Abstract
Bio-ethanol production from lignocellulosic raw materials could serve as a sustainable potential for improving the supply of liquid fuels in face of the food-to-fuel competition and the growing energy demand. Xylose is the second abundant sugar of lignocelluloses hydrolysates, but its commercial-scale conversion to ethanol by fermentation is challenged by incomplete and inefficient utilization of xylose. Here, we use a coupled strategy of simultaneous maltose utilization and in-situ carbon dioxide (CO2) fixation to achieve efficient xylose fermentation by the engineered Saccharomyces cerevisiae. Our results showed that the introduction of CO2 as electron acceptor for nicotinamide adenine dinucleotide (NADH) oxidation increased the total ethanol productivity and yield at the expense of simultaneous maltose and xylose utilization. Our achievements present an innovative strategy using CO2 to drive and redistribute the central pathways of xylose to desirable products and demonstrate a possible breakthrough in product yield of sugars.
Fossil fuel depletion and environmental concerns drive the worldwide development of renewable fuel sources, with high focus on biofuels and biochemicals produced from alternative and sustainable raw materials1,2. Bio-ethanol, as typical example, can be produced by first, second and third generation processes depending on the use of hexoses, biomass cellulose-released sugars, or algae starch and cellulose, respectively3,4,5. Whereas first generation bio-ethanol processes are widely applied, they currently suffer from high production costs, low fossil fuel prices, and food-to-fuel competition6. The development of lignocellulosic raw materials is hence recognized as essential for a sustainable potential of bio-ethanol production7. Third generation bio-ethanol processes are at early stages of investigation, and only the algae left-over cake (after lipids extraction) should be considered, hence reversing into a second generation raw material of starch and cellulose. Xylose is the second most abundant sugar and consists of up to 35% of cellulosic sugars present in lignocellulosic biomass8. However, incomplete and inefficient conversion of xylose into bio-ethanol has hindered its commercial-scale processes.
Although xylose-fermenting microbes exist in nature, their ethanol production rates and tolerances are inferior to Saccharomyces cerevisiae9. Its endogenous pathway can catalyze xylulose to ethanol, but it cannot convert xylose to xylulose10. Metabolic engineering has thus been used to develop xylose-utilizing S. cerevisiae11,12,13. Generally, the xylose catabolism in engineered yeasts is mediated by a heterologously expressed fungal pathway consisting of xylose reductase (XR) and xylitol dehydrogenase (XDH)14, or by the bacterial xylose isomerase (XI)15 to covert xylose into xylulose. Compared with the XI-carrying yeasts, the XR-XDH-carrying strains had greater strengths on xylose consumption rate and ethanol productivity16. Moreover, co-expression of a xylulokinase (XK)17,18 and the key enzymes involved in the pentose phosphate pathway (PPP)19,20 with XR and XDH was able to further improve the utilization of xylose.
However, XR prefers nicotinamide adenine dinucleotide phosphate (NADPH) to nicotinamide adenine dinucleotide (NADH), while XDH uses only nicotinamide adenine dinucleotide (NAD+) as a cofactor14. Compared with the redox-neutral glycolysis21 for ethanol production from glucose, the engineered XR-XDH pathway in S. cerevisiae may lead to cofactor imbalance22. On the one hand, NADPH for XR is supplied by glucose-6-phosphate (G6P) metabolism through oxidative branch of PPP, but gluconeogenesis pathway for conversion of xylose to G6P is limited when S. cerevisiae grows on xylose23,24. On the other hand, catalysis of NAD+ to NADH by XDH usually facilitates the formation of by-products such as glycerol and xylitol14,22. Many efforts have been devoted to relieve the cofactor imbalance. Use of the mutant XR (mXR) that exhibits higher preference for NADH25 or the mutant XDH (mXDH) using nicotinamide adenine dinucleotide phosphate (NADP+)26 showed positive effects on xylose utilization. Besides, by co-expressing a heterogeneous acetate-consumption pathway27 or a carbon dioxide (CO2)-fixation pathway28 in the yeasts with XR and XDH, a decrease of by-products and an increase of ethanol from xylose were also observed since acetate acid and CO2 could be used as electric acceptor for NADH oxidation.
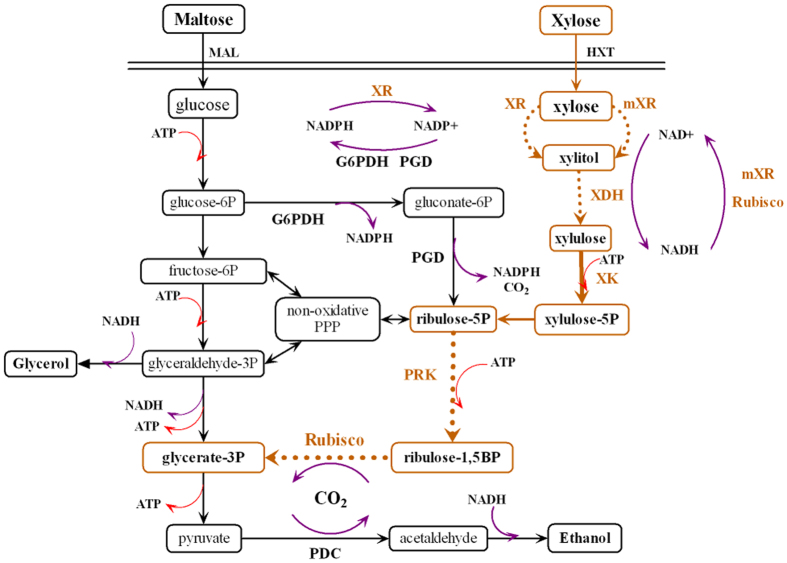
Therefore, the present research constructed a PRK-Rubisco module together with a mXR-XDH module into S. cerevisiae. Then, the resulted yeast strains were applied for xylose fermentation with simultaneous utilization of maltose and CO2 (Fig. 1). This strategy may have three major benefits: (i) The mXR-XDH module containing a mutant XR (R276H) preferring NADH, a XR, a XDH and a XK could improve the ethanol productivity and decrease the by-product accumulation. (ii) Catabolism of maltose could simultaneously supply energy and cofactors to meet the requirement for xylose metabolism. (iii) The CO2 fixation pathway may bypass the glucose repression on xylose catabolism inside the cell. The CO2 produced during fermentation can also form an appropriate atmosphere for Rubisco activity and a redox sink for improving the cofactor balance.
Figure 1. Engineered S. cerevisiae for co-utilization of xylose, maltose and CO2.
MAL: maltose transporter; HXT: hexose transporter; XR: xylose reductase; mXR: xylose reductase with the mutant of R276H; XDH: xylitol dehydrogenase; XK: xylulokinase; PRK: phosphoribulokinase; Rubisco: ribulose bisphosphate carboxylase-oxygenase; PPP: pentose phosphate pathway; G6PDH: glucose 6-phosphate dehydrogenase; PGD: 6-phosphogluconate dehydrogenase; PDC: pyruvate decarboxylase; P: phosphate; BP-bisphosphate; ATP: adenosine triphosphate.
Results
Construction of xylose metabolic pathway in S. cerevisiae
We obtained a xylose-fermenting yeast strain YSX4 (Table 1) with co-expression of wild-type XR, mutant XR (R276H), XDH and XK in S. cerevisiae YS58. In previous work, the same xylose metabolic pathway has been used in an engineered S. cerevisiae DA2429. XR (R276H) was found to exhibit much higher preference for NADH whereas XR showed two-fold higher preference for NADPH25. Theoretically, co-expression of wild-type XR and mutant XR (R276H) could enable YSX4 with comparable XR activities towards NADPH and NADH.
Table 1. Plasmids and strains used in this work.
| Plasmids and strains | Description |
|---|---|
| Plasmids | |
| pRS425 | LEU2, a multi-copy plasmid |
| pPS425-mXYL1-XYL2-XKS1-XYL1 | Expression of XR (R276H), XDH, XK and XR through pPS425 |
| YCplac33 | URA3, a single-copy plasmid |
| YCplac33-cbbM-sPRK | Expression of cbbM and sPRK through YCplac33 |
| YCplac33-cbbM-cfxP1 | Expression of cbbL1-cbbS1 and PRK through YCplac33 |
| YCplac33-cbbM-sPRK-GroEL-GroES | Expression of cbbM, sPRK and GroEL-GroES through YCplac33 |
| YCplac33-cbbL1-cbbS1-cfxP1-HSP60-HSP10 | Expression of cbbL1-cbbS1, PRK and HSP60-HSP10 through YCplac33 |
| Strains | |
| YS58 | MATa, leu2, his3,ura3, trp1 |
| YSX4 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 |
| YSC000 | YS58 harboring YCplac33 |
| YSC110 | YS58 harboring YCplac33-cbbM-sPRK |
| YSC111 | YS58 harboring YCplac33-cbbM-sPRK-GroEL-GroES |
| YSC220 | YS58 harboring YCplac33-cbbM-cfxP1 |
| YSC222 | YS58 harboring YCplac33-cbbM-cfxP1- HSP60-HSP10 |
| YSX4C000 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 and YCplac33 |
| YSX4C110 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 and YCplac33-cbbM-sPRK |
| YSX4C111 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 and YCplac33-cbbM-sPRK-GroEL-GroES |
| YSX4C220 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 and YCplac33-cbbL1-cbbS1-cfxP1 |
| YSX4C222 | YS58 harboring pPS425-mXYL1-XYL2-XKS1-XYL1 and YCplac33-cbbL1-cbbS1-cfxP1-HSP60-HSP10 |
The fermentation performance of the resulted YSX4 was evaluated under an oxygen-limited condition with xylose and glucose, or xylose and maltose as carbon source respectively (Table 2). We found that the xylose consumption rate of YSX4 (0.57 g/L/h) was similar with that of DA24 (0.53 g/L/h) on a glucose-xylose mixture, although the key enzymes involved in the xylose metabolic pathway in YSX4 were expressed by using a multi-copy plasmid, while in DA24, they were integrated in genome. Interestingly, compared with the glucose-xylose mixture, YSX4 exhibited a higher efficiency of xylose utilization (0.70 g/L/h) when grown on xylose and maltose, which is in agreement with previous results using cellobiose and xylose as carbon source29,30. Evidences showed that intracellular hydrolysis of cellobiose minimized glucose repression on xylose uptake, and simultaneous fermentation of cellobiose and xylose was capable of improving ethanol productivity when compared to fermentation with xylose only30. For both of maltose and cellobiose, disaccharides are formed from two glucose molecules. The difference is that glucose units are joined with an α (1 → 4) bond in maltose and a β (1 → 4) bond in cellobiose respectively. Therefore, we suspected that YSX4 could also co-ferment maltose and xylose, thus consumption of xylose by YSX4 was enhanced when cells were grown on maltose-xylose mixture.
Table 2. Fermentation performances of the engineered yeasts in this work.
| Sugars (g/L) | Strains | YEth | VEth | RXyl | RGlc/Mal | RTotal | Reference |
|---|---|---|---|---|---|---|---|
| Xylose (40) | YSX4 | 0.35 | 0.12 | 0.33 | — | — | This study |
| Glucose/Xylose | DA24 | 0.39 | 0.74 | 0.53 | — | 1.5 | 33 |
| (70/40) | YSX4 | 0.38 | 0.60 | 0.57 | 3.8 | 1.6 | This study |
| Maltose/Xylose (70/40) | YSX4 | 0.42 | 0.88 | 0.76 | 3.2 | 2.1 | This study |
| Glucose (70) | YSX4 | 0.45 | 1.8 | — | 3.9 | — | This study |
| Maltose (70) | YSX4 | 0.44 | 1.7 | — | 3.5 | — | This study |
| Maltose/Xylose (70/40) | YSX4C000 | 0.41 | 0.9 | 0.69 | 2.9 | 1.9 | This study |
| YSX4C110 | 0.41 | 1.0 | 0.73 | 2.8 | 2.0 | This study | |
| YSX4C111 | 0.46 | 1.3 | 0.97 | 2.4 | 2.8 | This study | |
| YSX4C222 | 0.47 | 1.5 | 1.1 | 2.2 | 3.1 | This study | |
| Maltose/Xylose (30/30) | YSX4C000 | 0.37 | 0.32 | 0.54 | 2.1 | 0.86 | This study |
| YSX4C110 | 0.39 | 0.38 | 0.58 | 2.0 | 0.93 | This study | |
| YSX4C111 | 0.43 | 0.50 | 0.68 | 1.8 | 1.2 | This study | |
| YSX4C222 | 0.46 | 0.57 | 0.70 | 1.6 | 1.3 | This study |
YEth: ethanol yield (g ethanol/g sugar): VEth: ethanol productivity (g/L/h); RXyl: xylose consumption rate (g/L/h); RGlc/Mal: glucose or maltose consumption rate (g/L/h); RTotal: total sugar consumption rate (g/L/h).
Functional expression of CO2-fixation pathway in S. cerevisiae
As a heterotrophic microorganism, S. cerevisiae intracellularly converts ribulose-5-phosphate (Ru5P) to glycerate-3-phosphate (G3P) through the non-oxidative branch of PPP without consumption of CO2. In contrast, many autotrophic microbes achieve this conversion by a two-step reaction using ribulose bisphosphate carboxylase-oxygenase (Rubisco) and phosphoribulokinase (PRK)31,32. PRK catalyzes Ru5P to ribulose-1,5-bisphosphate (Ru1,5BP), while Rubisco converts Ru1,5BP to G3P with CO2 fixation. There are four known forms of Rubisco in nature33. Form-I Rubisco (L8S8) is composed of eight large subunits and eight small subunits, of which the small subunits are able to concentrate surrounding CO2 to improve the reactivity of the large subunits. Form-II (L8) only contains eight large subunits. In previous works, the gene of cbbM from Rhodospirillum rubrum or Thiobacillus denitrificans coding for Form-II Rubisco and the gene of sPRK from Spinacia oleracea coding for PRK have already been successfully expressed in S. cerevisiae28,34. Interestingly, it was found that functional expression of cbbM in yeast depended on co-expression of a chaperone (GroEL-GroES) from Escherichia coli. In this work, we engineered S. cerevisiae YS58 with a Form-II based CO2 fixation system by co-expression of cbbM, sPRK, and GroEL-GroES, resulting in a strain named YSC111. Similarly, in another yeast strain called YSC222, cbbL1-cbbS1 (Form-I Rubisco gene) and cfxP1 (PRK gene) from Ralstonia eutropha H1635 constructed a Form-I based CO2 fixation system. To ensure the functionality of Form-I Rubisco, an endogenous chaperone (Hsp60-HSP10) of S. cerevisiae YS58 was simultaneously over-expressed.
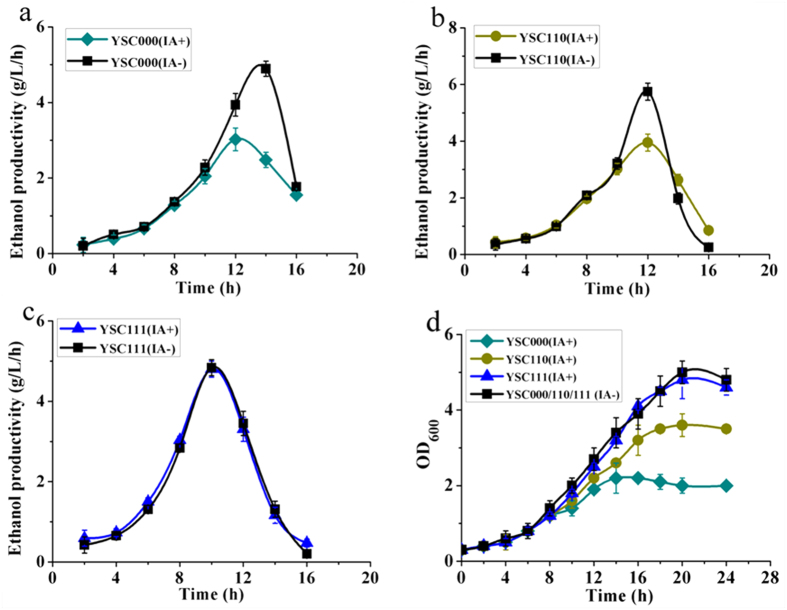
To test whether the CO2-fixation pathway was workable in the engineered S. cerevisiae, iodoacetate (IA) was used to inhibit the activity of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), thus repressing the cellular glycolysis. After addition of IA to 0.19 g/L (~0.001 mol/L), the ethanol productivity of YSC000 or YSC110 decreased sharply (Fig. 2a,b), but no negative effects on YSC111were observed (Fig. 2c). YSC111 also showed similar growth profiles with or without IA addition (Fig. 2d). The ethanol yields of YSC222 and YSC111 were slightly affected by IA, but the ethanol yields of YSC220 and YSC110 lacking of chaperones decreased by 27% (Table S1), indicating that chaperones were essential for functional expression of both Form-I and Form-II Rubisco. We also found that the performance of the engineered CO2-fixation pathway would be enhanced with increase of glucose concentrations (Fig. S1), which was probably due to more surrounding CO2 present.
Figure 2. Fermentation profiles of YSC000, YSC110 and YSC111 in YP medium containing 70 g/L glucose with IA (IA+) or without IA (IA-) addition.
Effects of CO2 fixation on xylose utilization
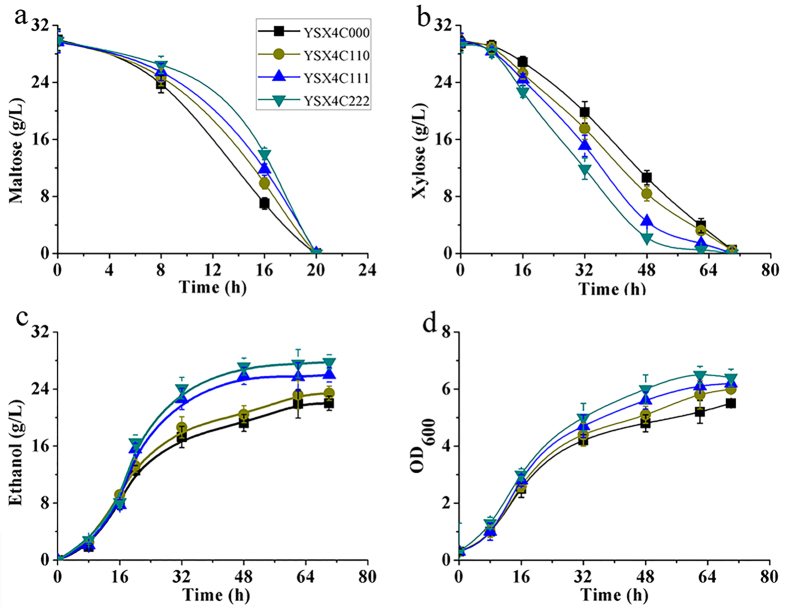
The functional CO2-fixation pathway was constructed in the engineered S. cerevisiae YSX4, resulting in YSX4C111 and YSX4C222. YSX4C222 exhibited the highest xylose consumption rate, ethanol productivity and strain growth rate (Fig. 3). Compared with YSX4C000, the rate of xylose consumption elevated from 0.54 g/L/h to 0.70 g/L/h by using YSX4C222 (Table 3), while both of the final ethanol level and cell density got a over 25% increase (Fig. 3c,d).
Figure 3. Fermentation profiles of YSX4C000, YSX4C110, YSX4C111 and YSX4C222 in YP medium containing 30 g/L maltose and 30 g/L xylose.
Table 3. The CO2-fixation rates of engineered YSX4C000, YSX4C111 and YSX4C222.
| Sugars (g/L) | Strains | CEth | CXyl | CGly | CXylit | CAcet | DCW | RCO2 |
|---|---|---|---|---|---|---|---|---|
| Maltose/Xylose (70/40) | YSX4C000 | 38.9 ± 0.5 | 15.2 ± 0.5 | 3.2 ± 0.3 | 2.8 ± 0.1 | 0.2 ± 0.1 | 5.2 ± 0.2 | — |
| YSX4C111 | 48.2 ± 0.5 | 5.2 ± 0.5 | 2.5 ± 0.2 | 2.4 ± 0.2 | 0.1 ± 0.1 | 5.7 ± 0.3 | 336.6 ± 1.5 | |
| YSX4C222 | 51.9 ± 0.5 | 0.3 ± 0.3 | 1.6 ± 0.2 | 2.0 ± 0.1 | 0.1 ± 0.1 | 6.4 ± 0.3 | 436.3 ± 3.0 |
CEth: ethanol concentration (g/L); CXyl: Residual xylose concentration (g/L); CGly: glycerol concentration (g/L); CXylit: xylitol ethanol concentration (g/L); CAcet: acetate concentration (g/L); DCW: Dry Cell Weight (g/L); RCO2: CO2-fixation rate (mg CO2/L/h).
Then, YSX4C111 and YSX4C222 were applied in the medium containing 70 g/L maltose and 40 g/L xylose. As shown in Table 2, the xylose consumption rates of YSX4C111 and YSX4C222 arrived to 0.97 and 1.1 g/L/h respectively. The total sugar consumption rate and the ethanol yield of YSX4C222 reached 3.1 g/L/h and 0.47 g/g sugars, which was 63% and 15% higher than YSX4C000. YSX4C222 cell extract also showed the highest carboxylation activity (Fig. S2). These findings suggested that the Form-I based CO2 fixation system was preferred in facilitation of the xylose metabolism.
Relative quantification of heterotrophic CO2 fixation
To evaluate the strength of the CO2 flux in YSX4C111 and YSX4C222, a reported metabolic flux index, MFIh-CO236, was employed to indicate the metabolic flux ratio between the CO2-fixation and the PPP-based pathways. The G3P generated by these two pathways was differentiated by using 13C-labeled CO2 and unlabeled sugars to obtain the MFIh-CO2 value.
As illustrated in Fig. S3, we assumed that a mole of glyceraldehyde-3-phosphate (GADP) was metabolized to G3P, while b mole of 13CO2 from NaH13CO3 and 12CO2 from sugars was fixed to G3P within a given time period. Moreover, we assumed that c mole of unlabeled G3P and d mole of 13C-G3P were channeled into the downstream metabolism. Fermentation experiments were performed using 10 g/L maltose and 20 g/L xylose with 100 mM NaH13CO3 and cell density of OD600 = ~0.1. The amounts of 13C-labeled and unlabeled G3P were measured by LC-MS/MS. S. cerevisiae YS58 was cultivated in a medium free of any carbon isotope to determine the ratio of 13C-G3P to the unlabeled G3P as the basal isotopic level, which was about 3.1%. The actually detected molar amount of 13C-G3P (y) can be calculated by Eq. (1), while the actually detected unlabeled G3P (x) can be calculated by Eq. (2).
 |
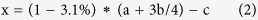 |
Under a metabolic steady-state, the relationship of d, c, x and y is shown in Eq. (3).
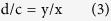 |
Therefore, the value of MFIh−CO2 can be calculated by Eq. (4).
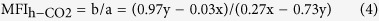 |
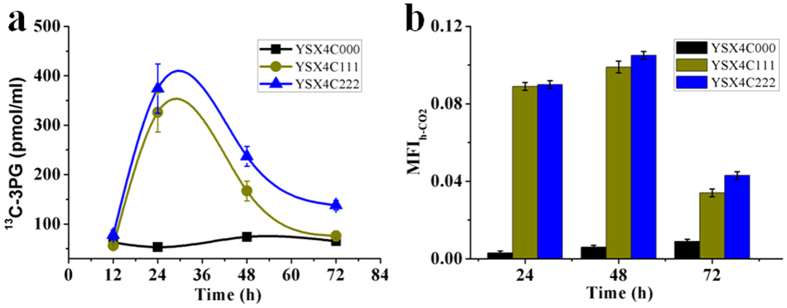
As shown in Fig. 4a, 13C-G3P amount in YSX4C000 did not change during fermentation. In contrast, a significant increase of 13C-G3P was observed in YSX4C111 and YSX4C222. These results demonstrated that CO2 was able to be incorporated into G3P through the engineered CO2-fixation pathways. The MFIh-CO2 values of these strains at different times were calculated to evaluate the relative CO2 flux (Fig. 4b), and the results indicated that over 8% of Ru5P from xylose was converted to G3P through the CO2-fixation pathway in YSX4C111 or YSX4C222. More interestingly, the higher levels of 13C-G3P and MFIh-CO2 in YSX4C222 than in YSXC111 suggested that the Form-I based system was more efficient than Form-II in S. cerevisiae.
Figure 4.
Intracellular 13C-G3P contents (a) and MFIh-CO2 values (b) of YSX4C000, YSX4C111 and YSX4C222.
Determination of the CO2-fixation rate in the engineered yeasts
The carbon balance of wild-type S. cerevisiae37 indicated that 96% carbon of the consumed sugars (Cconsumed sugars) was incorporated into biomass (Cbiomass), CO2, ethanol (Cethanol), xylitol (Cxylitol), glycerol (Cglycerol) and Acetate (Cacetate). Therefore, the carbon of CO2 fixed by YSX4C111 or YSX4C222 (C fixed CO2) can be calculated using Eq. (5). CCO2 with biomass and CCO2 with ethanol represent the CO2 from cell respiration and from ethanol biosynthesis respectively.
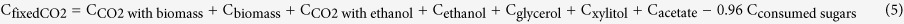 |
We assume that CCO2 with biomass is in direct proportion to Cbiomass. Therefore, Eq. (5) can be transformed into Eq. (6).
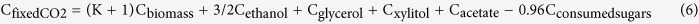 |
The carbon balance of YSX4C000 is shown as Eq. (7).
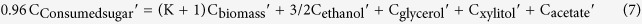 |
Substitution of K in Eq. (6) into Eq. (7), thus
 |
Fermentation data of YSX4C000, YSX4C111 or YSX4C222 on 70 g/L maltose and 40 g/L xylose are summarized in Table 3. The results showed that our yeasts were able to fix CO2 at a rate of 336.6–436.3 mg CO2/L/h, significantly exceeding the natural or the engineered microbes (5.8 to 147.0 mg CO2/L/h) in previous reports36 (Table S2).
Discussion
Currently, xylose fermentation with the engineered S. cerevisiae is inferior to glucose fermentation by the wild-type S. cerevisiae. Besides cofactor imbalance of the engineered XR-XDH pathway, supply of NADPH was limited because expression of the enzymes involved in gluconeogenesis in S. cerevisiae cannot be upregulated on xylose. Some rate-limiting enzymes in the non-oxidative branch of PPP need to be over-expressed to enhance the xylose downstream metabolic flux. Here, we engineered S. cerevisiae to achieve efficient bio-ethanol production from maltose and xylose with CO2 fixation. NADH could be reduced through the mXR and CO2-fixation pathway, while the NADPH for the wild-type XR and the adenosine triphosphate (ATP) for activating Ru5P to Ru1,5BP used in Rubisco reaction were provided by co-fermentation of maltose and xylose.
Xylose can enter the cell by facilitated diffusion through some yeast hexose transporters (HXTs)38. For engineered XR-XDH-carrying yeasts, intracellular xylose conversion is only slightly affected by catabolism of intracellular glucose, but xylose transport is strongly inhibited by extracellular glucose39. The engineered xylose transporters by non-rational mutagenesis or rational design have been used to improve simultaneous utilization of xylose and glucose40,41. However, the specific xylose transporters without glucose inhibition have still not been developed42. On the other hand, introduction of a cellodextrin transport (CDT-1) from Neurospora crassa into the yeast with intracellular β-glucosidase (GH1-1) achieved simultaneous utilization of cellobiose and xylose and showed synergistic effects due to sufficient NADPH supply29,30. But in yeast, β-glucosidase (GH1-1) can convert cellobiose to cellodextrins (cellotriose, cellotetraose) by its transglycosylation activity, while cellodextrin transport (CDT-1) can transport cellodextrins and glucose into the medium. This problem resulted in low ethanol yield and productivity from cellobiose and xylose. In this study, we employed another disaccharide, maltose, which can be utilized as efficiently as glucose by the wild-type yeast, to facilitate xylose metabolism and achieved the synergistic effects.
Heterotrophic Form-I and Form-II based CO2 fixation systems were successfully expressed in S. cerevisiae. We observed that the engineered yeast YSX4C111 and YSX4C222 grew better and exhibited higher ethanol productivity than YSX4C000. The results suggest that additional ATP consumption by implementing the CO2-fixation pathway had a less negative effect on cell growth than the beneficial effect that the pathway might bring during co-fermentation of xylose and maltose. With increase of sugar concentrations in fermentation, the engineered yeasts expressing CO2 fixation pathway showed better ethanol productivity. The possible reason was more CO2 production from sugars increased environmental CO2 and HCO3− concentrations so that improved the carboxylation activity of Rubisco under oxygen-limited condition. The same effects were also observed by supplementation of external CO234. The activity of Rubisco could further be optimized through expressing heterogeneous carbonic anhydrase (CA)36, which catalyzes the reversible conversion of CO2 and HCO3−, or constructing a microcompartment for Rubisco43, similar to the semi-permeable carboxysome in cyanobacteria.
At present, low efficiency of photosynthetic processes and electro-synthetic processes using photoautotrophic and chemoautotrophic microbes are limited in their growths and productivities32,44. Differing from autotrophic microbes, heterotrophic microbes such as S. cerevisiae, which has very short doubling time, have been widely applied in industrial production of bio-chemicals. The CO2 fixation at the expense of sugars in heterotrophic S. cerevisiae presented high CO2-fixation rates and product yields. The construction of a functional CO2-fixation pathway for efficient ethanol production from xylose and maltose in S. cerevisiae would create a foundation for other biofuels and chemicals production from lignocellulosic biomass.
Methods
Strains, plasmids and media
Escherichia coli Trans10 (TransGen Biotech) was used for genetic manipulation. The plasmids and yeasts used in this work were summarized in Table 1. Synthetic complete (SC)-Ura-Leu minimal medium contains 0.67% yeast nitrogen base with ammonium sulfate and without amino acids (YNB), 2% glucose, 0.005% histidine and 0.01% tryptophan. SC-Leu or SC-Ura minimal medium contains 0.67% YNB, 2% glucose, 0.005% histidine, 0.01% tryptophan, 0.005% leucine or 0.01% Uracil. SC-Ura-Leu, SC-Ura and SC-Leu medium contains 0.67% YNB, 2% glucose, 0.01% (adenine, arginine, cysteine, lysine, threonine, tryptophan) and 0.005% (aspartic acid, histidine, isoleucine, methionine, phenylalanine, proline, serine, tyrosine, valine), while appropriate leucine and uracil addition when required. YP medium contains 1% yeast extract and 2% peptone.
Construction of xylose and CO2-fixaiton pathway
The gene expression cassettes of xylose pathway were constructed. The genes of XYL1 (Gene ID: 4839234, coding for XR) and XYL2 (Gene ID: 4852013, coding for XDH) from S. stipitis CBS6054 (Laboratory-stored), and XKS1 (Gene ID: 853108, coding for XK) from S. cerevisiae YS58 (laboratory-stored) were amplified by using the primers in Table S3. Obtained gene fragments were inserted between promoters and terminators individually in the recombinant pUC19 as shown in Table S4. The mutant XYL1 (A826C, G827A, A828C) coding for XR (R276H) was amplified from pUC19-TEF2p-XYL1-TPI1t using point mutant primers (R276H-F and R276H-R) (Table S3), and then mutant XYL1 fragment was cloned and inserted into pUC19-TEF1p-mXYL1-PGIt (Table S4).
The genes of sPRK (GenBank: X07654.1) and cfxP1 (Gene ID: 4456348), cbbM (GenBank: L37437.2) and cbbL1-cbbS1 (GenBank: U20584.1), and GroEL-GroES (Gene ID: 948665) and HSP60-HSP10 (Gene ID: 850963, no mitochondrion signal peptide) were chosen for construction of the CO2-fixation pathway. Except HSP60-HSP10, other genes were codon-optimized by Jcat45 (http://www.jcat.de/) and synthesized in KLSBE (Key Laboratory of Systems Bioengineering, Ministry of Education, China). The synthetic gene fragments were linked into pEASY-Blunt with the promoters and terminators (Table S4). The gene expression cassettes were linked into pRS425 or YCplac33 vectors to generate yeast expression plasmids (Table 1). S. cerevisiae transformants were selected on SC-Ura-Leu, SC-Leu or SC-Ura minimal medium.
Fermentation experiments
The engineered yeast YSX4 was pre-cultured in SC-Leu medium. YSC000, YSC110, YSC111 and YSC222 were pre-cultured in SC-Ura medium. YSX4C000, YSX4C110, YSX4C111 and YSX4C222 were pre-cultured in SC-Ura-Leu medium. After washing with distilled water, the cells were inoculated into 200 mL YP medium with appropriate sugars (maltose, glucose or xylose) in 500 mL flasks. All fermentation experiments were carried out at 30 °C and 200 rpm under oxygen-limited conditions. The initial cell density was adjusted to an OD600 (optical density at 600 nm) of ~0.5. Rubisco activity was determined through Rubisco Carboxylation Activity Assay Kit (GENMED, Shanghai) by UV-visible Spectrophotometer SU-2000.
IA inhibition assay
The cells of YSC000, YSC110, YSC111 and YSC222 were inoculated into 200 mL YP medium with 70 g/L glucose in 500 mL flasks. The initial cell density was adjusted to an OD600 of ~0.1. IA was added to the media at 8 h. Ethanol concentration and OD600 were detected in whole fermentation process.
Analytical methods
Cell growth was monitored using UV-visible Spectrophotometer SU-2000 (OnLab Instruments). Maltose, glucose, xylose, xylitol, glycerol, acetate and ethanol were quantified by high performance liquid chromatography (Agilent 1200 Series HPLC system) equipped with a refractive index detector (Shimadzu, Japan) and an Bio-Rad Aminex HPX-87H organic acid analysis column (7.8 × 300 mm) which was maintained at 50 °C and used 0.05 mM sulfuric acids as mobile phase. The sample injection volume was 10 μL and the flow rate was 0.6 mL/min. Metabolites was detected by liquid chromatography-mass spectrometry/mass spectrometry system (Agilent 6460 series LC-MS/MS system) with Agilent XDC18 column (5 uM, 150 mm × 4.6 mm)36. Di-n-butylammonium acetate (DBAA) and NaH13CO3 were purchased from Sigma-Aldrich. Methanol was purchased from Fisher Scientific. The mobile phase was the mixture of solution A (water with 5 mM DBAA) and solution B (methanol with 5 mM DBAA) at the gradient shown in Table S6. The flow rate was 0.6 mL/min. The injection volume was 50 μL and the column temperature was 40 °C. The negative ion and selected multiple reactions monitoring (MRM) mode were used for MS detection. All experiments were conducted at least in triplicate, and the error bars in the figures denote the standard deviation from the means of independent experiments.
Additional Information
How to cite this article: Li, Y.-J. et al. Engineered yeast with a CO2-fixation pathway to improve the bio-ethanol production from xylose-mixed sugars. Sci. Rep. 7, 43875; doi: 10.1038/srep43875 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Basic Research Program of China (973 program, 2013CB733600), the National Nature Science Foundation of China (21390202) and the National Natural Science Foundation of China (21376023). We acknowledge Prof. Ying-Jin Yuan (Key Laboratory of Systems Bioengineering, Tianjin University) for help in gene synthesis, Prof. Yin Li and Dr. Fu-Yu Gong (Institute of Microbiology, Chinese Academy of Sciences) for help in determination of metabolites by LC-MS/MS, and Jan Baeyens (University of Warwick, School of Engineering, Coventry, UK) for his help in revising the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.-J.L., L.-H.F. and T.-W.T. conceived of the study, designed experiments, performed the statistical analysis, wrote and revised the manuscript. Y.-J.L., M.-M.W., Y.-W.C. and M.W. performed experiments. All authors read and approved the final manuscript.
References
- Baeyens J. et al. Challenges and opportunities in improving the production of bio-ethanol. Prog. Energ. Combust. 47, 60–88 (2015). [Google Scholar]
- Ragauskas A. J. et al. The path forward for biofuels and biomaterials. Science 311, 484–489 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang H. L., Baeyens J. & Tan T. W. The bubble-induced mixing in starch-to-ethanol fermenters. Chem. Eng. Res. Des. 90, 2122–2128 (2012). [Google Scholar]
- Kang Q., Appels L., Tan T. & Dewil R. Bioethanol from lignocellulosic biomass: current findings determine research priorities. The Scientific world J. 2014, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C. E. D. F. & Bertucco A. Bioethanol from microalgae and cyanobacteria: a review and technological outlook. Process Biochem. 51, 1833–1842 (2016). [Google Scholar]
- Tilman D. et al. Beneficial biofuels-the food, energy, and environment trilemma. Science 325, 270–271 (2009). [DOI] [PubMed] [Google Scholar]
- Brethauer S. & Studer M. H. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ. Sci. 7, 1446–1453 (2014). [Google Scholar]
- Jeffries T. W. & Jin Y. S. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63, 495–509 (2004). [DOI] [PubMed] [Google Scholar]
- Jeffries T. W. Emerging technology for fermenting D-xylose. Trends Biotechnol. 3, 208–212 (1985). [Google Scholar]
- Toivari M. H., Salusjärvi L., Ruohonen L. & Penttilä M. Endogenous xylose pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70, 3681–3686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L. H. et al. Engineering yeast with bifunctional minicellulosome and cellodextrin pathway for co-utilization of cellulose-mixed sugars. Biotechnol. Biofuels 9, 1–11 (2016). Jeffries, T. W. & Jin, Y. S. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63, 495–509 (2004).
- Nielsen J., Larsson C., van Maris A. & Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr. Opin.Biotechnol. 24, 398–404 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang G. C., Liu J. J., Kong I. I., Kwak S. & Jin Y. S. Combining C6 and C5 sugar metabolism for enhancing microbial bioconversion. Curr. Opin. Chem. Biol. 29, 49–57 (2015). [DOI] [PubMed] [Google Scholar]
- Walfridsson M., Anderlund M., Bao X. & Hahn-Hagerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl. Microbiol. Biotechnol. 48, 218–224 (1997). [DOI] [PubMed] [Google Scholar]
- Kuyper M. et al. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res. 4, 69–78 (2003). [DOI] [PubMed] [Google Scholar]
- Karhumaa K., Sanchez R. G., Hahn-Hagerdal B. & Gorwa-Grauslund M. F. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell. Fact. 6, 1–10 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivari M. H., Aristidou A., Ruohonen L. & Penttilä M. Conversion of xylose to ethanol by recombinant saccharomyces cerevisiae: importance of xylulokinase (xks1) and oxygen availability. Metab. Eng. 3, 236–249 (2001). [DOI] [PubMed] [Google Scholar]
- Kim B., Du J., Eriksen D. T. & Zhao H. Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl. Environ. Microbiol. 79, 931–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson M., Hallborn J., Penttilä M., Keränen S. & Hahnhägerdal B. Xylose-metabolizing saccharomyces cerevisiae strains overexpressing the tkl1 and tal1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61, 4184–90 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer L. N. et al. Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab. Eng. 25, 20–29 (2014). [DOI] [PubMed] [Google Scholar]
- Bar-Even A., Flamholz A., Noor E. & Milo R. Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat. Chem. Biol. 8, 509–17 (2012). [DOI] [PubMed] [Google Scholar]
- Ho N. W. Y., Chen Z. & Brainard A. P. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64, 1852–1859 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R. E. et al. Saccharomyces cerevisiae engineered for xylose metabolism requires gluconeogenesis and the oxidative branch of the pentose phosphate pathway for aerobic xylose assimilation. Yeast 28, 645–660 (2011). [DOI] [PubMed] [Google Scholar]
- Runquist D., Hahn-Hägerdal B. & Bettiga A. M. Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb. Cell. Fact. 8, 49–49 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. et al. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153, 3044–54 (2007). [DOI] [PubMed] [Google Scholar]
- Watanabe S. et al. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+ -dependent xylitol dehydrogenase. J. Biotechnol. 130, 316–9 (2007). [DOI] [PubMed] [Google Scholar]
- Wei N., Quarterman J., Kim S. R., Cate J. H. & Jin Y. S. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat. Commun. 4, 2580–2580 (2013). [DOI] [PubMed] [Google Scholar]
- Xia P. F. et al. Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. doi: 10.1021/acssynbio.6b00167 (2016). [DOI] [PubMed] [Google Scholar]
- Ha S. J. et al. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. USA 108, 504–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Ha S. J., Wei N., Oh E. J. & Jin Y. S. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends biotechnol. 30, 274–282 (2012). [DOI] [PubMed] [Google Scholar]
- Wolf M. R. & Lenz L. L. The evolution of the calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Current Genetics 32, 1–18 (1997). [DOI] [PubMed] [Google Scholar]
- Li H. & Liao J. C. Biological conversion of carbon dioxide to photosynthetic fuels and electrofuels. Energy Environ. Sci. 6, 2892–2899 (2013). [Google Scholar]
- Tabita R. S. S., Hanson T. E., Kreel N. E. & Scott S. S. Distinct form i, ii, iii, and iv rubisco proteins from the three kingdoms of life provide clues about rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524 (2008). [DOI] [PubMed] [Google Scholar]
- Víctor G. M. et al. Carbon dioxide fixation by calvin-cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 6, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann A. et al. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24, 1257–62 (2006). [DOI] [PubMed] [Google Scholar]
- Gong F. et al. Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation. Biotechnol. Biofuels 8, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick O. & Wittmann C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13c flux analysis. Microb. Cell. Fact. 4, 1–16 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher T., Becker J., Gárdonyi M., Hahnhägerdal B. & Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148, 2783–8 (2002). [DOI] [PubMed] [Google Scholar]
- Subtil T. & Boles E. Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 5, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M. et al. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5, 925–934 (2005). [DOI] [PubMed] [Google Scholar]
- Young E. M., Tong A., Bui H., Spofford C. & Alper H. S. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc. Natl. Acad. Sci. USA 111, 131–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M. & Boles E. Engineering of yeast hexose transporters to transport d-xylose without inhibition by d-glucose. Proc. Natl. Acad. Sci. USA 111, 5159–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci W. et al. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA 109, 478–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S. H. & Atsumi S. Photosynthetic approaches to chemical biotechnology. Curr. Opin. Biotech. 24, 1031–1036 (2013). [DOI] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Hempel D. C. & Jahn D. Jcat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, 526–531 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.