Significance
The clinical development of targeted therapies has been hampered by their limited intrinsic antitumor activity and the rapid emergence of resistance, highlighting the need to identify highly active and synergistic drug combinations. However, empirical synergistic drug-screening approaches are challenging, and elucidating the mechanisms that underlie such drug interactions is typically complex. Here, we performed an expression-based screen and network analyses to identify drugs amplifying the antitumor effects of NOTCH inhibition in T-cell acute lymphoblastic leukemia (T-ALL). These studies uncovered a druggable synthetic lethal interaction between suppression of protein translation and NOTCH inhibition in T-ALL. Our results illustrate the power of expression-based analyses toward the identification and functional characterization of antitumor drug combinations for the treatment of human cancer.
Keywords: leukemia, T-ALL, NOTCH1, protein translation, synergy
Abstract
The Notch1 gene is a major oncogenic driver and therapeutic target in T-cell acute lymphoblastic leukemia (T-ALL). However, inhibition of NOTCH signaling with γ-secretase inhibitors (GSIs) has shown limited antileukemic activity in clinical trials. Here we performed an expression-based virtual screening to identify highly active antileukemic drugs that synergize with NOTCH1 inhibition in T-ALL. Among these, withaferin A demonstrated the strongest cytotoxic and GSI-synergistic antileukemic effects in vitro and in vivo. Mechanistically, network perturbation analyses showed eIF2A-phosphorylation–mediated inhibition of protein translation as a critical mediator of the antileukemic effects of withaferin A and its interaction with NOTCH1 inhibition. Overall, these results support a role for anti-NOTCH1 therapies and protein translation inhibitor combinations in the treatment of T-ALL.
T-cell acute lymphoblastic leukemias (T-ALL) are immature lymphoid tumors characterized by the diffuse infiltration of the bone marrow by malignant lymphoblasts expressing immature T-cell markers (1). Clinically, T-ALL patients typically present with elevated white cell counts in peripheral blood and frequently show mediastinal thymic masses and meningeal infiltration of the central nervous system at diagnosis (1). In the early days of combination chemotherapy, T-ALL was recognized as a high-risk leukemia group; however, current cure rates with intensified therapy have improved to about 80% in children (2) and 60% in adults (3). Despite this progress, the prognosis of primary resistant and relapsed T-ALL remains very poor (4). In this context, the identification of activating mutations in the NOTCH1 gene has created major interest in the development of γ-secretase inhibitors (GSI), which block a proteolytic cleavage of NOTCH1 receptor at the membrane required for the activation of NOTCH1 signaling, as potential targeted therapy in T-ALL (5). However, the clinical development of GSIs as anti-NOTCH1 therapy has been hampered by a paucity of therapeutic responses in early trials (6–8). Thus, the identification of highly effective and synergistic GSI drug combinations capable of eliciting strong and synergistic cytotoxic antileukemic effects has become a major priority toward the development of effective anti-NOTCH1 therapies in the clinic.
Here, we implemented and integrated a systems biology approach toward the identification of active drugs synergistic with GSIs for the treatment of NOTCH1-driven T-ALL. These analyses identified eIF2A-mediated translation inhibition as therapeutic target for the development of synergistic drug combinations. Our results uncover highly active drug combinations for the treatment of T-ALL and identify a targetable synthetic lethality interaction between anti-NOTCH1 therapies and eIF2A-mediated translation inhibition.
Results
Expression-Based Screen of T-ALL Antileukemic Drugs.
Transcriptomic studies have linked inhibition of NOTCH1 signaling with gene expression signatures related to down-regulation of anabolic pathway genes and up-regulation of genes associated with catabolic functions (9, 10). Significantly, these metabolic effects are antagonized by activation of the PI3K-protein kinase B (AKT)–signaling pathway upon either Pten deletion or via expression of a constitutively active form of AKT (10, 11). Here we hypothesize that pharmacologic perturbations converging on this core transcriptional response could yield drugs and drug targets with synergistic antileukemic effects in T-ALL when combined with NOTCH1 inhibition. Toward this goal we searched for positive associations between gene sets generated by drug treatments in the Connectivity Map (cMAP) (12) and the gene expression signatures induced by NOTCH1 inhibition and reversed by PI3K-AKT activation in T-ALL (SI Appendix, Fig. S1). To generate a NOTCH1 inhibition signature, we profiled mouse Notch1-induced T-ALL cells treated with vehicle only or a GSI [((S)-2-(2-(3,5-difluorophenyl)acetamido)-N-((S)-5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)propanamide), DBZ] in vivo (SI Appendix, Fig. S1). Genes differentially expressed in vehicle-only versus GSI-treated Notch1-induced mouse T-ALL cells identified 16 positive cMAP associations (P < 0.01) indicative of candidate drugs potentially enhancing the effects of NOTCH1 inhibition in T-ALL (Fig. 1A). These included drugs with known mechanisms of action and redundant activities, such as histone deacetylase inhibitors (vorinostat, valproic acid, and trichostatin A); phenothiazine-derivative antipsychotic compounds (trifluoperazine, thioridazine, and prochlorperazine); and antimalarial drugs (astemizole and mefloquine); together with rapamycin, an mTOR protein complex 1 (mTORC1) inhibitor; geldanamycin, a heat shock protein 90 (HSP90) inhibitor; resveratrol, an antioxidant sirtuin agonist; parthenolide, an nuclear factor KB (NFKB) inhibitor with leukemia stem cell suppressor activity; withaferin A, a steroidal lactone natural compound with antiinflamatory and antiangiogenic activities; phenoxybenzamine, an antiadrenergic alpha receptor antagonist; pyrvinium pamoate, an antihelmintic compound with antitumor activity and preferential cytotoxicity following glucose starvation; and lanatoside C, a cardiac glycoside ion channel inhibitor used in the treatment of congestive heart failure and cardiac arrhythmias. Notably, and most reassuringly of this approach, rapamycin (13), vorinostat (14), and different antipsychotic phenothiazine drugs (15) have been recently described to have antileukemic effects in T-ALL and to increase the activity of GSIs. Loss of Pten rescues the metabolic and antileukemic effects of NOTCH inhibition with GSI (10). Thus, we also investigated negative associations between cMAP gene sets and the expression signatures driven by Pten loss following tamoxifen treatment of Ptenf/f CreERT2 Notch1-induced T-ALL cells (SI Appendix, Fig. S1). Here, genes differentially expressed in Pten-positive vs. Pten-knockout Notch1-induced T-ALL cells identified five negative cMAP associations (P < 0.01) indicative of candidate drugs antagonizing the effects of Pten loss. These included two PI3K-mTOR inhibitor drugs (rapamycin and wortmannin), the trichostatin A histone deacetylase inhibitor, and two antipsychotic drugs (trifluoperazine and thioridazine) (Fig. 1A). Notably, and consistent with the antagonistic effects of NOTCH1 inhibition and Pten inactivation in T-ALL, our cMAP analyses of drugs potentially enhancing the effects of NOTCH1 inhibition and compounds antagonizing the effects of Pten loss identified rapamycin, thioridazine, trifluoperazine, and trichostatin A as redundant hits in both categories (Fig. 1A). Finally, and consistent with these results, cMAP analysis of the gene expression signatures induced by NOTCH1 inhibition in Pten WT cells, but no longer present upon GSI treatment of Pten-deleted Notch1-induced leukemias (SI Appendix, Fig. S1), identified three of these drugs (rapamycin, trifluoperazine, and trichostatin A), as well as parthenolide, as candidate agents to abrogate the prosurvival effects of Pten loss in the context of NOTCH1 inhibition (Fig. 1A).
Fig. 1.
Gene expression-based identification and characterization of candidate drugs antagonizing NOTCH and PI3K-AKT oncogenic programs in T-ALL. (A) cMAP top-scoring drugs positively associated with NOTCH inactivation signatures and negatively associated with Pten deletion signatures in Notch1-induced leukemias and with Pten deletion signatures in Notch1-induced tumors treated with the DBZ GSI. (B) Circos plot representation of pairwise relationships between the gene expression signatures induced by drug treatments in CUTLL1 cells.
Following on these results we next explored the functional relationships between these cMAP hits in CUTLL1, a NOTCH1-dependent T-cell lymphoblastic cell line (16), analyzing their gene expression profiles and measuring the pairwise relationship between their gene expression signatures using gene set enrichment analysis (GSEA) (17). Circos plot (Fig. 1B) and MANTRA plot (Mode of Action by NeTwoRk Analysis) (SI Appendix, Fig. S1) representations of the similarities between the transcriptional signatures induced by each of these compounds identified drugs with highly interrelated transcriptional programs (wortmannin, valproic acid, trifluoperazine, thrichostatin A, thioridazine, rapamycin, phenoxybenzamine, and parthenolide), suggestive of a common downstream effector mechanism. In contrast, pyrvinium pamoate, resveratrol, withaferin A, astemizole, mefloquine, and vorinostat showed the least number of associations with other drugs, suggesting more distinct mechanisms of action.
Analysis of Antileukemic Effects and Interaction with NOTCH1 Inhibition.
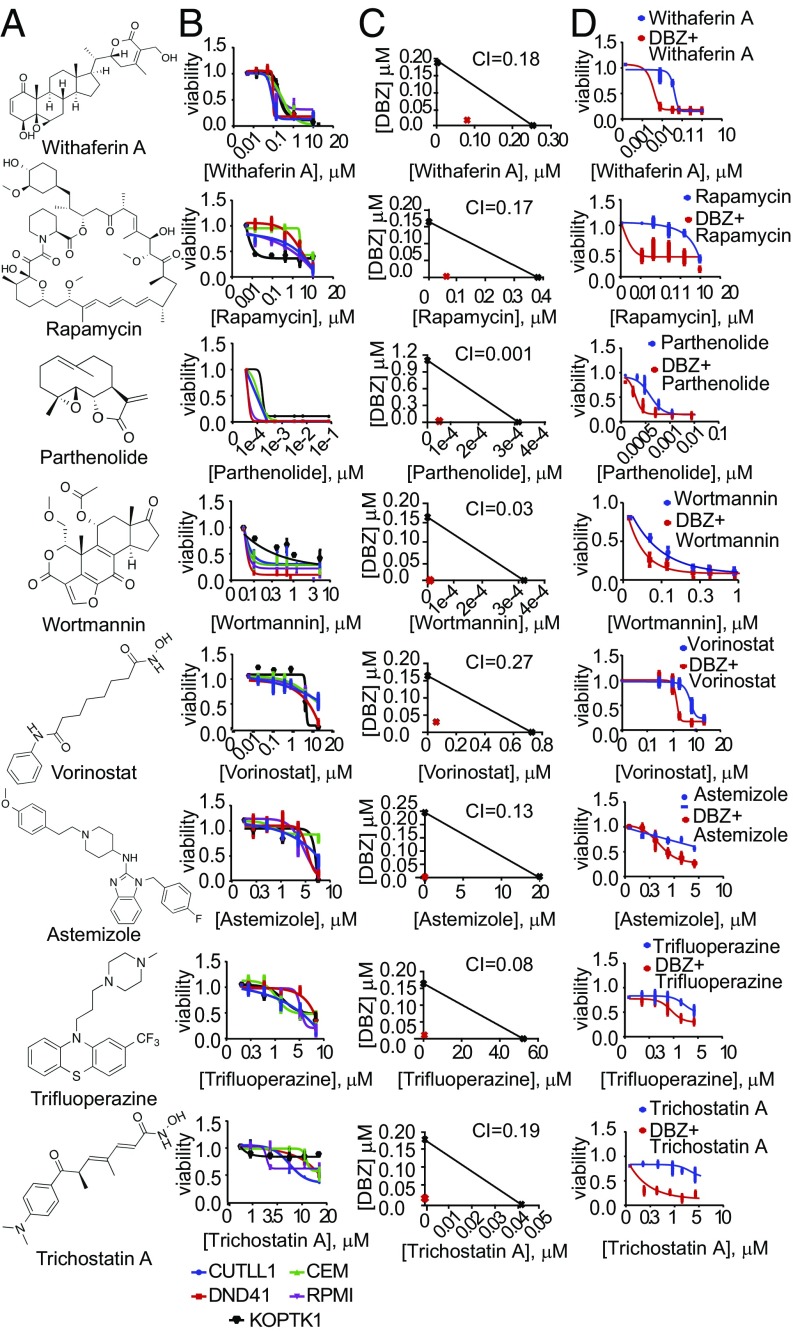
To evaluate the antileukemic effects of these drugs and their potential interaction with NOTCH1 inhibition, we first analyzed the response of CUTLL1 cells to each of our cMAP hits alone and in combination with the DBZ GSI. These analyses identified 11 compounds with strong synergism with GSI treatment (Combination Index < 0.4), 8 of which showed high (IC50 < 0.5 µM) (withaferin A, rapamycin, vorinostat, parthenolide, and wortmannin) or moderate (IC50 0.5–5 µM) (astemizole, trifluoperazine, and trichostatin A) intrinsic antileukemic activities based on reduced cell viability (Fig. 2 and SI Appendix, Figs. S2–S4). These results were verified in a broader panel of T-ALL lines, including PTEN WT (DND41 and KOPTK1) and PTEN mutant (RPMI8402 and CCRF-CEM) leukemias (SI Appendix, Table S1and SI Appendix, Figs. S2–S5).
Fig. 2.
Antileukemic effects of cMAP drugs that antagonize NOTCH1 and PI3K-AKT expression programs in T-ALL. (A) Withaferin A, rapamycin, parthenolide, wortmannin, vorinostat, astemizole, trifluoperazine, and trichostatin A structures. (B) Dose–response cell-viability curves relative to vehicle-treated controls in T-ALL (72 h treatment). (C) Isobologram analysis of the effects of fixed molar ratio combinations of each cMAP drug with the DBZ GSI in CUTLL1 cells during 6 d of treatment. (D) Cell-viability curves relative to vehicle-treated controls of cMAP drugs alone and in combination with a fully inhibitory concentration (250 nM) of the DBZ GSI (72 h treatment). All treatments were performed in triplicate and were repeated at least twice. CUTLL1 cells were used in C and D. Data in B and D represent mean ± SD.
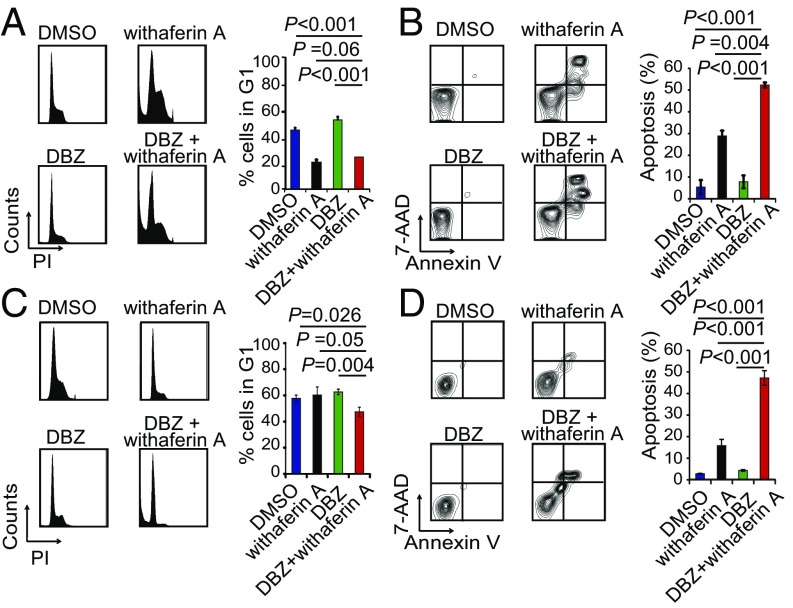
To elucidate the antileukemic effects of these drugs, we next analyzed their effects on cell cycle progression and apoptosis alone and in combination with DBZ (Fig. 3 and SI Appendix, Figs. S6–S8). These analyses demonstrated a major proapototic activity for withaferin A and parthenolide as single agents and markedly increased apoptotic responses for withaferin A, rapamycin, and astemizole when used in combination with DBZ (Fig. 3 and SI Appendix, Figs. S6 and S7). In contrast, vorinostat, wortmannin, trichostatin A, and trifluoperazine showed a strong and synergistic cytostatic activity in combination with GSI treatment, but with limited apoptosis (SI Appendix, Figs. S6 and S7). Analyses of primary T-ALL cells from three independent NOTCH1-mutant leukemia samples demonstrated strong antileukemic effects for the combination of withaferin A and DBZ (Fig. 4A). Consistently, and despite its short in vivo half-life (18), withaferin A induced a marked enhancement of the antitumor effects of Notch inhibition with DBZ in vivo in a mouse model of Notch1-induced T-ALL (10) (Fig. 4 B and C). In this experiment, we allografted mice with NOTCH1 HD-ΔPEST mouse-induced ALL cells and upon full leukemia development treated them with vehicle only, withaferin A, DBZ, or the combination of withaferin A plus DBZ, observing a marked and significant reduction in tumor burden by in vivo bioimaging after 6 d of treatment in animals treated with withaferin A plus DBZ (Fig. 4B), which translated in significant extension in survival from 20 d in control mice to 60 d in the combination treatment group (Fig. 4C). Finally, we evaluated the efficacy of this treatment against two independent Notch1-mutant human primary T-ALL xenografts (SI Appendix, Table S2) in vivo. These analyses demonstrated variable response to withaferin A alone, but robust responses to withaferin A plus DBZ in combination (Fig. 4 D–G, SI Appendix, Fig. S9).
Fig. 3.
Antitumor effects of withaferin A in combination with the DBZ GSI in T-ALL cells in vitro. (A) Cell cycle and (B) apoptosis analysis of CUTLL1 cells treated with DMSO, withaferin A (200 nM), the DBZ GSI (250 nM), and withaferin A (200 nM) plus DBZ (250 nM) in combination. (C) Cell cycle and (D) apoptosis analysis of CCRF-CEM cells treated with DMSO, withaferin A (200 nM), the DBZ GSI (250 nM), and withaferin A (200 nM) plus DBZ (250 nM) in combination. Cells were treated with drug alone or in combination per triplicate during 72 h. Bar graphs in A, B, C, and D indicate mean ± SD.
Fig. 4.
Characterization of the antileukemic effects of withaferin A in combination with the DBZ GSI in vivo. (A) In vitro analyses of the antileukemic responses of three independent primary T-ALL samples treated with DMSO control, DBZ (250 nM), withaferin A (200 nM), or with the same doses of DBZ plus withaferin A in combination during 72 h. (B) Representative images and quantitative analyses of changes in tumor burden in isogenic Notch1-positive, Pten-positive murine leukemia, upon treatment with vehicle only, the DBZ GSI, withaferin A ,and withaferin A plus DBZ in combination (n = 5 C57BL6 mice per group). (C) Survival analysis of mice harboring NOTCH1 L1601P Δ-PEST induced T-ALL upon treatment with vehicle only, DBZ (10 mg kg−1), withaferin A (10 mg kg−1), or withaferin A + DBZ (withaferin A + DBZ vs. all other groups P ≤ 0.001 Gehan–Breslow–Wilconson test; n = 10 per group). (D, E, F, and G) Luciferase images and quantitative analyses of changes in tumor burden in two independent human primary T-ALL xenografts upon treatment with vehicle only, the DBZ GSI, withaferin A, and withaferin A plus DBZ in combination (n = 5 per group). Horizontal bars in B, E, and G indicate mean luciferase levels.
Of note, therapy with withaferin A plus DBZ in combination was well tolerated clinically. Analysis of toxicity showed increased goblet cell numbers in the intestine of mice treated with DBZ, a phenotype linked with systemic inhibition of NOTCH signaling, and similar changes with no signs of increased toxicity were noted in animals treated with withaferin A plus DBZ (SI Appendix, Fig. S10). We observed no changes in the weight of C57BL/6 mice treated for 6 d with vehicle, DBZ, withaferin A, or both drugs in combination (SI Appendix, Fig. S11). Moreover, hematologic analyses revealed only a mild decrease of white blood cells at the expense of lymphocytes and monocytes in animals treated with the combination of DBZ and withaferin A (SI Appendix, Fig. S12).
eIF2A Translation Inhibition Mediates the Antileukemic Effects of Withaferin A.
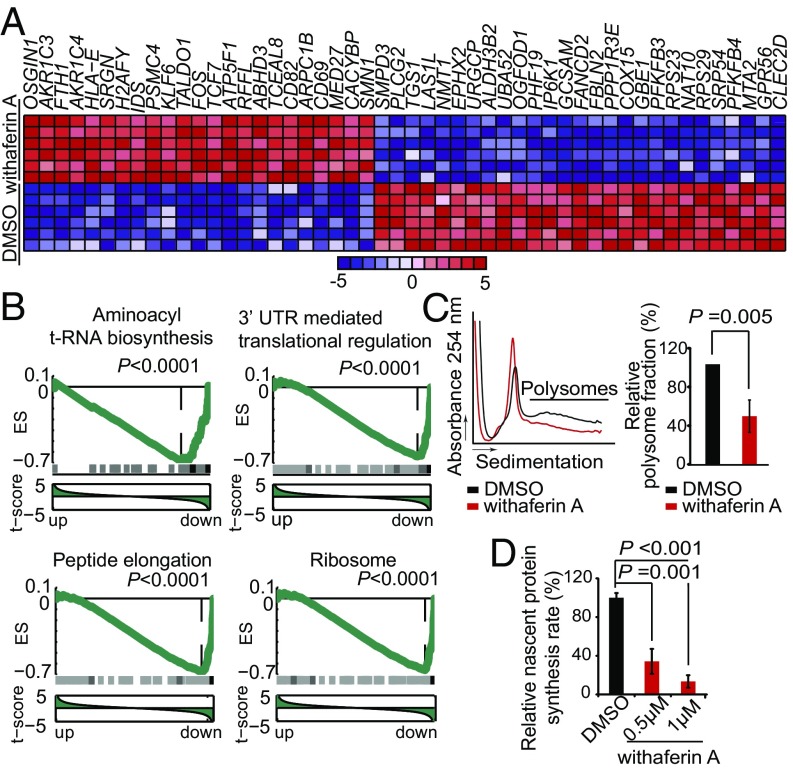
Withaferin A, a bioactive steroidal lactone originally isolated from Withania Somnifera, has shown antitumor effects against colorectal and breast cancer cell lines (19, 20). However, the mechanisms of action of this natural compound remain incompletely understood. To explore the potential effector mechanisms mediating the antileukemic activities of withaferin A in T-ALL and its interaction with NOTCH1 inhibition we further analyzed the transcriptional signatures induced by this drug. In CUTLL1 cells, withaferin A treatment induced broad changes in the gene expression profile with 433 up-regulated and 424 down-regulated genes (fold change 1.3, P < 0.001) (Fig. 5A). Notably, this signature was markedly enriched in genes and pathways implicated in protein translation, including translation (GO: 0006412), translational elongation (GO: 0006414), ribosome (GO: 0005840), and protein biosynthesis (SP_PIR_KEYWORD: protein biosynthesis). Moreover, gene set enrichment analysis (GSEA) revealed significant down-regulation of translation-related pathways upon withaferin A treatment, including aminoacyl transfer RNA (t-RNA) biosynthesis, 3′UTR translation regulation, peptide elongation, and ribosome (Fig. 5B). Consistently with our cMAP analysis results, genes down-regulated by GSI in T-ALL, including NOTCH1 direct targets (e.g., DTX1 and MYC), are also significantly down-regulated by GSEA in CUTLL1 cells treated with withaferin A (SI Appendix, Fig. S13). However, the transcriptional effects of DBZ and withaferin A are not completely overlapping, as evidenced by the lack of HES1 down-regulation upon withaferin A treatment (SI Appendix, Fig. S13). Further analysis of withaferin A-induced gene expression programs using DeMAND, a regulatory network algorithm for the identification of potential drug effector mechanisms as deregulated nodes induced by a drug treatment, identified 21 components of the translation machinery among the top 50 nodes perturbed by withaferin A (SI Appendix, Table S3). Notably, these included two subunits of eIF2A (eIF2S1 and eIF2S2), a protein complex that mediates the recruitment of the first Met-coupled t-RNA to the 40S ribosome subunit (21) and the inhibition of protein synthesis under conditions of cellular stress (22) (SI Appendix, Fig. S14). In all, these results suggest translation inhibition and the eIF2A complex as effectors of the antileukemic activities of withaferin A.
Fig. 5.
Inhibition of protein translation by withaferin A in T-ALL. (A) Heat map representation of top differentially expressed genes in CUTLL1 cells treated with vehicle only and withaferin A (24 h). Up-regulated transcripts are shown in red, and down-regulated transcripts are shown in blue. Bar at bottom indicates differential expression levels in SD units. (B) Representative examples of gene set enrichment plots corresponding to GSEA analysis of MSigDB C2 data sets enriched in the expression signature associated with withaferin A treatment samples compared with vehicle-treated controls. (C) Polysome profiling analysis of JURKAT cells treated with withaferin A or vehicle only (DMSO). (D) Quantitative analysis of nascent protein synthesis measured by ClickIT on JURKAT cells treated with DMSO or withaferin A. Bars in C and D represent mean ± SD.
To test this hypothesis, we analyzed the effects of withaferin A in the control of protein synthesis in polysome profiling and nascent protein synthesis assays. These studies revealed reduction of polysome numbers and a significant abrogation of nascent protein synthesis in withaferin A-treated T-ALL cells compared with vehicle-only–treated controls (Fig. 5 C and D). Moreover, and consistently with our network analyses prediction, withaferin A treatment of T-ALL cells induced dose-dependent phosphorylation of eIF2S1 at residue S51 (Fig. 6A and SI Appendix, Fig. S15), a posttranslational modification responsible for blocking the formation of eIF2A Met–t-RNA complexes in conditions of amino acid starvation and in response to oxidative and endoplasmic reticulum stress (21–23). Moreover, and concomitant with eIF2S1 S51 phosphorylation, we observed increased expression of ATF4, a transcription factor specifically activated by alternative translation in the context of eIF2A-mediated translation inhibition (Fig. 6B). Notably, expression of a phosphomimic mutant form of eIF2S1 (eIF2S1-S51D) in JURKAT and CUTLL1 cells impaired leukemia cell viability and proliferation (Fig. 6 C and D and SI Appendix, Fig. S15), whereas expression of a nonphosphorylatable form of eIF2S1 (eIF2S1-S51A) abrogated the antileukemic effects of withaferin A (Fig. 6 E and F and SI Appendix, Fig. S15) and of DBZ plus withaferin A in combination (Fig. 6 G and H and SI Appendix, Fig. S15). These results demonstrate a direct role for eIF2S1 phosphorylation and inhibition of eIF2A-dependent translation as a critical mediator in the antileukemic effects of withaferin A in T-ALL and support the role of therapies inhibiting protein translation in combination with NOTCH inhibition for the treatment of T-ALL. Consistent with this model, inhibition of protein translation with silvestrol, an inhibitor of eIF4A-cap–mediated translation, induced synergistic antileukemic effects with NOTCH inhibition by GSI in T-ALL (SI Appendix, Fig. S16).
Fig. 6.
eIF2A-dependent inhibition of protein translation mediates the antitumor effects of withaferin A in T-ALL. (A) Western blot analysis of eIF2S1-S51 phosphorylation in JURKAT cells treated with vehicle only or withaferin A. (B) Western blot analyses of ATF4 expression in JURKAT cells treated with vehicle only or withaferin A (0.5 µM) (C) Cell growth analysis of JURKAT cells after infection with empty-vector control lentiviruses and lentiviral constructs expressing eIF2S1 WT or the phosphomimic form of eIF2S1 (eIF2S1-S51D). (D) Cell-viability analysis (annexin-V APC exclusion) of JURKAT cells after infection with empty-vector control lentiviruses and lentiviral constructs expressing either WT or the phosphomimic form of eIF2S1 (eIF2S1-S51D) (72 h). (E) Differential cell growth of JURKAT cells treated with withaferin A relative to vehicle-treated controls after infection with empty-vector control lentiviruses and lentiviral constructs expressing eIF2S1 WT or the nonphosphorylatable form of eIF2S1 (eIF2S1-S51A) (72 h). (F) Cell-viability analysis (annexin-V APC exclusion) of JURKAT cells after infection with empty-vector control lentiviruses and lentiviral constructs expressing eIF2S1 WT or the nonphosphorylatable form of eIF2S1 (eIF2S1-S51A) treated with vehicle only or withaferin A (72 h). (G) Nascent protein synthesis in JURKAT cells infected with empty-vector control lentiviruses and lentiviral constructs expressing WT eIF2S1 or the nonphosphorylatable form of eIF2S1 (eIF2S1-S51A) upon treatment with withaferin A or withaferin A plus DBZ (250 nM) relative to vehicle-treated controls. (H) Cell-viability analysis of JURKAT cells infected with empty-vector control lentiviruses and lentiviral constructs expressing WT eIF2S1 or the nonphosphorylatable form of eIF2S1 (eIF2S1-S51A) upon treatment with withaferin A or withaferin A plus DBZ (250 nM) relative to vehicle-treated controls (72 h). Graphs in C, D, E, F, G, and H indicate mean ± SD. Asterisks in C and E indicate P < 0.001.
Discussion
There is an urgent need to identify drugs that synergistically enhance the antileukemic effects of anti-NOTCH1 therapies in T-ALL. However, empirical screening approaches to the identification of synergistic drug combinations are cumbersome and often do not directly inform on the mechanisms mediating drug interactions. To overcome these obstacles we implemented an expression-based discovery strategy that capitalizes on accurately engineered mouse models of NOTCH1-induced T-ALL with conditional loss of the Pten tumor suppressor gene. Reassuringly, this approach recovered inhibitors of the mTOR/PI3K/AKT pathway (rapamycin and wortmannin), histone deacetylase inhibitors (vorinostat, trichostatin A, and valproic acid), an NFKB inhibitor (parthenolide), and several phenothiazines (trifluoperazine, prochlorperazine, and thioridazine). However, the most active cytotoxic and synergistic compound identified in this screen was withaferin A, a natural compound with antitumor, antiinflammatory, antibacterial, and immunomodulary properties. Mice treated with DBZ plus withaferin A showed markedly reduced tumor loads, low levels of minimal residual disease, and increased survival in vivo. Moreover, the broad response of multiple T-ALL cell models tested here, including GSI-sensitive (CUTLL1, DND41, KOPT-K1) and GSI-resistant (CCRF-CEM, RPMI 8402, JURKAT) lines, to withaferin A plus GSI in combination supports that this combination can overcome resistance to anti-NOTCH1 therapies. It is unclear at this point if relapsed tumors following GSI plus withaferin A combination therapy would respond to retreatment. However, we have not observed development of resistance in vitro after sustained treatment with DBZ plus withaferin A in any of our models, suggesting that T-ALL cells need to overcome significant barriers to develop resistance to this combination.
Mechanisms proposed for the antitumor effects of withaferin A include proteasome (24), HSP90 (25), NFKB (26) inhibition, and induction of reactive oxygen species (ROS) (27). However, these mechanisms seem to be cell-type specific, as functional assays revealed decreased ROS levels and no or minimal impact of withaferin A in HSP90, proteasome, or NFKB activity in T-ALL cells (SI Appendix, Fig. S17). Functional annotation of transcriptomic data coupled with regulatory network analysis (DeMAND) pointed toward a prominent role of withaferin A in eIF2A translation inhibition. Consistently, withaferin A treatment induced decreased protein synthesis in T-ALL cells, which was mediated by eIF2S1 phosphorylation.
Key oncogenes and signaling pathways involved in the pathogenesis of T-ALL, including NOTCH1, MYC, and the PI3K-AKT-mTOR pathway, participate in the regulation of ribosome biogenesis and translation (9, 28, 29). Consistently, gene expression signatures induced by NOTCH inhibition in T-ALL show marked down-regulation on translation-related genes (SI Appendix, Fig. S18). Moreover, forced expression of the eIF4A translation initiation factor has been shown to accelerate NOTCH1-induced T-ALL in mice, and inhibition of eIF4A cap-mediated translation induces apoptosis in T-ALL (30). Our results shown here further highlight a central role of protein translation in T-ALL homeostasis and suggest a therapeutic role for targeting eIF2A-mediated translation in combination with GSIs.
Materials and Methods
T-ALL Human Cell Lines.
CCRF-CEM, RPMI-8402, KOPT-K1, and JURKAT T-ALL cell lines were obtained from ATCC, and HPBALL and DND41 cell lines were obtained from DSMZ (The Leibniz Institute). The CUTLL1 NOTCH-dependent T-cell lymphoblastic cell line has been previously described (16).
Human Primary Leukemia Samples.
T-ALL primary samples were obtained from the ECOG-ACRIN Cancer Research Group (ECOG-ACRIN) tumor bank. Clinical leukemia samples were obtained with informed consent at local institutions and used under the supervision of the Columbia University Medical Center Institutional Review Board Committee.
In Vitro Studies.
We analyzed cell viability and proliferation with the Cell Proliferation Kit I (Roche) and cell cycle analysis by flow cytometry after Propidium Iodide (Sigma Aldrich) DNA staining. We quantified cell viability and apoptosis by flow cytometry after Annexin V-allophycocyanin (APC) and 7AAD markers (AnnexinV BD Pharmigen). Western blot was performed using standard methods. Antibodies against NOTCH1 (Val1744) (#4147), GAPDH (#5174S), eIF2S1 (#9722), phospho-eIF2S1 S51 (#9721S), and ATF4 (#11815) were purchased from Cell Signaling Technologies; CDK4 (sc-260) and FYN (sc-271294) antibodies were obtained from Santa Cruz Biotechnology.
Microarray Gene Expression Profile Analysis.
We performed cMAP analyses on gene expression signatures derived from Pten conditional-inducible knockout NOTCH1-induced mouse T-ALLs upon treatment with the DBZ GSI; upon Pten deletion induced by tamoxifen treatment; and upon NOTCH1 inhibition with DBZ as before after tamoxifen-induced deletion of Pten. We applied Drug Mode of Action through Network Dysreguation (DeMAND) to withaferin A signature to investigate potential effectors of its antileukemic activity. A detailed description of analytical parameters is given in SI Appendix, Materials and Methods.
Mice and Animal Procedures.
We maintained animals in the animal facility at the Irving Cancer Center at Columbia University Medical Campus. All animal procedures were approved by the Columbia University Institute for Animal Care and Use Committee (IACUC). A detailed description of experimental therapeutic procedures is provided in SI Appendix, Materials and Methods.
Statistical Analyses.
We performed statistical analysis by Student’s t test. We considered results with P < 0.05 as statistically significant. We analyzed drug synergism using the median-effect method developed by Chou and Talalay (31, 32) and used the CalcuSyn software (Biosoft) to calculate the combination index (CI) and perform isobologram analysis of drug interactions. We used GraphPad Prism 4.0 software (GraphPad Software) for determination of drug IC50s using nonlinear regression analysis of dose–response curves. Statistical comparison of survival curves in pairs was performed using Gehan–Breslow–Wilconson test (GraphPad Software).
Supplementary Material
Acknowledgments
We thank the ECOG-ACRIN Cancer Research Group for clinical specimens; J. Aster for the MigR1-NOTCH1 L1601P ΔPEST vector; D. Ron for the pCDNA3 eIF2S1, pCDNA3 eIF2S1-S51A and pCDNA3 eIF2S1-S51D plasmids; P. P. Pandolfi for the Ptenfl conditional knockout mouse; T. Ludwig for the ROSA26Cre-ERT2/+ mouse; S. Indraccolo for xenograft T-ALL cells, R. Baer for helpful discussions and revision of the manuscript; and L. Xu for technical assistance in mouse experiments. This work was supported by the National Institute of Health (Grants R01CA129382 and CA120196 to A.A.F.; Grants CA180827 and CA196172 to E.P.; and CA180820, CA189859, CA14958, CA180791, and CA17145 to ECOG-ACRIN), the Stand Up to Cancer Innovative Research Award (to A.A.F.), the Swim Across America Foundation (to A.A.F.), the William Lawrence & Blanche Hughes Foundation (to A.A.F.), Fonds Wetenschappelijk Onderzoek Vlandereen (G084013) and European Research Council Starting Grant 334946 (to K.D.K.). D.H. is supported by the US National Institutes of Health Grant K99/R00 CA197869 and an Alex Lemonade Stand Foundation Young Investigator grant. M.S.-M. is a postdoctoral fellow funded by the Rally Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.A.C. is a Guest Editor invited by the Editorial Board.
Data deposition: Expression data are accessible at Gene Expression Omnibus using accession codes: GSE71087, GSE71089, GSE78189, and GSE5827.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611831114/-/DCSupplemental.
References
- 1.Pui CH, et al. Heterogeneity of presenting features and their relation to treatment outcome in 120 children with T-cell acute lymphoblastic leukemia. Blood. 1990;75(1):174–179. [PubMed] [Google Scholar]
- 2.Möricke A, et al. German-Austrian-Swiss ALL-BFM Study Group Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111(9):4477–4489. doi: 10.1182/blood-2007-09-112920. [DOI] [PubMed] [Google Scholar]
- 3.Stock W, et al. Dose intensification of daunorubicin and cytarabine during treatment of adult acute lymphoblastic leukemia: Results of Cancer and Leukemia Group B Study 19802. Cancer. 2013;119(1):90–98. doi: 10.1002/cncr.27617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton R, et al. Persistent MRD before and after allogeneic BMT predicts relapse in children with acute lymphoblastic leukaemia. Br J Haematol. 2015;168(3):395–404. doi: 10.1111/bjh.13142. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.Deangelo D, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. 2006;24(18S):6585. [Google Scholar]
- 7.Wei P, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9(6):1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 8.Takebe N, Nguyen D, Yang SX. Targeting notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol Ther. 2014;141(2):140–149. doi: 10.1016/j.pharmthera.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herranz D, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21(10):1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 13.Cullion K, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113(24):6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanda T, et al. Interconnecting molecular pathways in the pathogenesis and drug sensitivity of T-cell acute lymphoblastic leukemia. Blood. 2010;115(9):1735–1745. doi: 10.1182/blood-2009-07-235143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez A, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest. 2014;124(2):644–655. doi: 10.1172/JCI65093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palomero T, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil D, et al. Determination of withaferin A and withanolide A in mice plasma using high-performance liquid chromatography-tandem mass spectrometry: Application to pharmacokinetics after oral administration of Withania somnifera aqueous extract. J Pharm Biomed Anal. 2013;80:203–212. doi: 10.1016/j.jpba.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Das T, Roy KS, Chakrabarti T, Mukhopadhyay S, Roychoudhury S. Withaferin A modulates the Spindle assembly checkpoint by degradation of Mad2-Cdc20 complex in colorectal cancer cell lines. Biochem Pharmacol. 2014;91(1):31–39. doi: 10.1016/j.bcp.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Nagalingam A, Kuppusamy P, Singh SV, Sharma D, Saxena NK. Mechanistic elucidation of the antitumor properties of withaferin a in breast cancer. Cancer Res. 2014;74(9):2617–2629. doi: 10.1158/0008-5472.CAN-13-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6(4):318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 22.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 23.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci. 2013;70(19):3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, et al. Withaferin A inhibits the proteasome activity in mesothelioma in vitro and in vivo. PLoS One. 2012;7(8):e41214. doi: 10.1371/journal.pone.0041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu M, et al. Structure-activity relationship (SAR) of withanolides to inhibit Hsp90 for its activity in pancreatic cancer cells. Invest New Drugs. 2014;32(1):68–74. doi: 10.1007/s10637-013-9987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyninck K, Lahtela-Kakkonen M, Van der Veken P, Haegeman G, Vanden Berghe W. Withaferin A inhibits NF-kappaB activation by targeting cysteine 179 in IKKβ. Biochem Pharmacol. 2014;91(4):501–509. doi: 10.1016/j.bcp.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Hahm ER, et al. Withaferin A-induced apoptosis in human breast cancer cells is mediated by reactive oxygen species. PLoS One. 2011;6(8):e23354. doi: 10.1371/journal.pone.0023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell KJ, White RJ. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med. 2014;4(5):a018408. doi: 10.1101/cshperspect.a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iadevaia V, Liu R, Proud CG. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol. 2014;36:113–120. doi: 10.1016/j.semcdb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516):65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 32.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.