S1). Interestingly, the size of the mitochondrial genome strongly correlates with the amount of plastid sequences in angiosperm and gymnosperm mtDNAs, considering the total MTPT length (rho = 0.57, P = 1.05 × 10−07) or the total number of MTPTs (rho = 0.64, P = 6.57 × 10−10), but not with the MTPT mitochondrial coverage (rho = 0.16, P = 0.1693). In general, larger mtDNAs give shelter to more MTPTs (Figure S1). This observation agrees with previous studies on MTPTs and also on organelle-to-nucleus DNA transfers24,25, suggesting that genomes with extensive non-coding regions could harbor more alien sequences, but these alien insertions are not solely responsible for plant mitochondrial genome expansion26.
Foreign MTPTs are frequent among flowering plants
MTPTs can be derived from the plastid genome of the same species by IGT (termed native MTPTs) or from an unrelated species by HGT (termed foreign MTPTs). To determine the origin of the 1,385 MTPTs mentioned above (Table S1), all MTPTs with highest similarity to the ptDNA of an unrelated lineage were considered putatively foreign and were analyzed phylogenetically to confirm its origin and to determine the donor lineage. In addition to the 31 previously described cases8,15,16,17,27, 15 new foreign MTPTs were identified in this work (Table 1). MTPTs were considered foreign when phylogenetic analyses showed unexpected relationships with bootstrap support (BS) higher than 70% (Figure S2). In all cases, donor lineages were identified as members of the flowering plants, indicating angiosperm-to-angiosperm HGT events (Table 1, Figure S2). Out of the 72 angiosperm mtDNAs analyzed, 14 (19.4%) had at least one foreign MTPT (Table S1). That is, one out of five plant mtDNAs received plastid sequences by HGT. Sampling the >99.9% unexamined angiosperms may reveal that thousands of species bear foreign MTPTs in their mitochondria. These results are comparable to those of the cox1 intron horizontal transfers among angiosperms, in which 20% of the sequenced species had the invasive cox1 intron28. Among all alien DNA acquired by the mitochondrion, MTPTs and cox1 introns have the highest probability of being detected as they carry strong phylogenetic signal to corroborate their foreign origin. Therefore, these markers could speak for the underlying rate of DNA transfer among angiosperms.
Table 1. Information on foreign plastid sequences identified in flowering plant mitochondrial genomes.
| # Foreign MTPTa | Recipient speciesb | Recipient lineage | mtDNA genBank accession number | Start (nt) | End (nt) | Length of MTPT (bp) | Gene content of MTPT | BSc (%) | Putative donor lineaged | Evidence for mt-to-mt HGT in flanking regions of MTPT (+/−1000 bp)e |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||||||
| 1 | Amborella trichopoda1 | basal Magnoliophyta; Amborellales | KF754801 | 124144 | 127171 | 3028 | psbC; psbD | — | Santalales | no1 | no3 |
| 2 | KF754803 | 3077656 | 3078925 | 1270 | rbcL | — | Santalales | yes5 | no3 | ||
| 3 | 474251 | 476207 | 1957 | psaA | — | rosids; fabids; Oxalidales | no3 | no5 | |||
| 4 | KF754799 | 57856 | 61814 | 3959 | rps7; rps12; trnV; rrnS | — | rosids; fabids; Fagales; Fagaceae | yes5 | no3 | ||
| 5 | Asclepias syriaca | asterids; lamiids; Gentianales | NC_022796 | 29093 | 29432 | 340 | ndhB-1 | 100 | rosids; fabids; Fabales | yes4 | no1 |
| 6 | NC_022797 | 40124 | 42206 | 2083 | ndhB-1; rps7; rps12 | 100 | rosids; fabids; Fabales | no1 | no2 | ||
| 7 | NC_022798 | 310073 | 311353 | 1281 | non-coding region | 100 | rosids; fabids; Fabales | no3 | yes5 | ||
| 8 | NC_022796 | 497713 | 497990 | 278 | ndhD | 100 | rosids; malvids; Malvales; Malvaceae | no1 | no2 | ||
| 9 | Cucurbita pepo | rosids; fabids; Cucurbitales | NC_014050 | 252522 | 253546 | 1025 | cemA; petA | 100 | asterids; lamiids; Lamiales; Orobanchaceae | no1 | yes4 |
| 10 | NC_014050 | 848217 | 849548 | 1332 | rps7 | 93 | rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae | no2 | no1 | ||
| 11 | Erythranthe guttata | asterids; lamiids; Lamiales | NC_018041 | 469539 | 469886 | 348 | rps7 | 90 | rosids; fabids; Fabales; Fabaceae | no3 | no3 |
| 12 | Geranium brycei2 | rosids; malvids; Geraniales | KP974317 | 47781 | 49416 | 1636 | rbcL | — | asterids; lamiids; Solanales; Convolvulaceae; Cuscuta | no3 | no3 |
| 13 | KP974313 | 55290 | 56417 | 1128 | psaA | — | no1 | yes5 | |||
| 14 | KP974311 | 98551 | 100870 | 2320 | psaB; rps14 | — | no3 | yes5 | |||
| 15 | KP974317 | 42601 | 44151 | 1551 | atpB; atpE | — | no3 | no3 | |||
| 16 | KP974311 | 343448 | 347166 | 3719 | rpl2 intron; rpoB 3'; rpoC1 exon 1; ndhJ | — | rosids; fabids; Malpighiales; Euphorbiaceae; Acalyphoideae | no3 | yes6 | ||
| 17 | KP974311 | 87980 | 89010 | 1031 | petB | — | yes6 | yes6 | |||
| 18 | KP974317 | 19534 | 20704 | 1171 | psbD | — | no1 | no3 | |||
| 19 | KP974312 | 105427 | 106239 | 813 | matK | — | asterids; lamiids; Gentianales; Rubiaceae; Rubioideae | no3 | yes6 | ||
| 20 | Geranium maderense | rosids; malvids; Geraniales | NC_027000 | 454189 | 454564 | 376 | trnA; trnG | 98 | asterids; lamiids; Lamiales | no3 | yes4 |
| 21 | Glycine max3 | rosids; fabids; Fabales | NC_020455 | 230204 | 230895 | 692 | rbcL | — | asterids; lamiids; Gentianales; Apocynaceae | yes4 | yes4 |
| 22 | Gossypium harknessii | rosids; malvids; Malvales | NC_027406 | 365728 | 365950 | 223 | psbD | 82 | rosids; fabids; Malpighiales; Euphorbiaceae | yes4 | no3 |
| Gossypium hirsutum | NC_027407 | 368914 | 369136 | 223 | |||||||
| 23 | Helianthus annuus | asterids; campanulids; Asterales | NC_023337 | 107850 | 108495 | 646 | infA; rps8; rps11 | 98 | rosids; Saxifragales; Penthoraceae | no5 | yes5 |
| 24 | Hyoscyamus niger | asterids; lamiids; Solanales | NC_026515 | 351599 | 353572 | 1974 | rps12 | 98 | rosids; Rosales; Cannabaceae | yes4 | yes4 |
| 25 | 354959 | 355152 | 194 | non-coding region | 99 | rosids; Rosales; CannabaceaeΩ | yes4 | yes4 | |||
| 26 | 355156 | 356418 | 1263 | petB | 100 | rosids; Rosales; Cannabaceae; CannabisΩ | yes4 | yes4 | |||
| 27 | Lophophytum mirabile4 | Santalales | KU992322 to KU992380 | — | — | 245 | rpl2 | 100 | rosids; fabids; Fabales; Mimosoideae; Acacia ligulataΩ | yes5 | yes4 |
| 28 | — | — | 726 | rrn23 | 80 | rosids; fabids; Fabales; Mimosoideae; Acacia ligulataΩ | no3 | yes5 | |||
| 29 | — | — | 638 | psbA | 100 | rosids; fabids; Fabales; Mimosoideae; Acacia ligulataΩ | yes4 | yes5 | |||
| 30 | — | — | 520 | rpl16 | 72 | rosids; fabids; Fabales; Mimosoideae | no3 | no3 | |||
| 31 | — | — | 673 | petG; trnW | 76 | rosids; fabids; Fabales; Mimosoideae; Acacia ligulataΩ | yes5 | yes4 | |||
| 32 | — | — | 269 | rbcL | 99 | rosids; fabids; Fabales; Fabaceae | no3 | no5 | |||
| 33 | — | — | 771 | rpl2 | 90 | rosids; fabids; Fabales; Fabaceae | yes4 | no1 | |||
| 34 | Lotus japonicus3 | rosids; fabids; Fabales | NC_016743 | 307253 | 308275 | 1023 | rbcL | — | asterids; lamiids; Solanales; Convolvulaceae; Cuscuta | yes4 | no3 |
| 35 | Phoenyx dactylifera3 | Liliopsida; Arecaceae | NC_016740 | 179688 | 180534 | 847 | trnI; trnA | — | rosids; fabids; Fagales; Fagaceae | no3 | no3 |
| 36 | Rhazya stricta | asterids; lamiids; Gentianales | NC_024293 | 236269 | 237891 | 1623 | trnI; ycf2 | 87 | asterids; lamiids; Lamiales; Oleaceae; Oleeae; HesperelaeaΩ | no3 | yes4 |
| 37 | Salvia miltiorrhiza | asterids; lamiids; Lamiales | NC_023209 | 209571 | 209920 | 350 | psbA | 100 | rosids; fabids; Fabales; Fabaceae | yes4 | yes5 |
| 38 | Sapria himalayana5 | rosids; fabids; Malpighiales | — | — | — | 1737 | psbC; psbD | — | rosids; Vitales; TetrastigmaΩ | no1 | yes5 |
| 39 | — | — | — | 395 | psbA | — | rosids; Vitales; TetrastigmaΩ | no1 | no3 | ||
| 40 | — | — | — | 504 | ndhB | — | rosids; Vitales; Tetrastigma | yes4 | no3 | ||
| 41 | — | — | — | 477 | atpB | — | rosids; Vitales; TetrastigmaΩ | no1 | yes5 | ||
| 42 | — | — | — | 3703 | rpoC1; rpoC2 | — | rosids; Vitales; Tetrastigma | yes4 | yes4 | ||
| 43 | — | — | — | 2595 | rps12 | — | rosids; Vitales | NA | yes5 | ||
| 44 | — | — | — | 436 | rbcL | — | rosids; Vitales | no3 | no3 | ||
| 45 | — | — | — | 360 | psaB | — | rosids; Vitales; TetrastigmaΩ | no1 | no3 | ||
| 46 | — | — | — | 457 | atpA | — | asterids; campanulids; Apiales; Daucus | no3 | no1 | ||
aBold indicates MTPTs involving a host-parasite relationship.
b(1) Rice et al.15 (2) Park et al.17; (3) Sloan et al.8; (4) Sanchez-Puerta et al.27 and this study, Figure S2; (5) Xi et al.16.
cBS, bootstrap support value.
dΩ, the phylogenetic analysis showed the donor MTPT as sister to the MTPT of the recipient mitochondria.
e(1) no hit; (2) all hits related to the recipient lineage (putative native sequence); (3) hits to lineages unrelated to the donor or the recipient (unconclusive origin); (4) all hits related to the donor lineage; (5) hits to diverse lineages, phylogenetic analyses of flanking regions are shown in Figure S3; (6) Park et al.17; NA: not applicable, sequence not available for testing.
Foreign MTPTs flanking regions strengthen the mt-to-mt transfer hypothesis
The identification of foreign plastid-derived sequences in plant mtDNAs raises questions about the trajectory taken by these sequences until their arrival into the mitochondria. Foreign MTPTs could have originated through horizontal transfer from two different sources: (i) directly from the foreign ptDNA; or (ii) indirectly from the foreign mtDNA once the latter acquired the plastid sequences by IGT (Fig. 1). We favor hypothesis #2 for the following reasons: (i) all angiosperm mtDNAs analyzed contain native MTPTs, indicating that the initial acquisition of the plastid sequence by the native mitochondria is a trivial event8,24; and (ii) relatively frequent mitochondrion-to-mitochondrion HGT events among plants have been reported15,28,29,30,31,32,33,34,35. Here, we searched for evidence to test this hypothesis by analyzing each MTPT in detail. We reasoned that under hypothesis #2, foreign MTPTs should be embedded within foreign mitochondrial tracts, which were transferred as a whole via mt-to-mt HGT. Assuming the mt-to-mt transfer, we expect that foreign flanking mitochondrial sequences will have the same origin, i.e. they are related to the same donor lineage, as the foreign MTPT.
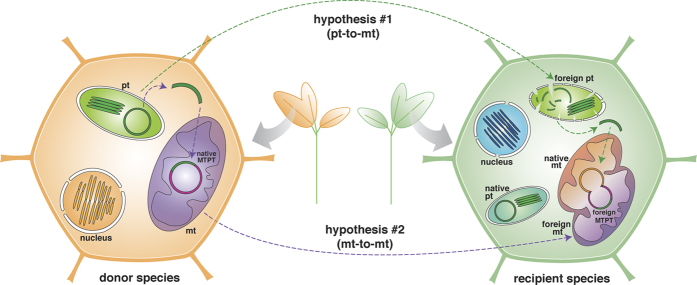
Figure 1. Hypotheses on the origin of foreign plastid sequences located in plant mtDNA (MTPTs).
Hypothesis #1: Plant-to-plant interactions (direct contact or via vector intermediates) enable the transfer of entire plastids (pt) whose genomic sequences are freed into the recipient cell and then captured by the native mitochondria (mt). Hypothesis #2: Plastid sequences are transferred by intracellular gene transfer from the plastid to the mitochondria within the donor plant; later, plant-to-plant interactions enable the transfer of entire foreign mitochondria into the recipient cell, both mitochondria (foreign and native) fuse and their genomes recombine.
We analyzed both flanking sequences (1 kb at each side) of the 46 foreign plastid insertions known to date (Table 1). BLAST and phylogenetic analyses of these regions revealed the presence of foreign mitochondrial sequences from the same donor lineage as the MTPT in 30 of the 46 cases (65%) (Table 1, Figure S3). Therefore, most of the foreign MTPTs were first integrated into the donor mitochondrial genome by IGT, and later horizontally transferred to the recipient mitochondria. The delivery of the MTPT from the donor mitochondria could follow the fusion-compatibility model15, in which the entire foreign mitochondria is captured by the recipient cell where the two mitochondria would fuse and their genomes recombine (Fig. 1).
Besides the 65% of the foreign MTPTs that showed evidence for mt-to-mt HGT, several MTPTs could not be fairly tested given the lack of mitochondrial genomic sequences from the donor lineages. For example, mitochondrial data from members of the family Fagaceae are not yet available, preventing the analyses of MTPTs found in Amborella trichopoda and Phoenix dactylifera. However, the upcoming sequencing of more plant mitochondrial genomes may uncover additional proof for the acquisition of other MTPTs via mitochondrial HGT.
Alternatively, a pt-to-mt horizontal transfer (hypothesis #1) is also conceivable. For example, it has been shown that plastids can be transferred through grafting between species36. Once in the recipient cell, the ptDNA can be freed and enter the mitochondria in the same way as the native ptDNA (Fig. 1). It is also possible that plastid DNA were horizontally transferred into the recipient cell and imported by the mitochondria37,38. Even though less likely than mt-to-mt HGT, pt-to-mt HGT may be responsible for some of the foreign MTPTs.
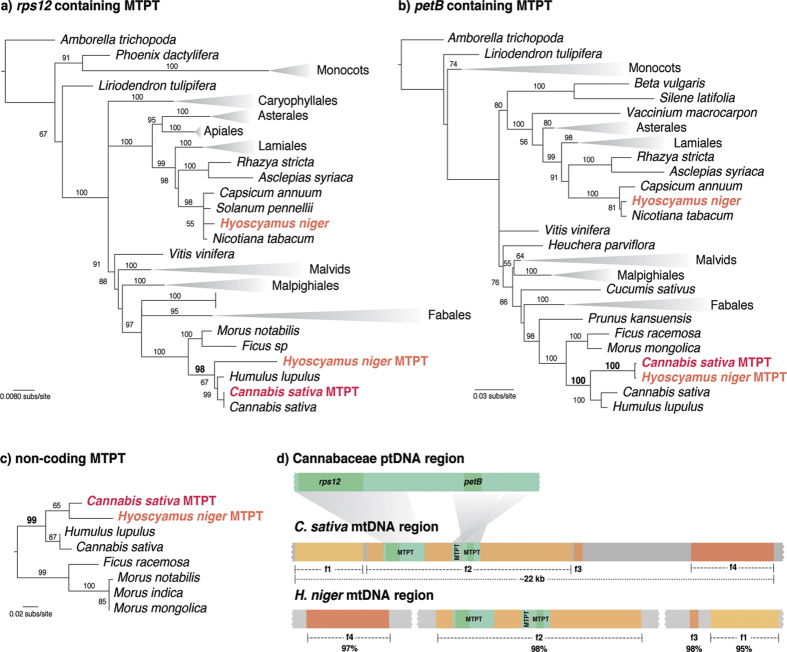
In only a few cases, mitochondrial genomic sequences were available from both donor and recipient lineages of the MTPTs, enabling more powerful comparisons. We found strong evidence for mt-to-mt HGT of foreign MTPTs in the angiosperm Hyoscyamus niger (Solanaceae). The three foreign plastid regions located in the H. niger mtDNA were confirmed to belong to the family Cannabaceae with strong phylogenetic support (BS ≥ 98%) (Fig. 2). Moreover, two of them, one containing the plastidial gene petB and the other a non-coding plastid region, were sister to the native MTPT found in Cannabis sativa mtDNA (Fig. 2b and c). The region containing the gene rps12 showed a different genealogical history, given that the C. sativa MTPT was more closely related to the C. sativa plastome than to the H. niger MTPT (Fig. 2a). However, the three plastid-derived regions were embedded within a mitochondrial region of C. sativa mtDNA that is also present in the mtDNA of H. niger (f2 in Fig. 2d), pointing to a single mt-to-mt HGT event. Therefore, the most plausible scenario is that after the mt-to-mt HGT event from C. sativa to H. niger, a second pt-to-mt intracellular gene transfer was experienced by the C. sativa rps12 MTPT8. To evaluate the extent of the mitochondrial HGT between those two species, we performed comparative analyses of both mitochondrial genomes. The analyses revealed the presence of four mitochondrial fragments (f1 to f4 in Fig. 2d) in H. niger mtDNA with high similarity (~95–98%) to sequences of C. sativa mtDNA (Fig. 2). Moreover, these mitochondrial sequences were only shared by H. niger and C. sativa. The four mitochondrial fragments, including the three MTPTs, were found within a 22 kb stretch of C. sativa mtDNA. This whole region was likely subjected to mt-to-mt transfer from a member of the family Cannabaceae to H. niger (Fig. 2d) and was slightly disrupted once integrated in the H. niger mtDNA.
Figure 2. HGT from the family Cannabaceae to Hyoscyamus niger mtDNA.
(a–c) Evidence of HGT from Cannabis sativa to H. niger mtDNA. Maximum likelihood trees of the plastid sequences rps12 (a), petB (b), and a non-coding region (c) are shown and include sequences located in the plastid or mitochondrial (MTPT) genomes of angiosperms. Several branches are collapsed and shown as triangles for clarity; the full trees are shown in Figure S2. Bootstrap support values >50% are shown above the branches. (d) Plastid (ptDNA) and mitochondrial (mtDNA) genomic comparisons of C. sativa and H. niger. The percent identity between mitochondrial homologous regions (f1–f4) found in C. sativa and H. niger are shown below the H. niger mtDNA.
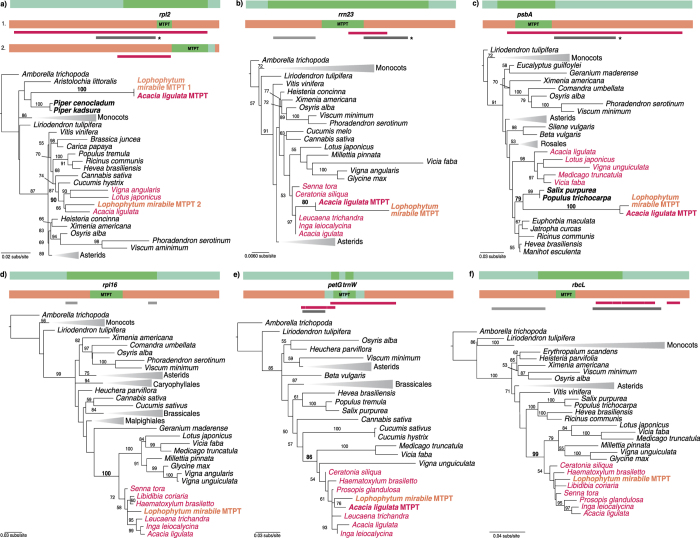
A remarkable number of HGT events among plants took place between hosts and parasites16,17,27,31,32,34. The haustorial connection that parasitic plants establish with their hosts provides a direct cell-to-cell contact, and a putative pathway for DNA transfers39. In agreement to this, we found that 24 of the 46 foreign MTPTs (52%) involved members of a parasitic relationship (shaded in grey in Table 1). Seven cases implicated the holoparasitic plant Lophophytum mirabile27 (Table 1, Fig. 3). Plants of the genus Lophophytum infect exclusively members of the tribe Mimosoideae (family Fabaceae)40 and phylogenetic analyses showed that five MTPTs of L. mirabile were acquired from its host27. However, two MTPTs were related to magnoliids and Salicales, respectively27. Here, we reanalyzed the MTPTs found in L. mirabile including recently available partial data from the mtDNA of the mimosoid Acacia ligulata41. Our results confirmed that five MTPTs were sister or nested within the tribe Mimosoideae (Fig. 3). In addition, the two MTPTs of L. mirabile (rpl2 and psbA) with odd relationships were now found sister to MTPTs of A. ligulata mtDNA with high bootstrap support (Fig. 3, Figure S2). In those two cases, both plastid sequences found in L. mirabile and A. ligulata mtDNAs were misplaced in the tree and the direction of the transfer could not be inferred from these data. However, the unparalleled acquisition of mitochondrial sequences from the mimosoids by L. mirabile27 suggests that these were also the result of transfers from the host to the parasite. Under such assumption, A. ligulata mtDNA must have received plastid sequences from Piperales (rpl2) and Salicales (rrn23), respectively, before the HGT to L. mirabile. Blast and phylogenetic analyses showed that flanking sequences of five of the seven MTPTs of L. mirabile were only similar (e.g. rpl2) or highly related (e.g. rrn23) to A. ligulata mtDNA (Table 1, Figure S3). These findings support transfers from the mimosoids via mt-to-mt HGT for most MTPTs in L. mirabile. A deeper inspection of the complete sequence of the A. ligulata mtDNA should reveal the extent of the HGT in this host-parasite relationship.
Figure 3. Analyses of foreign MTPTs in Lophophytum mirabile and their flanking regions.
Green rectangles represent a fragment of the donor plastid genome depicting coding sequences in dark green. Orange rectangles represent fragments of the L. mirabile mtDNA denoting the MTPT in green. BLAST results of the flanking regions are indicated with lines in fuchsia (hits to Acacia ligulata; donor lineage), grey (hits unrelated to the donor or the recipient lineages), and dark grey (hits to diverse lineages). When hits to diverse lineages were found, they were aligned and analyzed by phylogenetic analyses (Figure S3). In most cases, sequences of Acacia mtDNA were sister to the flanking regions of L. mirabile (depicted with an *). Best ML trees of plastid and MTPT fragments are shown. Members of the Fabaceae are shown in fuchsia. Several branches are collapsed and shown as triangles for clarity; the full trees are shown in Figure S2. Bootstrap support values >50% are shown above the branches.
Mitochondrion-to-plastid DNA transfers are rare
In contrast to the universally present MTPTs in angiosperm mtDNAs, mitochondrial sequences in plastid genomes (PTMTs) are rare. Since the first PTMT described by Iorizzo et al.19 in the carrot ptDNA, only three more cases have been published for angiosperm plastids20,21,22 (Table S2). To evaluate the frequency of PTMTs in plant plastids, we analyzed a total of 1,232 land plant ptDNAs using BLAST (Table S3). In addition to the four cases already described, we found three further PTMTs (Table S2). Unfortunately, we cannot rule out assembly errors for these novel cases because the original reads were not available in the public databases or shared by the authors. Surprisingly, the PTMT found in the obligate root holoparasite Orobanche californica has 93% identity to a mitochondrial sequence of one of its various hosts, Capsicum annuum42, becoming the first putative case of HGT within a plastome (Table S2). Among the eight complete plastid genomes of non-photosynthetic parasites of the family Orobanchaceae43, O. californica is the only one that showed the aforementioned insertion in the ptDNA, suggesting a recent transfer event.
Materials and Methods
To identify potential MTPTs we analyzed a total of 136 complete mitochondrial genomes of the green lineage (Table S1) available in the NCBI Organelle Genome Database as of April 2016 that have at least a plastid genome of the same order for comparison purposes. We blasted each mitochondrial genome against 1,232 plants plastid genomes available in the NCBI Organelle Genome Database using BLASTN v.2.4.0+ algorithm44 with the following settings: –task blastn –word_size 20 –e-value 1e-10. BLAST hits associated with ancient transfers8,9 or hits of ancient homology (atp1, rrn18, and rrn26)12 were excluded from further analysis. Mitochondrial sequences with blast hits to plastid genomes (named MTPTs) larger than 200 bp and with sequence identity >70% were further studied. MTPTs >200 bp separated by gaps <100 bp were taken together as one.
To detect MTPTs of foreign origin we searched for MTPTs with hits showing higher similarity to plastid sequences from a lineage unrelated to the one containing the MTPT. For each potential foreign MTPT, a set of homologous plastid sequences encompassing diverse plant species were extracted from NCBI databases and aligned using MUSCLE v3.745. To confirm the identity of the donor lineage, Maximum Likelihood analyses (1,000 rapid bootstrapping replicates) under a GTR+G substitution model were performed with RAxML v.8.0.046 (settings: -f a –m GTRGAMMA –k –N 1000 –x 67840 –p 7593029 –T 2). The presence of foreign MTPTs in the published mitochondrial assemblies was confirmed by paired-end read information, when available, also, in most cases, the library insert size was longer than the MTPTs. Flanking regions (1 kb at each side) of foreign MTPTs were analyzed with BLASTN using Unipro UGENE software47 to identify their origin. When BLAST hits included diverse angiosperms, we performed evolutionary analyses of the regions flanking the MTPT with RAxML, as described above.
To identify PTMTs we parsed a total of 1,232 complete plastid genomes that were available in the NCBI Organelle Genome Database as of August 2016 against all land plant mitochondrial genomes using BLASTN v.2.4.0+ algorithm44,48 with the following settings: –task blastn –word_size 7 –evalue 1e-10. For each hit, we fetched the subject mitochondrial features and excluded from further analyses all hits that were annotated as plastid-derived sequences or hits that held ancient homology between plastid and mitochondrial genomes (atp1, rrn18, and rrn26)12. The relevant regions were blasted against NCBI nr databases to corroborate their mitochondrial origin. We selected as potential PTMTs those sequences in which the bitscore value was higher for mitochondrial hits than for plastids.
Additional Information
How to cite this article: Gandini, C. L. and Sanchez-Puerta, M. V. Foreign Plastid Sequences in Plant Mitochondria are Frequently Acquired Via Mitochondrion-to-Mitochondrion Horizontal Transfer. Sci. Rep. 7, 43402; doi: 10.1038/srep43402 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank J.P. Mower, Z. Xi and I. Small for sharing assembled DNA sequences of Bartsia, Sapria, and Acacia, respectively. We are also grateful to A.A. Edera for helping with the scripts.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.L.G. collected the data, performed the analysis and the interpretation of the data and wrote the manuscript. M.V.S.-P. performed the interpretation of the data, gave technical support and conceptual advice and wrote the manuscript. Both authors discussed the results and implications and commented on the manuscript at all stages.
References
- Timmis J. N., Ayliffe M. A., Huang C. Y. & Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- Adams K. L., Daley D. O., Qiu Y. L., Whelan J. & Palmer J. D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357 (2000). [DOI] [PubMed] [Google Scholar]
- Stegemann S., Hartmann S., Ruf S. & Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 100, 8828–8833 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J., Hu X., Sun H., Yang Y. & Huang J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun 3, 1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci 15, 11–22 (2010). [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V. Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Soc Bot Pol 83, 317–323 (2014). [Google Scholar]
- Ellis J. Promiscuous DNA–chloroplast genes inside plant mitochondria. Nature 299, 678–679 (1982). [DOI] [PubMed] [Google Scholar]
- Sloan D. B. & Wu Z. History of plastid DNA insertions reveals weak deletion and at mutation biases in angiosperm mitochondrial genomes. Genome Biol Evol 6, 3210–3221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. et al. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol Biol Evol 24, 2040–2048 (2007). [DOI] [PubMed] [Google Scholar]
- Nakazono M., Nishiwaki S., Tsutsumi N. & Hirai A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mgg Mol Gen Genetics 252, 371–378 (1996). [DOI] [PubMed] [Google Scholar]
- Miyata S., Nakazono M. & Hirai A. Transcription of plastid-derived tRNA genes in rice mitochondria. Curr Genet 34, 216–220 (1998). [DOI] [PubMed] [Google Scholar]
- Hao W. & Palmer J. D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 106, 16728–16733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Rousseau-Gueutin M. & Timmis J. N. Plastid sequences contribute to some plant mitochondrial genes. Mol Biol Evol 29, 1707–1711 (2012). [DOI] [PubMed] [Google Scholar]
- Woloszynska M., Bocer T., Mackiewicz P. & Janska H. A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus. Plant Mol Biol 56, 811–820 (2004). [DOI] [PubMed] [Google Scholar]
- Rice D. W. et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342, 1468–1473 (2013). [DOI] [PubMed] [Google Scholar]
- Xi Z. et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet 9, e1003265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. et al. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol 208, 570–583 (2015). [DOI] [PubMed] [Google Scholar]
- Smith D. R. Mitochondrion‐to‐plastid DNA transfer: it happens. New Phytol 202, 736–738 (2014). [DOI] [PubMed] [Google Scholar]
- Iorizzo M. et al. Against the traffic: The first evidence for mitochondrial DNA transfer into the plastid genome. Mob Genetic Elements 2, 261–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. C. K., Cronn R. C., Edwards C., Fishbein M. & Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol Evol 5, 1872–1885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P.-F., Zhang Y.-X., Guo Z.-H. & Li D.-Z. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci. Rep. 5, 11608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke S. V. et al. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biol 16, 140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P. J. & Bateman R. M. Developmental bases for key innovations in the seed-plant microgametophyte. Trends Plant Sci 12, 317–326 (2007). [DOI] [PubMed] [Google Scholar]
- Smith D. R. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol 3, 743–748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazkani-Covo E., Zeller R. M. & Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6, e1000834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. B. et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10, e1001241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V., García L. E., Wohlfeiler J. & Ceriotti L. F. Unparalleled replacement of native mitochondrial genes by foreign homologs in a holoparasitic plant. New Phytol, doi: 10.1111/nph.14361 (2016). [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V., Cho Y., Mower J. P., Alverson A. J. & Palmer J. D. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol 25, 1762–1777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U., Adams K. L., Thomason B. & Palmer J. D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (2003). [DOI] [PubMed] [Google Scholar]
- Bergthorsson U., Richardson A. O., Young G. J., Goertzen L. R. & Palmer J. D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. USA 101, 17747–17752 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. C., Anderson W. R. & Wurdack K. J. Gene transfer from a parasitic flowering plant to a fern. Proy Soc Lond B Bio 272, 2237–2242 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J. P., Stefanović S., Young G. J. & Palmer J. D. Plant genetics: gene transfer from parasitic to host plants. Nature 432, 165–166 (2004). [DOI] [PubMed] [Google Scholar]
- Hao W., Richardson A. O., Zheng Y. & Palmer J. D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA 107, 21576–21581 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J. P. et al. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol 8, 150 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z. et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 13, 227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S. & Bock R. Exchange of genetic material between cells in plant tissue grafts. Science 324, 649–651 (2009). [DOI] [PubMed] [Google Scholar]
- Koulintchenko M., Konstantinov Y. & Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 22, 1245–1254 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov Y. M. et al. DNA import into mitochondria. Biochemistry Moscow 81, 1044–1056 (2016). [DOI] [PubMed] [Google Scholar]
- Davis C. C. & Xi Z. Horizontal gene transfer in parasitic plants. Curr Opin Plant Biol 26, 14–19 (2015). [DOI] [PubMed] [Google Scholar]
- Hansen B. Balanophoraceae. 23, 1–80 (Flora Neotrópica, 1980). [Google Scholar]
- Williams A. V., Boykin L. M., Howell K. A., Nevill P. G. & Small I. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. Plos One 10, e0125768–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasem J. R. Parasitic weeds of the Orobanchaceae family and their natural hosts in Jordan. Weed Biol Manag 9, 112–122 (2009). [Google Scholar]
- Wicke S. et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M. & team U. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.