Significance
Acquired aromatase inhibitor (AI) resistance is a major clinical issue in the treatment of estrogen receptor alpha (ERα)-positive breast cancer. Most AI-resistant breast tumors still express ERα and rely on its signaling for growth. The current study shows that serum- and glucocorticoid-inducible kinase 3 (SGK3), a kinase transcriptionally regulated by ERα in breast cancer, sustains ERα signaling and drives the acquired AI resistance by protecting against endoplasmic reticulum (EnR) stress-induced ERα downregulation and cell death through preserving sarcoplasmic/EnR calcium ATPase 2b (SERCA2b) function. Our study reveals regulation of ERα expression mediated by the EnR stress response and highlights SGK3 inhibition as a potential effective treatment of acquired AI-resistant breast cancer.
Keywords: SGK3, aromatase inhibitor, endoplasmic reticulum stress, estrogen receptor, SERCA2
Abstract
Many estrogen receptor alpha (ERα)-positive breast cancers initially respond to aromatase inhibitors (AIs), but eventually acquire resistance. Here, we report that serum- and glucocorticoid-inducible kinase 3 (SGK3), a kinase transcriptionally regulated by ERα in breast cancer, sustains ERα signaling and drives acquired AI resistance. SGK3 is up-regulated and essential for endoplasmic reticulum (EnR) homeostasis through preserving sarcoplasmic/EnR calcium ATPase 2b (SERCA2b) function in AI-resistant cells. We have further found that EnR stress response down-regulates ERα expression through the protein kinase RNA-like EnR kinase (PERK) arm, and SGK3 retains ERα expression and signaling by preventing excessive EnR stress. Our study reveals regulation of ERα expression mediated by the EnR stress response and the feed-forward regulation between SGK3 and ERα in breast cancer. Given SGK3 inhibition reduces AI-resistant cell survival by eliciting excessive EnR stress and also depletes ERα expression/function, we propose SGK3 inhibition as a potential effective treatment of acquired AI-resistant breast cancer.
About 70% of breast cancers express estrogen receptor alpha (ERα). Selective ER modulators (i.e., tamoxifen) and aromatase inhibitors (AIs) are first-line drugs for ERα+ breast cancers. Tamoxifen antagonizes estrogen-ERα binding, whereas AIs block estrogen biosynthesis by inhibiting aromatase, a key enzyme that converts androgens to estrogens. Because of their superior efficacy over tamoxifen, the third-generation AIs (i.e., exemestane, anastrozole, and letrozole) have largely replaced tamoxifen as the preferred treatment for ERα+ breast cancer in postmenopausal women (1). However, many patients develop resistance to AIs during treatment.
The mechanisms for acquired AI resistance have been proposed to be associated with constitutively active ERα mutations, altered activity of ERα coregulators, deregulation of oncogenic kinase signaling, or loss of ERα expression/function (1, 2). The majority of AI-resistant tumors still express ERα (3, 4). In those tumors, ERα is either hypersensitive to low levels of estrogens or activated in a ligand-independent manner, up-regulating genes that promote cell survival and growth.
The endoplasmic reticulum (EnR) is an organelle responsible for protein folding and the biosynthesis of secretory or membrane proteins, cholesterol, steroids, and lipids. The EnR is also the major intracellular calcium storage site. A high concentration of Ca2+ in EnR is maintained by the Ca2+ pump sarcoplasmic/EnR calcium ATPase (SERCA) and is required for the proper function of enzymes and chaperons involving protein folding (5). Suppressing SERCA activity by inhibitor thapsigargin depletes the EnR Ca2+ pool, triggering EnR stress and cell death (6). Of the three SERCA family members (SERCA1, SERCA2, and SERCA3), SERCA1 and SERCA3 are expressed in muscle cells or nonmuscle cells in a tissue-specific manner, whereas SERCA2 is ubiquitously expressed (7). SERCA2 exists in two isoforms: SERCA2a is expressed in cardiac and slow-twitch muscles, whereas SERCA2b, with a C-terminal extension, is the housekeeping isoform expressed in smooth muscles and nonmuscle cells.
EnR stress resulting from the accumulation of unfolded or misfolded proteins triggers unfolded protein response (UPR), which consists of three sensors: protein kinase RNA-like EnR kinase (PERK), inositol-requiring enzyme 1α (IRE1α), and activating transcription factor 6 (ATF6). Activation of these sensors of the UPR contributes to EnR homeostasis by reducing EnR protein loading and inducing expression of chaperones that increase protein folding capacity. Mild EnR stress response or UPR is protective, but hyperactive UPR converts it from cytoprotective to cytotoxic (8, 9).
Serum- and glucocorticoid-inducible kinases (SGKs) that have three isoforms, SGK1–SGK3, are AGC protein kinases and act in parallel to Akt downstream of phosphatidylinositol 3-kinase (PI3K) (10). SGK3 has been shown to play a pivotal role in Akt-independent signaling downstream of oncogenic PIK3CA mutations in some human cancers, including breast cancer (11). Most recently, Bago et al. reported that SGK3 counteracts the inhibition of PI3K/Akt signaling and stimulates tumor growth (12). SGK3 is amplified and hyperactivated in breast cancer (13), and its expression is regulated by ERα and positively correlates with ERα levels in breast tumors (14, 15). Here, we report a mechanism with which SGK3 sustains ERα signaling and drives acquired AI resistance by maintaining EnR homeostasis through preserving SERCA2b function in breast cancer.
Results
SGK3 Expression Is Up-Regulated During Development of Acquired AI Resistance.
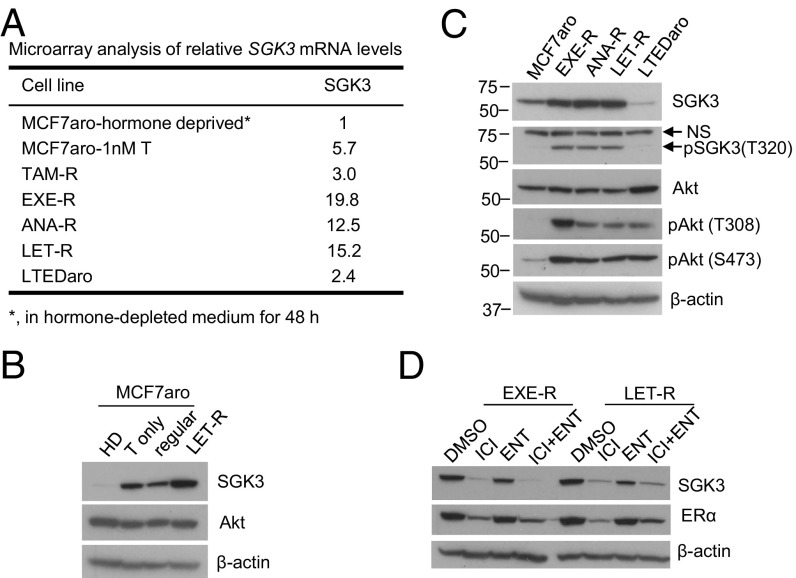
Three acquired AI-resistant cell lines designated EXE-R, ANA-R, and LET-R were generated by long-term exposure of aromatase-overexpressing ERα+ breast cancer cell line MCF7aro to testosterone (T) plus exemestane, anastrozole, and letrozole, respectively (16). Resistance to these AIs developed after an initial phase of suppression of T [T is converted to 17β-estradiol (E2) by aromatase]-dependent cell proliferation and survival. As one of the top up-regulated genes in all three AI-resistant cell lines, SGK3 mRNA levels were more than 12-fold and two- to fourfold greater in all three AI-resistant cell lines than in parental MCF7aro cells grown in hormone-depleted medium and T-containing medium, respectively (Fig. 1A). Elevated SGK3 protein levels in AI-resistant cells were confirmed by Western blot analysis (Fig. 1 B and C). Similar to LET-R cells derived from MCF7aro, HCC1428aro/LET-R cells had much higher levels of total and phosphorylated SGK3 than parental HCC1428aro cells (SI Appendix, Fig. S1A), suggesting increased SGK3 expression in acquired AI-resistant cells is not a single-cell-type phenomenon.
Fig. 1.
SGK3 expression is up-regulated in ERα+ breast cancer cells during development of acquired AI resistance. (A) Relative SGK3 mRNA levels in different cell lines determined by microarray. MCF7aro-1nM T, MCF7aro cells were cultured in hormone-depleted medium and treated with 1 nM T for 48 h; TAM-R, tamoxifen-resistant MCF7aro cells, generated by long-term culture of MCF7aro cells in hormone-depleted medium added with T plus tamoxifen (>6 mo); LTEDaro, long-term estrogen-deprived MCF7aro cells, which were generated by long-term culture of MCF7aro cells in hormone-depleted medium (>6 mo), and estrogen was not required for proliferation of the resulting cells. (B) Western blotting analysis of MCF7aro cells cultured in different conditions and LET-R cells. HD, cultured in hormone-depleted medium for 48 h; T only, cultured in hormone-depleted medium and treated with 1 nM T for 48 h; regular, cultured in the normal growth medium [MEM medium with 10% (vol/vol) FBS]. LET-R cells were cultured in hormone-depleted medium and treated with 1 nM T plus 200 nM letrozole. (C) Western blotting analysis of MCF7aro cells, AI-resistant cells, and LTEDaro cells. All the cells were cultured in their normal growth media, as described in Materials and Methods. (D) Western blotting analysis of levels of SGK3 and ERα in EXE-R and LET-R cells after being treated with 100 nM ICI 182,780, 5 μM enzalutamide, or their combination for 48 h.
SGK3 levels were up-regulated in MCF7aro-derived xenografts treated with T or T plus exemestane and were greater in exemestane-resistant xenografts than in exemestane-responsive xenografts (treated with T) measured by microarray and RT-qPCR analysis (SI Appendix, Fig. S2 A–C), demonstrating that SGK3 expression can be up-regulated in vivo during development of acquired AI resistance in ERα+ tumors.
Previously, we reported that SGK3 is transcriptionally regulated by estrogens and androgens (15, 17). We examined the effect of selective ER degrader ICI182,780 (fulvestrant) and anti-androgen enzalutamide on SGK3 expression in AI-resistant cells. ICI182,780 dramatically decreased SGK3 expression in both EXE-R and LET-R cell lines, whereas enzalutamide moderately reduced SGK3 expression (Fig. 1D), suggesting SGK3 is still mainly up-regulated by ERα, and androgen receptor (AR) may also contribute to increased SGK3 expression in AI-resistant cells.
SGK3 Is Critical for AI-Resistant Cell Proliferation and Survival.
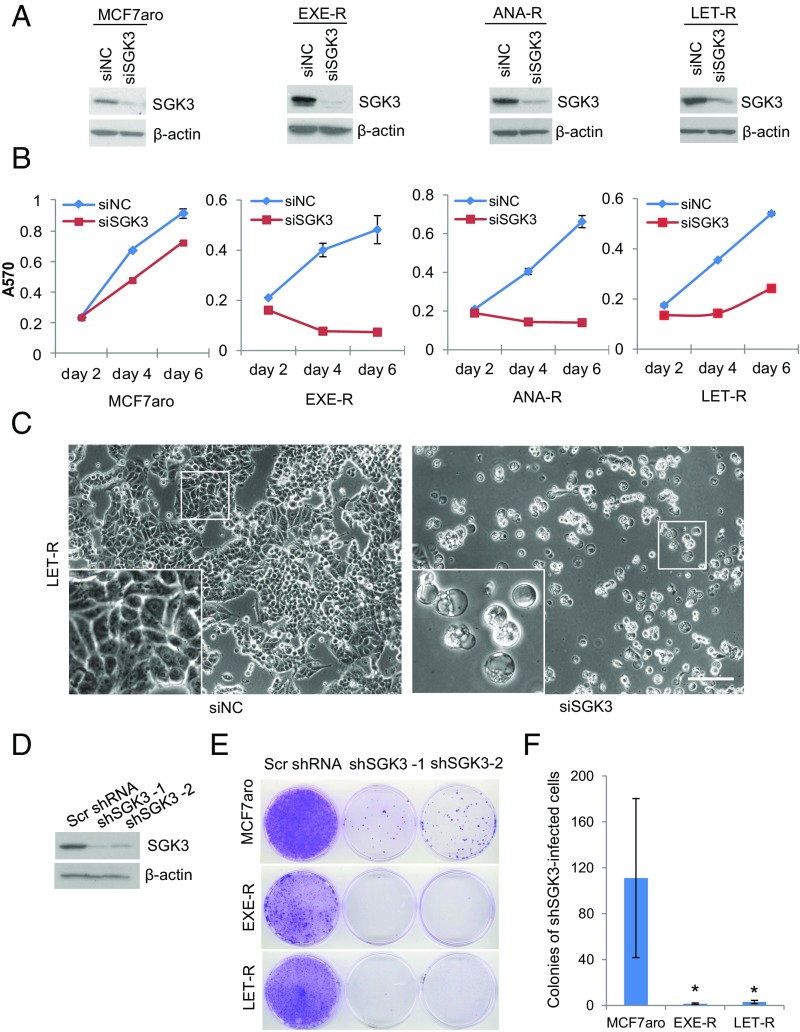
To explore the functional significance of increased SGK3 expression, we investigated the effects of SGK3 suppression on cell proliferation and survival of AI-resistant and AI-sensitive cells. Silencing SGK3 using siRNA severely impaired cell proliferation and survival of all three AI-resistant cell lines; the effect was greater in AI-resistant cells than in parental AI-sensitive MCF7aro cells (Fig. 2 A–C and SI Appendix, Fig. S3). Interestingly, massive cytoplasmic vacuoles were found in resistant cells after transfection with SGK3 siRNA (Fig. 2C and SI Appendix, Fig. S3). To further confirm that SGK3 is critical for AI-resistant cell proliferation and survival, we knocked down SGK3 using two distinct SGK3 shRNAs. Both SGK3 shRNAs dramatically reduced colony formation in AI-resistant cells and MCF7aro cells compared with scrambled shRNA, and AI-resistant cells were more vulnerable to the suppression of SGK3 expression (Fig. 2 E and F). Knockdown of SGK3 also significantly inhibited cell proliferation/survival of HCC1428aro/LET-R cells (SI Appendix, Fig. S1B), demonstrating that SGK3 is important for proliferation and survival of HCC1428aro/LET-R cells (Fig. 2F). These results imply that AI-resistant cells with increased SGK3 expression depend on this kinase for proliferation and survival.
Fig. 2.
SGK3 is critical for AI-resistant cell proliferation and survival. (A) Western blotting analysis of SGK3 levels in AI-sensitive and AI-resistant cells transfected with siRNA negative control or SGK3 siRNA for 72 h. (B) The effect of SGK3 knockdown on cell proliferation/survival in MCF7aro, EXE-R, ANA-R, and LET-R cells measured by MTT. (C) Light microscopy of LET-R cells transfected with siRNA negative control or SGK3 siRNA for 4 d. (Magnification: 100×.) (Scale bar: 50 μm.) (D) Western blotting analysis of SGK3 levels in MCF7aro cells transduced with scramble shRNA or two distinct SGK3 shRNA viral vectors. (E) Effect of knockdown of SGK3 by shRNA on colony formation. MCF7aro, EXE-R, and LET-R cells were cultured in 6-cm dishes and transduced with scrambled shRNA or two distinct SGK3 shRNA lentiviral vectors, respectively. Two days after transduction, cells were treated with 5 μg/mL puromycin to eliminate untransducted cells. Once the scrambled shRNA-infected cells reached confluency, both the scrambled shRNA-transduced cells and the SGK3 shRNA-transduced cell counterpart from the same cell line were fixed and stained with crystal violet simultaneously. (F) Quantization of colonies of the SGK3 shRNA-transduced cells. Both the scrambled shRNA-transduced cells and the SGK3 shRNA-transduced cell counterpart from the same cell line were fixed and stained with crystal violet simultaneously when the former reached confluency. Colonies of the SGK3 shRNA-transduced cells were counted and analyzed. *P < 0.05, by Student t test.
GSK650394 is a pan-SGK inhibitor and blocks SGK1 activity in the nanomolar range, but inhibits SGK3 activity at higher concentrations (18). As shown in SI Appendix, Fig. S4 A–C; 10 or 20 μmol/L GSK650394 inhibited phosphorylation of SGK3, but not Akt that is highly homologous to SGKs, and reduced resistant cell proliferation/survival. In contrast, 1 μmol/L GSK650394 had little effect on SGK3 phosphorylation and AI-resistant cell proliferation/survival, but inhibited SGK1 activity, as shown by blocking NDRG1 phosphorylation at Thr346, which is specifically phosphorylated by SGK1 (19), implying that SGK1 does not play a major role in AI-resistant cell proliferation and survival. Compared with SGK3 silencing, SGK1 silencing confirmed by RT-qPCR had a mild inhibitory effect on AI-resistant cell proliferation and survival (SI Appendix, Fig. S5 A–C), which rules out the possibility that GSK650394-induced AI-resistant cell death results from SGK1 inhibition (we did not examine SGK2 because of its tissue-specific expression).
SGK3 Mediates AI Resistance.
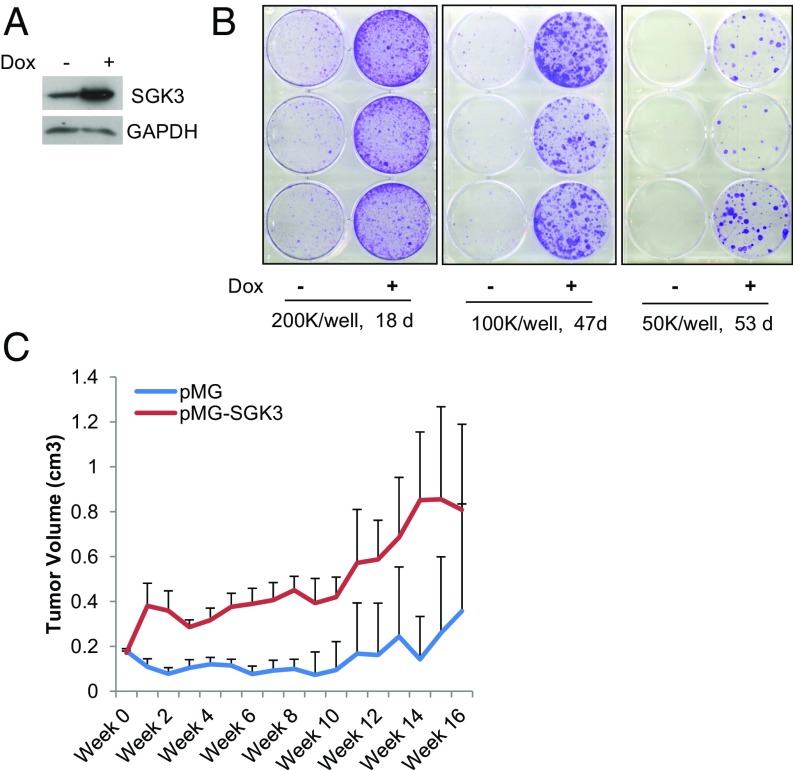
As expected, suppressing SGK3 using either inhibitor GSK650394 or SGK3 shRNA dramatically reduced surviving colonies of MCF7aro cells grown in hormone-depleted medium added with T plus letrozole, suggesting SGK3 is crucial for acquiring letrozole resistance (SI Appendix, Fig. S6 A and B). To test whether forced SGK3 overexpression in AI-sensitive cells promotes AI resistance, doxycycline (DOX)-inducible SGK3 expression cell line MCF7aro/TO/SGK3 was generated and cultured in hormone-depleted medium with T plus letrozole in the presence or absence of DOX. Blocking estrogen synthesis by letrozole caused the majority of cells to die, and induction of SGK3 expression by addition of DOX greatly increased surviving colonies (Fig. 3 A and B). To further test whether overexpression of SGK3 in AI-sensitive cells confers survival advantage on AI treatment, we generated MCF7aro cells stably infected with pTomo lentiviral control vector or pTomo-SGK3 and cocultured them in either regular medium or hormone-depleted medium with T plus letrozole. The cells stably infected with these two viral vectors can be easily distinguished under a fluorescence microscope, because pTomo vector will express both GFP and RFP fluorescent proteins (20), whereas pTomo-SGK3 will express GFP but not RFP, as RFP coding region was replaced with SGK3 coding region during vector construction. SI Appendix, Fig. S6C showed that SGK3-overexpressing cells MCF7aro/pTomo-SGK3 became dominant over vector control cells MCF7aro/pTomo after being cocultured in hormone-depleted medium with T plus letrozole. Western blotting analysis supported a dramatic reduction in MCF7aro/pTomo control cells, as shown by a marked decrease in RFP levels in similarly cultured mixed cells (SI Appendix, Fig. S6E). Using MCF7aro cells stably transfected with nonviral SGK3 expression vector or empty vector pMG, we showed that MCF7aro/pMG-SGK3 cells were resistant to anastrozole (SI Appendix, Fig. S6F), and MCF7aro/pMG-SGK3 cell-derived xenografts had less response to anastrozole treatment than MCF7aro/pMG control cell-derived xenografts in vivo, as measured by the xenograft growth (Fig. 3C).
Fig. 3.
SGK3 promotes AI resistance. (A) Western blotting analysis of SGK3 levels in MCF7aro/TO/SGK3 cells cultured with or without DOX. (B) Colony formation of MCF7aro/TO/SGK3 cells grown in hormone-depleted medium with T plus letrozole in the presence and absence of DOX. MCF7aro/TO/SGK3 cells seeded in three six-well plates at 200,000 cells per well, 100,000 cells per well, and 50,000 cells per well, respectively, were cultured in hormone-depleted medium with T plus letrozole and treated with or without 250 ng/mL DOX. After the indicated time, the cells were fixed and stained with crystal violet. (C) The growth curve of breast cancer xenografts derived from MC7aro/pMG or MCF7aro/pMG-SGK3 cells in ovariectomized mice inserted with T tablets and treated with anastrozole. The tumor size was measured every week.
SGK3 Is Essential for EnR Homeostasis of AI-Resistant Cells.
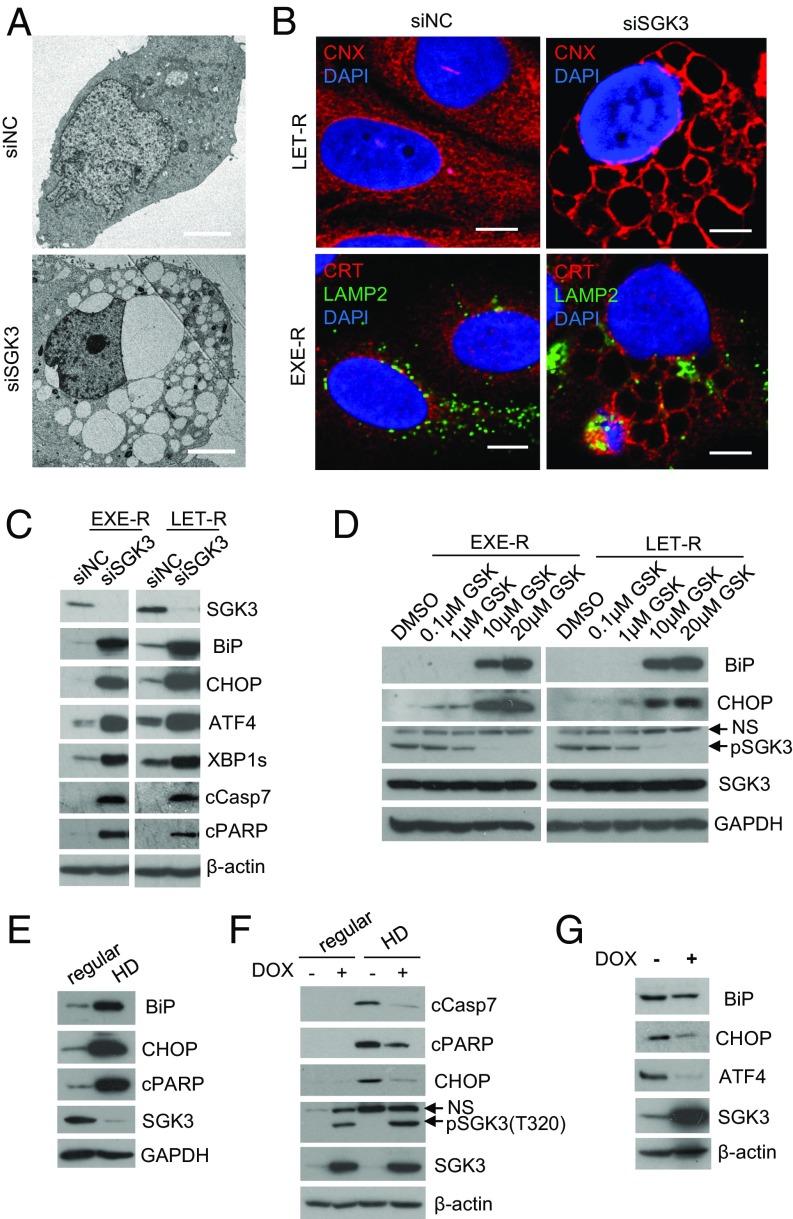
Because suppressing SGK3 using either SGK3 siRNA or inhibitor GSK650394 induced massive cytoplasmic vacuoles in AI-resistant cells, we investigated the origin of the vacuoles. Transmission electron microscopy showed that vacuoles in SGK3 siRNA-transfected LET-R cells varied in size, but were clear and lacked any visible cytoplasmic materials, thus ruling them out as autophagosomes (Fig. 4A). By immunostaining with EnR-specific markers calnexin and calreticulin, as well as the late endosomal and lysosomal marker lysosome-associated membrane protein 2 (LAMP2), we found that the vacuoles induced by SGK3 inhibition were EnR origin (Fig. 4B), suggesting that vacuoles result from EnR dilation and that SGK3 is critical for EnR healthiness of AI-resistant cells. Given that EnR vacuolization may result from excessive EnR stress, we tested whether suppressing SGK3 induces EnR stress in AI-resistant cells. The levels of EnR stress markers immunoglobulin heavy chain-binding protein (BiP, also named GRP78) and the CCAAT/enhancer binding protein homologous protein (CHOP) were dramatically increased in both EXE-R and LET-R cell lines after SGK3 suppression, using SGK3 siRNA or inhibitor GSK650394 (Fig. 4 C and D), suggesting SGK3 protects against EnR stress. Moreover, activating transcription factor 4 (ATF4) and the spliced X-box binding protein 1 (XBP1s), markers of the PERK and IRE1 arms of EnR stress response, respectively, were robustly induced (Fig. 4C). Excessive or prolonged EnR stress leads to apoptosis (21). We detected an increase of apoptotic markers cleaved PARP and cleaved caspase-7 in EXE-R cells and LET-R cells after SGK3 silencing (Fig. 4C). Silencing SGK3 also induced EnR stress and apoptosis in HCC1428aro/LET-R cells (SI Appendix, Fig. S1C).
Fig. 4.
SGK3 maintains EnR homeostasis. (A) Transmission electron microscopy of LET-R cells transfected with siRNA negative control or SGK3 siRNA for 72 h. (Magnification: 1,100×.) (Scale bar: 4 μm.) (B) Immunofluorescence microscopy of AI-resistant cells transfected with SGK3 siRNA or siRNA negative control. EXER-R and LET-R cells were transfected with siRNA negative control or SGK3 siRNA for 72 h and then fixed and immunostained with anti-calnexin (CNX) or anti-calreticulin (CRT) plus anti-LAMP2. After immunostaining, the cells were mounted in DAPI solution and imaged under a confocal microscope. DAPI stained nuclei blue. CNX and CRT were shown in red, and LAMP2 was shown in green. (Scale bar: 5 μm.) LAMP2 is a membrane protein of lysosome and late endosome, and thus serves as a marker of lysosome and later endosome. (C) Western blotting analysis of EXE-R and LET-R cells after being transfected with siRNA negative control or SGK3 siRNA for 72 h. (D) Western blotting analysis of EXE-R and LET-R cells after being treated with the increasing concentrations of GSK650394 for 48 h. (E) Western blotting analysis of MCF7aro cells cultured in regular growth medium or in hormone-depleted medium for 3 d. (F) Western blotting analysis of MCF7aro/TO/SGK3 cells grown in different conditions. MCF7aro/TO/SGK3 cells were cultured in normal growth medium or hormone-depleted medium and treated with or without 250 ng/mL DOX for 3 d. (G) Western blotting analysis of MCF7aro/TO/SGK3 cells grown in the medium with T plus letrozole in the presence or absence of 250 ng/mL DOX for 14 d.
RNA sequencing analysis of LET-R cells transfected with SGK3 siRNA or siRNA negative control also revealed that SGK3 silencing induced EnR stress (SI Appendix, Fig. S7A). The levels of EnR stress markers including HSPA5 (BiP), DDIT3 (CHOP), ERN1 (IRE1α), EIF2AK3 (PERK), and GADD45A were significantly elevated after SGK3 silencing (SI Appendix, Fig. S7B).
Estrogen withdrawal induced EnR stress and cell death, as shown by increased expression of BiP, CHOP, and cleaved PARP in MCF7aro cells (Fig. 4E). Induction of SGK3 expression significantly attenuated estrogen withdrawal-induced EnR stress and cell death, as shown by a decrease in expression of CHOP, cleaved caspase-7, and cleaved PARP in MCF7aro/TO/SGK3 cells (Fig. 4F). Moreover, induction of SGK3 expression significantly decreased the levels of BiP, ATF4, and CHOP in MCF7aro/TO/SGK3 cells grown in hormone-depleted medium added with T plus letrozole (Fig. 4G). Together, these data suggest SGK3 is important for maintaining EnR homeostasis in AI resistance acquisition.
SGK3 Maintains EnR Homeostasis Through Interaction with SERCA2b.
Using 372 differentially expressed genes in our microarray data of EXE-R cells transfected with SGK3 siRNA, or siRNA negative control [>1.5-fold change, false discovery rate (FDR) < 0.05], we queried the Connectivity Map, a collection of gene-expression signatures from cultured human cells treated with 1,309 small compounds (22) (www.broadinstitute.org/cmap/). It came out that the differentially expressed gene profile of EXE-R cells transfected with SGK3 siRNA vs. siRNA negative control mostly resembled the one of MCF-7 cells treated with thapsigargin vs. DMSO (SI Appendix, Table S1). Thapsigargin is a SERCA-specific irreversible inhibitor that induces EnR stress by depleting EnR calcium stores. The same as SGK3 suppression, thapsigargin also induced massive EnR vacuolization and EnR stress in AI-resistant cells (Fig. 5 A and B). In contrast, two other widely used EnR stress inducers tunicamycin (which induces EnR stress through inhibition of glycosylation) and brefeldin A (which induces EnR stress through blocking protein transportation from EnR to Golgi apparatus) did not induce EnR vacuolization, although they induced EnR stress (SI Appendix, Fig. S8 A and B), suggesting EnR vacuolization is not simply a result of EnR stress. Using SERCA2b siRNA, we showed that knockdown of SERCA2b expression induced massive EnR vacuolization and cell death in AI-resistant cells (SI Appendix, Fig. S8 C and D), which is the same as knockdown of SGK3.
Fig. 5.
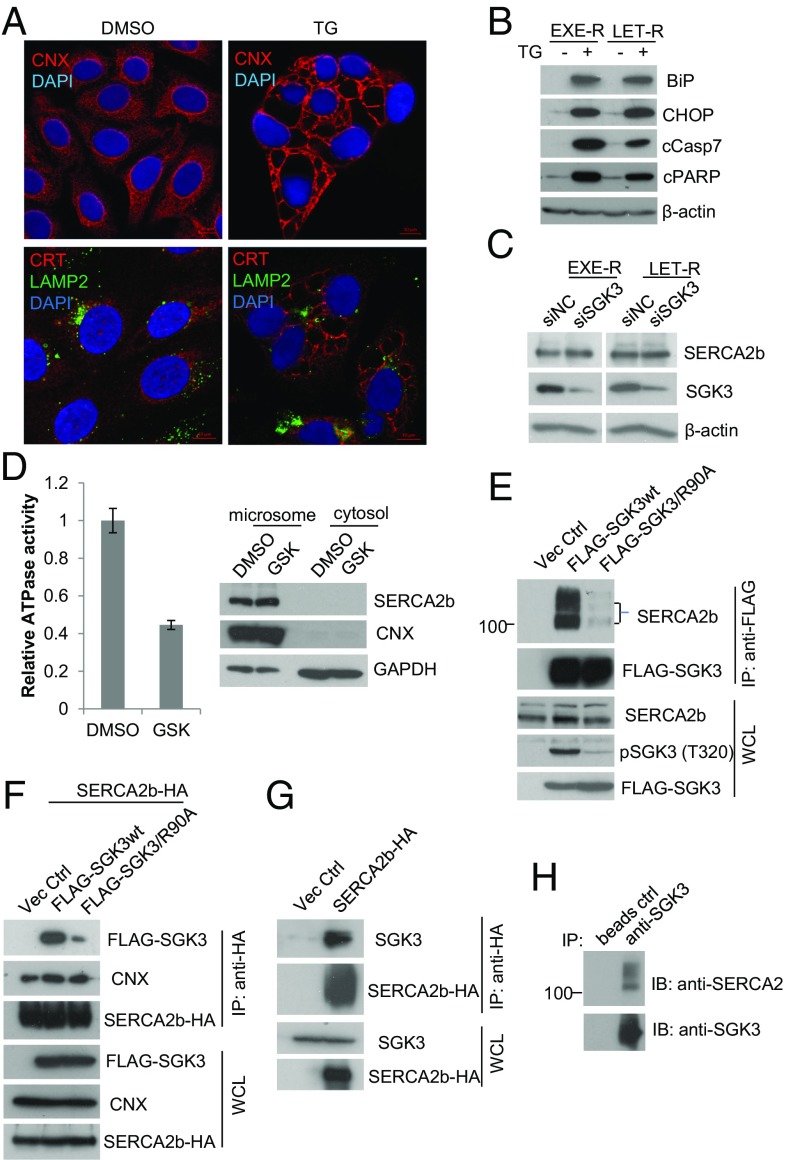
SGK3 maintains EnR homeostasis through interaction with SERCA2b. (A) Confocal microscopy of EXE-R cells treated with 0.5 μM thapsigargin (TG) for 48 h. Cells were immunostained with anti-CNX or anti-CRT plus anti-LAMP2 and mounted in DAPI solution. DAPI stained nuclei blue. CNX and CRT were shown in red, and LAMP2 was shown in green. (Scale bar: 10 μm.) (B) Western blotting analysis of EXE-R cells and LET-R cells after being treated with 0.5 μM TG for 72 h. (C) Western blotting analysis of EXE-R and LET-R cells after being transfected with SGK3 siRNA or siRNA negative control for 72 h. (D) Effect of SGK3 inhibition on SERCA2b ATPase activity. EXE-R cells were treated with DMSO or 20 μM GSK650394 overnight and harvested for microsome isolation. Five micrograms microsomes were used for analysis of SERCA2b ATPase activity in vitro (Left). The same amount of proteins from microsomal and cytosolic fractions was loaded for Western blotting analysis (Right). (E) Immunoprecipitation analysis of the interaction between SGK3 and SERCA2b. 293T cells were transfected with empty vector or vectors expressing either FLAG-wtSGK3 or FLAG-SGK3/R90A. An equivalent amount of cell lysate was immunoprecipitated with anti-FLAG resin, and the precipitated proteins were detected by Western blotting, using the relevant antibodies. (F) 293T cells were cotransfected with SERCA2b-HA expression vector and empty vectors or vector expressing FLAG-SGK3 wt or FLAG-SGK3/R90A. The cell lysates were immunoprecipitated with anti-HA resin, and precipitated proteins were detected by Western blotting. (G) MCF7aro cells were transfected with empty vector or SERCA2b-HA expression vector for 30 h. Cell lysates were immunoprecipitated with anti-HA resin, and precipitated proteins were detected by Western blotting analysis. (H) Immunoprecipitation analysis of the association between endogenous SGK3 and SERCA2b. The same amount of EXE-R cell lysates were incubated with beads alone or beads coated with anti-SGK3, and the precipitated proteins were detected by Western blotting using anti-SERCA2 or anti-SGK3.
The results prompted us to hypothesize that SGK3 might target SERCA2b. Because silencing SGK3 did not significantly affect SERCA2b levels in both EXE-R and LET-R cell lines (Fig. 5C), it suggests that SGK3 does not regulate SERCA2b expression. However, we noticed that FLAG-SGK3 pulled down SERCA2b in our previous proteomics study (23), implying that SGK3 might regulate SERCA2b function. SERCA2b is a membrane of P-type ATPase family, and the ATPase activity is indispensable for SERCA2b function. As shown in Fig. 5D, SERCA2b ATPase activity was significantly impaired after inhibition of SGK3 by GSK650394 in EXE-R cells, suggesting that SGK3 preserves SERCA2b function. To confirm the physical interaction between SGK3 and SERCA2b, we performed a serial of immunoprecipitation assays. FLAG-SGK3 pulled down SERCA2b in 293T cells. In contrast, mutation of the phox homology domain (SGK3/R90A) that is required for SGK3 activation (24) dramatically reduced the interaction of SGK3 with SERCA2b (Fig. 5 E and F). HA-SERCA2b was able to pull down endogenous SGK3 in MCF7aro cells (Fig. 5G). Moreover, we showed that SGK3 was associated with SERCA2b in nontransfected EXE-R cells (Fig. 5H).
SGK3 Sustains ERα Signaling in AI-Resistant Cells.
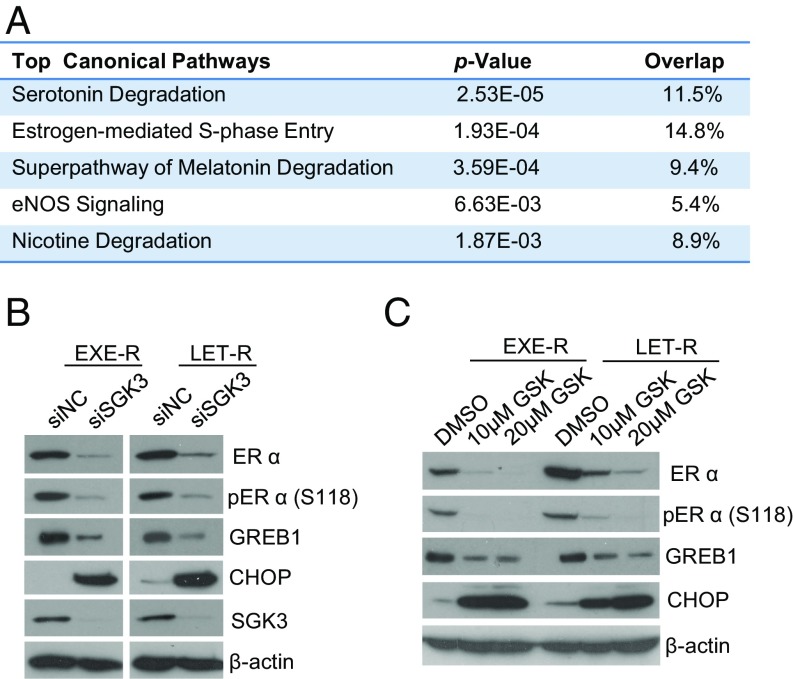
SGK3 has been shown to enhance ERα transactivation activity through phosphorylation of coactivator Flightless-I (25). Using RNA sequencing analysis, we found that knockdown of SGK3 significantly down-regulated ERα signaling in the LET-R cells (Fig. 6A). Interestingly, ERα levels were significantly decreased after SGK3 silencing. Expression of ERα target genes (e.g., IGFBP3, PGR, GREB1L, KIAA0101, CCND1, and MYBL1) was also significantly down-regulated (SI Appendix, Fig. S9A). Western blot analysis confirmed that silencing SGK3 dramatically reduced protein levels of the phosphorylated and total ERα, as well as the well-known ERα target Growth Regulation by Estrogen in Breast Cancer 1 (GREB1) in both EXE-R and LET-R cell lines (Fig. 6B). Pharmacological inhibition of SGK3 by GSK650394 also depleted ERα and reduced GREB1 expression (Fig. 6C), further confirming that SGK3 is required for ERα expression and signaling in AI-resistant cells. Because silencing SGK1 did not decrease ERα expression levels (SI Appendix, Fig. S9B), it is unlikely that down-regulation of ERα expression by GSK650394 results from SGK1 inhibition.
Fig. 6.
SGK3 retains ERα expression and signaling in AI-resistant cells. (A) The five most significantly down-regulated canonical pathways in siSGK3-transfected LET-R cells vs. siNC-transfected LET-R cells analyzed by IPA. LER-R cells were transfected with siRNA negative control or SGK3 siRNA for 72 h and then subjected to RNA-seq analysis. Differentially expressed genes from RNA-seq data (>1.5-fold change in siSGK3 cells vs. siNC cells; P < 0.05) were analyzed using IPA. (B) Western blotting analysis of EXE-R and LET-R cells after being transfected with siRNA negative control or SGK3 siRNA for 72 h. (C) Western blotting analysis of EXE-R and LET-R cells after being treated with 10- or 20 μM GSK650394 for 72 h.
SGK3 Retains ERα Expression by Protecting Against EnR Stress-Induced ERα Down-Regulation via SERCA2b.
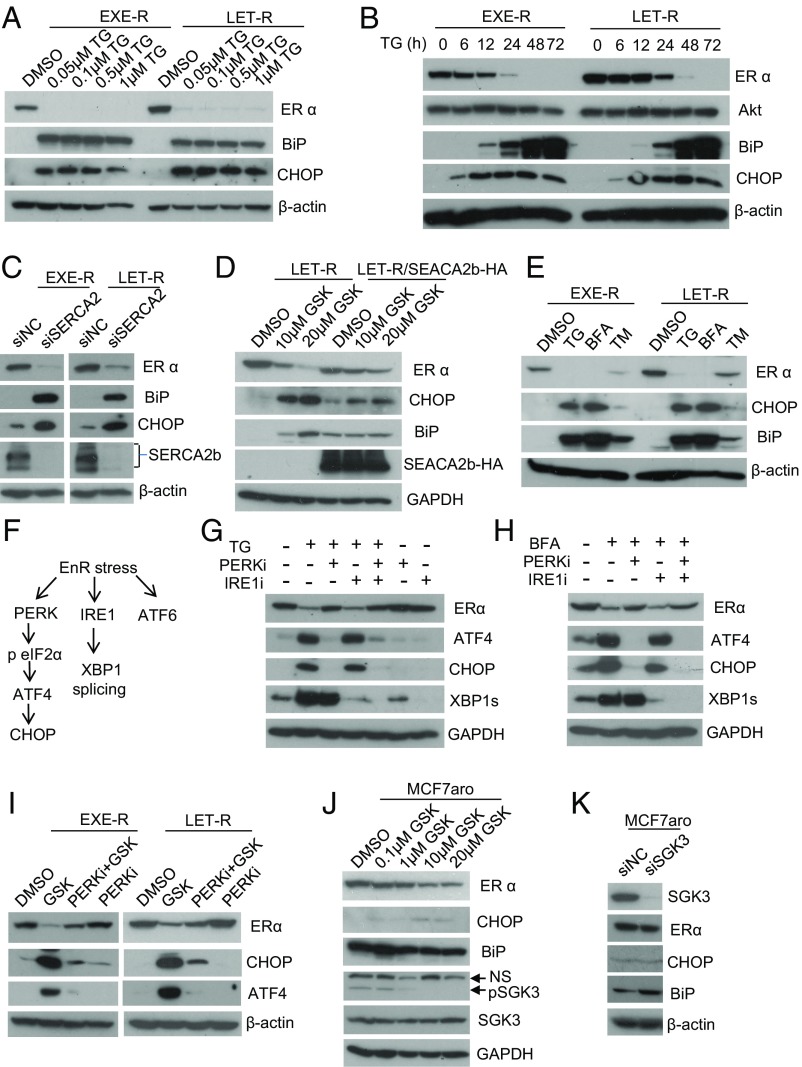
Given that SGK3 functionally associates with SERCA2b, we asked whether SERCA2b is involved in SGK3 regulation of ERα expression. Suppressing SERCA2b using thapsigargin or SERCA2b siRNA depleted ERα with a concomitant induction of EnR stress in EXE-R, LET-R, and HCC1428aro/LET-R cell lines (Fig. 7 A–C and SI Appendix, Fig. S1D), suggesting SERCA2b is required for ERα expression. We further generated LET-R cells stably expressing SERCA2b-HA (SI Appendix, Fig. S10A). Ectopic expression of SERCA2b-HA did not increase ERα expression, but it significantly attenuated SGK3 inhibition-induced ERα down-regulation and EnR stress (Fig. 7D), supporting that SGK3 maintains EnR homeostasis through SERCA2b, and that SERCA2b is involved in SGK3 regulation of ERα expression.
Fig. 7.
SGK3 retains ERα expression through maintaining EnR homeostasis via SERCA2b. (A) Western blotting analysis of EXE-R and LET-R cells after being treated with different concentrations of TG, as indicated, for 48 h. (B) Western blotting analysis of EXE-R and LET-R cells after being treated with 0.1 μM TG for different time, as indicated. (C) Western blotting analysis of EXE-R and LET-R cells after being transfected with siRNA negative control or SERCA2b siRNA for 72 h. (D) Western blotting analysis of LET-R and LET-R/SERCA2b-HA cells after being treated with different concentrations of GSK650394, as indicated, for 48 h. (E) Western blotting analysis of EXE-R and LET-R cells after being treated with 1 μM TG, 5 μg/mL brefeldin A (BFA), and 4 μg/mL tunicamycin (TM) for 48 h. (F) Schematic diagram of the three arms of EnR stress response or UPR. (G) Western blotting analysis of MCF7aro cells treated with 0.5 μM TG alone or plus 1 μM GSK2656157 (PERK inhibitor) or 25 μM 4μ8C (IRE1 inhibitor) or two in combination for 24 h. (H) Western blotting analysis of MCF7aro cells treated with 2.5 μg/mL BFA alone or plus 1 μM GSK2656157 (PERK inhibitor) or 25 μM 4μ8C (IRE1 inhibitor) or two in combination for 24 h. (I) Western blotting analysis of EXE-R cells and LET-R cells after being treated with 20 μM GSK650394, 2 μM PERK inhibitor (GSK2656157), alone or in combination for 24 h. (J) Western blotting analysis of MCF7aro cells treated with different concentrations of GSK650394, as indicated, for 48 h. (K) Western blotting analysis of MCF7aro cells transfected with siRNA negative control or SGK3 siRNA for 72 h.
Because ERα down-regulation was always correlated with EnR stress induction in our study, we asked whether EnR stress results in ERα down-regulation. Thapsigargin, tunicamycin, and brefeldin A all decreased ERα expression at both protein and mRNA levels, and the extent of ERα down-regulation was correlated with the extent of EnR stress induction (Fig. 7E and SI Appendix, Fig. S10 B and C), supporting that EnR stress response down-regulates ERα expression through transcriptional mechanisms. Moreover, PERK inhibitor GSK2606414, which inhibits the PERK arm of EnR stress response, blocked thapsigargin- or brefeldin A-induced ERα down-regulation (Fig. 7 G and H), suggesting that activation of the PERK arm is responsible for EnR stress-induced ERα down-regulation. PERK inhibitor was also able to attenuate SGK3 inhibition-induced ERα down-regulation (Fig. 7I), indicating that SGK3 retains ERα expression at least partially through preventing excessive activation of the PERK arm of EnR stress response.
As shown earlier, AI-resistant cells were more vulnerable to SGK3 suppression than parental AI-sensitive cells, and SGK3 suppression caused massive EnR vacuolization in resistant cells, but not in AI-sensitive cells. We thus compared the effects of SGK3 inhibition on ERα expression and EnR stress induction in AI-resistant cells and in MCF7aro cells. In contrast to the effects in AI-resistant cells, inhibition of SGK3 using inhibitor GSK650394 only slightly decreased ERα levels and had little effect on EnR stress induction in MCF7aro cells (Fig. 7J). Knockdown of SGK3 did not induce CHOP expression nor decrease ERα levels (Fig. 7K), suggesting that cells acquire a dependency on SGK3 for maintaining EnR homeostasis and ERα signaling during the development of AI resistance.
Discussion
PI3K pathway is frequently activated in breast cancer (26). SGK3 is a PI3K downstream kinase functioning in parallel to Akt (10). Activation of Akt has been shown to confer tamoxifen resistance and de novo AI resistance of breast cancer (27–29). The current study shows that up-regulation of SGK3 is crucial for acquiring AI resistance.
ERα is still engaged in advanced breast cancer, contributes to disease pathogenesis, and remains a viable therapeutic target. One of the major mechanisms by which ERα+ breast cancer cells become AI-resistant is the acquisition of estrogen hypersensitivity. After AI exposure, most estrogen-dependent cancer cells stop proliferation and go through apoptosis and/or autophagy, but some cancer cells gain estrogen hypersensitivity by ERα mutations or the cross-talk of ERα with growth factor signaling pathways, so that cancer cells can survive and proliferate at low levels of estrogen. Previously, we and others have reported that SGK3 is an ERα direct target, and SGK3 levels positively correlate with ERα levels in breast tumors (14, 15). SGK3 can enhance ERα transactivation through phosphorylation of coactivator Flightless-I (25). Here we show that SGK3 sustains ERα expression and signaling through a mechanism of EnR homeostasis maintenance in AI-resistant cells. On the basis of these findings, we propose that the feed-forward regulation between ERα and SGK3 promotes acquisition of estrogen hypersensitivity of ERα+ breast cancer cells.
Using long-term estrogen deprivation (LTED) as an in vitro model of AI treatment, Dowsett’s group documented that during adaption to LTED, MCF7 cells pass through three distinct phases: quiescent phase (LTED-Q), followed by an estrogen hypersensitivity phase (LTED-H), and finally an estrogen-independent phase (LTED-I) (30, 31). So far, most studies focus on LTED-I, not LTED-H. Our AI-resistant model provides an opportunity to study ERα hypersensitivity, because these acquired AI-resistant cells grow in the presence of T and AI (a small amount of E2 can be converted in this situation) and are sensitive to fulvestrant (16). LTED cells do not respond to any AI, whereas our AI-resistant cells acquire resistance to one AI and will respond to another AI (16, 32), which resembles a true clinical situation (33). Moreover, LTED cells are less responsive to fulvestrant compared with our AI-resistant cells (16), suggesting LTED cells are less dependent on ERα signaling than our resistant cell lines. Therefore, LTED cells and our acquired AI-resistant cells may represent different stages of AI resistance. SGK3 is extremely important for our acquired AI-resistant cells, because SGK3 expression requires ERα and in turn sustains ERα expression and signaling in these resistant cells.
SGK3 can be transcriptionally up-regulated by both ERα and AR, but AR regulation of SGK3 expression requires the presence of ERα (15, 17). Because AI blocks the conversion of androgens to estrogens, the levels of intratumoral androgens are elevated after treatment with AIs (34), which might up-regulate SGK3 and thus promote AI resistance. Recent studies have shown that AR expression and activity are increased in AI-resistant breast cancer cells, and AR collaborates with ERα to promote AI resistance (35–37), which is in favor of our thoughts.
Estrogen triggers intracellular Ca2+ release from EnR, which activates anticipatory UPR and is required for estrogen-mediated cell proliferation (9, 38). Estrogen also regulates expression and activity of SERCA (39–43). Here we showed that SERCA2b is required for ERα expression, and SGK3 preserves SERCA2b function in AI-resistant cells, although the detailed mechanism still needs to be resolved. Recently, Schmid et al. reported that SGK3 regulates Ca2+ entry in dendritic cells through up-regulation of STIM2 expression (44). Our study suggests SGK3 maintains EnR Ca2+ homeostasis through preserving SERCA2b function, and thus protects against excessive EnR stress, which results in ERα depletion and cell death in AI-resistant cells.
Raina et al. reported that mild EnR stress up-regulates estrogen signaling (45). In the current study, we showed that EnR stress inducers (i.e., thapsigargin and brefeldin A) deplete ERα expression through activation of the PERK arm. Thus, EnR stress has double-edged effects on estrogen signaling, and the effects depend on its severity or the extent of activation of PERK arm. Our study reveals a regulatory mechanism of ERα expression by EnR stress response previously unknown to modulate ERα expression.
EnR stress response or UPR is usually an adaptive response preventing further damage to cells. However, EnR stress triggers cell death if it remains unresolved or it is excessive (21, 46). Estrogen up-regulates expression of BiP and XBP1 through ERα, and activates anticipatory UPR that has been shown to contribute estrogen-induced cell proliferation in breast cancer cells (9, 47). UPR is activated in endocrine-resistant cells and contributes to the resistance in breast cancer (47–49). AIs block estrogen biosynthesis and induce EnR stress. Although the mechanisms are not well understood, EnR calcium levels drop during EnR stress (50). If EnR calcium levels drop too low, it will trigger extensive EnR stress, leading to cell death. We propose that up-regulation of SGK3 is crucial for preventing extensive EnR stress through preserving EnR calcium pump SERCA2b function during acquiring AI resistance, while retaining ER expression and its transactivation activity by SGK3 drives the resistance. Because SGK3 inhibition elicits massive EnR stress and depletes ERα, SGK3 could be a potential target for effective treatment of acquired AI-resistant breast tumors that still rely on ERα signaling for growth.
Materials and Methods
Additional methods are in SI Appendix, Materials and Methods.
Cell Culture, Plasmid Constructs, Lentivirus Production and Transduction, Immunofluorescence Microscopy, and Western Blotting.
Methods are described in SI Appendix, Materials and Methods.
Xenografts.
All animal experiments were approved by the institutional animal care and use committee at City of Hope. Xenografts were generated in female BALB/c Nu-Nu, athymic, ovariectomized mice through s.c. injections of MCF7aro cells (4 mice/group). Exemestane-resistant xenografts were established through daily injection of exemestane (250 µg/day) to mice with T tablets. The negative and positive controls were those mice inserted with placebo and T tablets, respectively. The positive control and exemestane-resistant tumors were collected when their volume reached 1,000 mm3. For the anastrazole treatment experiment, xenografts were generated in female BALB/c Nu-Nu, athymic, ovariectomized mice through s.c. injections of MCF7aro/pMG cells or MCF7aro/pMG-SGK3 cells in Matrigel (BD Biosciences) (six mice/group). The mice were inserted with T tablets, and administrated with daily injection of anastrozole (250 µg/day). Tumor volumes were measured every week. Body weights were monitored weekly as an indicator of overall health.
siRNA Transfection.
siRNA transfection was performed using siPORT NeoFX transfection agent (Ambion), according to manufacturer’s instruction on reverse transfection, and carried out in 6-cm dishes or 24-well plates. Briefly, cells were trypsinized and diluted in the medium at 1 × 105/mL For each 6-cm dish, 10 µL siPORT NeoFX transfection agent and 15 µL 10 µM siRNA negative control (Ambion) or siRNA against SGK3, SGK1, or SERCA2 (all from Santa Cruz) were diluted in 250 µL OPTI-MEM I medium, respectively. For each well of a 24-well plate, 1 µL siPORT NeoFX transfection agent and 1.5 µL 10 µM siRNA were introduced. After being mixed and incubated for 10 min, the mixture of siRNA and transfection agent was dispersed into 6-cm dishes or 24-well plates. Cell suspensions (1 × 105/mL) were overlaid onto the transfection complexes at 4 mL cells/dish or 0.5 mL cells/well.
RNA Microarray.
RNA from cells or xenografts was sent to City of Hope Integrative Genomic Core for microarray analysis, as previously described (16). The Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays were used. Ingenuity pathway Analysis (IPA, from QIAGEN) was used for analysis of microarray data. For SGK3 knockdown experiment, EXE-R cells were transfected with siRNA negative control or SGK3 siRNA, using the reverse transfection method for 48 h, and then harvested for RNA extraction and subsequent microarray analysis. The experiment was carried out in triplicate.
RNA Sequencing.
LET-R cells were transfected with siRNA negative control or SGK3 siRNA, using the reverse transfection method for 72 h, and were then harvested for RNA extraction. RNA was extracted using TRIZOL reagent (Invitrogen) and sent to City of Hope Integrative Genomic Core for RNA sequencing analysis. The sequencing type was Hiseq2500 SE40. The result was carried out in triplicate and analyzed by IPA.
Cell Proliferation Assay.
Cell proliferation was measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. At the indicated time, the medium in each well of a 24-well plate was removed from cells and replaced with 0.5 mL fresh phenol red-free medium containing 0.5 mg/mL MTT; the cells were then incubated at 37 °C for 1 h. The medium was discarded, and 0.5 mL DMSO was added to each well to dissolve the formazan dye trapped in the living cells. One hundred microliters of the supernatant was then transferred into a 96-well plate and read in a SpectraMax M5 plate reader (Molecular Devices) at A570.
Colony Survival Assay.
For shRNA knockdown experiments, an equal number of cells were seeded into 6-cm dishes, cultured in their normal growth media overnight, and then infected with the same amount of the scramble shRNA or the SGK3 shRNA viral suspensions. Two days after infection, cells were added with 5 μg/mL puromycin to eliminate the uninfected cells and continued to culture until the scramble shRNA-transduced cells reached confluency. Medium in each dish was replaced every 3 d. Once the scramble shRNA-transduced cells reached confluency, both the scramble shRNA-transduced cells and the SGK3 shRNA-transduced cell counterpart from the same cell line were fixed and stained with crystal violet simultaneously. For SGK3 induction experiments, MCF7aro/TO/SGK3 cells were hormone starved for 2 d and then seeded into six-well plates at different cell densities (200,000 per well, 100,000 per well, and 50,000 per well) and cultured in phenol red free MEM medium with 10% (vol/vol) CD FBS. The cells were added with or without DOX (final concentration, 250 ng/mL), and treated with 1 nM T plus 200 nM letrozole. Every 3 d, the cells were replaced with fresh medium and under the same treatments until the time indicated. For fixation and staining, we followed the protocol as described in ref. 51.
Microsome Isolation.
Two T150 flasks of cells were harvested and washed once with PBS. The cell pellets were washed with 1 mL 0.9% NaCl and resuspended in a fresh prepared ice-cold IBcell-1 buffer (225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA, and 30 mM Tris⋅HCl at pH 7.4). Cells were disrupted by passing through blunt-ended needles (26 gauge) 15 times. The lysates were centrifuged at 1,000 × g for 10 min at 4 °C twice to pellet cell debris, nuclei, and unbroken cells. The suspensions were centrifuged at 6,000 × g for 10 min at 4 °C to pellet mitochondria, and then centrifuged at maximum speed (≥20,000 × g) for 30 min at 4 °C. The suspension was cytosolic fraction, and the pellet was microsomal fraction. The microsomal pellets were washed with IBcell-1 buffer once, and suspended in a solution containing 0.25 M sucrose, 0.15 M KCI, 3 mM 2-mercaptoethanol, 20 μM CaC12, 10 mM Tris⋅HCl at pH 7.5.
SERCA2b ATPase Activity Assay.
The SERCA2b ATPase activity was assessed by measurement of microsomal Ca2+ ATPase activity (52), using a modified procedure, as reported in the reference (53). ATPase activities were determined by a colorimetric method by measuring the amount of Pi release, using an ATPase assay kit according to the manufacturer's protocol (Innova Biosciences), with a modification. Briefly, 5 μg of microsomes were mixed with the buffer containing 0.5 M assay buffer, 0.1 M MgCl2, 2 mM CaCl2, and 10 mM ATP for 30 min at 37 °C. Then 50 μL Gold mix was added to stop reactions. After 2 min, 20 μL stabilizer solution was added and the absorbance was read at 620 nm 30 min later. The experiment was performed in duplicate.
Coimmunoprecipitation.
Coimmunoprecipitaion assay was performed as described previously (23). Briefly, cells cultured in 6- or 10-cm dishes were lysed in 800 µL or 1 mL IP lysis buffer (50 mM Tris⋅HCl at pH 7.4, with 150 mM NaCl, 1 mM EDTA, 1% TRITON X-100 and complete protease inhibitor mixture) on ice for 50 min. The supernatants were collected by centrifugation at 16,300 × g for 5 min. After preclearing with 30 µL protein A or G agarose by incubation for 3 h at 4 °C on a rocking platform, equal amount of each lysate was incubated with 50 µL anti-FLAG resin or anti-HA resin overnight or incubated with anti-SGK3 antibodies for 1 h, followed by the addition of 50 µL protein A agarose for immunoprecipitation overnight at 4 °C with rotation. Immunoprecipitates were washed four times with 900 µL IP lysis buffer and boiled in 2× SDS loading buffer for subsequent Western blot analysis.
Supplementary Material
Acknowledgments
We thank Dr. Zheng Liu for assistance with analyzing microarray data, Dr. Brian Armstrong for assistance with confocal microscopy, Dr. Zhou Li for assistance with transmission electron microscopy, Dr. Jinhui Wang for assistance with RNA-Seq, and Lucy Brown for flow cytometry analysis. We also thank Dr. Cynthia Wong and Dr. Guoqiang Sun for some technical support and Dr. Chu-Yu Liu for discussions about the manuscript. This work was supported by a Carr Baird grant (to Y.W.) and Hope Idol 2012 (to S.C.), the ThinkCure grant (to S.C.) and NIH Grant CA44735 (to S.C.). The Analytical Cytometry Core, Bioinformatics Core, Electron Microscope Core, Light Microscope Core, and Integrative Genomics Core were supported by the National Cancer Institute under Award P30CA33572.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612991114/-/DCSupplemental.
References
- 1.Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer. 2015;15(5):261–275. doi: 10.1038/nrc3920. [DOI] [PubMed] [Google Scholar]
- 2.Chen S, et al. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol. 2006;102(1-5):232–240. doi: 10.1016/j.jsbmb.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson JF, et al. Endocrine treatment options for advanced breast cancer--the role of fulvestrant. Eur J Cancer. 2005;41(3):346–356. doi: 10.1016/j.ejca.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2(2):101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 5.Krebs J, Agellon LB, Michalak M. Ca(2+) homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochem Biophys Res Commun. 2015;460(1):114–121. doi: 10.1016/j.bbrc.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Denmeade SR, Isaacs JT. The SERCA pump as a therapeutic target: Making a “smart bomb” for prostate cancer. Cancer Biol Ther. 2005;4(1):14–22. doi: 10.4161/cbt.4.1.1505. [DOI] [PubMed] [Google Scholar]
- 7.Vangheluwe P, Raeymaekers L, Dode L, Wuytack F. Modulating sarco(endo)plasmic reticulum Ca2+ ATPase 2 (SERCA2) activity: Cell biological implications. Cell Calcium. 2005;38(3-4):291–302. doi: 10.1016/j.ceca.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro DJ, Livezey M, Yu L, Zheng X, Andruska N. Anticipatory UPR activation: A protective pathway and target in cancer. Trends Endocrinol Metab. 2016;27(10):731–741. doi: 10.1016/j.tem.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andruska N, Zheng X, Yang X, Helferich WG, Shapiro DJ. Anticipatory estrogen activation of the unfolded protein response is linked to cell proliferation and poor survival in estrogen receptor α-positive breast cancer. Oncogene. 2015;34(29):3760–3769. doi: 10.1038/onc.2014.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344(Pt 1):189–197. [PMC free article] [PubMed] [Google Scholar]
- 11.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bago R, et al. The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 2016;35(17):1902–1922. doi: 10.15252/embj.201693929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasser JA, et al. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell. 2014;56(4):595–607. doi: 10.1016/j.molcel.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, et al. SGK3 is associated with estrogen receptor expression in breast cancer. Breast Cancer Res Treat. 2012;134(2):531–541. doi: 10.1007/s10549-012-2081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Mol Endocrinol. 2011;25(1):72–82. doi: 10.1210/me.2010-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masri S, et al. Genome-wide analysis of aromatase inhibitor-resistant, tamoxifen-resistant, and long-term estrogen-deprived cells reveals a role for estrogen receptor. Cancer Res. 2008;68(12):4910–4918. doi: 10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Zhou D, Chen S. SGK3 is an androgen-inducible kinase promoting prostate cancer cell proliferation through activation of p70 S6 kinase and up-regulation of cyclin D1. Mol Endocrinol. 2014;28(6):935–948. doi: 10.1210/me.2013-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherk AB, et al. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res. 2008;68(18):7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray JT, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384(Pt 3):477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marumoto T, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15(1):110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 22.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Xu W, Zhou D, Neckers L, Chen S. Coordinated regulation of serum- and glucocorticoid-inducible kinase 3 by a C-terminal hydrophobic motif and Hsp90-Cdc37 chaperone complex. J Biol Chem. 2014;289(8):4815–4826. doi: 10.1074/jbc.M113.518480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessier M, Woodgett JR. Role of the Phox homology domain and phosphorylation in activation of serum and glucocorticoid-regulated kinase-3. J Biol Chem. 2006;281(33):23978–23989. doi: 10.1074/jbc.M604333200. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, et al. Identification of Flightless-I as a substrate of the cytokine-independent survival kinase CISK. J Biol Chem. 2009;284(21):14377–14385. doi: 10.1074/jbc.M807770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29(33):4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell RA, et al. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: A new model for anti-estrogen resistance. J Biol Chem. 2001;276(13):9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 28.Clark AS, West K, Streicher S, Dennis PA. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol Cancer Ther. 2002;1(9):707–717. [PubMed] [Google Scholar]
- 29.Wong C, Wang X, Smith D, Reddy K, Chen S. AKT-aro and HER2-aro, models for de novo resistance to aromatase inhibitors; molecular characterization and inhibitor response studies. Breast Cancer Res Treat. 2012;134(2):671–681. doi: 10.1007/s10549-012-2105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin LA, et al. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278(33):30458–30468. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- 31.Chan CM, Martin LA, Johnston SR, Ali S, Dowsett M. Molecular changes associated with the acquisition of oestrogen hypersensitivity in MCF-7 breast cancer cells on long-term oestrogen deprivation. J Steroid Biochem Mol Biol. 2002;81(4-5):333–341. doi: 10.1016/s0960-0760(02)00074-2. [DOI] [PubMed] [Google Scholar]
- 32.Masri S, Phung S, Wang X, Chen S. Molecular characterization of aromatase inhibitor-resistant, tamoxifen-resistant and LTEDaro cell lines. J Steroid Biochem Mol Biol. 2010;118(4-5):277–282. doi: 10.1016/j.jsbmb.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lønning PE. Lack of complete cross-resistance between different aromatase inhibitors; a real finding in search for an explanation? Eur J Cancer. 2009;45(4):527–535. doi: 10.1016/j.ejca.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Takagi K, et al. Increased intratumoral androgens in human breast carcinoma following aromatase inhibitor exemestane treatment. Endocr Relat Cancer. 2010;17(2):415–430. doi: 10.1677/ERC-09-0257. [DOI] [PubMed] [Google Scholar]
- 35.Rechoum Y, et al. AR collaborates with ERα in aromatase inhibitor-resistant breast cancer. Breast Cancer Res Treat. 2014;147(3):473–485. doi: 10.1007/s10549-014-3082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujii R, et al. Increased androgen receptor activity and cell proliferation in aromatase inhibitor-resistant breast carcinoma. J Steroid Biochem Mol Biol. 2014;144(Pt B):513–522. doi: 10.1016/j.jsbmb.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Hanamura T, et al. Androgen metabolite-dependent growth of hormone receptor-positive breast cancer as a possible aromatase inhibitor-resistance mechanism. Breast Cancer Res Treat. 2013;139(3):731–740. doi: 10.1007/s10549-013-2595-x. [DOI] [PubMed] [Google Scholar]
- 38.Divekar SD, et al. The role of calcium in the activation of estrogen receptor-alpha. Cancer Res. 2011;71(5):1658–1668. doi: 10.1158/0008-5472.CAN-10-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill BJ, Muldrew E. Oestrogen upregulates the sarcoplasmic reticulum Ca(2+) ATPase pump in coronary arteries. Clin Exp Pharmacol Physiol. 2014;41(6):430–436. doi: 10.1111/1440-1681.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YH, Li W, Song WH. Effects of oestrogen on sarcoplasmic reticulum Ca2+-ATPase activity and gene expression in genioglossus in chronic intermittent hypoxia rat. Arch Oral Biol. 2009;54(4):322–328. doi: 10.1016/j.archoralbio.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Liu CG, et al. 17Beta-oestradiol regulates the expression of Na+/K+-ATPase beta1-subunit, sarcoplasmic reticulum Ca2+-ATPase and carbonic anhydrase iv in H9C2 cells. Clin Exp Pharmacol Physiol. 2007;34(10):998–1004. doi: 10.1111/j.1440-1681.2007.04675.x. [DOI] [PubMed] [Google Scholar]
- 42.Thor D, Uchizono JA, Lin-Cereghino GP, Rahimian R. The effect of 17 beta-estradiol on intracellular calcium homeostasis in human endothelial cells. Eur J Pharmacol. 2010;630(1-3):92–99. doi: 10.1016/j.ejphar.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kooptiwut S, et al. Estrogen reduces endoplasmic reticulum stress to protect against glucotoxicity induced-pancreatic β-cell death. J Steroid Biochem Mol Biol. 2014;139:25–32. doi: 10.1016/j.jsbmb.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Schmid E, et al. SGK3 regulates Ca(2+) entry and migration of dendritic cells. Cell Physiol Biochem. 2012;30(6):1423–1435. doi: 10.1159/000343330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raina K, et al. Targeted protein destabilization reveals an estrogen-mediated ER stress response. Nat Chem Biol. 2014;10(11):957–962. doi: 10.1038/nchembio.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajapaksa G, Thomas C, Gustafsson JA. Estrogen signaling and unfolded protein response in breast cancer. J Steroid Biochem Mol Biol. 2016;163:45–50. doi: 10.1016/j.jsbmb.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 48.Clarke R, Cook KL. Unfolding the Role of Stress Response Signaling in Endocrine Resistant Breast Cancers. Front Oncol. 2015;5:140. doi: 10.3389/fonc.2015.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke R, et al. Endoplasmic reticulum stress, the unfolded protein response, and gene network modeling in antiestrogen resistant breast cancer. Horm Mol Biol Clin Investig. 2011;5(1):35–44. doi: 10.1515/hmbci.2010.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eletto D, Chevet E, Argon Y, Appenzeller-Herzog C. Redox controls UPR to control redox. J Cell Sci. 2014;127(Pt 17):3649–3658. doi: 10.1242/jcs.153643. [DOI] [PubMed] [Google Scholar]
- 51.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 52.Saborido A, Delgado J, Megías A. Measurement of sarcoplasmic reticulum Ca2+-ATPase activity and E-type Mg2+-ATPase activity in rat heart homogenates. Anal Biochem. 1999;268(1):79–88. doi: 10.1006/abio.1998.3043. [DOI] [PubMed] [Google Scholar]
- 53.Sahoo SK, et al. The N terminus of sarcolipin plays an important role in uncoupling sarco-endoplasmic reticulum Ca2+-ATPase (SERCA) ATP hydrolysis from Ca2+ transport. J Biol Chem. 2015;290(22):14057–14067. doi: 10.1074/jbc.M115.636738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.