The extensive evolution of intracellular compartmentalization requires highly selective mechanisms for protein targeting to distinct membrane systems. One of the fundamental processes in protein targeting is the insertion of proteins into biological membranes. The efficient and accurate insertion of membrane proteins is an important step for their proper function in different organelles, and any targeting error may lead to mislocalization of these proteins with detrimental cellular effects. Posttranslational insertion is required for a class of tail-anchored (TA) proteins, which are characterized by a transmembrane domain (TMD) near their C terminus, for their correct targeting to the destined membrane (1). In PNAS, Xing et al. uncover a pathway for TA protein insertion into the endoplasmic reticulum (ER) in the model plant Arabidopsis thaliana, which plays an unexpected role in root hair growth (2). The authors identify several key components in the guided entry of tail-anchored protein (GET) complex that has a conserved function in regulating TA protein insertion. However, in contrast to yeast and animals, the core GET system in Arabidopsis involves a distinct GET3 clade, suggesting an ancient evolution of the GET3 paralogs in plants and which may function divergently as plant-specific organelle chaperones.
The GET pathway is the most extensively studied system for shuttling TA proteins to distinct organelles in yeast and mammalian cells. Structural and biochemical investigations in yeast have provided insights into the molecular mechanism of the GET complex (1). In the yeast GET pathway, newly synthesized TA proteins are initially recognized by a cytosolic pretargeting complex (PTC), comprising SGT2 and GET4–GET5, and then transferred to the ATPase GET3. GET3 loaded with a TA protein will then bind to its receptor: the ER-localized GET1–GET2 complex, which subsequently promotes the insertion of the TA protein into the ER membrane (Fig. 1A). The recognition and insertion processes mediated by the GET complex give rise to the membrane specificity for TA proteins, as overexpression of a subset of TA proteins in get mutants results in the redistribution of the aberrant protein into the yeast mitochondrial membrane (3, 4). Alternatively, TA proteins may use other chaperones/receptors for insertion into the ER, as defects in the GET pathway are only conditionally lethal in yeast (4). In contrast, loss of the GET3 homolog in mammals leads to embryonic lethality (5), indicating a divergent role of GET components in different organisms.
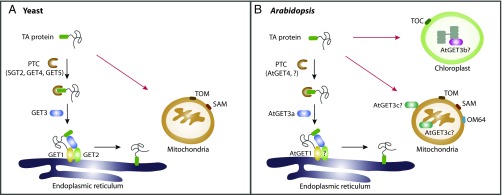
Fig. 1.
Working model for the GET system. (A) In yeast, some TA proteins targeted to the ER membrane are GET dependent (black arrows), whereas targeting to mitochondria is GET independent and uses receptors including TOM and SAM complexes (red arrows). In the GET pathway, the newly synthesized protein is initially recognized by the PTC complex (SGT2 and GET4–GET5), and then is transferred to the ATPase GET3. Subsequently, cytosolic GET3 guides the TA protein to the ER membrane via interacting with the ER-localized GET1–GET2 complex, and finally promotes the insertion of the TA protein into the ER membrane. (B) In Arabidopsis, the AtGET3a-dependent pathway is involved in TA protein insertion into the ER (black arrows). TA protein transport to mitochondria and chloroplast is likely GET independent (red arrows) and requires distinct receptors, including TOM and SAM complexes, as well as a plant-specific receptor OM64 for mitochondria and TOC complex for chloroplast. For TA proteins targeted to the ER, the PTC complex (AtGET4 and possibly other unidentified components?) binds to the nascent proteins and recruits the cytosolic AtGET3a. Then, AtGET3a interacts with AtGET1 and a possible coreceptor (?) to transfer the TA protein to AtGET1 for insertion into the ER. In contrast, AtGET3b is distributed in chloroplast stroma, whereas AtGET3c is found in the mitochondrial matrix or on the mitochondrial outer membrane. The biological function of both AtGET3b and AtGET3c in chloroplast or mitochondrial remains elusive.
By contrast, the underlying mechanism for TA protein targeting in plants remains obscure, although increasing evidence supports the existence of a similar regulatory pathway. Bioinformatic screens have predicted over 500 TA proteins in the Arabidopsis thaliana genome that are directed to distinct membrane compartments, including the ER, mitochondria, and chloroplast (6). However, genetic evidence for a functional GET pathway guiding the TA protein insertion process in plants is currently lacking, thus hindering our understanding of TA protein targeting in plants. Previous sequence alignment analyses indicate that, except for GET3, most GET homologs are missing in the Arabidopsis genome. However, whether GET3 isoforms are able to mediate protein insertion independently of the other GET components or whether divergent proteins have evolved to function similarly in plant cells, is an unresolved question.
To address this issue, Xing et al. have performed an in silico sequence comparison and identified several core GET homologs, including AtGET1, AtGET3a-c, and AtGET4 in Arabidopsis. Consistent with their yeast homologs, AtGET1 and AtGET4 are found to be localized on the ER and in the cytosol, respectively. Strikingly, the three AtGET3 paralogs display different subcellular distributions (AtGET3a, cytosol; AtGET3b, chloroplast stroma; AtGET3c, mitochondria matrix) (Fig. 1B). Among these three AtGET3 paralogs, only AtGET3a was shown to be involved in TA protein insertion into the ER membrane. AtGET3a, but neither AtGET3b nor AtGET3c, underwent homodimerization and could interact with AtGET4 as well as AtGET1, demonstrating that the AtGET3a protein interactome resembles the yeast GET complex.
Further investigations on the physiological function of the GET complex in plants using different get mutants has uncovered a role for the GET complex in root hair growth. Loss of AtGET1, AtGET3a, and AtGET4 leads to reduced growth of root hairs, and this defect in get mutants can be rescued by transforming back the corresponding GET components. However, mutation of the ATPase binding site in AtGET3a failed to recover the atget3a mutant phenotype, implying that a functional ATPase activity of AtGET3a is responsible for root hair growth. In addition, several TA protein substrates have been identified as interacting with AtGET3a, whereas other AtGET3a-independent TA proteins underpin that additional posttranslational pathways are required in plant. The authors used the root hair-specific Qa-SNARE SYP123, which targets to the PM, and showed that loss of AtGET components impacts on the protein abundance with a reduction in both transcript and protein levels of SYP123, rather than its subcellular localization. These findings suggest that an alternative pathway for TA protein insertion exists in plants as SYP123 could still be targeted to the PM in Atget mutants. This conclusion is also supported by the observation that Atget mutants are viable, whereas overexpression of AtGET3a in Atget1 causes severe growth defects. The authors suggest that overexpressed AtGET3a may saturate the binding sites of some essential TA protein substrates, thus preventing their recognition by other receptors and subsequent targeting.
Xing et al. thus uncovered a conserved GET pathway for posttranslational regulation of TA proteins in Arabidopsis, which allows for the efficient insertion of TA proteins to the prescribed compartment (Fig. 1B). Another recent related study (7) using an in vitro assay showed that yeast GET1/2 is able to promote the insertion of the Arabidopsis SNARE SYP72 by the aid of GET3, which further supports the conserved nature of the GET pathway for TA protein insertion in eukaryotes. Although most of the GET counterparts have been identified in Arabidopsis, it remains to be seen whether other GET orthologs such as GET2 and GET5 exist in plants. Because CAML, the mammalian GET2, shows no significant sequence similarity to its yeast ortholog (8), it would appear that different organisms might have evolved sequence divergency or other counterparts in this pathway.
The GET system in plants may contribute to specific plant developmental processes such as root hair growth and ER stress response, through the alteration of abundance of SYP123 (2) or subcellular localization of SYP72 (7). It was recently noted that multiple selection filters have been used to distinguish correct and incorrect substrates based on minor differences of the TMD and C-terminal elements of TA proteins (9). Because several TA proteins have been identified as the AtGET3a substrates, future studies will aim at elucidating the underlying molecular mechanisms for plant TA protein recognition in plants to understand why and how the plant GET complex impacts the substrates to different extents (e.g., abundance or mislocalization), and their other specific physiological roles during plant growth and development.
Gene duplication may also have given rise to the evolution of novel functions in plants, such as the plant-specific organelle chloroplast. Plants contain the conserved translocase of the inner membrane (TOM) and sorting and assembly machinery (SAM) components for TA protein delivery to mitochondria, but also use the plant-unique receptor, OM64 (10). On the other hand, targeting to chloroplast requires the translocon at the outer envelope membrane of chloroplasts (TOC) complex as the general entry gate for most of the chloroplast TA proteins (11). However, only a few chloroplast chaperones including AKR2 and HSP70 have been reported (12). The study by Xing et al. showed that Arabidopsis has another two GET3 paralogs (AtGET3b and AtGET3c) with distinct functions to AtGET3a, because (i) both AtGET3b and AtGET3c do not contain a GET1-binding motif and show lack of interaction with AtGET1, (ii) both AtGET3b and AtGET3c reside on distinct compartments (chloroplast and mitochondria, respectively), and (iii) both cytosolic-localized truncated forms of AtGET3b and AtGET3c fail to rescue the root hair growth defect in AtGET3a. Therefore, it is very likely that the gene duplication event of AtGET3 evolves as an adaptation in plant cells to function as plant-specific organelle chaperons, either within the chloroplast stroma or mitochondria matrix (Fig. 1B). In addition, because AtGET3c was previously shown to localize on the mitochondria outer membrane in transient-expressed protoplasts (13), alternatively, AtGET3c might also be involved in mitochondria TA protein targeting. Further in-depth investigations are required to explore the biological functions of the AtGET3b/c paralogs in plant distinct organelles.
Footnotes
The authors declare no conflict of interest.
See companion article on page E1544.
References
- 1.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12(12):787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing S, et al. Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc Natl Acad Sci USA. 2017;114:E1544–E1553. doi: 10.1073/pnas.1619525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci USA. 2014;111(22):8019–8024. doi: 10.1073/pnas.1405755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134(4):634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Kriechbaumer V, et al. Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic. 2009;10(12):1753–1764. doi: 10.1111/j.1600-0854.2009.00991.x. [DOI] [PubMed] [Google Scholar]
- 7.Srivistava R, Zalisko BE, Keenan RJ, Howell SH. The GET system inserts the tail-anchored SYP72 protein into endoplasmic reticulum membranes. Plant Physiol. 2016;2016:pp-00928. doi: 10.1104/pp.16.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48(3):387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Rao M, et al. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. eLife. 2016;5:21301. doi: 10.7554/eLife.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan O, Murcha MW, Whelan J. Unique components of the plant mitochondrial protein import apparatus. Biochim Biophys Acta. 2013;1833(2):304–313. doi: 10.1016/j.bbamcr.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14(12):787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 12.Bae W, et al. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol. 2008;10(2):220–227. doi: 10.1038/ncb1683. [DOI] [PubMed] [Google Scholar]
- 13.Duncan O, et al. Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol. 2011;157(3):1093–1113. doi: 10.1104/pp.111.183160. [DOI] [PMC free article] [PubMed] [Google Scholar]