Significance
Introduction of new anti-hepatitis C virus (HCV) agents, so-called direct-acting antivirals (DAAs), has greatly changed treatment for HCV, and a variety of choices for anti-HCV drug combinations are available. For better management and control of this worldwide infectious disease with anti-HCV agents, it is critical to develop a method for precisely profiling the antiviral efficacy of possible combination drug regimens and seek the “best treatment” based on scientific evidence. In this study, we showed how cell culture data can be combined with a mathematical model and computer simulation to quantify the anti-HCV drug efficacy of different drug concentrations and combinations in a preclinical setting, and hence develop a quantitative basis for selecting drug combinations prior to costly clinical trials.
Keywords: HCV, antiviral, mathematical model, replicon, instantaneous inhibitory potential
Abstract
With the introduction of direct-acting antivirals (DAAs), treatment against hepatitis C virus (HCV) has significantly improved. To manage and control this worldwide infectious disease better, the “best” multidrug treatment is demanded based on scientific evidence. However, there is no method available that systematically quantifies and compares the antiviral efficacy and drug-resistance profiles of drug combinations. Based on experimental anti-HCV profiles in a cell culture system, we quantified the instantaneous inhibitory potential (IIP), which is the logarithm of the reduction in viral replication events, for both single drugs and multiple-drug combinations. From the calculated IIP of 15 anti-HCV drugs from different classes [telaprevir, danoprevir, asunaprevir, simeprevir, sofosbuvir (SOF), VX-222, dasabuvir, nesbuvir, tegobuvir, daclatasvir, ledipasvir, IFN-α, IFN-λ1, cyclosporin A, and SCY-635], we found that the nucleoside polymerase inhibitor SOF had one of the largest potentials to inhibit viral replication events. We also compared intrinsic antiviral activities of a panel of drug combinations. Our quantification analysis clearly indicated an advantage of triple-DAA treatments over double-DAA treatments, with triple-DAA treatments showing enhanced antiviral activity and a significantly lower probability for drug resistance to emerge at clinically relevant drug concentrations. Our framework provides quantitative information to consider in designing multidrug strategies before costly clinical trials.
Hepatitis C virus (HCV) affects ∼170 million people worldwide (1–4) and is a major cause of liver cirrhosis and hepatocellular carcinoma. The standard treatment has long been a combination of IFN, IFN-α or pegylated IFN-α (peg–IFN-α), with ribavirin (RBV), with a sustained virological response (SVR) rate of around 50% (5). Improvements in the SVR rate have been made by using anti-HCV agents that inhibit viral-derived factors or cellular factors that are essential for viral replication. Agents inhibiting viral proteins, called direct-acting antivirals (DAAs), typically target HCV nonstructural (NS)3 protease, NS5A, and NS5B polymerase (3). Anti-HCV molecules that target cellular factors, so-called host-targeting antivirals (HTAs), include those HTAs inhibiting cyclophilins and microRNA-122, which are required for HCV replication (3). These agents have been evaluated in clinical trials. In 2011, the protease inhibitors (PIs) telaprevir (TPV) and boceprevir were approved by the US Food and Drug Administration for use in combination with peg-IFN and RBV. These drug combinations achieved significantly improved clinical outcomes, attaining more than a 70% SVR rate (5). The second-generation PI simeprevir (SMV) was approved in 2013 and has been widely used as one of the first choices of PIs in combinations such as SMV&peg–IFN-α&RBV and SMV&sofosbuvir (SOF) (4). SOF is a nucleoside polymerase inhibitor (NI) that was approved in 2013 and is or has been used in combination with RBV, SMV, and ledipasvir (LDV) (4). NS5A inhibitors (NS5AIs) that are already approved include daclatasvir (DCV) and LDV, which can be used in combinations such as DCV-SOF; DCV-asunaprevir (ASV), a PI; and, most importantly, SOF-LDV. Other treatment choices include a combination of paritaprevir (PI), ombitasvir (NS5AI), dasabuvir [DAS; nonnucleoside polymerase inhibitor (NNI)], and ritonavir (6). Additional drugs have just been approved, and others will eventually be approved for adding new combination choices (7). Anti-HCV treatment with triple-DAA regimens has also been in clinical trials (8–10).
In an era of rapid progress for anti-HCV treatments, patients and clinicians select one combination treatment from the available approved choices based on clinical trial results and practical issues, such as insurance company reimbursement policies. Toward better management and control of HCV infection, it is important to understand the intrinsic characteristics of each drug, including its antiviral activity, drug resistance profile, and adverse effects when used both singly and in combination to determine the “best” available combination treatment. Although the intrinsic antiviral activity is the most fundamental factor for treatment, there have been no data available that systematically evaluate and compare the intrinsic anti-HCV activity of drugs that are currently available or that will be available in the future. Here, by combining experimental and mathematical approaches, we evaluated the intrinsic antiviral activity and the theoretical emergence of drug-resistant viruses upon treatment with clinically available and developmental-phase anti-HCV agents for single drugs and double- and triple-drug combinations.
Results and Discussion
Evaluation of Intrinsic Antiviral Activity of Single and Double-Combination HCV Drugs.
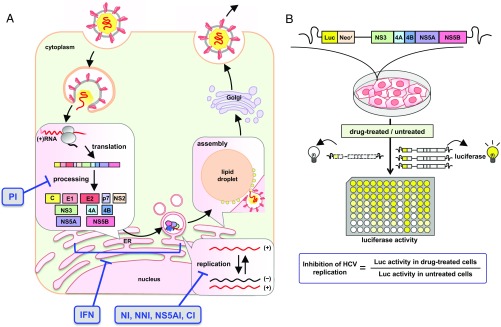
We evaluated the intrinsic antiviral activity of anti-HCV agents of different classes (Fig. 1A) in a cell culture model for HCV genotype 1, the most prevalent HCV genotype worldwide. The replicon system enables one to evaluate the efficacy of these drugs to inhibit HCV replication in a highly sensitive and high-throughput manner (11). We treated an HCV subgenomic replicon (strain-NN) (12) with each drug for 72 h and measured the HCV replication activity (Fig. 1B, Methods, and SI Appendix). The antiviral activity of a drug can be expressed as the instantaneous inhibitory potential (IIP) (13–18):
| [1] |
Here, is the fraction of infection events unaffected by the drug, is the drug concentration, is the drug concentration that inhibits 50% inhibition of the activity, and is the slope parameter reflecting the steepness of the dose–response curve (SI Appendix, Supplementary Note 1). If a drug reduces HCV replication by 1 log, then and its , whereas if it reduces replication by 2 logs (i.e., 100-fold), its . Note that the IIP incorporates all three parameters of the dose–response curve: , , and . Eq. 1 indicates that the higher the of the drug, the higher is the IIP at a given and .
Fig. 1.
Schematics of the anti-HCV drug targets and the experimental system. (A) HCV life cycle and drug targets. After entry into the host cell, HCV genomic RNA is translated into viral precursor polyprotein and processed into functional proteins (C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). HCV RNA replicates inside the isolated membrane compartments derived from the endoplasmic reticulum (ER), and assembles into viral particles on lipid droplets, which traffic through the Golgi and are released outside of the cell. PIs (TPV, DPV, ASV, and SMV) inhibit the processing step, and drugs such as NI (SOF), NNIs (VX, DAS, NSV, and TGV), NS5AIs (DCV and LDV), and CIs (CsA and SCY) target HCV RNA replication. IFNs (IFN-α and IFN-λ1) supposedly inhibit at least the step(s) of translation and replication. (B) HCV replication activity was evaluated using an HCV subgenomic replicon (genotype 1b, strain NN) carrying a fusion of the firefly luciferase gene (Luc) with the neomycin phosphotransferase (Neor). The replicon autonomously and persistently replicates in Huh-7 cells. Cells treated with drugs were incubated for 72 h and then harvested for luciferase assay. Inhibition of HCV replication was measured by the luciferase activity in drug-treated cells, relative to activity in DMSO-treated cells.
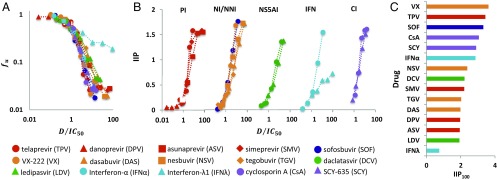
By profiling the anti-HCV activity of 15 clinically available and currently developmental-phase drugs (PIs [TPV, danoprevir (DPV), ASV, and SMV], NI [SOF], NNIs [VX-222 (VX), DAS, nesbuvir (NSV), and tegobuvir (TGV)], NS5AI [DCV and LDV], IFNs [IFN-α and IFN-λ1], and cyclophilin inhibitors (CIs) [cyclosporin A (CsA) and SCY-635 (SCY)] (Fig. 1A), we found that the dose–response curve slope, and thus the IIP value, varied among drugs (Fig. 2 A and B and SI Appendix, Supplementary Note 1). By extrapolation (Fig. 2C), we also determined the IIP100, defined as the IIP when , to estimate the effects of high drug concentrations, because clinical doses can range between 10- and 100-fold above the (19). We found that previous or current first-line drugs against HCV infection, such as IFN-α, TPV, SMV, and SOF, as well as CIs, can inhibit more than 99% of HCV replication in this concentration range ().
Fig. 2.
Quantification of the IIP of single-HCV drugs. (A) Log–log plots of dose–response curves normalized by , determined from the replicon assay, of PIs (TPV, DPV, SMV, and ASV; red), the NI (SOF; blue), NNIs (VX, DAS, NSV, and TGV; orange), NS5AIs (DCV and LDV; green), IFNs (IFN-α and IFN-λ1; cyan), and CIs (CsA and CSY; purple). Each point represents the mean of three experiments. (B) IIP of classes or subclasses of antiviral drugs, normalized by , calculated from the experimentally measured by Eq. 1. (C) IIP values at drug concentration (IIP100) determined by extrapolation.
Although the precise molecular basis for determining IIP value remains to be understood, IIP is likely to be governed by the subclass and target of a drug (SI Appendix, Supplementary Note 1). High IIPs were achieved by agents that included SOF and also HTAs, implying that these drugs inhibit the largest number of HCV replication events when administered at doses above their (Fig. 2 B and C and SI Appendix, Supplementary Note 1). This result adds a favorable characteristic to the already-known advantages of HTAs; pan-genotypic antiviral effect, high barrier to drug resistance, and relatively low cost (7), although the antiviral efficacy in patients may be affected by cellular conditions and other in vivo factors, such as the pharmacodynamics, local tissue environment, and targeted cellular compartment. However, given the current trends in anti-HCV therapy, the replacement of IFN-α–based regimens by all-oral, IFN-free therapies, evaluating DAA-only combinations is a timely issue. Among the DAA double combinations in this study, SOF combinations yielded desirably high IIP of the combination (IIPcom) values (Fig. 3A and SI Appendix, Supplementary Notes 2 and 3). SOF is one of the strong candidates for a constituent in the current standard-of-care multidrug treatment (7). Our IIP and IIPcom analyses show that even a small increase in the concentration of SOF can present a dramatic gain of antiviral effect, and the potential antiviral effect of SOF combinations is much higher compared with other drug combinations that show low IIPcom values.
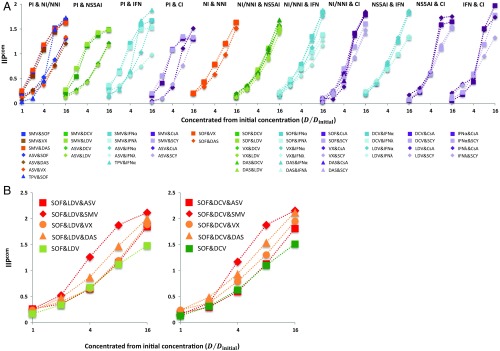
Fig. 3.
Quantification of inhibitory potential of anti-HCV drug multicombinations. (A) IIPcom of antiviral drug double combinations was calculated from the measured by Eq. 1. Fifty-two double combinations of interclass (or subclass) antiviral drugs were analyzed using the HCV replicon assay. Each point represents the mean of three experiments. Drugs were concentrated at a constant ratio from their initial concentrations , where the values were determined in separate single-drug experiments. (B) IIPcom of antiviral drug triple combinations was calculated from the measured by Eq. 1. Eight triple combinations of antiviral drugs were analyzed using the HCV replicon assay. Each point represents the mean of three experiments. Drugs were concentrated at a constant ratio from their initial concentrations , where the values were determined in separate single-drug experiments.
Profiling of Triple-Combination Anti-HCV Drugs.
Triple-DAA IFN-free combinations are being clinically evaluated to seek more rapid and efficacious elimination of HCV, including NI&NS5AI&PI and NI&NS5AI&NNI (8, 20–22). However, it is not yet understood how much advantage triple-DAA treatment can provide over double-DAA treatment, and which triple-DAA combination will give the best treatment outcome (8, 20–22). Here, we quantified the anti-HCV activity of eight candidate combinations of triple-DAA treatment; SOF with NS5AI (DCV, LDV) plus PI (SMV, ASV) or NNI (VX, DAS) (Fig. 3B and SI Appendix, Fig. S9 and Table S3). Interestingly, we found that these triple-DAA combinations greatly enhanced antiviral activity (i.e., increased the IIPcom by as much as twofold) compared with double-DAA treatments in Fig. 3B ( by Welch’s t test for all combinations), and that these drug combinations exhibited an intermediate activity compared with Loewe additivity and Bliss independence (SI Appendix, Fig. S10). Especially among the tested combinations, SOF&LDV&SMV and SOF&DCV&SMV achieved the highest IIPcom values (also SI Appendix, Table S3). At its clinical concentration (23, 24) (SI Appendix, Table S4), adding SMV to SOF&DCV increased the IIP of the combination, IIPcom, from 5.4 to 9.4 (i.e., it increased the antiviral activity by 4 logs) (Fig. 4 A and B). A proof-of-concept clinical trial of triple-DAA treatment (10) also showed that SOF&DCV&SMV can achieve a high SVR rate, while reducing the treatment period from 12 wk to only 3 wk using a response-guided protocol. Our analysis clearly supports a clinical advantage for triple-DAA–based IFN-free treatments as discussed elsewhere (8, 10, 20–22).
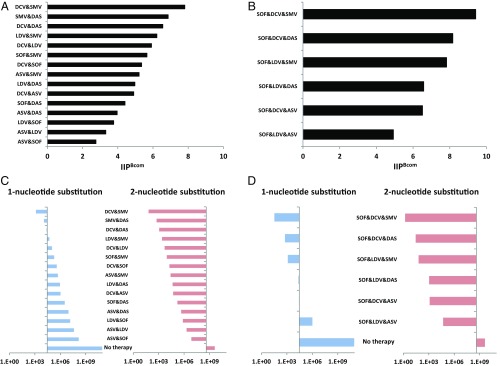
Fig. 4.
Quantification of the risk of HCV drug resistance. The fraction of unaffected HCV replication events of each double-drug (A) and triple-drug (B) combination at clinical concentrations is shown. The expected number of newly produced mutants with one-nucleotide (blue) and two-nucleotide (red) substitutions after the first day of double-drug (C) and triple-drug (D) combination treatment is shown. Each number is calculated by multiplying the number of newly produced mutants per day and the fraction of production events unaffected by a drug combination as follows: and , where and are the probability of one and two mutations occurring in the HCV genome after one replication event. The y axis shows the number of all possible one-nucleotide and two-nucleotide mutants ( and , respectively). Thus, if the bar faces to the left for a drug combination, it means that the expected number of newly produced mutants is below the number of all possible mutants under the corresponding treatment, suggesting drug-resistant mutants are unlikely to occur.
Calculation of Risk for HCV Drug Resistance Emergence.
With some DAA combination treatments, the emergence of drug-resistant HCV is one of the major causes leading to treatment failure (4, 7, 25). As reported by Rong et al. (26), because the number of newly produced virions per day is higher than the number of all possible single and double mutants of a drug-sensitive viral strain, all possible one-nucleotide and two-nucleotide drug-resistant mutants are predicted to be produced multiple times each day and may happen after 1 d of single-drug treatment (Fig. 4 C and D and SI Appendix, Supplementary Note 4). To minimize the emergence or selection of drug-resistant virus during treatment, multidrug combinations are the key treatment strategy. Using the mutation-estimating approach developed previously (26), we calculated the risk of emergent drug resistance against clinically important multidrug combinations for the drug concentrations in clinical use.
Given the clinical concentration of each drug reported (23, 24) (SI Appendix, Table S4) and applying a drug combination theory, Bliss independence (27), we estimated the fractions of unaffected production events (SI Appendix, Fig. S11 and Supplementary Note 4) and the Bliss-estimated IIP of double- and triple-DAA combinations (IIPBcom), shown in Fig. 4 A and B. According to our results, most of multidrug combinations show anti-HCV activity intermediate between Loewe additivity and Bliss independence (SI Appendix, Figs. S7 and S10 and Tables S2 and S3). Thus, we here assumed the anti-HCV effects of drug combinations calculated by Bliss independence to be the upper limit of their effectiveness. Our analyses indicated that SOF&DCV&SMV achieved the highest IIPBcom among the eight triple combinations composed of SOF&NS5AI&PI or SOF&NS5AI&NNI (Fig. 4B). Based on the estimated antiviral activity of these multidrug combinations, we calculated the expected number of newly produced virions carrying one-nucleotide or two-nucleotide mutations after 1 d of multiple-drug treatment in Fig. 4 C and D (also SI Appendix, Supplementary Note 4). Interestingly, even with suppression of viral replication events by most double-DAA combinations, except for DCV&SMV and SMV&DAS, there is still a chance for all of the possible one-nucleotide mutants to be generated (Fig. 4C), although many of those mutants are expected to be lethal (or unable to grow under double-combination treatment) and have lower fitness than wild-type virus. In contrast, triple combinations, except for SOF&LDV&ASV, showed a lower probability for allowing the emergence all of the possible one-nucleotide mutations. For example, SOF&LDV&SMV, a clinical choice for triple DAAs, showed an 11,000-fold lower risk of emergent mutants compared with SOF&LDV. Thus, our analysis clearly showed an advantage of triple-DAA combinations over double-DAA combinations, which greatly reduced the possible emergence of mutant viruses (Fig. 4 C and D). Our analysis was conservative in that it did not take into account the possible lower replication fitness of mutant virus, as seen with SOF resistance mutations (28). Hence our calculations may underestimate the barrier to resistance.
This high genetic barrier is especially important in cases where resistance-associated HCV variants preexist in patients before antiviral treatment, because the acquisition of drug resistance against double-DAA treatment requires only one additional nucleotide substitution, which can be easily introduced even under antiviral treatment, but the acquisition of drug resistance against triple-DAA treatment needs two additional nucleotide substitutions, which are much less frequent. It is known that PI-resistant variants are generally seen with low frequency (0.1–3%) in untreated patients; however, the Q80K mutation in NS3, which generates weak resistance to SMV, has been observed in 9–48% of patients infected with HCV genotype 1a, but at a much lower frequency in patients treated with genotype 1b (29–31). L31M and Y93H in NS5A, conferring resistance to NS5AIs, have high frequency in ∼30% of treatment-naive patients infected with HCV genotype 1b (32, 33). Preexistence of these resistant variants against anti-HCV agents, such as SMV, DCV, or LDV, limits treatment efficacy (34). Our analysis showed the advantage of triple-DAA treatments for universal clearance of HCV independent of individual viral genotypes and quasispecies. The analysis also suggested that SOF&DCV&SMV would have the highest barrier to resistance of any combination tested.
Conclusions
Because a series of HCV drugs have recently been or will soon be approved for clinical use, the clinical outcome of HCV treatment has been dramatically improved. To achieve better management and control of HCV infection worldwide, it is essential to understand the characteristics of each drug and to choose the optimal drug combination based on scientific evidence. The practical choice of drug depends on many factors: side effects of the drug, the genotype of HCV, patient characteristics, the presence of resistance-associated variants of HCV in the patient, and the patient’s treatment history. Among these factors, the primary and fundamental factors to be considered for treatment optimization are the magnitude of antiviral activity and the potential for emergence of drug resistance. Until now, however, the intrinsic anti-HCV activity achieved by monotreatment and combination treatments has not been systematically quantified, and the difference in the characteristics of each anti-HCV drug has not been tabulated. In this study, we evaluated the anti-HCV activity in an HCV genotype 1 replicon cell culture system (Fig. 1B). Although some anti-HCV drugs block multiple steps, including viral assembly/secretion (35), the primary target of all of the drugs used in this study is viral replication, which prompted us to use the replicon system to evaluate drug effectiveness. This system supports efficient replication of genotype 1 HCV, and thus enables one to measure the intrinsic antiviral effects of any drug combination and at any concentration of the component drugs in a highly sensitive manner and with high throughput. The experimental data were analyzed by calculating the IIP, which is the log reduction in HCV replication caused by drugs singly and in combination at a particular concentration (13–18). From the calculated IIP of 15 anti-HCV drugs, we found that IFN-α and the NI SOF had the largest potential to inhibit viral replication events. Profiling of 52 double-combination treatments indicated that combinations using a PI, SMV, achieved high IIP. By taking into account clinical concentrations, different SMV-based double-DAA combinations under clinical development showed the most desirable IIP score. Further, quantitative analysis showed that triple DAA combinations greatly enhanced the antiviral activity and reduced the probability of emergence of drug-resistant virus compared with double-DAA treatments.
Quantifying antiviral activity based on the IIPs was originally developed (14, 15, 17, 18) for quantifying the anti-HIV effect of combination antiretroviral therapy (cART). Interestingly, these approaches provided a quantitative basis for determining cART efficacy and for predicting drug resistance in patients (36). Likewise, our experimental evidence-based mathematical analysis is useful for optimizing drug use because it computes the antiviral activity for various combinations and drug concentrations in a preclinical setting, thereby providing basic information for designing more efficacious and cost-effective drug treatments with a high barrier to drug resistance. Given that the antiviral efficacy of most DAAs varies among the HCV genotypes, optimizing drug combinations that target other genotypes should be possible using other replicon or infectious virus cell culture systems.
Methods
In this study, HCV replication was evaluated in the HCV replicon system. It should be noted that the anti-HCV activity (IIP) of drugs, including PIs, NS5AIs, NIs, and NNIs, based on the replicon assay showed significant correlations with the anti-HCV activity obtained in the HCV infectious cell culture system, suggesting that the replicon system can be used to characterize the essential anti-HCV activity of drugs (SI Appendix, Fig. S12 and Supplementary Note 5). In addition, a Spearman’s rank-order correlation analysis showed that the anti-HCV activity was not significantly affected by the adaptive mutations in our replicon system (SI Appendix, Fig. S13 and Supplementary Note 5). We used LucNeo#2 cells, which carry an HCV subgenomic replicon that includes ORFs for a fusion protein of firefly luciferase-neomycin phosphotransferase and the NS3–NS5B region of an HCV of genotype 1b (strain NN) (12). LucNeo#2 cells were seeded at cells per well, incubated for 24 h, and treated with each compound at the indicated concentration. After incubation for 72 h, the cells were lysed and their luciferase activity was measured with a Luciferase Assay System according to the manufacturer’s protocol (Promega) (12). Simultaneously, cell viability was measured at 72 h posttreatment with a Cell Proliferation Kit II (XTT), as recommended by the manufacturer (Roche) (37). This replicon’s advantage for high-throughput assays enables one to produce the large scale of data that is required for quantifying the antiviral activity of drugs and drug combinations.
In the monotreatment study, we evaluated the intrinsic anti-HCV activity of 15 anti-HCV drugs (Fig. 1): DAAs that directly inhibit a viral-derived factor and HTAs that inhibit HCV replication by targeting cellular factors. The DAAs included PIs (TPV, DPV, ASV, and SMV), an NI (SOF), NNIs (VX, DAS, NSV, and TGV), and NS5AIs (DCV and LDV). The HTAs comprised IFNs (IFN-α and IFN-λ1) and CIs (CsA and SCY). In the cotreatment experiment, we treated cells with the indicated combinations of drugs and measured their HCV replication activity as described above. We confirmed that no toxicity was observed in any of drug combinations. SMV, ASV, DAS, NSV, TGV, and LDV were purchased from MedChem Express. TRV, DPV, SOF, VX, and DCV were from Selleckchem. IFN-α was obtained from MSD. IFN-λ1 was purchased from R&D Systems. CsA was purchased from Sigma–Aldrich, and SCY was kindly provided by Scynexis, Inc.
Supplementary Material
Acknowledgments
We thank Dr. Kunitada Shimotohno (National Center for Global Health and Medicine) for providing LucNeo#2 cells; Scynexis, Inc. for SCY; and Dr. Mohsan Saeed and Dr. Charles M. Rice (The Rockefeller University) for HCV Con1 replicon in SEC14L2-overexpressing cells. We also thank Dr. Senko Tsukuda (Department of Virology II, National Institute of Infectious Diseases) for editorial assistance. This work was supported, in part, by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to T.W. and K.W.); the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (T.W. and K.W.); NIH Grants R01-AI028433, R01-AI078881, and R01-OD011095 (to A.S.P.); the Japan Science and Technology Agency (JST) PRESTO program (S.I.); the JST Research Institute of Science and Technology for Society (RISTEX) program (S.I.); the Commissioned Research Program of the Ministry of Health, Labour and Welfare, Japan (S.I.); Japan Society for the Promotion of Science (JSPS) KAKENHI [Grants-in-Aid for Scientific Research; 16H04845, 16K13777, 15KT0107, and 26287025 (to S.I.) and 26460565 (to K.W.)]; the Mitsui Life Social Welfare Foundation (S.I.); the Shin-Nihon of Advanced Medical Research (S.I.); GlaxoSmithKline plc (GSK) Japan Research Grant 2016 (to S.I.); the JST CREST program (S.I. and K.W.); and a Grant-in-Aid from the Ministry of Health, Labor, and Welfare, Japan (to K.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.F.S. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610197114/-/DCSupplemental.
References
- 1.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19(7):837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368(20):1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11(7):482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 4.Pawlotsky JM. Hepatitis C treatment: The data flood goes on-an update from the liver meeting 2014. Gastroenterology. 2015;148(3):468–479. doi: 10.1053/j.gastro.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Cheng R, Tu T, Shackel N, McCaughan GW. Advances in and the future of treatments for hepatitis C. Expert Rev Gastroenterol Hepatol. 2014;8(6):633–647. doi: 10.1586/17474124.2014.909725. [DOI] [PubMed] [Google Scholar]
- 6.Feld JJ, et al. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370(17):1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 7.Pawlotsky JM, Feld JJ, Zeuzem S, Hoofnagle JH. From non-A, non-B hepatitis to hepatitis C virus cure. J Hepatol. 2015;62(1) Suppl:S87–S99. doi: 10.1016/j.jhep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Kohli A, et al. Virological response after 6 week triple-drug regimens for hepatitis C: a proof-of-concept phase 2A cohort study. Lancet. 2015;385(9973):1107–1113. doi: 10.1016/S0140-6736(14)61228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang SS, Kao JH. Daclatasvir-containing all-oral regimens for the treatment of hepatitis C virus infection. Hepatol Int. 2016;10(2):258–266. doi: 10.1007/s12072-015-9668-3. [DOI] [PubMed] [Google Scholar]
- 10.Lau G, et al. Efficacy and safety of 3-week response-guided triple direct-acting antiviral therapy for chronic hepatitis C infection: A phase 2, open-label, proof-of-concept study. Lancet Gastroenterol Hepatol. 2016;1(2):97–104. doi: 10.1016/S2468-1253(16)30015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann V, Bartenschlager R. On the history of hepatitis C virus cell culture systems. J Med Chem. 2014;57(5):1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 12.Goto K, et al. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem Biophys Res Commun. 2006;343(3):879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 13.Sampah ME, Shen L, Jilek BL, Siliciano RF. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc Natl Acad Sci USA. 2011;108(18):7613–7618. doi: 10.1073/pnas.1018360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen L, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14(7):762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen L, et al. A critical subset model provides a conceptual basis for the high antiviral activity of major HIV drugs. Sci Transl Med. 2011;3(91):91ra63. doi: 10.1126/scitranslmed.3002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskey SB, Siliciano RF. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat Rev Microbiol. 2014;12(11):772–780. doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- 17.Jilek BL, et al. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med. 2012;18(3):446–451. doi: 10.1038/nm.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen L, Rabi SA, Siliciano RF. A novel method for determining the inhibitory potential of anti-HIV drugs. Trends Pharmacol Sci. 2009;30(12):610–616. doi: 10.1016/j.tips.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy MB, et al. Pharmacokinetic/Pharmacodynamic predictors of clinical potency for hepatitis C virus nonnucleoside polymerase and protease inhibitors. Antimicrob Agents Chemother. 2012;56(6):3144–3156. doi: 10.1128/AAC.06283-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everson GT, et al. Efficacy of an interferon- and ribavirin-free regimen of daclatasvir, asunaprevir, and BMS-791325 in treatment-naive patients with HCV genotype 1 infection. Gastroenterology. 2014;146(2):420–429. doi: 10.1053/j.gastro.2013.10.057. [DOI] [PubMed] [Google Scholar]
- 21.Poordad F, et al. UNITY-1 Study Group Fixed-dose combination therapy with daclatasvir, asunaprevir, and beclabuvir for noncirrhotic patients with HCV genotype 1 infection. JAMA. 2015;313(17):1728–1735. doi: 10.1001/jama.2015.3860. [DOI] [PubMed] [Google Scholar]
- 22.Muir AJ, et al. Daclatasvir in combination with asunaprevir and beclabuvir for hepatitis C virus genotype 1 infection with compensated cirrhosis. JAMA. 2015;313(17):1736–1744. doi: 10.1001/jama.2015.3868. [DOI] [PubMed] [Google Scholar]
- 23.Friborg J, et al. In vitro assessment of re-treatment options for patients with hepatitis C virus genotype 1b infection resistant to daclatasvir plus asunaprevir. Infect Dis Ther. 2014;4(1):137–144. doi: 10.1007/s40121-014-0052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalezari J, et al. Ombitasvir/paritaprevir/r and dasabuvir plus ribavirin in HCV genotype 1-infected patients on methadone or buprenorphine. J Hepatol. 2015;63(2):364–369. doi: 10.1016/j.jhep.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Zoulim F, et al. Hepatitis C virus treatment in the real world: Optimising treatment and access to therapies. Gut. 2015;64(11):1824–1833. doi: 10.1136/gutjnl-2015-310421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2(30):30ra32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 28.Tong X, Kwong AD. Barrier to resistance: lessons from 2 direct-acting hepatitis C virus inhibitors, MK-5172 and Sofosbuvir. Clin Infect Dis. 2014;59(12):1675–1677. doi: 10.1093/cid/ciu700. [DOI] [PubMed] [Google Scholar]
- 29.Sarrazin C, et al. Prevalence of the hepatitis C virus NS3 polymorphism Q80K in genotype 1 patients in the European region. Antiviral Res. 2015;116:10–16. doi: 10.1016/j.antiviral.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson IM, et al. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): A phase 3, randomised, double-blind, placebo-controlled trial. Lancet. 2014;384(9941):403–413. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- 31.Forns X, et al. Simeprevir with peginterferon and ribavirin leads to high rates of SVR in patients with HCV genotype 1 who relapsed after previous therapy: A phase 3 trial. Gastroenterology. 2014;146(7):1669–1679.e1663. doi: 10.1053/j.gastro.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 32.McPhee F, et al. High sustained virologic response to daclatasvir plus asunaprevir in elderly and cirrhotic patients with hepatitis C virus genotype 1b without baseline NS5A polymorphisms. Adv Ther. 2015;32(7):637–649. doi: 10.1007/s12325-015-0221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimi S, et al. Long term persistence of NS5A inhibitor-resistant hepatitis C virus in patients who failed daclatasvir and asunaprevir therapy. J Med Virol. 2015;87(11):1913–1920. doi: 10.1002/jmv.24255. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki Y, et al. Dual oral therapy with daclatasvir and asunaprevir for patients with HCV genotype 1b infection and limited treatment options. J Hepatol. 2013;58(4):655–662. doi: 10.1016/j.jhep.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 35.Guedj J, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci USA. 2013;110(10):3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbloom DI, Hill AL, Rabi SA, Siliciano RF, Nowak MA. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med. 2012;18(9):1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukuda S, et al. Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis B virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression. J Biol Chem. 2015;290(9):5673–5684. doi: 10.1074/jbc.M114.602540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.