Significance
A key feature of plant cells is rapid cytoplasmic streaming that is driven by myosin motors. However, specific mechanisms of myosin-dependent streaming are poorly understood. Here, we characterize a dense network of plant myosins and their receptors and adaptors that, jointly with myosins, appear to mediate cytoplasmic streaming through distinct endomembrane compartments. We additionally present data suggestive of a myosin-dependent nucleocytoplasmic trafficking pathway. The myosin network is an ancient functional module that was already present in the common ancestor of green algae and land plants but underwent a major expansion in the latter, probably contributing to land colonization by plants.
Keywords: myosins, receptors, adaptors, cytoplasmic streaming, nuclear transport
Abstract
We investigate the myosin XI-driven transport network in Arabidopsis using protein–protein interaction, subcellular localization, gene knockout, and bioinformatics analyses. The two major groups of nodes in this network are myosins XI and their membrane-anchored receptors (MyoB) that, together, drive endomembrane trafficking and cytoplasmic streaming in the plant cells. The network shows high node connectivity and is dominated by generalists, with a smaller fraction of more specialized myosins and receptors. We show that interaction with myosins and association with motile vesicles are common properties of the MyoB family receptors. We identify previously uncharacterized myosin-binding proteins, putative myosin adaptors that belong to two unrelated families, with four members each (MadA and MadB). Surprisingly, MadA1 localizes to the nucleus and is rapidly transported to the cytoplasm, suggesting the existence of myosin XI-driven nucleocytoplasmic trafficking. In contrast, MadA2 and MadA3, as well as MadB1, partition between the cytosolic pools of motile endomembrane vesicles that colocalize with myosin XI-K and diffuse material that does not. Gene knockout analysis shows that MadB1–4 contribute to polarized root hair growth, phenocopying myosins, whereas MadA1–4 are redundant for this process. Phylogenetic analysis reveals congruent evolutionary histories of the myosin XI, MyoB, MadA, and MadB families. All these gene families emerged in green algae and show concurrent expansions via serial duplication in flowering plants. Thus, the myosin XI transport network increased in complexity and robustness concomitantly with the land colonization by flowering plants and, by inference, could have been a major contributor to this process.
For half of a century, it had been assumed that rapid organelle trafficking and cytoplasmic streaming in plant cells rely on myosin motors (1) and, in particular, class XI myosins that are homologous to fungal and animal class V myosins (2). Over the past decade, a massive amount of information on specific functions of myosins and mechanisms of myosin XI-dependent processes in plants has been obtained (3–5). The first genetic evidence of myosin function in cell expansion involved the demonstration of a dramatic reduction of the polarized root hair growth upon inactivation of the myosin XI-K and XI-2 genes (6, 7). Subsequently, it has been shown that myosin-dependent trafficking is required for the growth of multiple cell types, as well as normal plant growth and development (4, 8–10). Among the 13 paralogous myosins XI encoded in the Arabidopsis thaliana genome, the highly expressed myosins XI-K, XI-1, and XI-2 have been shown to provide the most pronounced, functionally redundant contributions to each of these processes. In contrast, another highly expressed paralog, myosin XI-I, plays only limited roles in cell and plant growth but has a specialized function in nuclear repositioning through binding nuclear envelope protein WIT2 (11).
Studies on subcellular localization of the myosin XI-K, the dominant contributor to most of the myosin-dependent processes in plants, show that, contrary to the long-standing beliefs, the bulk of the intracellular pool of this myosin is associated not with the larger organelles, such as Golgi, mitochondria, and peroxisomes, but with a vesicle-like compartment that rapidly moves in dense, “beads-on-a-string” arrays along F-actin bundles (12). Furthermore, although the endoplasmic reticulum (ER) flow in plant cells has been demonstrated to rely on myosins XI-K, XI-1, and XI-2 (13), the appearance of the XI-K–associated membrane compartment was distinct from the appearance of the ER network (12). These findings imply that the observed motility of the ER is not caused by direct association with the myosin motors, but is rather elicited indirectly, perhaps via the cytosolic flow driven by vesicle-associated myosins.
The latter hypothesis was further boosted by the discovery of the bona fide myosin receptors, the MyoB family proteins, which colocalized with myosin XI-K to the distinct vesicular compartment described above (14). The MyoB protein family is characterized by the universally conserved, myosin-binding, coiled-coil domain (previously known as DUF593); most of the MyoBs also contain a predicted transmembrane helix (TM) that is sufficient for MyoB anchoring to endomembrane vesicles in a myosin-dependent manner (14). In addition, it has been shown that the MyoB compartment belonged neither to the ER-to-Golgi-to-plasma membrane secretory pathway nor to several other known exo- or endocytic compartments, with the implication that the myosin–MyoB complex defines a previously undescribed, specialized transport compartment, a potential driver of cytoplasmic streaming (14). The MyoB family is conserved in all land plants and some green algae, but not in other eukaryotes (14). Despite the advanced status of the yeast model, analogous bona fide, membrane-anchored, vesicular myosin receptors remain to be identified (15), although a variety of fungal and animal myosin V adaptors and regulators have been characterized in distinct cell types (16). For the sake of clarity, we henceforth reserve the term “receptors” for TM-containing proteins that provide a direct link between myosin and its membrane cargo, whereas the term “adaptor” will be used for myosin-binding proteins that lack a TM and are indirectly associated with a target membrane.
A correlation between myosin-driven transport and plant cell expansion has been established by the demonstration that inactivation of the four highly expressed myosins results in virtual abolishment of the intracellular motility, as well as a dramatic reduction in cell growth (9). Conversely, expression of a faster moving, engineered myosin XI resulted in an increase in organelle trafficking velocity and, consequently, faster cell and plant growth (17). Although it has been proposed that these growth effects depend on acceleration of cytoplasmic streaming, it remained unclear which specific myosin cargo was responsible for driving the streaming. Moreover, the streaming itself has been traditionally measured as the velocity of undefined cytoplasmic particles or organelles that could be either drivers or passive cargoes of streaming (17, 18).
This conundrum was largely resolved with the introduction of an inert tracer of cytoplasmic streaming, namely, fluorescently labeled, heterologous protein inclusion bodies incapable of myosin XI binding (19). It has been shown that the velocity distribution of these tracer bodies was virtually indistinguishable from the velocity distributions for Golgi, mitochondria, peroxisomes, or secretory vesicles, with a mean velocity of ∼1 μm⋅s−1. In contrast, the velocity distribution of the MyoB1 vesicles was significantly skewed toward higher velocities, up to 6 μm⋅s−1, with a mean velocity of 3.2 μm⋅s−1 (19). These findings suggest that the larger organelles and secretory vesicles, as well as the inert tracer, all moved in the same passive manner, following cytoplasmic streaming driven by the myosin-MyoB compartment. This notion is strongly supported by negative dominant inhibition experiments: Overexpression of either the globular tail domain (GTD) of myosin responsible for cargo binding or the DUF593 domain of MyoB responsible for myosin attachment to vesicles resulted in nearly identical extents of reduction in the velocities of Golgi, mitochondria, peroxisomes, and secretory vesicles, as well as, most importantly, cytoplasmic streaming itself as attested by the inert tracer (19). Furthermore, inactivation of four of the 16 MyoB genes present in the Arabidopsis genome resulted in a statistically significant 24% reduction in cytoplasmic streaming velocity. Taken together, these findings validate the mechanistic connection between myosin XI motors, their vesicular cargo defined by the MyoB receptors, and the process of cytoplasmic streaming. This type of connection between myosins and their receptors is yet to be established in animal models of cytoplasmic streaming (20). It also has been shown that myosins XI and MyoBs genetically interact, in agreement with a model in which the myosin-MyoB compartment drives cytoplasmic streaming that carries along organelles and vesicles, boosts cells’ metabolic status, and enables cell growth (19).
Despite the substantial recent progress made in understanding myosin transport in plants, many critical questions remain unanswered. With only three Arabidopsis MyoBs characterized in any detail, what are the functions, myosin-binding activity, and subcellular localization of the remaining 13 MyoBs? Could it be the case that some of the MyoBs are associated with the organelles, ER, or secretory vesicles, thus directly contributing to streaming, which would require an amendment to the above model? Are there other myosin-binding proteins unrelated to MyoB, in addition to the nuclear WIT2 receptor of myosin XI-I?
To address these questions, we investigate a wider transport network including five distinct myosins XI and 11 MyoB receptors, and show that diverse MyoBs localize to typical MyoB vesicles rather than organelles. We identify two previously uncharacterized families of myosin-binding proteins unrelated to MyoB and show that most of these putative myosin adaptors are associated with distinct vesicular compartments. Surprisingly, one of these adaptors localizes to the nucleoplasm, suggestive of a function in nuclear export. We also reveal shared phylogenomic patterns for myosins XI, MyoBs, and the two myosin adaptor families, indicating concerted evolution of the myosin-driven transport network in plants. The proliferation of the myosin network components in flowering plants implies an important contribution to their evolutionary success during land colonization.
Results
Myosins XI Interact with Distinct Subsets of MyoB Receptors.
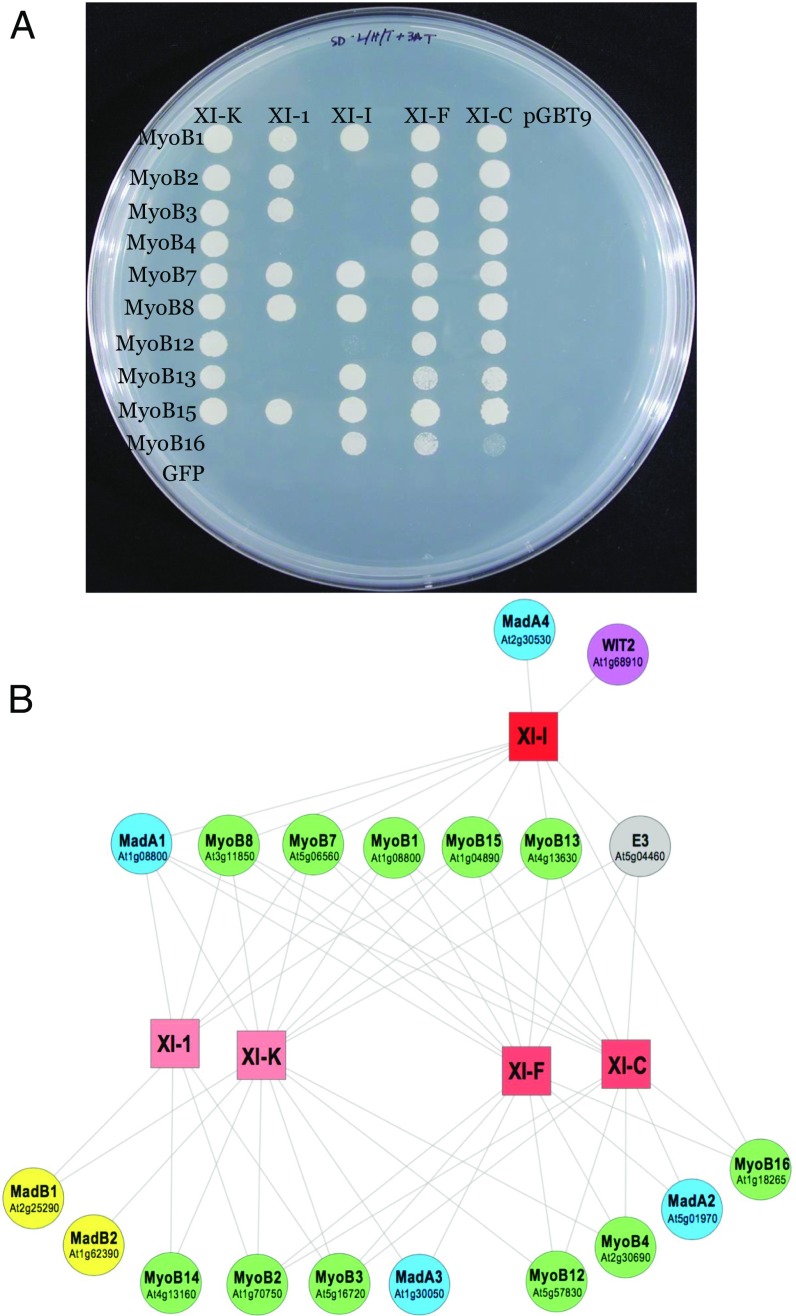
To gain detailed insight into the network of myosin–MyoB interactions, we conducted deep Y2H screens using an Arabidopsis cDNA library and five distinct myosin GTDs as bait. These screens included GTDs of myosins XI-K, XI-1, XI-I, XI-F, and XI-C, representing four diverse myosin XI groups (21). We found that each myosin interacted with one to four MyoBs (Table 1). Reciprocally, distinct MyoBs exhibited interactions with one to four myosins XI. Because the frequency with which the cDNA clones are identified in Y2H screens depends on their library representation, these screens are prone to bias, as indicated by the fact that the most abundant hits were observed with MyoB1 and MyoB2, which are expressed to the highest levels throughout Arabidopsis tissues. To overcome this bias, we conducted directed, pairwise Y2H assays that involved the same five myosins and 11 MyoBs representing each of the six MyoB subfamilies (14). Seven of these MyoBs that produced no hits in the previous library screens were included to determine whether myosin binding is indeed a common property of MyoB family proteins. Although screening of 10 MyoBs yielded a rich set of interaction data (Fig. 1A; discussed below), a prey plasmid harboring MyoB14 failed to show any myosin binding in these experiments. Thus, we changed the experimental design by using MyoB14 as bait for screening the Arabidopsis cDNA library. The resulting hits with myosins XI-K and XI-1 validated myosin binding by MyoB14 (SI Appendix, Table S1).
Table 1.
Top hits of the Y2H screens using five distinct myosin XI GTDs as bait
| Gene ID | XI-K | XI-1 | XI-I | XI-F | XI-C | Total | Protein description |
| At1g08800 | 14 | 7 | 18 | 5 | 44 | MyoB1, myosin receptor | |
| At1g70750 | 5 | 13 | 3 | 21 | MyoB2, myosin receptor | ||
| At5g16720 | 2 | 1 | 1 | 4 | MyoB3, myosin receptor | ||
| At2g30690 | 2 | 2 | MyoB4, myosin receptor | ||||
| At5g06560 | 1 | 2 | 3 | MyoB7, myosin receptor | |||
| At4g02800 | 4 | 9 | 5 | 7 | 3 | 28 | MadA1; two CCs, NLS |
| At5g01970 | 2 | 16 | 18 | MadA2; two CCs | |||
| At1g30050 | 2 | 1 | 3 | MadA3; two CCs | |||
| At2g30530 | 1 | 1 | MadA4; two CCs | ||||
| At5g04460 | 6 | 15 | 4 | 4 | 29 | E3 ubiquitin ligase | |
| At1g68910 | 11 | 11 | WIT2, nuclear envelope | ||||
| At2g25290 | 6 | 2 | 8 | MadB1; CC-TPR repeats |
Nos. of hits for each GTD are shown. CC, coiled coil; ID, identification.
Fig. 1.
Pairwise interactions between five myosins XI and 10 MyoB receptors assessed using the yeast two-hybrid system. (A) Yeast colony growth on triple-deficient medium. Negative controls: GFP, prey plasmid engineered to express GFP; pGBT9, bait vector harboring no inserted ORF. (B) Bipartite network showing the interactions between myosins XI (squares) and their partners (circles). Shades of red identify groups of myosins that share similar interaction patterns. Sets of homologous myosin receptors and adaptors are represented with distinct colors. Gene identifications are also shown for MyoB receptors and Mad adaptors.
All detected interactions between myosins and MyoB receptors were represented as a bipartite network, in which two classes of nodes correspond to the myosins and the receptors, respectively, and the edges denote the interactions between these partner proteins (myosin adaptors are also included; discussed below) (Fig. 1B). Beyond the convenient graphical representation of the data, the network facilitates quantitative analysis of the interaction patterns. Specifically, we assessed two key properties of the network, nestedness and modularity. The former parameter evaluates the node hierarchy that can be described in terms of generalists [high-degree nodes (i.e., broadly interacting proteins)] and specialists [low-degree nodes (i.e., proteins with a small number of partners)]. Modularity quantifies the existence of groups of nodes that preferentially interact with each other but not with the rest of the network (22).
Network randomization indicates that the overall nestedness and modularity of the myosin network are not significantly greater than expected in random networks of similar complexity, a result that could be expected, given the small network size. The lack of a clear modular structure in the network reflects the high connectivity of several receptors that interact with all five (MyoB1, MyoB7, MyoB8, and MyoB15) or four (MyoB2, MyoB3, and MyoB13) myosins. The rest of the network shows some degree of specificity, with three groups of myosins with their own distinct interaction patterns: (i) XI-K and XI-1, (ii) XI-F and XI-C, and (iii) XI-I (Fig. 1B).
Taken together, the Y2H assays and the network analysis that involved five myosins XI and 11 MyoB receptors demonstrated highly redundant interactions along with higher degrees of interaction specificity for certain nodes. This pattern is compatible with the functional redundancy that has been observed for both myosins and MyoBs in genetic analyses (9, 14, 19).
Subcellular Distributions of MyoB12, MyoB13, and MyoB14.
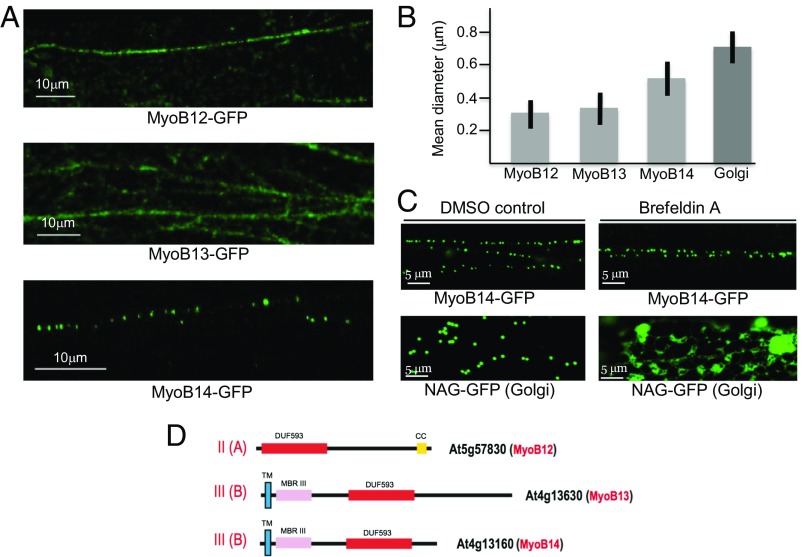
Previously, GFP-tagged myosin XI-K, MyoB1 (MyoB subfamily IA), MyoB2 (subfamily IB), and MyoB7 (subfamily IIB) have been localized to motile vesicles that appeared distinct from larger organelles and several exocytic or endocytic compartments (12, 14). To obtain further insight into the localization of the structurally and evolutionarily diverse MyoB receptors, we tagged MyoB12 (subfamily IIA), MyoB13 (subfamily IIIB), and MyoB14 (subfamily IIIB) with GFP and stably expressed these proteins in Arabidopsis under the control of the respective native promoters, aiming to obtain normal accumulation levels. As shown in Fig. 2D, MyoB13 and MyoB14 both contain an N-terminal TM followed by putative metal-binding domains, whereas MyoB12 lacks a predicted TM and a metal-binding domain, which is an exception among MyoB proteins and is suggestive of association with a nonmembrane cargo (14).
Fig. 2.
Analysis of MyoB12-GFP, MyoB13-GFP, and MyoB14-GFP localization and particle size in the leaf epidermal cells of transgenic Arabidopsis plants. (A) Distribution of the fluorescent bodies in linear vesicle-like arrays. (B) Mean diameters of the bodies associated with MyoB12-GFP, MyoB13-GFP, MyoB14-GFP, and Golgi stacks. (C) Distribution of the Golgi-specific marker NAG-GFP and MyoB14-GFP in leaf epidermal cells treated with DMSO (control) or BFA. (D) Domain architectures of the MyoB12, MyoB13, and MyoB14 that belong to subfamilies II(A) and III(B) as shown. CC, coiled coil; DUF593, a myosin-binding domain; MBRIII, predicted metal-binding domain III; TM, transmembrane α-helix.
The beads-on-a-string localization patterns of MyoB12-GFP and MyoB13-GFP (Fig. 2A and Movies S1 and S2) closely resembled the patterns of MyoB1-GFP and MyoB2-GFP (14), implying their association with analogous vesicular compartments. Unexpectedly, MyoB14-GFP labeled discrete bodies that also moved in linear patterns but were sparser and larger than those bodies formed by other MyoBs (Fig. 2A and Movie S3). To investigate the membrane association of the MyoB12–14 proteins, we isolated the microsome fraction of the cells and used denaturing agents that allow one to distinguish among distinct types of membrane-associated proteins. In particular, peripheral proteins are dissociated from membranes by treatments with 1 M NaCl, whereas integral membrane proteins are solubilized by 1% Triton X-100. This fractionation demonstrated that the bulk of MyoB12, MyoB13, and MyoB14 was present in microsomes and solubilized after Triton X-100 treatment (SI Appendix, Fig. S1B). None of these proteins was released after NaCl treatment, indicating that they are either integrated into or are tightly associated with the membranes.
Next, we confirmed the statistically significant difference in the mean size of the compartments containing MyoB12 and MyoB13, as well as the compartments containing MyoB14 (Fig. 2B). The larger size of the latter compartments potentially could imply that in contrast to the prototypic MyoB1 compartment, MyoB14 is associated with endomembranes along the ER-to-Golgi-plasma membrane secretory pathway. To test this possibility, we used Brefeldin A (BFA) treatment inducing the formation of a distinct BFA compartment, to which the markers that are normally targeted to the ER, Golgi, trans-Golgi network, or secretory vesicles, colocalize (23). As expected, the Golgi stack marker repartitioned into this compartment (Fig. 2C). However, BFA treatment had no effect on MyoB14-GFP vesicles, thus excluding an association of MyoB14 with the main secretory pathway (Fig. 2C).
Taken together, the findings on the three evolutionarily and structurally diverse MyoB receptors described here reveal features similar to previously characterized MyoB1, MyoB2, and MyoB7 (14), except for the larger size of the MyoB14-associated compartment. These results are compatible with the hypothesis that diverse MyoB receptors are associated with a distinct class of compartments that apparently specialize in rapid endomembrane transport.
Previously Uncharacterized Protein Partners of Myosins XI.
In addition to numerous hits to MyoB receptors, deep Y2H library screens baited with five distinct myosin GTDs identified four paralogous Arabidopsis proteins (Table 1). Among these proteins, a protein encoded by the At4g02800 gene interacted with each of the tested myosins, whereas the proteins encoded by the genes At5g01970, At1g30050, and At2g30530 showed hits with one or two myosins each (Table 1). None of these proteins contained a predicted TM that is typical of bona fide MyoB receptors. Therefore, we dubbed this previously undetected family of myosin-binding proteins putative myosin adaptors of family A (MadA1–4; Table 1).
To validate the physical interactions between myosins and these candidate adaptors, we conducted pull-down assays using myosin XI-K GTD fused to maltose-binding protein (MBP-GTD) and FLAG-tagged MadA1 and MadA3. As shown in SI Appendix, Fig. S2, each of these proteins specifically interacted with the myosin XI-K GTD. Given that the MadA family is not homologous to the MyoB family of myosin receptors, we hypothesize that MadA1–4 adaptors act either as a more specialized alternative to or in concert with MyoBs.
The next most common myosin interactor identified in this work was a protein encoded by the At5g04460 gene (Table 1), a previously uncharacterized member of a large family of proteins containing a RING domain typical of the E3 ubiquitin ligases (24). Recently, yeast E3 ligase Dma1 has been shown to control the release of the myosin V cargo (25). It seems likely that the Arabidopsis homolog of this ubiquitin ligase similarly functions as a regulator of myosin XI cargo attachment.
Our screens additionally identified WIT2 protein that interacted with only one myosin, XI-I. Fittingly, this outer nuclear membrane protein has been identified previously as a linker between the nucleus and myosin XI-I, an interaction required for slow nuclear relocation (11). This result further supports the specificity and functional relevance of our Y2H screens.
Finally, a previously uncharacterized protein encoded by the gene At2g25290 was confidently identified as a myosin interactor (Table 1). This protein, which we denoted MadB1, contains a PB1 domain and tetratricopeptide repeat, two structural elements that are typically involved in protein–protein interactions (26, 27). There are three paralogs of MadB1 in Arabidopsis, which implies potential differential myosin binding by the proteins of this family. To determine if myosin binding is a common property of the MadB family, we conducted pull-down assays with MadB1 and MadB2 (encoded by gene At1g62390), two divergent MadB family members. These assays validated physical interactions between myosin the XI-K GTD and each of these MadB proteins (SI Appendix, Fig. S2).
The adaptors, the newly added members of the bipartite myosin transport network, followed the familiar patterns of generalists (e.g., MadA1 that interacted with all five myosins) and specialists (e.g., WIT2 that interacted only with the most specialized myosin XI-I) (Fig. 1B).
Typical of the Y2H screens, there were also numerous single hits with diverse proteins. Because investigating all these hits was impractical, we did not explore the relevance of the corresponding proteins any further but, instead, focused on the two families of validated myosin interactors (adaptors), MadA and MadB.
The MadA Proteins Localize Either to the Nucleus or to the Cytoplasm.
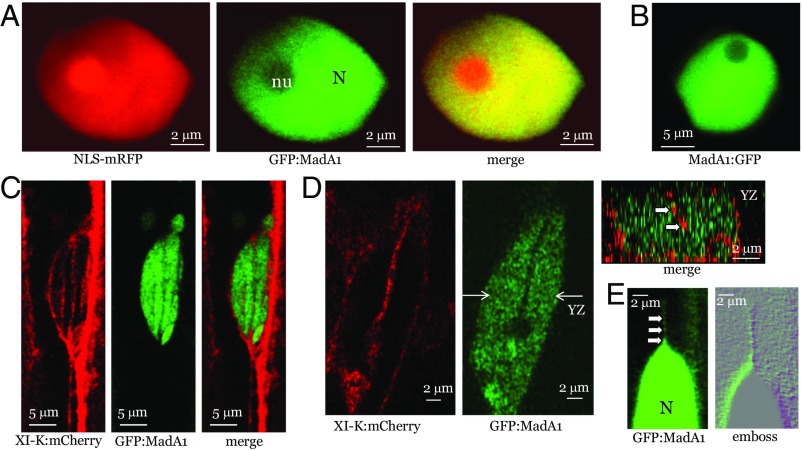
Bioinformatic analysis of the MadA proteins identified two putative coiled-coil domains in each of them. Intriguingly, MadA1, but not the other members of the MadA family, harbors a nuclear localization signal (NLS). To determine if MadA1 is indeed a nuclear protein, it was tagged by an N-terminal fusion to GFP. The resulting GFP:MadA was transiently coexpressed in Nicotiana benthamiana with NLS-tagged monomeric red fluorescent protein (mRFP). As expected, the NLS-mRFP was nuclear-localized (Fig. 3A); the GFP:MadA1 was also observed in the nucleus, with a preferential localization to the nucleoplasm rather than the nucleolus (Fig. 3A). To ensure that the N-terminal GFP fusion in GFP:MadA1 did not affect the protein localization, we showed that placing GFP at the C terminus in MadA1:GFP also resulted in nuclear localization (Fig. 3B).
Fig. 3.
Analysis of GFP:MadA1 localization relative to nucleus-specific marker NLS-mRFP (fused to NLS derived from SV40 T-antigen) or XI-K:mCherry (myosin XI-K tagged by mRFP Cherry). (A) Colocalization of NLS-mRFP and GFP:MadA1. (B) Nuclear localization of MadA1:GFP. (C and D) Colocalization of XI-K:mCherry and GFP:MadA1. In D, the nuclear cross-section along the y–z plane marked by white arrows (Center) is shown (Upper Right); white arrows point to red, XI-K:mCherry-tagged material present within the nucleus. (E) GFP:MadA1-containing material apparently exported from the nucleus (N) as a linear array of discrete bodies marked by white arrows.
Next, we stably transformed the GFP:MadA1-encoding gene under control of its native promoter into Arabidopsis and validated its nuclear targeting in the homologous system (Fig. 3C). Strikingly, we also observed rapid linear movement of the small green fluorescent bodies out of the nucleus, starting with apparent protrusions of nuclear material into the cytosol and continuing with its fragmentation into progressively smaller motile bodies that rapidly became invisible in the cytosol, either being dispersed or moving out of the focal plane (Fig. 3E and Movie S4). This phenomenon was transient and was detected only in ∼10% of the screened nuclei, possibly because it could only be observed when occurring precisely in the focal plane. Conceivably, such rapid trafficking of the GFP:MadA1-labeled nuclear material is due to active export mediated by MadA1–myosin interactions.
To address potential colocalization of MadA1 and myosin, we stably coexpressed GFP:MadA1 and myosin XI-K:mCherry in Arabidopsis using the corresponding native promoters for each gene. The latter protein was observed along thick cortical arrays corresponding to F-actin bundles (12, 14), as well as in thinner arrays surrounding the nuclear surface (Fig. 3D). Interestingly, similar linear arrays traversed the nucleus, as was validated by imaging the cross-section of the nucleus that clearly showed internal myosin XI-K:mCherry surrounded by, but not colocalized with, GFP:MadA1 (Fig. 3D). Although the exact organization of the intranuclear strands associated with myosin remains to be determined, such strands could be analogous to well-known transvacuolar strands, the F-actin– and myosin-containing cytosolic sleeves traversing the vacuolar interior (28). To our knowledge, transnuclear strands of this type have not been reported so far.
We were unable to determine whether the GFP:MadA1-tagged material exiting the nucleus (Fig. 3E) also colocalized with XI-K:mCherry. Nevertheless, it appears that the available data are best compatible with the existence of two distinct, myosin-driven, hypothetical nuclear transport pathways: (i) transnuclear trafficking and (ii) MadA1-mediated nuclear export.
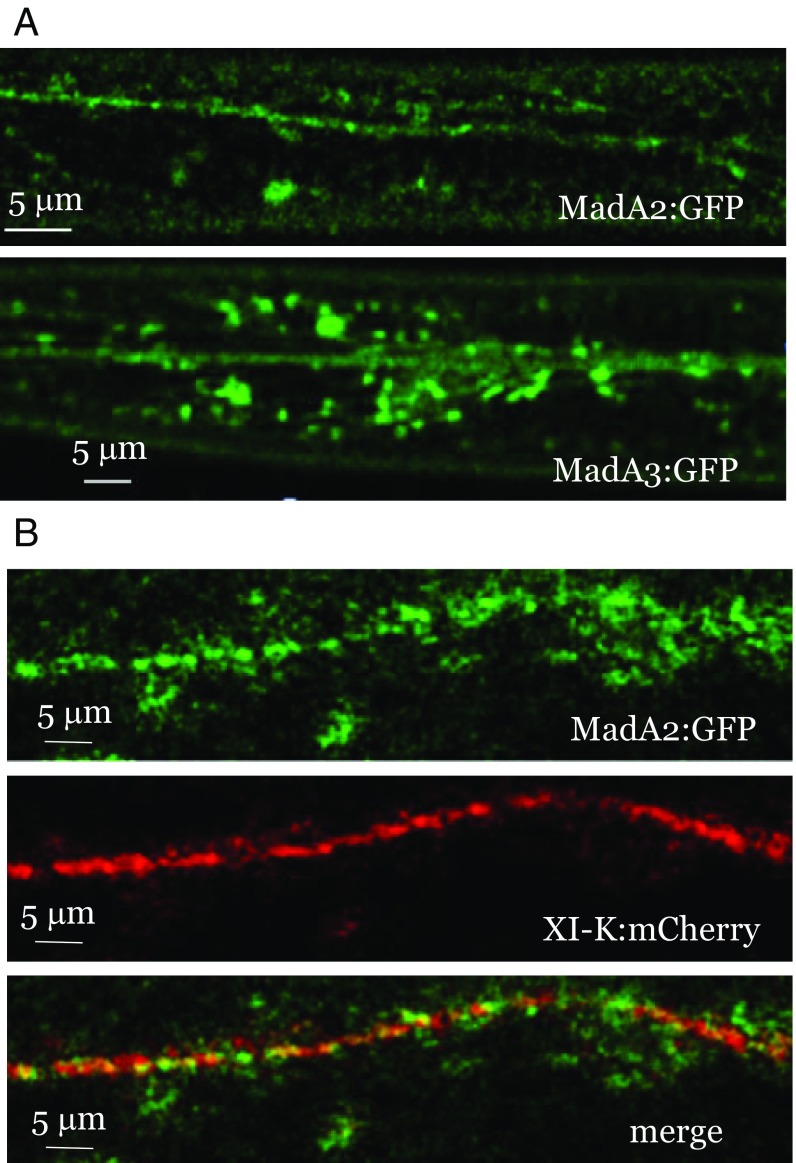
To investigate the subcellular localization of the MadA proteins that lack a predicted NLS, we tagged MadA2 and MadA3 with GFP and stably expressed the resulting constructs in Arabidopsis under control of their native promoters. Each of these proteins was found to localize to the cytosol, and specifically to two major types of fluorescent bodies: (i) those fluorescent bodies moving in linear patterns and (ii) those fluorescent bodies distributed diffusely in the cytosol (Fig. 4A and Movies S5 and S6). Stable coexpression of MadA2:GFP with XI-K:mCherry showed that most of the diffuse MadA2:GFP fluorescence did not colocalize with XI-K:mCherry. In contrast, a substantial fraction of Mad2:GFP moving in linear patterns did colocalize with XI-K:mCherry (Pearson’s coefficient of 0.58, corresponding to a statistically significant overlap; P < 0.001), indicative of its myosin-driven transport (Fig. 4B). However, a substantial fraction of the myosin did not colocalize with MadA2:GFP.
Fig. 4.
(A) Analysis of MadA2:GFP and MadA3:GFP localization stably expressed in leaf epidermal cells. (B) Colocalization of MadA2:GFP with XI-K:mCherry.
To determine the nature of the compartments associated with MadA2:GFP and MadA3:GFP, we analyzed subcellular fractions and found that each of these proteins was present primarily in the microsomal pellet, was not solubilized by 1 M NaCl, but was released from the membranes by Triton X-100 treatment (SI Appendix, Fig. S1). Thus, similar to myosin XI-K and MyoB receptors, both MadA2 and MadA3 appear to be tightly associated with vesicle-like compartments, but not with the larger organelles.
To assess the functions of the MadA proteins, we generated knockout mutants in which one, two, three, or all four of the corresponding genes were inactivated. Because none of the plant lines exhibited noticeable phenotypic differences from the Columbia control, we limited quantitative analysis to the quadruple-knockout (4KO) mutant: mada1 mada2 mada3 mada4. Neither the plant height nor the leaf rosette span of the mutant plants was significantly different from the Columbia plants (P > 0.05; n = 70; SI Appendix, Dataset S1). Analysis of the root hair length showed no significant difference between the 4KO mutant and Columbia plants either (the mean root hair lengths were 308 ± 104 μm for Columbia and 327 ± 111 μm for 4KO lines; P > 0.05; n = 365). Thus, it was concluded that the MadA protein family plays no major role in plant growth, at least under optimal conditions.
Membrane-Associated MadB1 Contributes to Polar Growth of Root Hairs.
The subcellular fractionation analysis showed that, similar to MadA2 and MadA3, MadB1 was tightly associated with membranes (SI Appendix, Fig. S1). Next, the MadB1 gene under control of its native promoter was tagged by inserting the GFP coding sequence, and the resulting MadB1:GFP was stably expressed along with myosin XI-K:mCherry. Again, the subcellular distribution of this protein was similar to MadA2:GFP and MadA3:GFP: The green fluorescence was observed in motile vesicles moving in linear patterns, as well as in diffuse cytosolic material (Fig. 5A and Movie S7). Colocalization analysis showed partial overlap with myosin XI-K:mCherry in linear arrays along F-actin bundles but not outside of these arrays (Fig. 5A). Quantitative analysis validated a statistically significant (P < 0.001) partial overlap between two fluorophores, with a Pearson’s coefficient of 0.69.
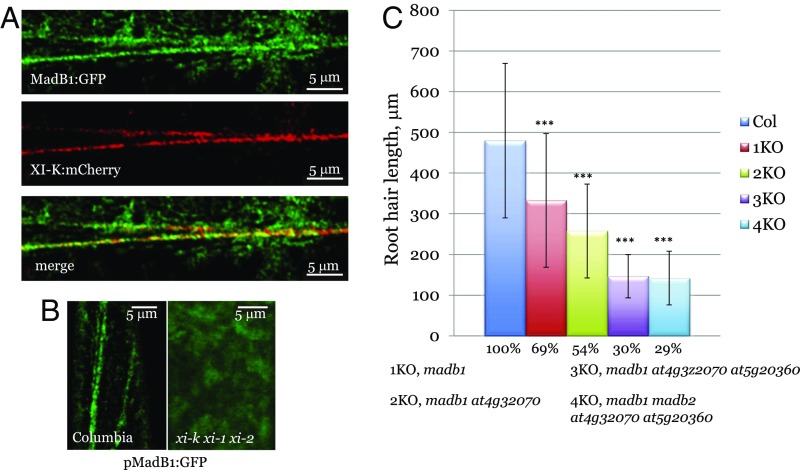
Fig. 5.
Analysis of MadB1:GFP localization and function of MadB proteins in root hair growth. (A) Colocalization of MadB1:GFP with XI-K:mCherry. (B) Localization of MadB1:GFP in Columbia and myosin triple-knockout (xi-k xi-1 xi-2) mutant plants. (C) Analysis of root hair length in Columbia (Col) plants and in gene knockout plants in which from one (1KO) to four (4KO) genes encoding pMad family B proteins were inactivated. The genotypes of the 1KO–4KO plants are shown below the graph. Asterisks (***) denote P < 0.001 for pairwise comparisons between Columbia and each of the mutant lines.
To evaluate the functional significance of the interactions between myosin XI-K and MadB1, we transformed the MadB1:GFP gene into the triple-myosin knockout mutant xi-k xi-1 xi-2 plants (9). As shown in Fig. 5B, the MadB1:GFP in such plants was no longer present in motile vesicles, but rather appeared as diffuse fluorescence, demonstrating that transport of MadB1:GFP-associated compartment is myosin-dependent.
To explore the functions of MadB1 and its three paralogs in Arabidopsis growth, we generated gene knockout lines with the following genotypes: madb1 (1KO), madb1 at4g32070 (2KO), madb1 at4g32070 at5g20360 (3KO), and madb1 at4g32070 at5g20360 madb2 (4KO). Phenotypic analysis of the plant height and leaf rosette diameter did not reveal any significant differences from the wild-type plants (SI Appendix, Dataset S1). In contrast, analysis of the root hair growth demonstrated statistically significant differences for each mutant (Fig. 5C). Importantly, the reduction in root hair growth was progressive: 69% of the wild type for 1KO, 54% for 2KO, and 30% for 3KO, suggesting additive functions of the MadB proteins in the same genetic pathway. Because the length of root hairs in 4KO was only marginally lower than in 3KO, the fourth member of the MadB family appears to be functionally redundant. Given that root hair growth is a sensitive functional indicator of the myosin activity, the finding that MadB proteins phenocopied myosin knockout mutants further supported the functional association of myosins with these putative myosin adaptors.
Evolution of the Myosin-Driven Transport Network in Plants.
To characterize the evolution of the emerging multicomponent, myosin-driven vesicular transport system, we updated our previous analysis of the major components of the myosin transport network, namely, myosins XI and MyoB receptors, as well as MadA1–4 and MadB1–4 adaptors, using the available databases of plant genomes (Materials and Methods). The patterns of representation of these families in major lineages of green plants (Viridiplantae) are shown in SI Appendix, Dataset S2.
In agreement with previous findings (21), class XI myosins are represented in all lineages of land plants and green algae. However, the burst of duplications that gave rise to the five myosin XI subclasses that are conserved in extant monocots and eudicots (XI F, G, I, J, and K) most likely occurred in their common ancestral lineage after the divergence from gymnosperms (Acrogymnospermae). This evolutionary scenario is fully compatible with the fact that Amborella trichopoda, a basal member of the flowering plant lineage Magnoliophyta, possesses a single myosin of each myosin XI subclass (SI Appendix, Datasets S2 and S3).
The distribution of the MyoB receptor family is somewhat more limited: At least two MyoB genes are present in the incompletely sequenced genome of Nitella mirabilis of Charophyta, the lineage of freshwater green algae that comprise the sister group to terrestrial plants (19), but not in Chlorophyta. In striking parallel to the myosin phylogeny, A. trichopoda has a single representative in most branches of the MyoB family (SI Appendix, Datasets S2 and S4). These findings indicate that the expansion of myosins in the common ancestor of monocots and dicots was accompanied by the concomitant expansion of the myosin receptors of the MyoB family.
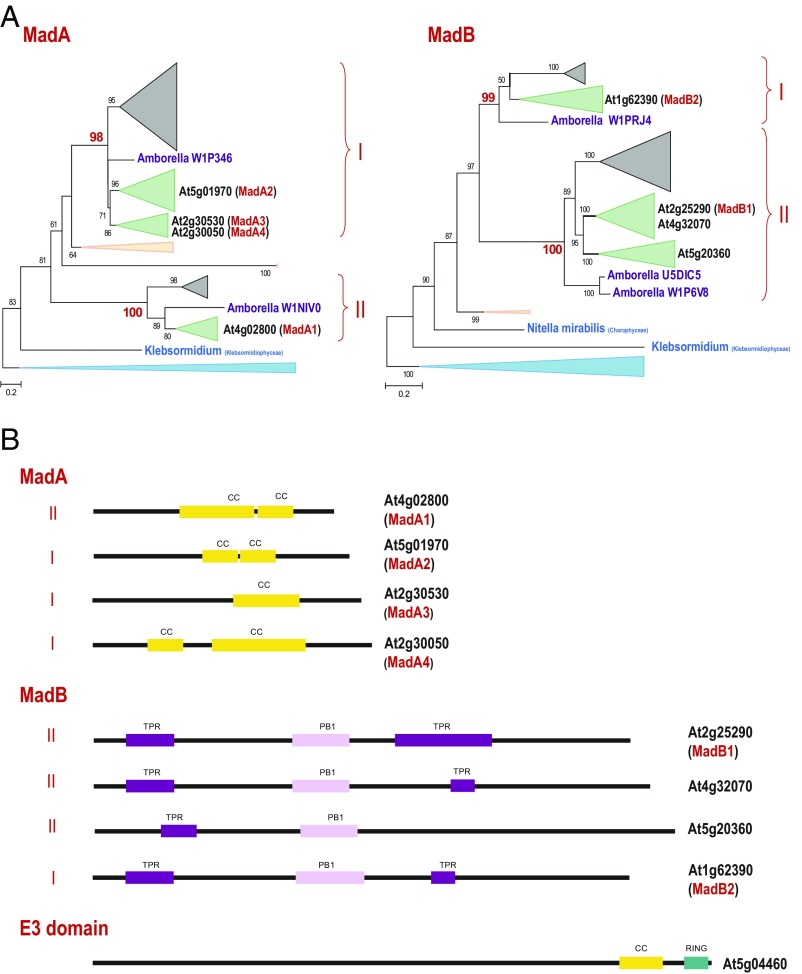
Another family of myosin interactors identified in this work, MadA, also shows a clear expansion in monocots and dicots, although not as pronounced as the expansion of the MyoB family. We detected representatives of this family in most lineages of terrestrial plants and green algae, both Chlorophyta and Charophyta (SI Appendix, Dataset S2). Phylogenetic analysis identified two major branches of this family that are represented by several paralogs in all Magnoliophyta (Fig. 6A and SI Appendix, Dataset S5). Because the nucleus-localized Arabidopsis MadA1 belongs to one major branch of the tree, whereas cytosolic MadA2–4 belong to another, it appears that the functional divergence between these branches occurred in the common ancestor of flowering plants. In agreement with this inference and recapitulating the pattern observed with the myosins and the MyoB family, A. trichopoda encodes a single protein in each branch (Fig. 6A and SI Appendix, Dataset S5). Apparently, branch II of the MadA family evolved faster than branch I, so that the grouping of branch I with Bryophyta is most likely due to short branch attraction. Notwithstanding the overall low sequence conservation, most of the MadA proteins share a similar organization, with the only conserved feature being the coiled-coil central region (Fig. 6B).
Fig. 6.
Phylogenies of the MadA and MadB families and sequence features of the plant myosin interactors. (A) Maximum-likelihood unrooted trees of the MadA and MadB families. Bootstrap values (percentage points) are shown for all internal branches. The key bootstrap values supporting subfamilies are shown in red. The collapsed branches are shown by triangles according to the respective plant taxa, as follows: moss and spikemoss in light peach; monocots in gray; dicots in green; Chlorophyta in light blue; Amborella, a basal Magnoliophyta plant, in purple; and Klebsormidium, a filamentous terrestrial alga, in teal blue. For A. thaliana, the gene identifiers are highlighted (bold). The myosin-interacting proteins are shown in red in parentheses. The complete trees in the Newick format are provided in SI Appendix as Datasets S5 (MadA) and S6 (MadB). (B) Sequence features of the putative myosin adaptor families. The lengths of the proteins and identified domains, presented as boxes, are drawn roughly to scale. CC, coiled-coil domain; PR1, PR1 domain; RING, RING finger (E3-like domain); TPR, TPR repeats.
The MadB family shows a distribution and phylogenetic tree topology that are closely similar to the MadA family, despite completely different domain architectures (Figs. 6 A and B). Indeed, we identified MadB proteins in the same set of plant clades as the MadA proteins (SI Appendix, Dataset S2). Furthermore, the MadB family also forms two major branches in Magnoliophyta, with branch II evolving faster and branch I accordingly grouping with Bryophyta and A. trichopoda, which, respectively, possess only one and two representatives in these branches (Fig. 6A and SI Appendix, Dataset S6). Thus, similar to the MadA family, the first duplication in the MadB family most likely occurred at the same stage in plant evolution, namely, in the common ancestor of Magnoliophyta.
Our phylogenetic analysis suggests that a primitive version of the myosin transport network, including myosins XI, MadA, and MadB adaptors, was already operative in the common ancestor of green algae and land plants. In contrast, the MyoB receptor family apparently emerged in the lineage that included the common ancestor of terrestrial plants and freshwater characean green algae. Regardless of the exact provenance of MyoB, the flourishing of plants during land colonization was marked by an extensive, concomitant expansion of myosins XI and the MyoB, MadA, and MadB families of myosin receptors and adaptors. This evolutionary scenario reflects the tight coevolution of myosins and their receptors and adaptors, which form a dense interaction network (Fig. 1B).
Discussion
The defining cellular feature of the green branch of eukaryotic life (Plantae or Archaeplastida) that includes glaucophytes, euglenids, green algae, and land plants is the presence of chloroplasts that were acquired through Cyanobacteria endosymbiosis over a billion years ago (29, 30). Arguably, the next most prominent feature of the plant cells are the remarkably vigorous intracellular dynamics paraded in cytoplasmic streaming that reach velocities of 4 μm⋅s−1 in higher plants and 50 μm⋅s−1 in green algae (17, 31). In contrast, only much slower cytoplasmic streaming (∼0.4 μm⋅s−1) has been described in certain animal cell types (e.g., mouse oocytes) (32).
The progress in understanding the mechanism of cytoplasmic streaming that was discovered over two centuries ago and later attributed to myosin XI motors can be roughly divided into the following phases: (i) biochemical and biophysical studies of myosins XI (3), (ii) elucidation of the roles of individual myosins in cell dynamics and expansion and plant development (6–10, 13, 17, 33–35), and (iii) identification of the MyoB receptors and their functions in cytoplasmic streaming and cell and plant growth (14, 19). The major findings of the present work seem to mark the beginning of the next phase, the system analysis of the entire transport network driven by myosins XI.
First, we identified all pairwise interactions between five myosins XI and 11 MyoB receptors, representing the evolutionary and structural diversity of these proteins in Arabidopsis (Fig. 1A and Table 1), and showed that myosin binding is indeed a common property of MyoBs. The myosin–MyoB interaction network is dominated by generalists (i.e., myosins, MyoBs) that each bind many of their respective protein partners (Fig. 1B). These results are compatible with the functional redundancies of myosins XI and MyoBs (8, 9, 14, 19), and they attest to the biological robustness of the myosin-driven transport network in plants.
Second, we showed that structurally diverse MyoB12–14 are each tightly associated with vesicle-like, motile compartments (Fig. 2), similar to those compartments associated with MyoB1, MyoB2, and MyoB7 (14). Among these myosins, MyoB13 and MyoB14 contain a predicted TM, whereas MyoB12 does not, suggesting that its tight membrane association occurs through anchoring to a distinct TM protein. Interestingly, although the mean diameter of the motile vesicles associated with MyoB14 was larger than the mean diameter of the motile vesicles formed by other MyoBs (Fig. 2B), these MyoB14 vesicles were unaffected by BFA, again similar to other MyoB compartments that function outside the ER-to-Golgi-to-plasma membrane secretory pathway (Fig. 2C). These and previously published (14) results imply that association with the specialized transport vesicles is a common property of diverse MyoBs. Importantly, none of the six MyoBs studied so far was targeted to larger organelles, in agreement with the model proposing that MyoB vesicles drive cytoplasmic streaming that carries passively moving organelles (14, 19). In addition to their general roles in intracellular motility and cell growth, certain MyoBs are involved in more specialized processes. Thus, a pollen-specific MyoB has been implicated in pollen tube growth in tobacco (36), whereas maize MyoBs have been found in association with protein bodies specific to maize endosperm (37, 38).
Third, we identified two previously uncharacterized families of myosin XI-binding proteins, putative myosin adaptors dubbed MadA and MadB (Table 1). Of these adaptors, MadA2, MadA3, and MadB1 were localized to vesicle-like compartments despite the absence of predicted TM (Figs. 4 and 5), further emphasizing the roles of myosins in endomembrane transport. However, unlike MyoB receptors, which are mostly present in motile vesicles (14), compartments associated with MadA2, MadA3, or MadB1 were partitioned between motile vesicles and diffuse material (Figs. 4 and 5). This difference implies that these adaptors are only transiently associated with myosins and are likely to perform functions distinct from the functions of the MyoB receptors.
Within the myosin transport network, MadA1–4 and MadB1, as well as myosin-binding E3 ligase and WIT2 (Table 1), exhibited the same generalist/specialist pattern as myosins XI and MyoB receptors (Fig. 1B). The network partitions into three groups of myosins. The first group consists of myosins XI-K and XI-1, which are generalists and closely related paralogs that share functions in cell growth and dynamics (8, 9, 12–14, 19). The second group includes myosins XI-F and XI-C, which are somewhat odd bed fellows, given that the latter is pollen-specific, whereas the former is not (21). However, these two myosins share highly similar network connectivity patterns and could perform analogous functions in distinct cell types. Finally, myosin XI-I is the most narrow specialist, in accord with its distinct position in the phylogenetic tree (21) and unusual functional and enzymatic properties (11, 39). Fittingly, the most specialized myosin interactor, WIT2, exclusively interacts with myosin XI-I. Thus, the network structure seems to show substantial biological relevance despite its relatively small size.
Fourth, we demonstrated that the NLS-containing myosin adaptor MadA1 is indeed nucleus-localized and apparently is extruded from the nucleus in rapidly moving particle arrays (Fig. 3 and Movie S4). This unexpected discovery suggests the existence of a previously unknown pathway of myosin-driven nuclear export in plants. This pathway might complement nuclear export via the nuclear pore complex (40) and nuclear envelope budding (41). Such MadA1-mediated export might involve MadA1 attachment to material that, upon emergence from the nuclear pore complex, would bind myosin and would be rapidly trafficked through the cytosol. The deep evolutionary conservation of the MadA1-like proteins (Fig. 6A) further strengthens the potential significance of this hypothetical plant nuclear export pathway.
Another intriguing aspect of the present results is the presence of an apparent myosin-driven transport route that traverses the nuclear interior. Despite the accumulating supportive evidence, the existence of the F-actin–myosin complexes in the nucleus remains controversial (42, 43). Although the presence of actins in the plant nucleus (44), as well as the contributions of F-actin and myosin XI-I to nucleus relocation and shape, has been described (11, 45), to our knowledge, myosin-driven transport through the nucleus has not. Although the molecular architecture of the linear arrays associated with myosin XI-K:mCherry (Fig. 3C) requires further investigation, it seems likely that these arrays reside within the transnuclear cytosolic sleeves surrounded by the membrane. If such is the case, these sleeves would be analogous to dynamic transvacuolar strands (28), the formation of which depends on actomyosin transport (13).
Fifth, we assessed the functionality of the MadA and MadB adaptors using gene knockout analysis. Unexpectedly, inactivation of MadA1–4 genes did not produce any observable phenotypic changes under optimal growth conditions (SI Appendix, Dataset S1), although their fitness under stress was not evaluated. In contrast, elimination of the MadB proteins resulted in a dramatic reduction in the polar elongation of root hairs, such that the root hair length in the triple- and quadruple-knockout plants was less than a third of the root hair length in the control (Fig. 5C). These root hair growth defects closely mimicked the defects in the myosin XI knockout mutants (8, 9), whereas analysis of MyoB gene knockouts showed little if any role of these receptors in root hair growth (19). Collectively, these findings suggest that myosins and the MadB adaptors constitute a distinct transport pathway that is specifically responsible for the root hair growth. Because the PB1 domain that is conserved in the MadB family (Fig. 6B) is typically found in the proteins involved in auxin signal transduction (27), the myosin-MadB transport pathway also might be regulated by this plant hormone.
Sixth, phylogenomic analysis of myosins XI and their receptors and adaptors reveals a near-perfect pattern of coevolution, whereby the primitive ancestral network dramatically expanded through serial duplications in the lineage, leading to the common ancestor of monocots and dicots. This extent of the network expansion implies that it was driven by adaptation of the plants to the growth on land. The functional underpinnings of this adaptive diversification of myosin-driven processes remain to be studied in detail.
Conclusions
Previous studies have identified the MyoB family of plant myosin receptors that, jointly with myosins XI, appear to mediate cytoplasmic streaming through distinct endomembrane compartments. The present work led to the identification of two distinct families of previously unknown myosin adaptors and reveals a far greater complexity of the myosin interactor network than previously suspected. The network is strikingly dense and redundant, indicative of high robustness, which is also manifest in gene knockout experiments. Unexpectedly, we identified a putative nuclear branch of myosin-driven transport that seems to rely specifically on the myosin adaptor MadA1 described here. The myosin network is clearly an ancient functional module that was already present in the common ancestor of green algae and land plants but underwent a major expansion in the latter, through several concordant duplications of the genes encoding myosins and their receptors and adaptors. Characterization of the full functional and mechanistic repertoire of this network is a major task for follow-up studies.
Materials and Methods
Yeast Two-Hybrid Assay.
The Arabidopsis sequences encoding GTDs of the myosin XI-F (I1112 to S1545) and XI-C (A1061 to G1538) were cloned into bait vector pGBKT7. Fragments of MyoB cDNA clones containing entire myosin-binding DUF593 domains but lacking TM were cloned into a prey vector pGADT7. The following amino acid sequences were selected: MyoB3 (At5g16720), G273 to A659; MyoB4 (At2g30690), G552 to N760; MyoB7 (At5g06560) D18 to V518; MyoB8 (At3g11850), E2 to V504; MyoB12 (At5g57830), V2 to L387; MyoB13 (At4g13630), V36 to A569; MyoB15 (At1g04890), T2 to S411; and MyoB16 (At1g18265), S54 to M280. A fragment of MyoB14 (At4g13160) between amino acids T37 and P282 was inserted into pGBKT7 as bait. Bait plasmids harboring myosin XI-K, XI-1, and XI-I GTDs; prey plasmids carrying fragments of the MyoB1 and MyoB2 cDNAs; and protocols for library screens have been described previously (14). Pairwise interactions between myosin GTDs and MyoBs were assayed by growth on triple-dropout selective medium minus Trp/Leu/His supplemented with 10 mM 3-amino-1,2,4-triazole. Yeast cells cotransformed with the empty pGBT9 vector and MyoB constructs, or a prey vector expressing GFP in combination with the GTD-carrying bait plasmids, were used as negative controls. Transformed yeast cells were grown in a liquid medium lacking Trp and Leu and diluted to OD600 = 0.1 before plating onto selective medium. The plates were incubated for 7 d at 30 °C before screening and imaging.
Subcellular Distribution of MyoB Receptors and Mad Adaptors.
Genomic fragments encoding MyoB8, MyoB12, MyoB13, MyoB14, MyoB15, and MyoB16, including their promoter regions, were PCR-amplified using KOD Hot Start high-fidelity DNA polymerase (Novagen) and cloned into a modified pMDC32 plasmid using the SbfI and AscI sites. The GFP cDNA was added downstream from the MyoB ORF between the AcsI and PacI sites. GFP-tagged MadA1, MadA2, MadA3, and MadB1 were also generated using the same approach. In addition, MadA1 was GFP-tagged at its N terminus by inserting a GFP ORF between the first and second codons using overlapping PCR assays. The resulting plasmids were mobilized into Agrobacterium GV3101 for floral dipping transformation of Arabidopsis plants. T1 seeds were plated on 0.6% Phytagar containing half-strength MS medium supplemented with 2% (wt/vol) sucrose and 50 μg/mL Hygromycin B. Seeds collected from the T1 plants were plated on the same medium supplemented with 25 μg/mL Hygromycin B, and the surviving T2 seedlings were transplanted into soil and grown for 2 wk. Rosette leaves from these plants were harvested and ground in 3 vol (wt/vol) of grinding buffer [20 mM Hepes (pH 7.2), 50 mM KOAc, 1 mM EDTA, 1 mM EGTA, 250 mM sorbitol, 2 mM DTT and 1× Plant Protease Inhibitor mixture (Amresco)] on ice. Homogenates were filtered through Miracloth (Calbiochem) and centrifuged at 3,000 × g for 10 min. Supernatants were loaded onto the 20% (wt/vol) sucrose cushion in the same buffer and centrifuged at 100,000 × g for 1 h. The 100,000 × g pellets containing microsomes were resuspended in the grinding buffer containing either 1 M NaCl or 1% Triton X-100. Samples were incubated for 30 min and centrifuged at 100,000 × g for 30 min to separate solubilized and treatment-resistant proteins. Samples for immunoblot assays were boiled in SDS sample buffer and analyzed using SDS/PAGE [10% (wt/vol) acrylamide]. The polyclonal antibodies used for immunodetection were specific to GFP (Rockland Immunochemicals), myosin XI-K (7), a cytosolic marker fructose-1,6 bisphosphatase (cFBPase; Agrisera), and ER lumen-resident binding protein (BiP).
BFA Treatment and Confocal Microscopy.
The T2 Arabidopsis lines expressing GFP-tagged proteins were imaged by confocal laser scanning microscopy. BFA treatment and imaging were done as described (14). To observe both myosin XI-K and its counterpart concurrently under physiological conditions, pMDC-based dual-expression cassettes harboring both mCherry-tagged myosin XI-K and a GFP-tagged myosin interactor were made as described (14). The resulting plasmids were used for transformation of the Arabidopsis homozygous xi-k knockout line SALK_067972 (7).
Pull-Down Assays.
Bacterial vectors engineered to express MBP fused either to β-glucuronidase (GUS) or myosin XI-K GTD were described previously (14). The pET expression vector (EMD Millipore) was modified to add a FLAG epitope tag at the C terminus of each expressed protein. The cDNA copies of the MadA1, MadA3, MadB1, MadB2, or GFP ORF were cloned into the modified vector, and the resulting constructs were mobilized into Rosetta(DE3)pLysS cells for protein expression in Escherichia coli. The liquid bacterial cultures were grown for 16 h at 28 °C in the presence of 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested by centrifugation, resuspended in a protein interaction buffer [20 mM Hepes-KOH, 50 mM potassium acetate, 1 mM EDTA, 1 mM EGTA, 5% (vol/vol) glycerol, 1% Triton X-100 (pH 7.2)], supplemented with 1 mg/mL lysozyme, and frozen/thawed once to initiate cell lysis. Cells were sonicated for 1 min, and the lysates were centrifuged for 10 min at 10,000 × g. All of the procedures were carried out on ice. Supernatants of the MBP::GUS or MBP::XIK-GTD were mixed with amylose resin (New England Biolabs) and incubated for 1 h. After washing the matrix with the protein interaction buffer, supernatants harboring the putative, myosin-interacting, recombinant proteins were added to the beads and incubated for 1 h. After washing with the protein interaction buffer, beads were boiled in SDS/PAGE loading buffer and analyzed by immunoblotting using FLAG-specific antibodies.
Generation and Phenotypic Characterization of the Gene Knockout Mutants.
Arabidopsis mutants used for this study are in the Columbia ecotype background. T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH); homozygous mutant plant lines were identified by PCR-based genotyping, and their null phenotypes were validated by end-point RT-PCR. The Arabidopsis lines SALK_108010 (AT4G02800; MadA1), SALK_014695 (AT5G01970; MadA2), SALK_144872 (AT1G30050; MadA3), and SK24834 (CS1007495; AT2G30530; MadA4), respectively, were used to construct a quadruple-knockout mutant mada1 mada2 mada3 mada4. Single-, double-, triple-, and quadruple-knockout lines of the MadB family were generated by obtaining and crossing the following Arabidopsis lines: CS864474 (AT2G25290; MadB1), SALK_001325 (AT4G32070), CS384293 (SALK_088673), and SALK_088673 (AT1G62390; MadB2). Plant heights and rosette diameters were measured as described by Peremyslov et al. (19) (n = 32). Root hair growth was analyzed in accord with the method of Peremyslov et al. (7). Typically, 200–300 root hairs were measured for each mutant.
Bioinformatics.
The modularity of the bipartite network comprising myosins and myosin-binding proteins was assessed using MODULAR software (46), which employs simulated annealing to assign nodes to modules in a way that maximizes the Barber’s bipartite modularity index. The nestedness was quantitatively evaluated as the magnitude of the leading eigenvalue of the adjacency matrix (47). The significance of the modularity and nestedness indices was determined by comparing the values obtained for the analyzed network with the values of 200 random bipartite networks with the same degree of distribution.
The PSI-BLAST program (48) with default parameters was used to search for myosin XI, MyoB, MadA, and MadB family proteins in the following databases: National Center for Biotechnology Information nonredundant database, UniProt, and the Joint Genome Institute database. The MUSCLE program (49) with default parameters was used to generate multiple sequence alignments. Incomplete sequences were discarded before further analysis. Conserved blocks of alignments were used for phylogenetic tree reconstruction. The maximum-likelihood phylogenetic tree was built using the FastTree program (50), with the Whelan and Goldman (WAG) substitution matrix and gamma-distributed evolutionary rates. The same program was used to compute bootstrap values. The search for conserved protein domains was performed using the CDD-search (51). The TMHMM (52) and Marcoil (53) programs with default parameters were used for prediction of TMs and coiled-coil regions, respectively.
Supplementary Material
Acknowledgments
We thank Prof. Viktor Zarsky for critical reading of the manuscript and helpful suggestions. V.V.D. is a Fellow, Scientific Visitors Program, National Center for Biotechnology Information, NIH. The work in the V.V.D. laboratory was supported, in part, by US-Israel Binational Science Foundation Award 2013084. The research in group of E.V.K. is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1756.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620577114/-/DCSupplemental.
References
- 1.Shimmen T. The sliding theory of cytoplasmic streaming: Fifty years of progress. J Plant Res. 2007;120(1):31–43. doi: 10.1007/s10265-006-0061-0. [DOI] [PubMed] [Google Scholar]
- 2.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103(10):3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tominaga M, Ito K. The molecular mechanism and physiological role of cytoplasmic streaming. Curr Opin Plant Biol. 2015;27:104–110. doi: 10.1016/j.pbi.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Ueda H, Tamura K, Hara-Nishimura I. Functions of plant-specific myosin XI: From intracellular motility to plant postures. Curr Opin Plant Biol. 2015;28:30–38. doi: 10.1016/j.pbi.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Geitmann A, Nebenführ A. Navigating the plant cell: Intracellular transport logistics in the green kingdom. Mol Biol Cell. 2015;26(19):3373–3378. doi: 10.1091/mbc.E14-10-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ojangu EL, Järve K, Paves H, Truve E. Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma. 2007;230(3-4):193–202. doi: 10.1007/s00709-006-0233-8. [DOI] [PubMed] [Google Scholar]
- 7.Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 2008;146(3):1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokhnevsky AI, Peremyslov VV, Dolja VV. Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA. 2008;105(50):19744–19749. doi: 10.1073/pnas.0810730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peremyslov VV, Prokhnevsky AI, Dolja VV. Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell. 2010;22(6):1883–1897. doi: 10.1105/tpc.110.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojangu EL, et al. Myosins XI-K, XI-1, and XI-2 are required for development of pavement cells, trichomes, and stigmatic papillae in Arabidopsis. BMC Plant Biol. 2012;12:81. doi: 10.1186/1471-2229-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, et al. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol. 2013;23(18):1776–1781. doi: 10.1016/j.cub.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Peremyslov VV, Klocko AL, Fowler JE, Dolja VV. Arabidopsis myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Front Plant Sci. 2012;3:184. doi: 10.3389/fpls.2012.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda H, et al. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA. 2010;107(15):6894–6899. doi: 10.1073/pnas.0911482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peremyslov VV, et al. Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell. 2013;25(8):3022–3038. doi: 10.1105/tpc.113.113704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell. 2011;20(1):47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer JA, 3rd, Sellers JR. Walking to work: Roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2011;13(1):13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 17.Tominaga M, et al. Cytoplasmic streaming velocity as a plant size determinant. Dev Cell. 2013;27(3):345–352. doi: 10.1016/j.devcel.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Stefano G, Renna L, Brandizzi F. The endoplasmic reticulum exerts control over organelle streaming during cell expansion. J Cell Sci. 2014;127(Pt 5):947–953. doi: 10.1242/jcs.139907. [DOI] [PubMed] [Google Scholar]
- 19.Peremyslov VV, Cole RA, Fowler JE, Dolja VV. Myosin-powered membrane compartment drives cytoplasmic streaming cell expansion and plant development. PLoS One. 2015;10(10):e0139331. doi: 10.1371/journal.pone.0139331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodhouse FG, Goldstein RE. Cytoplasmic streaming in plant cells emerges naturally by microfilament self-organization. Proc Natl Acad Sci USA. 2013;110(35):14132–14137. doi: 10.1073/pnas.1302736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peremyslov VV, et al. Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol. 2011;155(3):1191–1204. doi: 10.1104/pp.110.170720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weitz JS, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21(2):82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14(6):382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 25.Yau RG, et al. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev Cell. 2014;28(5):520–533. doi: 10.1016/j.devcel.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grove TZ, Cortajarena AL, Regan L. Ligand binding by repeat proteins: Natural and designed. Curr Opin Struct Biol. 2008;18(4):507–515. doi: 10.1016/j.sbi.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilfoyle TJ. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell. 2015;27(1):33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oda Y, Higaki T, Hasezawa S, Kutsuna N. Chapter 3. New insights into plant vacuolar structure and dynamics. Int Rev Cell Mol Biol. 2009;277:103–135. doi: 10.1016/S1937-6448(09)77003-0. [DOI] [PubMed] [Google Scholar]
- 29.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delwiche CF, Cooper ED. The evolutionary origin of a terrestrial flora. Curr Biol. 2015;25(19):R899–R910. doi: 10.1016/j.cub.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 31.Ito K, et al. Kinetic mechanism of the fastest motor protein, Chara myosin. J Biol Chem. 2007;282(27):19534–19545. doi: 10.1074/jbc.M611802200. [DOI] [PubMed] [Google Scholar]
- 32.Ganguly S, Williams LS, Palacios IM, Goldstein RE. Cytoplasmic streaming in Drosophila oocytes varies with kinesin activity and correlates with the microtubule cytoskeleton architecture. Proc Natl Acad Sci USA. 2012;109(38):15109–15114. doi: 10.1073/pnas.1203575109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 2008;146(3):1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avisar D, Abu-Abied M, Belausov E, Sadot E. Myosin XIK is a major player in cytoplasm dynamics and is regulated by two amino acids in its tail. J Exp Bot. 2012;63(1):241–249. doi: 10.1093/jxb/err265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto K, et al. Regulation of organ straightening and plant posture by an actin-myosin XI cytoskeleton. Nat Plants. 2015;1(4):15031. doi: 10.1038/nplants.2015.31. [DOI] [PubMed] [Google Scholar]
- 36.Stephan O, et al. RISAP is a TGN-associated RAC5 effector regulating membrane traffic during polar cell growth in tobacco. Plant Cell. 2014;26(11):4426–4447. doi: 10.1105/tpc.114.131078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, et al. Comprehensive proteomic analysis of developing protein bodies in maize (Zea mays) endosperm provides novel insights into its biogenesis. J Exp Bot. 2016;67(22):6323–6335. doi: 10.1093/jxb/erw396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holding DR, et al. The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell. 2007;19(8):2569–2582. doi: 10.1105/tpc.107.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haraguchi T, Tominaga M, Nakano A, Yamamoto K, Ito K. Myosin XI-I is mechanically and enzymatically unique among class-XI myosins in Arabidopsis. Plant Cell Physiol. 2016;57(8):1732–1743. doi: 10.1093/pcp/pcw097. [DOI] [PubMed] [Google Scholar]
- 40.Adams RL, Wente SR. Uncovering nuclear pore complexity with innovation. Cell. 2013;152(6):1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fradkin LG, Budnik V. This bud’s for you: Mechanisms of cellular nucleocytoplasmic trafficking via nuclear envelope budding. Curr Opin Cell Biol. 2016;41:125–131. doi: 10.1016/j.ceb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarshad AA, Percipalle P. New insight into role of myosin motors for activation of RNA polymerases. Int Rev Cell Mol Biol. 2014;311:183–230. doi: 10.1016/B978-0-12-800179-0.00004-0. [DOI] [PubMed] [Google Scholar]
- 43.Hendzel MJ. The F-act’s of nuclear actin. Curr Opin Cell Biol. 2014;28:84–89. doi: 10.1016/j.ceb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Kandasamy MK, McKinney EC, Meagher RB. Differential sublocalization of actin variants within the nucleus. Cytoskeleton. 2010;67(11):729–743. doi: 10.1002/cm.20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Groves NR, Meier I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus. 2015;6(2):144–153. doi: 10.1080/19491034.2014.1003512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquitti FMD, Guimaraes PR, Pries MM, Bittencourt LF. Modular: Software for the autonomous computation of modularity in large network sets. Ecography. 2014;37:221–224. [Google Scholar]
- 47.Staniczenko PP, Kopp JC, Allesina S. The ghost of nestedness in ecological networks. Nat Commun. 2013;4(1391):1391. doi: 10.1038/ncomms2422. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 53.Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18(4):617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.