Abstract
In the last 40 years only one new antitubercular drug has been approved, whilst resistance to current drugs, including rifampicin, is spreading. Here, we used the model organism Mycobacterium smegmatis to study mechanisms of phenotypic mycobacterial resistance, employing quantitative mass spectrometry-based proteomics to investigate the temporal effects of sub-lethal concentrations of rifampicin on the mycobacterial proteome at time-points corresponding to early response, onset of bacteriostasis and early recovery. Across 18 samples, a total of 3,218 proteins were identified from 31,846 distinct peptides averaging 16,250 identified peptides per sample. We found evidence that two component signal transduction systems (e.g. MprA/MprB) play a major role during initial mycobacterial adaptive responses to sub-lethal rifampicin and that, after dampening an initial SOS response, the bacteria supress the DevR (DosR) regulon and also upregulate their transcriptional and translational machineries. Furthermore, we found a co-ordinated dysregulation in haeme and mycobactin synthesis. Finally, gradual upregulation of the M. smegmatis-specific rifampin ADP-ribosyl transferase was observed which, together with upregulation of transcriptional and translational machinery, likely explains recovery of normal growth. Overall, our data indicates that in mycobacteria, sub-lethal rifampicin triggers a concerted phenotypic response that contrasts significantly with that observed at higher antimicrobial doses.
Mycobacterium tuberculosis is the causative agent of tuberculosis disease (TB), which accounts for ~1.5 million deaths and ~9 million new cases annually1. The global rise in these figures has begun to turn thanks to effective drug treatments and increased drive for diagnosis and treatment of infected individuals. However, the effective treatment of this disease relies upon anti-tubercular drugs in use since the 1950 s2 with only 1 new drug being licenced by the FDA in almost 40 years3. In the light of this slow introduction of new TB therapies - drug resistance, including multi-drug resistant TB (MDR-TB), has emerged as a major threat to the success of global TB management. As evidence of this, in 2013 there were an estimated 480 000 new cases of, and 210 000 deaths from, MDR-TB.
Exposure of M. tuberculosis to sub-lethal drug concentrations allows the bacterium to persist and, importantly, replicate in the presence of drug. Whilst the Darwinian selection of resistant mutants by lethal concentrations has received much attention over the years, there is now a growing awareness that sub-lethal drug exposure can bring about non-lethal selection and have large effects on bacterial biology4. Sub-lethal concentrations of anti-tubercular drugs can result from poor adherence to treatment schedules, incorrect dosing or drug regimen, irregular availability of drugs or poor penetration of the infected tissue by the drug. In illustration of this last mechanism, Dartois and Barry used mass spectrometry to demonstrate the poor penetration of human granulomas by rifampicin5.
High pressure liquid chromatography and modern mass spectrometry-based proteomics enables the rapid quantitation of thousands of proteins across many samples. This allows investigation, in an untargeted manner, into how the proteome as a whole changes in an organism in response to stimulus. Rifampicin binds to the RNA polymerase subunit RpoB and is one of the frontline anti-tubercular drugs; acquisition of resistance to rifampicin is a strong predictor for later development of MDR-TB6. Additionally, whilst one traditionally thinks of antimicrobial agents as foreign to the bacterial target, many antibiotic drugs, including rifampicin, are polyketides – a class of compound synthesised by bacteria and commonly employed in bacterial chemical warfare as a species seeks dominance in a nutrient rich environment. With this in mind, we reasoned that polyketide antibiotics such as rifampicin may in fact not be totally foreign to mycobacterial species and that ancient, broad specificity evolutionary mechanisms might therefore exist for mycobacteria to sense polyketide exposure and rapidly induce phenotypic resistance.
We further reasoned that the underlying mechanisms of phenotypic resistance to rifampicin might thus be revealed through proteomic analysis of mycobacterial spp. exposed to sub-lethal drug doses. Previously, Hu et al.7 used quantitative proteomics to study mycobacterial response to isoniazid in an M. bovis BCG strain, whilst Koul et al.8 studied the delayed M. tuberculosis response to bedaquiline. Just recently de Keijzer et al. reported a time-course, phospho-proteomic analysis of the effects of very high dose rifampicin treatment in M. tuberculosis9. This analysis provides good insight into the early, phenotypic responses of M. tuberculosis to strong rifampicin challenge. As rifampicin concentration in plasma experiences pulses of high concentration, followed by fading of the same, this may be useful and complementary to the present study where we consider the phenotypic adaptations of mycobacteria exposed to a relatively low, or sub-inhibitory/sub-lethal dosage which, for the reasons discussed above, may better represent the challenge experienced by the organism in situ. It is also more likely that signalling responses specific to rifampicin will be uncovered in a low-dose model than in a high-dose model where general stress response is likely to overpower the specific signalling response. The time-course used in the present study also examines later time points, relative to the generation-time of the mycobacterial species, allowing the examination of time points corresponding with early response, onset of bacteriostasis and early recovery of normal growth.
Here, we used M. smegmatis as a non-pathogenic model for M. tuberculosis, since it is considered to be a suitable model for the study of regulatory mechanisms of mycobacterial resistance10. In the current study we report the time-dependent, quantitative dysregulation of the M. smegmatis proteome upon exposure in vitro to sub-lethal rifampicin concentrations. Through this study, we gain insight into a series of rapid, temporal changes in the M. smegmatis proteome that collectively enable the bacilli to avoid cell death upon rifampicin exposure and instead to become phenotypically resistant to the antibiotic.
Materials and Methods
Culture Conditions, Colony Counts and Killing Curves
Wild Type Mycobacterium smegmatis (strain mc2155) glycerol stocks were streaked on 7H10 Middlebrook agar plates and single colonies were picked and inoculated in 7H9 Middlebrook broth supplemented with 10% OADC Enrichment Media and 0.2% glycerol and grown to A600nm 0.6 or A600nm 1.2 for synchronising initial inoculum in larger cultures. Cultures were grown to mid-log phase at 37 °C on a shaker at 120–130 rpm before inoculation into 150 mL 7H9, prepared as above with the addition of filter sterilised (0.2 µm cellulose acetate filter) Tween 20 to final concentration of 0.1% to prevent clumping of bacteria.
For determination of sub-lethal rifampicin drug concentration, triplicate flasks of 150 mL liquid cultures at mid-log phase (generated as above; A600nm 1.2) were treated with various concentrations of rifampicin dissolved in DMSO, or with DMSO only as a control. Cell density, and hence replication and death, were inferred from A600nm readings measured at 15 minute intervals for 2 hours, followed by 30 minute intervals for a further 2 hours (4 hours total monitoring) with a final measurement approximately 14 hours later using a Varian Cary 50 UV-visible spectrophotometer. At the highest concentrations used, rifampicin showed negligible absorbance and DMSO negligible impact on cell growth.
Colony counts for mid-log phase, A600nm 1.2, were determined using 7H10 agar plates and serial, ten-fold dilutions of bacteria with culture medium.
Cell Lysis and Protein Purification
Bacterial cultures were transferred to 50 mL centrifuge tubes and bacteria harvested by means of centrifugation at 4000 rpm and 4 °C (Heraeus Megafuge 1.0 R) for 10 minutes. The supernatant was discarded, the cells washed by resuspension in PBS and the process repeated for a second wash before snap freezing cells in liquid nitrogen and storing at −80 °C.
Frozen cell pellets were subsequently thawed by the addition of 700 μL lysis buffer consisting of 1.5% deoxycholate, 1% sodium dodecasulphate (SDS), 375 μg lysozyme, 1 tablet PhosSTOP and 1 tablet cOmplete ULTRA protease inhibitor (Roche) in 10 mL 500 mM Tris-HCl, pH 7. Once thawed the bacteria were subjected to 8 rounds of probe sonication (VirSonic UltraSonic Cell Disrupter 100 at setting 10) for 20–30 seconds, with several minutes on ice in between rounds, adapted from Chopra et al.11. The lysate was clarified by centrifugation and the supernatant was purified for protein by means of methanol/chloroform precipitation. Briefly, equal parts lysate and methanol and ¾ parts chloroform were mixed and centrifuged at 4000 rpm for 5 min. A white band of protein developed at the interface and the top layer was carefully removed before the addition of a further ¾ parts (of original lysate volume) methanol. Centrifugation caused the precipitated protein band to pellet and the remaining aqueous layer was removed.
In-Solution Digestion of Proteins and Clean-Up of Peptides
The protein pellet was solubilised by means of 6 M Urea, 2 M Thiourea in 10 mM Tris-HCl, pH 8. Bulk protein quantitation was done by means of a modified Bradford assay12 and digestion by means of the in-solution method reported by Borchert et al.13. Briefly, proteins were reduced by 1 mM dithiothreitol, alkylated with 5.5 mM iodoacetamide and, pre-digested with Lys-C (1:100 mass ratio) before dilution of the denaturing buffer with 4 volumes 20 mM ammonium bicarbonate. The diluted sample was then digested with Trypsin (1:50 mass ratio) overnight. Resulting peptides were desalted using STop And Go Extraction tips14 (STAGE tips) and solubilised in 2% acetonitrile in preparation for LC-MS/MS.
LC-MS/MS
Samples were fractionated in-line by means of a Dionex Ultimate 3500 RSLC Nano System (Thermo Fisher Scientific) running a reversed phase gradient over an in-house built 40 cm column (75 μm internal diameter; 3.6 μm Aeris Peptide C18 beads, Phenomenex 04A-4507) and maintained at 40 °C. Solvent A was 0.1% Formic Acid in HPLC grade water and solvent B 0.1% Formic Acid in Acetonitrile. Gradient consisted of holding 1% solvent B for 10 minutes, increasing to 6% B over 2 minutes and then increasing to 35% B over 118 minutes; washing with 80% B followed.
Tandem mass spectrometry analysis was performed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) operating in top 10 data-dependant acquisition mode. Precursor MS1 scan range was between 300 and 1,750 with resolution of 70,000, and automatic gain control (AGC) target of 3e6 and maximum fill time of 250 ms. Fragmentation of precursor ions was set to a normalised collision energy of 28. MS2 scans employed a resolution of 17,500 and an isolation window 2 Th. Scan range for MS2 was 200 to 2,000 Th, AGC target was set to 5e4 and maximum fill time was 80 ms. Sample injection volumes were adjusted so as to yield a total ion count of approximately 5e9 at the highest point in the peptide region for each sample.
Protein Identification/Quantitation
Raw data files from the Q-Exactive were processed in MaxQuant15 version 1.5.0.3. The M. smegmatis mc2155 reference proteome from Uniprot16 (6,600 entries) was used to define the search space for the built-in Andromeda search engine17. Methionine oxidation and N-terminal acetylation were set as variable modifications and carbamidomethylation of cysteine as a constant modification. The data mass measurement corrections were performed by means of a first search with 20 ppm accuracy tolerance which was followed by a main search on the re-calibrated data with 4.5 ppm tolerance. Missed cleavages were limited to at-most two and an empirically derived false discovery rate (FDR) of 1%, estimated using the reversed proteome in a target-decoy approach, was used to restrict identifications at both the peptide spectrum matching and protein inference levels. Protein inference required at least one unique or razor peptide for identification of a protein group. The label free quantitation (LFQ) was enabled through the MaxLFQ algorithm18. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium19 via the PRIDE20 partner repository with the dataset identifier PXD004197.
Data Handling and Statistical Treatment
The resulting protein quantifications were batch normalised using R21 and Combat22 (contained in the SVA package23), outlier samples were identified by means of hierarchical clustering and principle components analysis plots and were repeated. Subsequent data processing was done using Perseus (www.perseus-framework.org) and R: protein identifications were filtered in silico so as to consider only proteins with non-zero LFQ values for triplicate measures and mean values for each protein were compared between treated and control groups using a student’s t-test (assuming equal variance) with a cut-off value of p = 0.05. A second level of stringency was used for initial analysis wherein the absolute value of a protein’s log-2 transformed fold-change was required to be greater than twice the standard deviation of the same for all proteins in a given time-point. Those proteins passing the second criteria were referred to as being strongly dysregulated. Within each time point, those proteins entirely absent from either treatment group, whilst consistently observed in the other, were also considered to be dysregulated. Any proteins discussed in the text that were identified through presence/absence are explicitly labelled as having been so identified.
Gene Ontology Analysis
STRING24 (http://string-db.org/) and the STRINGdb25 package in R were used for Gene Ontology terms enrichment analysis. The full set of identified mycobacterial proteins was used as the background proteome by which to calculate enrichment. The resulting enriched terms were filtered for those containing fewer than 100 proteins so as to display fewer overly-general terms.
Results
Determination of Sub-Lethal Rifampicin Concentration
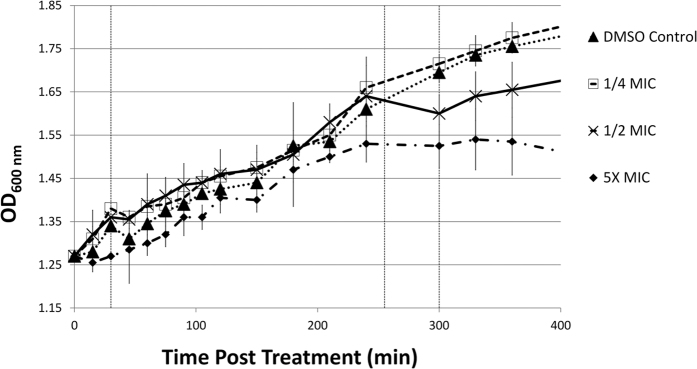
Figure 1 shows the various growth curves obtained for different concentrations of rifampicin. Bacteria were treated with various concentrations of rifampicin at mid-log phase which corresponded to A600nm of 1.2. Serial dilution plate counts determined this to correspond to 3.8e8 CFU/mL. The Minimal Inhibitory Concentration (MIC) was reported in the literature26 to be 20 μg/mL and this was confirmed by visual MIC assays. Various multiples of this concentration, including 1/4X, 1/2X, 1X, 2X and 5X this MIC were tested for growth effects through growth curves (1X and 2X in Supplementary Fig. S1). 5X MIC (100 μg/mL) resulted in a slight and immediate growth defect whilst all lower concentrations initially displayed growth similar to that of the DMSO only control until 240 minutes whereafter growth retardation was evident for all excepting the 1/4 MIC treated culture which showed no difference in growth relative to the DMSO control. After 300 minutes the 2X and 5X MIC (40, 100 μg/mL) concentrations appeared bactericidal whilst the 1/2 MIC and 1X MIC (10, 20 μg/mL) treated cultures showed recovery of growth after 300 minutes and optical densities rose parallel to that of the DMSO controls, only delayed. It was thus concluded that 1/2 MIC (10 μg/mL) induced bacteriostasis between 240 and 300 minutes post-treatment, with recovery evident after 300 minutes post-treatment. Thus the time points selected for proteomic investigations were 30, 255 and 300 minutes post-exposure corresponding with early exposure, onset of bacteriostasis and just prior to recovery for 1/2 MIC (10 μg/mL) – which was selected as the sub-lethal rifampicin concentration for this study.
Figure 1. Growth Curves for cultures treated with various rifampicin concentrations at mid-log phase.
MIC was 20 μg/mL and so 5X, 1/2 and 1/4 MIC indicate cultures treated with rifampicin at 100, 10 and 5 μg/mL respectively. DMSO control was treated with DMSO only. 1/4 MIC showed no difference in growth relative to DMSO control. 1/2 MIC onward showed growth defect relative to controls from 240 minutes until recovery after 300 minutes excepting 5X MIC which showed no signs of recovery. 1/2 MIC, 10 μg/mL, was selected for use as a sub-lethal concentration. Vertical dashed lines indicate time points 30, 255 and 300 minutes post-treatment which were used for time course experiment. Time points correspond to initial response, onset of bacteriostasis and early recovery respectively. Error bars indicate standard deviation.
Data Quality
Across 18 samples, using in-line reverse-phase chromatography of unfractionated samples, a total of 3,218 proteins were identified from 31,846 distinct peptides averaging 16,250 identified peptides per sample (see Supplementary Fig. S2), and 10 unique peptides per protein, with a total of 997,139 spectra submitted with high accuracy (average mass deviations less than 1 ppm). Figure 2 shows an example of the collected spectra annotated with b- and y-ions from the Andromeda search results.
Figure 2. Example of an annotated MS2 spectrum.
An exemplary spectrum showing high coverage of the matching peptide sequence and annotation of a high proportion of spectrum peaks.
Batch effects due to certain replicates having been processed separately were identified and batch correction applied through the ComBat algorithm22 found in the SVA package23 in R, after which samples did not cluster by batch (replicate number) as shown in Supplementary Fig. S3. Principle component 1, which accounts for the majority of the variation in the data, appeared to separate the data primarily on treatment group and this separation was larger at more advanced time points, or rather, the level of variation observed within time points, as pertaining to those proteins weighted most heavily in the PCA analysis, enlarged with later time points.
Those proteins that met the necessary criteria for application of the t-test (i.e. non-zero values for all three replicates within treatment groups) for each time point numbered 1,843; 1,951 and 1,972 for time points one, two, and three respectively. In accordance with the normalizing assumption that overall protein abundance should not change: the average log2-fold change was close to zero and the standard deviation thereof varied between 0.22 and 0.30. Thus total variation was similar between samples, whilst variation due to treatment increased with time of exposure.
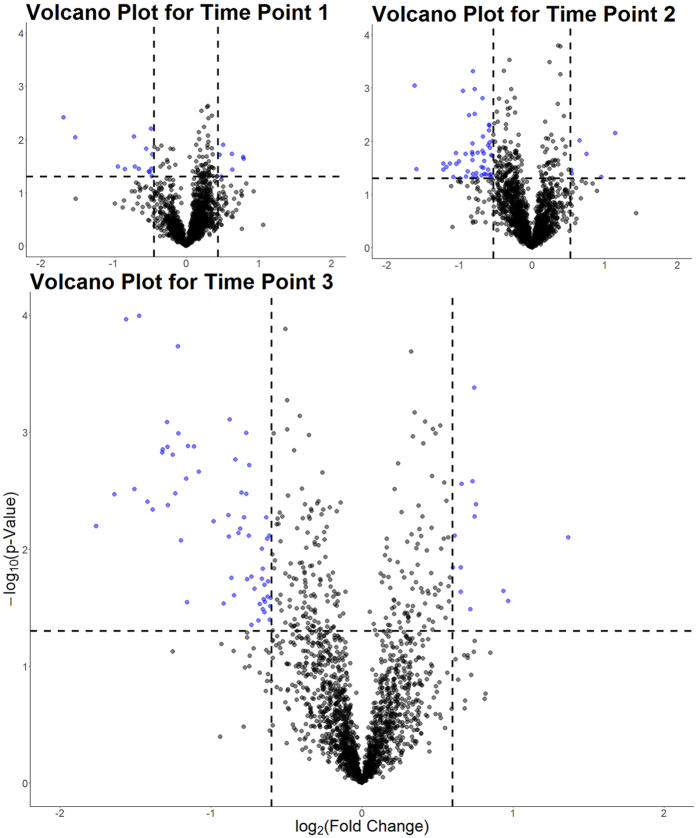
In keeping with this observation, there were 91, 205 and 296 dysregulated (p < 0.05) and 21, 57 and 75 strongly dysregulated (by more than twice the standard deviation for fold changes within that time point; p < 0.05) proteins for time points one, two and three respectively (see Fig. 3). The 10 most strongly dysregulated proteins for each time point can be seen in Table 1. More detail is available in Supplementary Spreadsheet S1, including a list of those proteins identified as dysregulated through a presence/absence analysis in which 17, 9 and 9 proteins were uniquely expressed in either treated or control samples for time points one, two and three respectively with 15/17, 6/9 and 4/9 respectively of those dysregulated proteins being downregulated (absent in treated samples).
Figure 3. Volcano plots for time points 1, 2 and 3.
Coloured points indicate those proteins significantly (p < 0.05, indicated by the horizontal dashed line) dysregulated by twice or more the standard deviation of the average fold changes for that time point (vertical dashed lines). Upper left and right panels display plots for time points one and two respectively with lower panel showing the volcano plot for the third time point.
Table 1. Ten most strongly dysregulated proteins/protein group after rifampicin exposure for each time point.
| Uniprot Protein ID* | Protein Name or Description | Gene Name | log2 (Fold Change) | p-Values for Fold Change | Sequence Coverage |
|---|---|---|---|---|---|
| Time point 1 | |||||
| A0R112 | Probable nicotinate-nucleotide adenylyltransferase | nadD | 0.79 | 2.33E-02 | 31.9 |
| A0R198 | Clp protease | clpP | 0.78 | 2.14E-02 | 69 |
| A0R0H6 | Coenzyme F420-dependent N5, N10-methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase | MSMEG_4389 | 0.64 | 3.67E-02 | 28.3 |
| A0R3H9 | Type I antifreeze protein | MSMEG_5479 | −0.65 | 3.55E-02 | 42.5 |
| A0QT09 | Succinate dehydrogenase hydrophobic membrane anchor protein SdhD | sdhD | −0.70 | 3.22E-02 | 21.8 |
| A0QWU1 | LemA protein | MSMEG_3063 | −0.71 | 8.75E-03 | 53.4 |
| A0R1D5 | HpcH/HpaI aldolase/citrate lyase family protein | MSMEG_4713 | −0.84 | 3.63E-02 | 29 |
| A0QTR3 | TetR-family protein transcriptional regulator | MSMEG_1935 | −0.94 | 3.26E-02 | 49.8 |
| A0R316 | Uncharacterized protein | MSMEG_5308 | −1.52 | 9.14E-03 | 34.2 |
| A0R722 | Glucanase | MSMEG_6752 | −1.69 | 3.82E-03 | 32.3 |
| Time Point 2 | |||||
| A0QVH8 | Zinc metalloprotease Rip1 | rip1 | 1.15 | 7.04E-03 | 11.1 |
| A0R2B2 | Major facilitator superfamily MFS_1 | MSMEG_5051 | −1.04 | 2.70E-02 | 12.4 |
| A0QUN8 | Hydrogenase assembly chaperone HypC/HupF | hypC | −1.05 | 1.69E-02 | 57 |
| A0QUN1 | Tetratricopeptide repeat domain protein | MSMEG_2267 | −1.08 | 4.76E-02 | 59.9 |
| A0QTK6 | S30AE family protein | MSMEG_1878 | −1.13 | 2.64E-02 | 50 |
| A0QUM7 | Hydrogenase-2, large subunit | hybC | −1.18 | 3.08E-02 | 58.9 |
| A0QXA8 | Cytochrome D ubiquinol oxidase subunit 1 | cydA | −1.22 | 2.65E-02 | 17.2 |
| A0QUM5 | Uncharacterized protein | MSMEG_2261 | −1.22 | 3.41E-02 | 81.5 |
| A0R3I6 | Peptidase S1 and S6, chymotrypsin/Hap | MSMEG_5486 | −1.59 | 3.36E-02 | 65.1 |
| A0R045 | NLP/P60 | MSMEG_4256 | −1.62 | 9.01E-04 | 27 |
| Time Point 3 | |||||
| A0QUE0 | Putative helicase | MSMEG_2174 | 1.37 | 7.92E-03 | 56.7 |
| A0QUM7 | Hydrogenase-2, large subunit | hybC | −1.32 | 1.50E-03 | 58.9 |
| A0R368 | Uncharacterized protein | MSMEG_5362 | −1.39 | 4.59E-03 | 51.2 |
| A0R2B2 | Major facilitator superfamily MFS_1 | MSMEG_5051 | −1.42 | 3.94E-03 | 12.4 |
| A0QUM8 | Hydrogenase maturation protease | MSMEG_2264 | −1.48 | 1.02E-04 | 57.7 |
| P0CH00/P0CG99 | Ribonucleoside-diphosphate reductase subunit alpha 1/alpha 2 | nrdE2 | −1.51 | 3.06E-03 | 78.8 |
| A0QUN2 | Rieske (2Fe-2S) region | MSMEG_2268 | −1.54 | 6.79E-05 | 50.2 |
| A0QXA8 | Cytochrome D ubiquinol oxidase subunit 1 | cydA | −1.56 | 1.08E-04 | 17.2 |
| A0R3I6 | Peptidase S1 and S6, chymotrypsin/Hap | MSMEG_5486 | −1.64 | 3.41E-03 | 65.1 |
| A0QTK6 | S30AE family protein | MSMEG_1878 | −1.76 | 6.38E-03 | 50 |
(*)Q-Values for Identifications were vanishingly small and reported as 0 for proteins shown.
Time Point 1: Initial Rifampicin Exposure is Characterised by Transient SOS Response
From the GO term enrichment analysis (see Supplementary Fig. S4) “SOS response” was, in the first time point, one of the most highly enriched biological processes with the DNA repair proteins UvrB, UvrC and RuvB all up regulated at this time point and related terms featuring in all four enrichment categories. The redox related Superoxide dismutase (Sod) was also up regulated (p < 0.05) in the first time point, consistent with reports that rifampicin causes metal catalysed ROS production27.
The SOS response was not found to be an enriched term in subsequent time points as significance of dysregulation for some proteins was lost. Exceptions to this were LexA, a repressor of the SOS DNA damage response, which was down regulated in the second time point, and UvrC that was observed to be again up regulated in the third time point. Thus it would seem that a transient SOS response was induced – possibly in response to oxidative stresses – and then mostly, but not entirely, silenced. Among the top 15 enriched KEGG pathways “Two-component system” features in all three time points with RegX3, PrrB, PstS and PhoP all dysregulated as early as the second time point; indicating the bacteria monitor and respond to their environment throughout the time course. It would thus appear the bacteria engage in careful sensing of the environment and, in the case of sub-lethal challenge, determine not to commit to dormancy whereas this would be expected with high dose rifampicin challenge9.
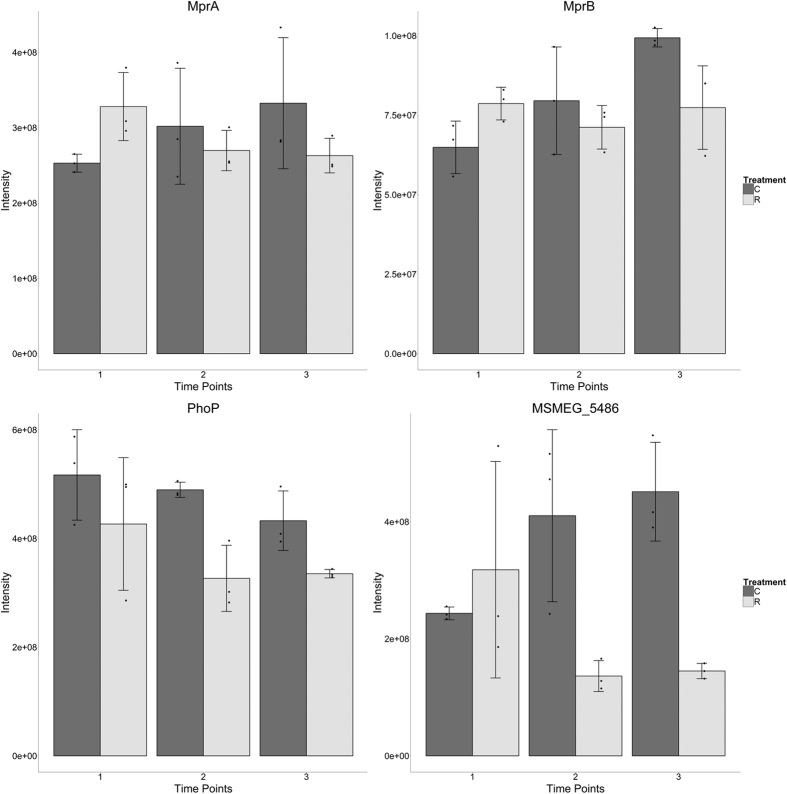
Important persistence regulating transcription factor28 MprA was also observed to be significantly (p < 0.05) up regulated in the first time point (see Fig. 4). MprA has been shown to protect against various perturbants of the cell wall including cell wall antibiotics29 and its up regulation deserves further discussion. External to the cell wall is another important virulence factor, namely the encasing polysaccharide capsule thought to help prevent association of M. tuberculosis with macrophages30,31. Capsule organisation was an enriched biological process in this early time point with DprE2 and galactofuranosyl transferase GlfT2, involved in creation of the arabinan and galactan of the arabinogalactan component of the cell wall respectively, each down regulated. Both Ms3580, a predicted mycolyltransferase, and FbpB a known antigen 85-C orthologue responsible for synthesising the virulence associated trehalose dimycolate (TDM - or chord factor) from trehalose monomycolate in M. tuberculosis, were also observed to be down regulated in time point 1. Thus there is clear evidence for an immediate dysregulation in important cell wall and capsule organising enzymes.
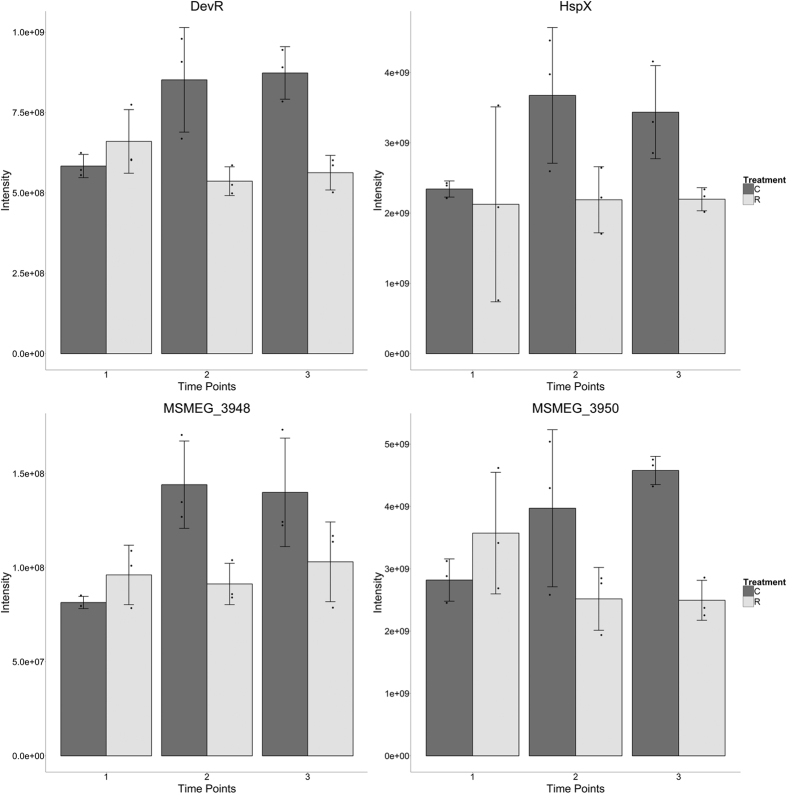
Figure 4. Protein expression profiles for several DevR related proteins.
Upper panel, effector (left) and sensor (right) proteins of two-component system MprAB coregulate dormancy alongside DevR. Lower panel, MSMEG_5486 (left) is an orthologue of the virulence related Mtb PepD protease. PhoP (right) of two-component system PhoPR is thought to be able to modulate the DevR stress response. Bars indicate mean expression with error bars indicating standard deviation and points indicating individual replicate values. Control samples shown in dark grey and rifampicin treated in light grey.
In the second and third time points several enzymes involved in phosphatidylinositol mannoside (PIM) and mycolic acid synthesis, two of the major lipid forms in the mycobacterial cell walls and essential for growth, are down regulated. This could be due to decreased demand for production of such lipids due to decreased cellular replication.
Time Point 2: Rifampicin Induces Dysregulation of DosR Response, TetR and MarR Transcriptional Regulators and Haeme Synthesis
DosR Response
Following on from the up regulation of MprA, the DevR (DosR) regulon was observed to be down regulated relative to controls from the second time point onward: with 13 of 33 genes known to be part of the regulon identified as dysregulated in at least one time point and all following similar expression profiles across time points. Figure 5 shows the expression profiles for 4 proteins in the regulon: DevR (the transcriptional activator of the regulon), HspX (a major stress response protein), Ms3950 (another universal stress protein; USP) and Ms3948 (an acyl-transferase representative of the non-USP proteins in the regulon). In all cases the expression in the controls rises steadily, or else rises and plateaus, whilst in the rifampicin treated cultures expression of the protein either remains constant or drops and plateaus.
Figure 5. Expression profiles for representative members of the DevR regulon.
Upper left panel: DevR (A0R2V2) is a major regulator in the dormancy response and is sensitive to oxidative and other stresses through DevS and DevT. Upper right panel: HspX is a stress response protein under the (activating) regulation of DevR. Lower left panel: Ms3950 is a universal stress response protein. Lower right panel: Ms3948 is an acyl-transferase. Points indicate individual replicate values measured, bars indicate mean expression with error bars indicating standard deviation. Control samples shown in dark grey and rifampicin treated in light grey.
Transcriptional regulators
In the second time point there was greater dysregulation of transcriptional regulators with 18 transcriptional regulators down regulated and just 4 up regulated. Those down regulated included five TetR family regulators, including Ms1380 identified by presence/absence, and the only MarR (Ms1492) identified as significantly dysregulated after data quality filters. TetR and MarR regulators are both typically repressors of gene transcription whose down regulation is commonly associated with multi-drug resistance and efflux pumps32,33. Accordingly, from the second to the third time points there were increasing numbers of dysregulated ABC transporters causing “ABC transporters” to appear among the top 15 enriched KEGG pathways in the second and third time points. Whilst diverse in their annotated functions and showing no obvious pattern in expression profiles, this increased dysregulation is notable as up regulation of ABC transporter drug efflux pumps is a common mechanism of MarR induced drug resistance. Also, of the 4 proteins identified as down regulated (absent in treated samples) in time point three, two are predicted transporters of the major facilitator superfamily. A recent study identified Ms4022 (MSMEG_4022) as a novel TetR-family regulator that directly activates the expression of seven transport related genes and enhanced M. smegmatis resistance to anti-tuberculosis drugs including rifampicin34. In the current study however Ms4022 did not significantly differ between treatment and control groups across all three time points (see Supplementary Spreadsheet S1). Additionally, we have identified five of the related proteins of which four (Ms1448; Ms2727; Ms5278 and Ms6909) were not significantly different and one was down regulated (Ms5051); suggesting that under our experimental conditions M. smegmatis employed different transport resistance mechanisms in response to rifampicin challenge.
Haeme Synthesis
GltD and GltX feed into the “Porphyrin and chlorophyll synthesis” pathway which was found in the top 15 enriched KEGG pathways of time point two. The iron-dependant repressor IdeR was down regulated in the second time point with iron scavenging mycobactin synthases MbtA and MbtE up regulated in the second and third time points – dysregulation of the mycobactins indicating that the treated bacteria were possibly attempting to increase iron uptake. Increased iron uptake and abundance of siderophores have previously been linked to success of the M. tuberculosis Beijing strain and rifampicin tolerance9,35,36. Supplementary Fig. S5 shows the pathway from glutamate to haeme synthesis was dysregulated in a coordinated fashion. At several branch points along the pathway there was dysregulation observed that would favour increased haeme production: notably chelatase Ms2615 and HemD. Ms6306 and Ms2383, each named GltX, are up regulated in the second and third time points respectively (the former by presence/absence) and likely generating increased quantities of Glutamyl-tRNAGlu to feed into this pathway while the down regulation of HemA may have served a conflicting purpose to increase the availability of charged tRNA for translation.
Time Point 3: Growth Recovery is Preceded by Up Regulation of Transcriptional and Translational Machinery and Mediated by Drug Inactivation
Redox
By time point three, both the dehydrogenase type 2a subunits, HybA and HybC, and almost the full complement of dehydrogenase-2 accessory/chaperone proteins, HypB-HypF, were down regulated. This is notable as the HybA and HybC subunits are part of the [NiFe] hydrogenase Hyd1 shown to constitutively catalyse the oxidation of hydrogen and aid growth in M. smegmatis37. The down regulation of this protein and its related accessories may offer further evidence of attempts to limit metal catalysed ROS production.
Growth Recovery
Time point 3 represents the time at which proteomic changes that enable recovery of growth were expected to be observable, prior to actual resumption of growth. However, in terms of bacterial replication machinery, “DNA replication” remained an enriched term and there was little other evidence in the expression profiles for replicative recovery in time point three. Indeed, NrdE2 and NrdF2 of the ribonucleoside-diphosphate reductase (responsible for reducing RNA to DNA) were down regulated further in time point three and several other cell division related proteins, such as CwsA, FtsH and FtsZ, were also down regulated in the third time point. It would thus appear that recovery of cellular replication machinery, and replicative recovery occurs closer to the subsequent time point on the curve than anticipated (see Fig. 1).
Transcription/Translation
Supplementary Fig. S4 shows a strong cluster of dysregulated proteins at time point three, including both transcriptional RNA polymerase subunits and translational ribosomal subunits. Supplementary Fig. S6 shows that the expression levels of RpoA, RpoB and RpoC all gradually decreased in control samples across the time-course, but steadily increased in rifampicin treated samples. Expression levels of the sigma factor SigF generally increased in both treated and control samples across the time-course, but notably SigF showed a marked down regulation at the first time point in rifampicin-treated cells and did not subsequently recover to the same levels observed in the control samples at any time point.
Supplementary Fig. S7 shows a similar general trend of reduced ribosomal expression with time in the control samples, and expression in the treated samples generally increased. In the majority of cases the protein expression of the treated and control samples were approximately equal in the first time point (with RplF being the exception) but unlike with the transcriptional machinery in Supplementary Fig. S6 some ribosomal proteins did not follow a monotonically increasing pattern of expression in the treated samples. These proteins instead showed a general increase relative to the controls but with a dip in expression in time point two that rose again in the third time point. From the GO enrichment analysis “RNA polymerase” and related terms “RNA polymerase activity” and “Primosome complex” appeared among the top 15 KEGG pathways, molecular functions and cellular compartments from the second time point onward and “Ribosome” in the top 15 most enriched cellular compartment terms in the third time point along with “Translation” which appeared in the top 15 enriched biological processes in the third time point. This, together with the expression profiles of those proteins observed to be dysregulated, argues that up regulation of both transcriptional and translational machinery was necessary to overcome the inhibitory effects of rifampicin on protein production.
Arr
Ms1221, or rifampin ADP-ribosyl transferase, is a broad specificity enzyme that is reported to inactivate rifampicin through ADP-ribosylation on C23, with inversion of configuration at C1 of the NAD+ -derived ribose, and is thought to account for the larger rifampicin MIC values for M. smegmatis as compared with those of M. tuberculosis38. This protein was consistently missing in control samples (absent in 8 out of 9 samples) and consistently present (8 out of 9 samples) in treated cultures and, as shown in Supplementary Fig. S8, showed steadily increasing expression in the treated samples over time. It would thus appear the most straight-forward explanation for recovery of growth in M. smegmatis lies in the gradual accumulation of this rifampicin-deactivating enzyme.
Discussion
A great deal of our current knowledge regarding bacterial acquired antibiotic resistance is based on assays using high or lethal concentrations of antibiotics that would necessarily favour the selection of rare mutants with strong, pre-existing phenotypes or genotypes39,40. However phenotypically resistant bacteria can also be selected at antibiotic concentrations several hundred-fold below the lethal concentration for susceptible cells4,40,41. This argues that the most relevant mycobacterial responses to antibiotic exposure include those observed at sub-MIC antibiotic concentrations, which are sub-lethal and allow continued growth of the pathogen40. Within this context, our data shed new light on numerous molecular mechanisms which contribute to the acquisition by mycobacteria of phenotypic resistance to rifampicin – including in M. smegmatis the eventual up regulation of an M. smegmatis-specific, broad specificity chromosomal ADP-ribosyl transferase – and contrast with previous studies that selected and characterised pre-existing persister cell populations at higher antimicrobial doses9,42,43,44.
Two component signal transduction systems a primary control mechanism mediating mycobacterial adaptive responses to sub-MIC rifampicin
It has been suggested that at sub-MIC concentrations several antibiotics act as signalling molecules inducing changes associated with gene expression, quorum sensing, biofilm formation and virulence4. However the circumstances and respective temporal dynamics of such responses remain largely elusive. Here, we employed an in vitro experimental design that allowed us to examine the antibiotic-induced proteomic changes that take place in the model organism M. smegmatis at different time points after an initial insult using a sub-MIC concentration of rifampicin. Our analyses focused on quantifying the temporal dynamics of differential protein expression across early response, stasis and regrowth phases post-antibiotic exposure (Fig. 1), in order to gain insight into the induction of phenotypic antibiotic resistance in mycobacteria. Amongst others changes, our data revealed a differential regulation of different two-component systems across the three time points, including the MprA/MprB and LuxR systems (Figs 4 and 5). Two-component systems are well known signal transduction devices responsible for important bacterial adaptive responses to environmental stimulus, mostly by modulating gene expression45. Concordant with our findings, a recent study showed that several two component systems are up regulated in multidrug resistant M. tuberculosis clinical isolates and could be induced by first line antitubercular agents46. Here, we highlight the differential regulation of a stress responsive two component system, comprising a sensor kinase MprB and response regulator MprA which act as a signal transduction pair in vitro and in vivo47 and which are required for persistence during host infection48. In M. tuberculosis the activation of this two-component signal transduction system directly regulates the expression of sigma factors sigB and sigE - which in turn regulates pepD48. This is of particular interest since, according to our data, the induction of MprA at time point 1 (Fig. 4) was followed by a later (time point 3) down regulation of its cognate kinase MprB with the pattern of PepD abundance mirroring that of MprA and showing a significant and strong decrease in abundance in the latter time points (Fig. 4). Our data therefore suggest MprA/B as an important and versatile regulatory mechanism, in which the respective components can be regulated separately such that by the up regulation of the transcriptional regulator MprA enables mycobacteria to achieve a rapid response to initial rifampicin insult, whilst its activity can later be decreased by the down regulation of the sensor component MprB.
In another two-component system, the observed dysregulation of the LuxR family response regulator DevR deserves further attention. DevR-DevS/DosR-DosS is one of the best characterised two component systems of M. tuberculosis and responds rapidly to oxidative stress signals, including hypoxia and nitric oxide49,50,51, inducing the DevR dormancy programme in mycobacteria by regulating the expression of an ~48 gene regulon49,52. A recent proteomic study on M. tuberculosis Beijing B0/W148 exposed to high doses of rifampicin revealed the induction of DosR regulon proteins upon antibiotic exposure9. By contrast, our results show two LuxR family two-component response regulators (A0R2V2 and A0QZ95) being down regulated at time points 2 and 3 (A0R2V2 seen in Fig. 5). Furthermore, whilst our data provides no direct evidence for DosR induction, within our dataset we observed 13 of the 33 proteins in the DevR regulon being down regulated (Regulon as defined in the manually curated RegPrecise; http://regprecise.lbl.gov/RegPrecise/index.jsp). It is of course possible that, in our study, DosR was in fact rapidly up regulated following rifampicin insult and then down regulated again to initial baseline by time point 1. However, it is also possible that exposure of mycobacteria to sub-lethal rifampicin challenge may not induce the DevR operon. Interestingly, an earlier quantitative proteomic analysis involving two M. bovis strains showed that increased expression of BCG_2013 (latency protein belonging to DosR regulon) leads to a decrease in MprA expression followed by up regulation of KatG and consequently increased INH susceptibility53 (Hu et al.7). By contrast, in our study MprA was up regulated at time point 1, supporting the idea that sub-MIC rifampicin does not necessarily trigger a dormancy response. This is further supported by the findings of Bartek et al. who observed that induction of the dormancy response was not required for drug tolerance in mice54. It has also been shown that induction of DevR alone is not sufficient to alter growth dynamics55 and moreover W-Beijing lineages of M. tuberculosis constitutively express this regulon56. Overall then, our data suggest that there may be an immediate preparatory stress response by the bacteria to the initial rifampicin insult, but that the bacteria continue to monitor their environment through “Two-component system[s]” (enriched in all three time points) and other sensing mechanisms and subsequently quell any early dormancy response, allowing the later recovery of growth once phenotypic resistance has been attained.
Sub-MIC levels of rifampicin induce SOS response and virulence determinants
Sub MIC antibiotic concentrations have been reported to increase mutagenesis, in particular for those drugs known to interfere with DNA/RNA synthesis40. Our data indicate that after rifampicin insult, M. smegmatis primarily uses an SOS system - specifically uvrABC (which is involved in nucleotide-excision repair) - as a rapid means by which to deal with possible DNA damage. Additionally the down regulation of LexA, (a repressor of SOS DNA damage response) and the up regulation of UvrD/REP family (an inhibitor of RecA- mediated DNA strand exchange57) implies that SOS system was in fact activated. To our knowledge this is the first report of mycobacteria associating the induction of the SOS response system with presence of sub-lethal antibiotic concentrations, despite the induction of SOS response by sub-MIC antibiotic concentrations being well established in other bacterial species (e.g. Escherichia coli58,59; Staphylococcus aureus60; Vibrio cholerea61). For example, in E. coli fluoroquinolones are known to induce an SOS response, whilst other antibiotics, such as aminoglycosides, tetracycline and chloramphenicol, do not62. It will therefore of interest to explore in the future which and under what conditions different antibiotics induce SOS response in mycobacteria.
There is an increasing body of evidence indicating that several antibiotics stimulate the production of reactive oxygen species in bacteria63,64 and that sub lethal antibiotic treatment leads to multi-drug resistance, through a radical based mutagenic mechanism65. In line with this, our data revealed an up regulation of several oxidative stress responsive proteins across all time points, including proteins such as superoxide dismutase, oxidoreductases, thioredoxin and others. In addition to the important role of such detoxification proteins during antibiotic challenge, these proteins also represent important potential virulence determinants that may play a crucial role during bacterial survival inside macrophages, in which pathogens are thought to be primarily killed by production of ROS and NO species66,67. Our data may therefore imply that, in a scenario like that described by Dartois and Barry68, sub-lethal concentrations of antibiotic at the site of TB disease may have the undesired effect of potentiating pathogenic mycobacteria with tools of survival and perhaps also directly enhance the evolution of acquired drug resistance.
The quantitative proteomics analyses indicate an evident differential abundance (mainly induction) of several proteins associated with iron up take and sequestration, including MbtA and MbtE. A possible explanation for this could be the fact that, as mentioned earlier, mycobacteria respond to low concentrations of rifampicin via ancestral sensing mechanisms triggered by polyketide natural products. As mentioned above, the latter are commonly employed in bacterial chemical warfare as a species seeks dominance in a natural environment69. Since iron is essential for bacterial growth, it is therefore reasonable to speculate that M. smegmatis perceives rifampicin as a signal molecule indicating the presence of other bacterial competitors, which would immediately lead to iron uptake/sequestration as means to compete and avoid imminent iron depletion. Importantly, in the context of host infection by mycobacterial pathogens, iron uptake and sequestration have been reported to be tightly linked to virulence70,71. Thus, if similar proteomic responses are observed in M. tuberculosis to those reported here, sub-MIC dosing of rifampicin could have multiple detrimental effects, directly inducing a more virulent mycobacterial phenotype. Further experiments on pre-rifampicin-stressed mycobacteria are now underway to test this hypothesis.
Identification of proteomic signatures resulting from rifampicin pre-exposure
Our growth curve data shows that after 300 mins post-rifampicin challenge (corresponding to ~2 doublings of M. smegmatis) the bacterial cells resume growth. Taking in to account the observed changes in the M. smegmatis proteome throughout the time course assay, this prompted us to investigate whether re-growing cells at time point 3 preserved markers of pre-exposure to rifampicin. Interestingly, in our data, time point 3 is characterized by the dysregulation of several ABC transporters (PstS and LpqZ were up regulated while CydD and Ms0553 were down regulated, amongst others) and, whilst neither were observed to be dysregulated in our data, we observed an inverse correlation between one MarR and one of its controlled ABC transporters, supporting the findings of Zhang et al.72 who previously identified a novel marRAB operon in which up regulation of the regulated ABC transporters resulted in increased, not decreased, susceptibility of M. smegmatis to rifampicin. Amongst the other ABC transporters found to be up regulated at time point 3 in our data, the up regulation of proteins such as Ferric Iron-binding periplasmic protein of ABC transporter – which is also implicated in bacterial virulence73,74 – is notable.
We also observed that oxidative stress responsive protein such as oxidoreductases and thioredoxin, as well proteins involved in iron uptake (e.g. Fe III-dicitrate-binding periplasmic Lipoprotein FecB; FeS assembly protein SufD) were up regulated at time point three. Interesting here we have also detected the up regulation of a number of additional virulence factors with a known role during infection such as zinc metalloprotease75 and membrane-anchored mycosin76 trigger factor77. In sum, although several stress associated proteins were down-regulated at time point 3, it is evident that even shortly prior to resumption of growth cells conserve markers of rifampicin pre-exposure, suggesting that those cells are no longer “naive” and instead continue to dysregulate certain stress responsive proteins and virulence factors that may confer a longer-lasting phenotypic advantage.
It is thus tempting to speculate overall that the proteomic responses of M. smegmatis to sub-lethal concentrations of rifampicin observed in the present work might be part of a more ancient, conserved, broad-specificity mechanism for mycobacteria to selectively regulate, for example, ABC transporters and the chromosomal rifampicin ADP-ribosyl transferase in order to respond rapidly to exogenous polyketide exposure; further efforts to identify the possible upstream signalling processes responsible for this phenomenon are now in hand. In summary our data demonstrates that despite M. smegmatis having a chromosomal resistance factor the response of the organism to sub lethal rifampicin exposure is far more complex than simple up regulation of that factor. This may reflect the fact that the resistance marker is undetectable in the absence of rifampicin, so the organism perhaps requires other more rapid stress responses to provide time for phenotypic adaption. Whether chemical blockade of these specific early stress responses will sensitise mycobacteria to lower doses of rifampicin remains to be seen.
Additional Information
Accession Codes: PRIDE Accession Number: PXD004197.
How to cite this article: Giddey, A. D. et al. A temporal proteome dynamics study reveals the molecular basis of induced phenotypic resistance in Mycobacterium smegmatis at sub-lethal rifampicin concentrations. Sci. Rep. 7, 43858; doi: 10.1038/srep43858 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the National Research Foundation (NRF) for their financial support of this research through grant numbers 98963 and 95984 and for a PhD bursary to ADG. JMB thanks the NRF for a South African Research Chair grant and SG thanks the CSIR for a PhD bursary. NCS thanks the South African Medical Research Council for a research fellowship. Computations were performed using facilities provided by the University of Cape Town’s ICTS High Performance Computing Team (http://hpc.uct.ac.za).
Footnotes
The authors declare no competing financial interests.
Author Contributions A.G. carried out the experimental work, data analysis and co-wrote the manuscript. E.K. carried out the experimental work and data analysis. K.N. supervised the experimental work and assisted with data analysis. S.G. and A.N. contributed to the experimental work. N.S. and J.B. designed the experiments, analysed the data and co-wrote the manuscript. All authors reviewed the manuscript.
References
- World Health Organisation. Global tuberculosis report, doi: WHO/HTM/TB/2014.08 (2014).
- Steenken W. & Wolinsky E. Isoniazid in experimental tuberculosis. Trans. Annu. Meet. Natl. Tuberc. Assoc. 48, 425–30 (1952). [PubMed] [Google Scholar]
- Mahajan R. Bedaquiline: First FDA-approved tuberculosis drug in 40 years. Int. J. Appl. Basic Med. Res. 3, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I. & Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–78 (2014). [DOI] [PubMed] [Google Scholar]
- Prideaux B. et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat. Med. 21, 1223–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caws M. et al. Mutations Prevalent among Rifampin- and Isoniazid-Resistant Mycobacterium tuberculosis Isolates from a Hospital in Vietnam. J. Clin. Microbiol. 44, 2333–2337 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. et al. Quantitative Proteomics Reveals Novel Insights into Isoniazid Susceptibility in Mycobacteria Mediated by a Universal Stress Protein. J. Proteome Res. 14, 1445–1454 (2015). [DOI] [PubMed] [Google Scholar]
- Koul A. et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5, 3369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keijzer J. et al. Mechanisms of phenotypic rifampicin tolerance in Mycobacterium tuberculosis Beijing genotype strain B0/W148 revealed by proteomics. J. Proteome Res. acs.jproteome 5b01073, doi: 10.1021/acs.jproteome.5b01073 (2016). [DOI] [PubMed] [Google Scholar]
- Rao M., Liu H., Yang M., Zhao C. & He Z.-G. A Copper-responsive Global Repressor Regulates Expression of Diverse Membrane-associated Transporters and Bacterial Drug Resistance in Mycobacteria. J. Biol. Chem. 287, 39721–39731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra T. et al. Quantitative Mass Spectrometry Reveals Plasticity of Metabolic Networks in Mycobacterium smegmatis. Mol. Cell. Proteomics 13, 3014–3028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagli L. S. & Rodriguez L. V. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 6, 559–563 (1985). [Google Scholar]
- Borchert N. et al. Phosphoproteome of Pristionchus pacificus Provides Insights into Architecture of Signaling Networks in Nematode Models. Mol. Cell. Proteomics 11, 1631–1639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y. & Mann M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 75, 663–670 (2003). [DOI] [PubMed] [Google Scholar]
- Cox J. & Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- The Uniprot Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- Cox J. et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. Update of the PRIDE database and its related tools. Nucleic Acids Res. gkw880, doi: 10.1093/nar/gkw880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. at < http://www.r-project.org/>; accessed on 14 December 2016 (2012).
- Johnson W. E., Li C. & Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–27 (2007). [DOI] [PubMed] [Google Scholar]
- Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E. & Storey J. D. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. STRING v10 : protein – protein interaction networks, integrated over the tree of life. 43, 447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan S., Venter H. & Dabbs E. R. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 41, 2456–60 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccaro G., Pietraforte D., Giannoni F., Mustazzolu A. & Fattorini L. Rifampin Induces Hydroxyl Radical Formation in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 7527–7533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrt T. C. & Deretic V. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 98, 12706–12711 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. J., He H., Penoske R. M., Twining S. S. & Zahrt T. C. PepD Participates in the Mycobacterial Stress Response Mediated through MprAB and SigE. J. Bacteriol. 192, 1498–1510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywes L., Hoppe H. C., Daffé M. & Ehlers M. R. W. Nonopsonic binding of Mycobacterium tuberculosis to complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect. Immun. 65, 4258–4266 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffé M. & Etienne G. The capsule of Mycobacterium tuberculosis and its implications for pathogenicity. Tuber. Lung Dis. 79, 153–169 (1999). [DOI] [PubMed] [Google Scholar]
- Ramos J. L. et al. The TetR Family of Transcriptional Repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhana R. J. C., Singla A., Sikder M. H., Withers M. & Kendall S. L. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics 16, 479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yang M. & He Z.-G. Novel TetR family transcriptional factor regulates expression of multiple transport-related genes and affects rifampicin resistance in Mycobacterium smegmatis. Sci. Rep. 6, 27489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keijzer J., de Haas P. E., de Ru A. H., van Veelen P. a. & van Soolingen D. Disclosure of selective advantages in the ‘modern’ sublineage of the Mycobacterium tuberculosis Beijing genotype family by quantitative proteomics. Mol. Cell. Proteomics 2632–2645, doi: 10.1074/mcp.M114.038380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu B., Sarma G. R. & Venkatesan P. Effect of anti-tuberculosis drugs on the iron-sequestration mechanisms of mycobacteria. Indian J. Pathol. Microbiol. 38, 287–292 (1995). [PubMed] [Google Scholar]
- Greening C., Berney M., Hards K., Cook G. M. & Conrad R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc. Natl. Acad. Sci. 111, 4257–4261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysarowich J. et al. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc. Natl. Acad. Sci. 105, 4886–4891 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg E. et al. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog 7, e1002158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. & Andersson D. I. Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr Opin Microbiol 15, 555–560 (2012). [DOI] [PubMed] [Google Scholar]
- Sandegren L. Selection of antibiotic resistance at very low antibiotic concentrations. Ups. J. Med. Sci. 119, 103–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. & Marcotte E. M. The Proteomic Response of Mycobacterium smegmatis to Anti-Tuberculosis Drugs Suggests Targeted Pathways. J Proteome Res 7, 855–865 (2008). [DOI] [PubMed] [Google Scholar]
- Grant S. S., Kaufmann B. B., Chand N. S., Haseley N. & Hung D. T. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc. Natl. Acad. Sci. 109, 12147–12152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A. et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D. & Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9, 143–152 (2006). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. Transcriptional and proteomic analyses of two-component response regulators in multidrug-resistant Mycobacterium tuberculosis. Int J Antimicrob Agents 46, 73–81 (2015). [DOI] [PubMed] [Google Scholar]
- Zahrt T. C., Wozniak C., Jones D. & Trevett A. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect Immun 71, 6962–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Hovey R., Kane J., Singh V. & Zahrt T. C. MprAB Is a Stress-Responsive Two-Component System That Directly Regulates Expression of Sigma Factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188, 2134–2143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N. et al. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80, 141–159 (2000). [DOI] [PubMed] [Google Scholar]
- Voskuil M. I. & Schlesinger L. S. Toward Resolving the Paradox of the Critical Role of the DosR Regulon in Mycobacterium tuberculosis Persistence and Active Disease. Am. J. Respir. Crit. Care Med. 191, 1103–1105 (2015). [DOI] [PubMed] [Google Scholar]
- Kaur K., Kumari P., Sharma S., Sehgal S. & Tyagi J. S. DevS/DosS sensor is bifunctional and its phosphatase activity precludes aerobic DevR/DosR regulon expression in Mycobacterium tuberculosis. FEBS J. 283, 2949–2962 (2016). [DOI] [PubMed] [Google Scholar]
- Selvaraj S., Sambandam V., Sardar D. & Anishetty S. In silico analysis of DosR regulon proteins of Mycobacterium tuberculosis. Gene 506, 233–241 (2012). [DOI] [PubMed] [Google Scholar]
- Hu X. et al. Quantitative proteomics reveals novel insights into isoniazid susceptibility in mycobacteria mediated by a universal stress protein. J Proteome Res 14, 1445–1454 (2015). [DOI] [PubMed] [Google Scholar]
- Bartek I. L. et al. The DosR regulon of M. tuberculosis and antibacterial tolerance. Tuberculosis 89, 310–316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minch K., Rustad T. & Sherman D. R. Mycobacterium tuberculosis Growth following Aerobic Expression of the DosR Regulon. PLoS One 7, e35935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallow A., Domenech P. & Reed M. B. Strains of the East Asian (W/Beijing) Lineage of Mycobacterium tuberculosis Are DosS/DosT-DosR Two-Component Regulatory System Natural Mutants. J. Bacteriol. 192, 2228–2238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. et al. Mycobacterium tuberculosis UvrD1 and UvrA Proteins Suppress DNA Strand Exchange Promoted by Cognate and Noncognate RecA Proteins. Biochemistry 49, 4872–4883 (2010). [DOI] [PubMed] [Google Scholar]
- Gutierrez A. et al. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun 4, 1610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadlallah S. M., Rahal E. A., Sabra A., Kissoyan K. A. B. & Matar G. M. Effect of Rifampicin and Gentamicin on Shiga Toxin 2 Expression Level and the SOS Response in Escherichia coli O104:H4. Foodborne Pathog. Dis. 12, 47–55 (2014). [DOI] [PubMed] [Google Scholar]
- Nagel M., Reuter T., Jansen A., Szekat C. & Bierbaum G. Influence of ciprofloxacin and vancomycin on mutation rate and transposition of IS256 in Staphylococcus aureus. Int. J. Med. Microbiol. 301, 229–236 (2011). [DOI] [PubMed] [Google Scholar]
- Baharoglu Z., Krin E. & Mazel D. RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in Vibrio cholerae. PLoS Genet 9, e1003421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z. & Mazel D. Vibrio cholerae Triggers SOS and Mutagenesis in Response to a Wide Range of Antibiotics: a Route towards Multiresistance. Antimicrob Agents Chemother 55, 2438–2441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A. & Collins J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J. & Collins J. J. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8, 423–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A., DePristo M. A. & Collins J. J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37, 311–320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S. A. et al. The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic Biol Med 53, 1625–1641 (2012). [DOI] [PubMed] [Google Scholar]
- Weiss G. & Schaible U. E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartois V. & Barry C. E. A medicinal chemists’ guide to the unique difficulties of lead optimization for tuberculosis. Bioorganic Med. Chem. Lett. 23, 4741–4750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley C. P., Lee H. Y. & Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA 105, 4595–4600 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieracci F. M. & Barie P. S. Iron and the Risk of Infection. Surg. Infect. (Larchmt). 6, s41–s46 (2005). [DOI] [PubMed] [Google Scholar]
- Skaar E. P. The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLoS Pathog 6, e1000949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. A Novel marRAB Operon Contributes to the Rifampicin Resistance in Mycobacterium smegmatis. PLoS One 9, e106016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. S., Gilliland S. M., Ruiz-Albert J. & Holden D. W. Characterization of Pit, a Streptococcus pneumoniae Iron Uptake ABC Transporter. Infect Immun 70, 4389–4398 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarau A. et al. FutA2 Is a Ferric Binding Protein from Synechocystis PCC 6803. J. Biol. Chem. 283, 12520–12527 (2008). [DOI] [PubMed] [Google Scholar]
- Vemula M. H. et al. Mycobacterium tuberculosis Zinc Metalloprotease-1 Assists Mycobacterial Dissemination in Zebrafish. Front. Microbiol. 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave J. A., Gey van Pittius N. C., Beyers A. D., Ehlers M. R. W. & Brown G. D. Mycosin-1, a subtilisin-like serine protease of Mycobacterium tuberculosis, is cell wall-associated and expressed during infection of macrophages. BMC Microbiol 2, 30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot A., Botton E., Dubail I. & Charbit A. A Homolog of Bacillus subtilis Trigger Factor in Listeria monocytogenes Is Involved in Stress Tolerance and Bacterial Virulence. Appl Env. Microbiol 72, 6623–6631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.