Skeletal muscle is a plastic tissue that can increase in size (hypertrophy) in response to exercise or decrease in size (atrophy) with inactivity (1). Muscle atrophy occurs systemically in physiological conditions, such as fasting or aging, and in a variety of pathological conditions, including cancer cachexia, cardiac failure, renal failure, and chronic obstructive pulmonary disease. Focal atrophy of specific muscles or muscle groups can be induced by plaster cast immobilization or traumatic injury to peripheral nerves. An amazing aspect of muscle tissue loss in these conditions is the fact that muscle fibers maintain essentially their normal structure and function during atrophy, at least during the early stages. Consider fasting, when muscles undergo rapid protein degradation and release amino acids that are converted into glucose by the liver, thus allowing survival of the organism. However, despite the loss of muscle mass, contractile force normalized per muscle mass is preserved (2), and a fasted animal can stalk its prey or move to distant places for foraging, stressing the programmed nature of this evolutionarily conserved adaptive response. A study on atrophic fibers isolated from denervated muscles also showed that the force normalized per cross-sectional area is not reduced at 7 and 14 d after sciatic nerve section (3). How can the muscle machine lose pieces without losing function, something that man-made machines cannot do?
A distinctive feature of the contractile machine is a modular structure that contributes to its resilience after damage and allows preserving contractile function during atrophy. In muscle fibers, which are gigantic elongated cells containing hundreds of nuclei, the contractile material is organized into myofibrils arranged in parallel and composed of modular units arranged in series, the sarcomeres (Fig. 1). A single human muscle fiber, which in long muscles can be up to 10–20 cm long and 0.1 mm wide, may contain millions of sarcomeres, each ∼2 μm long and 1 μm wide. Sarcomeres consist of two sets of filaments, the thick (myosin) and thin (actin) filaments, which can slide one over the other thanks to the ATPase activity of a molecular motor, the myosin molecule, thus producing shortening and/or force generation. Thick and thin filaments are contained within the sarcomeric cytoskeleton made of rigid Z disks and elastic titin filaments that extend between the two Z disks. An extrasarcomeric cytoskeleton, composed of desmin intermediate filaments and other proteins, provides a transverse scaffold surrounding the myofibrils at the level of the Z disks and linking adjacent myofibrils, thus keeping in register the sarcomeres and generating the typical striated appearance of skeletal muscle fibers.
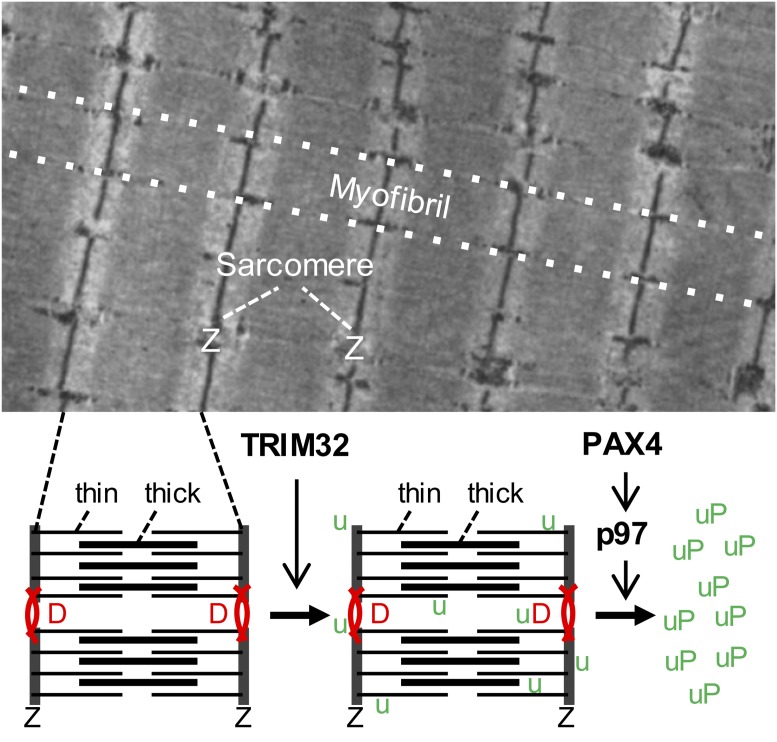
Fig. 1.
The upper panel shows a longitudinal section of a rat skeletal muscle fiber visualized by electron microscopy. The cytoplasm is filled with myofibrils, which consist of sarcomeres arranged in series and bordered by Z disks (Z). The lower panel shows a scheme of two adjacent sarcomeres with thick (myosin) and thin (actin) filaments. Z disks are connected by transversally oriented desmin filaments (D). Two crucial steps in the dismantling of the sarcomeres during muscle atrophy are illustrated: first, the TRIM32-induced ubiquitination (u) of desmin filaments, thin filaments, and Z-disk proteins, followed by the PAX4-dependent up-regulation of p97, which is responsible for the extraction of ubiquitinated proteins (uP) from the sarcomeres before their degradation in the proteasome.
The mechanism leading to the loss of contractile proteins during muscle atrophy is poorly understood, but recent studies, as in PNAS by Volodin et al. (4), are shedding light on this process. The ubiquitin–proteasome system, a major pathway of protein degradation present in all cells, has been implicated in the degradation of muscle contractile proteins (5). In this system, proteins are first conjugated with ubiquitin via a three-step process, with an E1 enzyme first binding ubiquitin that is subsequently transferred to an E2 enzyme and finally conjugated to the substrate by an E3 ligase. Ubiquitinated proteins are recognized by the proteasome receptors and degraded by the proteasome proteases. A large number of E3 ligases with distinct substrate specificities have been identified. In particular, two muscle-specific ubiquitin ligases, muscle RING-finger 1 (MuRF1) and Atrogin1/MAFbx, are rapidly induced in different types of muscle atrophy (6, 7), and their loss reduces muscle wasting (6). The expression of both MuRF1 and Atrogin1 is regulated by the transcription factor FoxO3, which causes dramatic muscle atrophy when overexpressed in adult muscle fibers (8). A common set of many genes, referred to as atrogenes, are induced by FoxO transcription factors in different muscle-wasting conditions (9).
Subsequent studies showed that thick and thin filaments are degraded by distinct mechanisms. MuRF1 ubiquitinates thick filament proteins, first myosin light chains and myosin binding protein C, and later myosin heavy chains, leading to their proteasomal degradation (10, 11). In contrast, a distinct ubiquitin ligase, tripartite motif-containing protein 32 (Trim32), is responsible for the degradation of thin filament proteins, actin, tropomyosin, and troponin, and of the Z-disk protein, α-actinin, whose loss is apparently preceded by the Trim32-dependent degradation of the desmin filaments (12). However, these analyses on thin-filament degradation were performed during the extremely rapid muscle atrophy induced by fasting; therefore, the precise sequence of events leading to muscle atrophy could not be clearly dissected. In PNAS, Volodin et al. (4) reinvestigate thin filament and desmin changes during the slower atrophy induced by denervation. They report that an early change during the first week after nerve section is the phosphorylation and ubiquitination of desmin, which is followed during the second week by the dissociation of desmin filaments and their ubiquitination followed by ubiquitination and degradation of contractile proteins. The findings indicate that the depolymerization of the desmin network, a process accelerated by electroporating denervated muscles with a dominant-negative inhibitor of desmin assembly, is a condition for myofibril breakdown, presumably because it makes thin-filament proteins more susceptible for ubiquitination. On the other hand, the total amount of desmin was not reduced 14 d after denervation, in contrast with the disappearance of desmin after fasting for 2 d (12).
A first important result of the present study (4) is the identification of two phases in the transcriptional program induced by denervation atrophy: an early phase characterized by the up-regulation of different atrogenes, which occurs a few days after nerve section under the control of FoxO transcription factors, and a second phase after 10–14 d, characterized by the up-regulation of other enzymes, including p97/VCP, which is not regulated by FoxO. Indeed, the level of p97 is not changed by overexpression of constitutively active FoxO3 in myotubes, although a dominant-negative p97 mutant blocks the increase in degradation of long-lived proteins by constitutively active FoxO3 (13). The AAA-ATPase p97/VCP is known to interact with ubiquitinated proteins associated to endoplasmic reticulum membranes, chromatin, or mitochondria with the help of ubiquitin-binding cofactors and is able to extract these target proteins from those structures to allow degradation by the proteasome (14). By analogy, it was suggested that p97, which in muscle forms complexes with cofactors Ufd1 and p47, associates with and extracts ubiquitinated proteins from the myofibrils before delivery to the proteasome (13). This interpretation is supported by the finding that the accumulation of p97, Ufd1, and p47 in atrophying muscle fibers coincides with the rapid breakdown of myofibrillar proteins at 10–14 d after denervation (4).
To identify the transcription factor(s) responsible for the expression of P97 and other transcripts up-regulated during the second week after nerve section, Volodin et al. (4) analyzed the promoters of these genes and found that they all contain binding motifs for the transcription factor PAX4. This finding is completely unexpected, first because PAX transcription factors are known for their role in
Volodin et al. provide a global, although certainly not yet complete, picture of the pathway involved in the controlled dismantling of cytoskeletal and myofibrillar structures during muscle atrophy.
organogenesis during development and are usually down-regulated in the adult, second because Pax4 controls the differentiation of insulin-producing beta cells in the pancreas (15), whereas two other members of the PAX family, Pax 3 and Pax7, regulate muscle progenitor cells (16). Volodin et al. (4) show that Pax4 is expressed in skeletal muscle and that p97 expression is markedly decreased when PAX4 levels are reduced by electroporation of fasted muscles with specific shRNAs. Under these conditions, ubiquitinated proteins were retained in the myofibrillar fraction, an effect similar to that produced by a dominant-negative p97, supporting a crucial role of the PAX4–p97 axis in the extraction and degradation of myofibrillar components. Interestingly, PAX4 also induces MuRF1 expression at 10 d after nerve section, thus promoting the degradation of thick filaments.
In conclusion, Volodin et al. (4) provide a global, although certainly not yet complete, picture of the pathway involved in the controlled dismantling of cytoskeletal and myofibrillar structures during muscle atrophy (Fig. 1). The first step is less well characterized, as the kinase responsible for desmin phosphorylation during atrophy in vivo has not yet been identified. This is followed by desmin ubiquitination and disassembly by the ubiquitin ligase TRIM32, which in turn promotes the subsequent ubiquitination of thin-filament and Z-disk proteins by TRIM32. At a later stage, the PAX4-dependent up-regulation of p97 allows the binding and extraction of myofibrillar proteins by p97, presumably followed by their transfer to the proteasome. This scheme has been validated both after food deprivation and nerve section, although the progression of the changes is much more rapid after fasting compared with denervation. It will be of interest to determine how the numerous other factors known to be involved in myofibril breakdown fit into this scheme, and whether a similar sequence of changes takes place in other models of muscle wasting, such as cancer cachexia or cardiac failure.
Another open issue is whether myofibrils and the cytoskeleton are affected homogeneously throughout the muscle fiber during atrophy or whether the degradation process occurs preferentially in specific regions. Early ultrastructural studies pointed out that, during the first 2 wk after denervation, peripheral myofibrils often appeared disrupted, whereas the inner myofibrils showed “good preservation and alignment of the bands” (17). It will be important to reinvestigate this issue by immunostaining with appropriate antibodies to map the areas of cytoskeleton and myofibril breakdown. For example, one may ask whether proteasome recruitment occurs in specific regions, for example, at the periphery of the fibers or at the periphery of the myofibrils. One might also try to visualize myofibril ubiquitination and breakdown in living muscle fibers, an approach previously used to visualize protein dislocation from the endoplasmic reticulum (18). Live-cell imaging would allow to distinguish whether contractile protein degradation is a homogeneously and slowly progressing process or is rather characterized by sporadic catastrophic events triggered by focal myofibrillar destabilization.
Footnotes
The author declares no conflict of interest.
See companion article on page E1375.
References
- 1.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280(17):4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 2.Nishio ML, Madapallimattam AG, Jeejeebhoy KN. Comparison of six methods for force normalization in muscles from malnourished rats. Med Sci Sports Exerc. 1992;24(2):259–264. [PubMed] [Google Scholar]
- 3.Raffaello A, et al. Denervation in murine fast-twitch muscle: Short-term physiological changes and temporal expression profiling. Physiol Genomics. 2006;25(1):60–74. doi: 10.1152/physiolgenomics.00051.2005. [DOI] [PubMed] [Google Scholar]
- 4.Volodin A, Kosti I, Goldberg AL, Cohen S. Myofibril breakdown during atrophy is a delayed response requiring the transcription factor PAX4 and desmin depolymerization. Proc Natl Acad Sci USA. 2017;114:E1375–E1384. doi: 10.1073/pnas.1612988114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271(43):26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 6.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 7.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 10.Clarke BA, et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Cohen S, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198(4):575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 2012;31(15):3334–3350. doi: 10.1038/emboj.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 15.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386(6623):399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 16.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 17.Pellegrino C, Franzini C. An electron microscope study of denervation atrophy in red and white skeletal muscle fibers. J Cell Biol. 1963;17(2):327–349. doi: 10.1083/jcb.17.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Y, Fang S. Live cell imaging of protein dislocation from the endoplasmic reticulum. J Biol Chem. 2012;287(33):28057–28066. doi: 10.1074/jbc.M112.381798. [DOI] [PMC free article] [PubMed] [Google Scholar]