Significance
Endoplasmic reticulum (ER) stress is linked to the development of complex metabolic diseases, including diabetes, obesity, and atherosclerosis. Irremediable ER stress can push the unfolded protein response (UPR) toward proinflammatory and proapoptotic signaling. The need to dissociate the adaptive UPR responses from its destructive outputs has become a major challenge for therapeutic strategies aimed at mitigating ER stress that is often observed in chronic diseases. Our findings show that inositol-requiring enzyme 1 (IRE1) plays a critical role in metaflammation and that administering IRE1-specific inhibitors to hyperlipidemic mice counteracts atherosclerosis progression.
Keywords: endoplasmic reticulum stress, unfolded protein response, metaflammation, lipotoxicity, atherosclerosis
Abstract
Metaflammation, an atypical, metabolically induced, chronic low-grade inflammation, plays an important role in the development of obesity, diabetes, and atherosclerosis. An important primer for metaflammation is the persistent metabolic overloading of the endoplasmic reticulum (ER), leading to its functional impairment. Activation of the unfolded protein response (UPR), a homeostatic regulatory network that responds to ER stress, is a hallmark of all stages of atherosclerotic plaque formation. The most conserved ER-resident UPR regulator, the kinase/endoribonuclease inositol-requiring enzyme 1 (IRE1), is activated in lipid-laden macrophages that infiltrate the atherosclerotic lesions. Using RNA sequencing in macrophages, we discovered that IRE1 regulates the expression of many proatherogenic genes, including several important cytokines and chemokines. We show that IRE1 inhibitors uncouple lipid-induced ER stress from inflammasome activation in both mouse and human macrophages. In vivo, these IRE1 inhibitors led to a significant decrease in hyperlipidemia-induced IL-1β and IL-18 production, lowered T-helper type-1 immune responses, and reduced atherosclerotic plaque size without altering the plasma lipid profiles in apolipoprotein E-deficient mice. These results show that pharmacologic modulation of IRE1 counteracts metaflammation and alleviates atherosclerosis.
Complex molecular interactions between environment, diet, and genetics that take place at the metabolic and immune interface provoke a low-grade, chronic inflammatory response—metaflammation—that engages cells of the immune system (macrophages, neutrophils, and lymphocytes) and metabolic tissues (adipocytes, hepatocytes, and pancreatic cells) (1). An important primer for metaflammation is chronic metabolic overloading of organelles, such as the endoplasmic reticulum (ER) and mitochondria, which results in impairment of their functions (2).
The ER serves essential cellular functions that include the synthesis and folding of secreted and transmembrane proteins, calcium storage, and lipid synthesis for membrane biogenesis or energy storage. Disruption of any of these functions leads to ER stress and the subsequent activation of an elaborate network of adaptive responses, collectively known as the unfolded protein response (UPR) (3). The UPR reestablishes homeostasis through both transcriptional and translational layers of control. The UPR signals through three mechanistically distinct branches that are initiated by the ER-resident protein folding sensors inositol-requiring enzyme 1 (IRE1), protein kinase RNA-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) (3).
IRE1 controls the phylogenetically most conserved branch of the UPR found from fungi to metazoans. It has an ER-lumenal sensor domain that recognizes unfolded peptides and cytosolic kinase and endoribonuclease (RNase) domains that relay the information to downstream effectors (3). On sensing unfolded proteins, IRE1 oligomerizes and trans-autophosphorylates, thereby activating its RNase function. Metazoan IRE1 possesses two functional outputs dependent on its RNase activity. (i) It initiates a nonconventional splicing reaction that processes the mRNA encoding X-box binding protein-1 (Xbp1) to allow the translation of its active form, XBP1s, a potent transcription factor that, together with ATF6, drives expression of numerous genes, including those encoding ER-resident chaperones and ER-associated protein degradation machinery (3). (ii) IRE1 selectively degrades ER-bound mRNAs in a process known as regulated inositol-requiring enzyme 1-dependent decay (RIDD) to alleviate ER load (4). By these mechanisms, UPR activation reinstates homeostasis.
Increased ER stress and activation of the UPR are well-documented in atherosclerosis (5). Many metabolic cardiovascular risk factors observed in obesity, including hyperglycemia, hypercholesterolemia, and elevated saturated fats, can induce ER stress in all stages of atherogenesis, the process leading to the development of atherosclerotic plaques. During atherogenesis, a maladaptive inflammatory response is initiated by the deposition of cholesterol-rich lipoproteins in the subendothelial layer of arterial walls (6). Signs of ER stress are most prominent in the atherosclerosis-prone regions of vascular lesions, such as the branching points of arteries, and typically observed in macrophages—among other immune cells—infiltrating these regions (7, 8). Chronic, irremediable ER stress triggers apoptosis in macrophages, contributing to the growth of the necrotic core that is observed in atherosclerotic plaques, which leads to a subsequent reduction in plaque stability, promoting their rupture (5).
Work over the last decade has pinpointed ER stress as a driving force for atherosclerosis progression (5, 7–10). For example, inhibiting the apoptotic signaling downstream of ER stress through genetic deletion of the proapoptotic transcription factor CCAAT box binding enhancer homologous protein (CHOP) or the signal transducer c-jun N-terminal kinase (JNK) blocks atherosclerosis progression (5, 11–13). Moreover, modulation of ER stress by using chemical chaperones alleviates atherosclerosis, further supporting the idea that therapeutic modulation of ER function is a promising avenue to combat atherosclerosis (14–16). Precedence for this notion is provided by pharmacological approaches to improve ER function—and thereby, limit ER stress—that include autophagy inducers, antioxidants, and regulators of ER calcium homeostasis (17–19).
Recent drug discovery efforts have opened the door to approaches that entail selective modulation of UPR signaling. This quest has led to the identification of several new small molecules that target the enzymatic activities of specific UPR regulators (19–23). Specifically, blocking IRE1 or XBP1s function has been shown to be beneficial for restraining tumor progression in mouse models (21, 24), highlighting that the specific targeting of the UPR can have beneficial impact in disease.
Several lines of evidence support the notion that selective pharmacological targeting of IRE1 is a desirable therapeutic approach for treatment of atherosclerosis. First, a profound increase in IRE1 phosphorylation and XBP1s expression is observed in atherosclerotic plaques of mice and humans (8, 10). Second, mechanical sheer stresses activate IRE1, whereas cardiovascular disease risk factors, such as oxidized phospholipids and homocysteine, induce both IRE1 and PERK (5, 25–29). Third, experimentally sustained Xbp1 mRNA splicing in the vessel wall promotes atherosclerosis, whereas its ablation ameliorates hypercholesterolemia in obese or apolipoprotein E-deficient (ApoE−/−) mice (10, 30). Two small molecules, STF-083010 and 4µ8C, which selectively inhibit IRE1’s RNase function, have been used in cells and animals to produce favorable therapeutic outcomes in other disease settings: STF-083010 reduced growth of multiple myeloma (21, 24, 31, 32), and 4µ8c suppressed inflammation in a murine arthritis model (33). The action of both compounds is well-understood mechanistically. Both form a Schiff base with a specific lysine positioned in the active site of the IRE1 RNase domain, blocking its function, and both show no overt toxicity when administered systemically (21, 32, 33). Thus, we reasoned that these drugs might have therapeutic applicability in atherosclerosis.
Here, we investigated the direct contribution of IRE1 to lipid-induced inflammation and atherosclerotic disease progression by administering these two IRE1 inhibitors to macrophages and ApoE−/− mice on a Western-type (high-fat and high-cholesterol) diet. Our results substantiate the concept that specific pharmacological modulation of IRE1 counteracts metaflammation and yields therapeutic gains in atherosclerotic disease, warranting additional validation in human disease.
Results
Proatherogenic Genes Are Regulated by IRE1.
To understand the contributions of IRE1 in atherogenesis, we first analyzed the impact of its inhibition on the transcriptome of macrophages using RNA sequencing (RNA-seq). ER poisons, such as tunicamycin, which inhibits protein N-linked glycosylation in the ER lumen, or thapsigargin, which disrupts ER calcium reuptake, activate all UPR branches simultaneously, impeding the dissection of the signaling contributions of individual UPR branches. By refraining from using these pleiotropic drugs, we aimed to identify the specific IRE1-regulated gene expression changes. To this end, we probed the transcriptional response to acute inhibition of IRE1 using STF-083010 in primary mouse bone marrow-derived macrophages (BMDMs) (34, 35). We analyzed differentially regulated mRNAs at early time points (6 hours) after IRE1 inhibition to distinguish immediately responsive genes from those with expression that may be altered as part of an adaptive response to chronic inhibition. Using an arbitrary cutoff of 1.5-fold, we observed increased expression of 169 genes and decreased expression of 135 genes on IRE1 inhibition (Fig. 1A, Dataset S1, and Tables S1 and S2) (P < 0.05). To categorize the affected genes functionally according to their association with disease processes, we used the Ingenuity Pathway Analysis (IPA) tool (36). IPA identified the down-regulation of many important proatherogenic genes, including cytokines, chemokines, and chemokine receptors, on inhibition of the steady-state IRE1 activity (Fig. 1B).
Fig. 1.
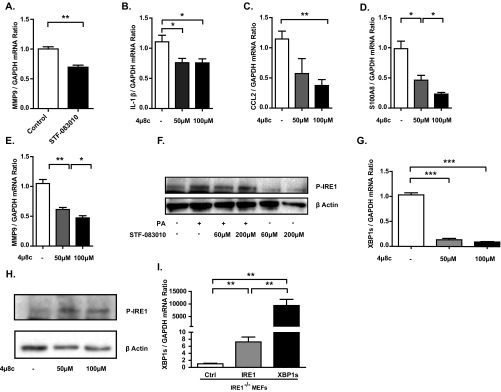
IRE1 regulates the expression of proatherogenic genes. (A) RNA-seq analysis in BMDMs treated with 60 μM STF-083010 or DMSO (control) for 6 hours. Volcano plot of differentially expressed mRNAs. (B) Analysis of atherosclerosis-related mRNAs using the IPA tool (details are in the text). (C–E) Confirmation of IRE1-dependent atherogenic gene regulation in mouse BMDMs treated with STF-083010 or DMSO (control) by qRT-PCR. (F–H) qRT-PCR analysis of atherogenic gene expression in IRE1−/− MEFs on forced expression of XBP1s or restoring IRE1’s function. Data: mean values ± SEM; n = 3. Student’s t test. Ctrl, control; ns, not significant. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Table S1.
Top 25 up-regulated genes with STF-083010 treatment in mouse BMDM cells
| Gene | Description | Fold change | P value |
| Gem | GTP binding protein (gene overexpressed in skeletal muscle) | 5.28 | 0.0197 |
| Aqp9 | Aquaporin 9 | 4.26 | 5.00E-05 |
| Slc7a11 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 | 4.25 | 5.00E-05 |
| Chac1 | ChaC, cation transport regulator-like 1 | 4.20 | 5.00E-05 |
| Trib3 | Tribbles homolog 3 | 4.14 | 5.00E-05 |
| Dclk1 | Doublecortin-like kinase 1 | 4.10 | 0.0061 |
| Slc1a4 | Solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 3.96 | 5.00E-05 |
| Stx1a | Syntaxin 1A (brain) | 3.96 | 0.03755 |
| Soat2 | Sterol O-acyltransferase 2 | 3.45 | 5.00E-05 |
| Ifrd1 | IFN-related developmental regulator 1 | 3.39 | 5.00E-05 |
| Trim46 | Tripartite motif-containing 46 | 3.37 | 0.00455 |
| Dnmt3l | Similar to DNA cytosine-5 methyltransferase 3-like protein | 3.33 | 0.00085 |
| Zfp827 | Zinc finger protein 827 | 3.32 | 0.0057 |
| Bcar1 | Breast cancer antiestrogen resistance 1 | 3.24 | 0.01765 |
| Fzd4 | Frizzled homolog 4 (Drosophila) | 3.24 | 0.04035 |
| Tmeff1 | Transmembrane protein with EGF-like and two follistatin-like domains 1 | 3.22 | 5.00E-05 |
| Slc7a5 | Similar to solute carrier family 7 | 3.17 | 5.00E-05 |
| Efcab5 | EF-hand calcium binding domain 5 | 3.15 | 0.03165 |
| Gm13889 | Predicted gene 13889 | 3.10 | 0.02935 |
| Matn2 | Matrilin 2 | 3.06 | 5.00E-05 |
| Aplp1 | Amyloid beta (A4) precursor-like protein 1 | 2.98 | 0.0086 |
| Asns | Asparagine synthetase | 2.96 | 5.00E-05 |
| Ddit3 | DNA damage-inducible transcript 3 | 2.94 | 5.00E-05 |
| Riiad1 | Regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 | 2.93 | 0.04555 |
| Map6 | Microtubule-associated protein 6 | 2.86 | 0.0004 |
Table S2.
Top 25 down-regulated genes with STF-083010 treatment in mouse BMDM cells
| Gene | Description | Fold change | P value |
| Plxnb3 | Plexin B3 | −4.45 | 0.03955 |
| Smpd3 | Sphingomyelin phosphodiesterase 3, neutral | −3.94 | 0.006 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | −3.83 | 5.00E-05 |
| Fpr2 | Formyl peptide receptor 2 | −3.76 | 0.0461 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | −3.72 | 5.00E-05 |
| Cd177 | CD177 antigen | −3.66 | 0.04935 |
| Cx3cr1 | Chemokine (C-X3-C) receptor 1 | −3.48 | 5.00E-05 |
| Chil3 | Chitinase-like 3 | −3.46 | 5.00E-05 |
| Rhobtb1 | Rho-related BTB domain containing 1 | −3.45 | 0.00245 |
| Hpgd | Hydroxyprostaglandin dehydrogenase 15 (NAD) | −3.28 | 5.00E-05 |
| Syne1 | Synaptic nuclear envelope 1 | −3.09 | 0.03485 |
| Gpr34 | G protein-coupled receptor 34 | −2.99 | 0.0171 |
| Ccr2 | Chemokine (C-C motif) receptor 2 | −2.99 | 5.00E-05 |
| Sult1a1 | Sulfotransferase family 1A, phenol-preferring, member 1 | −2.88 | 0.00875 |
| Ltf | Lactotransferrin | −2.84 | 5.00E-05 |
| Gm5086 | Predicted gene 5086 | −2.83 | 0.00595 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | −2.73 | 0.006 |
| Cd69 | CD69 antigen | −2.63 | 0.0091 |
| Idi1 | Isopentenyl-diphosphate delta isomerase | −2.61 | 0.02395 |
| Ms4a6b | Membrane-spanning 4 domains, subfamily A, member 6B | −2.60 | 5.00E-05 |
| Il1b | IL-1β | −2.59 | 0.0409 |
| Ccnd1 | Cyclin D1 | −2.58 | 5.00E-05 |
| Sesn1 | Sestrin 1 | −2.57 | 5.00E-05 |
| Rab3il1 | RAB3A interacting protein (rabin3)-like 1 | −2.57 | 5.00E-05 |
| Gm1966 | GTPase, very large IFN-inducible 1 | −2.57 | 0.0001 |
We next validated our findings using quantitative (q)RT-PCR. In these experiments, we observed a significant reduction in the mRNA levels of Il-1β, C-C motif chemokine ligand 2 (Ccl2), calgranulin A (S100a8), and matrix metalloprotease-9 (Mmp9) (P < 0.05, P < 0.001, P < 0.01, and P < 0.01, respectively) after IRE1 inhibition with either STF-083010 or 4µ8c (Fig. 1 C–E and Fig. S1 A–E). Both of these drugs were used at concentrations that do not compromise cell viability or induce any other toxicity (21, 34, 35, 37–40). Consistent with earlier reports, these IRE1 inhibitors had no effect on the kinase function of IRE1 (Fig. S1 F–H), confirming that the identified proatherogetic genes are regulated by IRE1’s RNase activity.
Fig. S1.
Modulation of IRE1 signaling by various methods: validation and impact on various proatherogenic genes. (A) qRT-PCR analysis of Mmp9 mRNA levels in mouse BMDMs treated with 60 μM STF-083010 or DMSO (control). (B–E) qRT-PCR analysis of Il-1β, Ccl2, S100a8, and Mmp9 mRNA levels in mouse BMDMs treated with 4µ8c (at the indicated doses) or DMSO (control). (F) Validation of results in A and Fig. 1 A–E. IRE1 phosphorylation was detected by Western blot in lysates of PA-stimulated BMDMs treated with STF-083010 or DMSO (control). (G and H) Validation of results in B–E. (G) qRT-PCR analysis of spliced Xbp1 mRNA levels in mouse BMDMs treated with increasing doses of 4µ8C. (H) Western blot analysis of IRE1 phosphorylation in BMDMs after IRE1 inhibition with 4µ8c. (I) Validation of results in Fig. 1 F–H. qRT-PCR analysis of spliced Xbp1 mRNA levels in IRE1−/− MEFs transfected with plasmids encoding IRE1 or XBP1s. Statistics are the same as in Fig. 1. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Because IRE1 is rate-limiting for the production of XBP1s, we next assessed if XBP1s regulates the expression of these proatherogenic genes. To this end, we forced expressed XBP1s or restored IRE1 function in inositol-requiring enzyme 1-deficient (IRE1−/−) mouse embryonic fibroblasts (MEFs) (Fig. S1I). Both experiments showed a marked induction of Il-1β, Ccl2, and S100a8 mRNA levels (Fig. 1 F–H). Together, our findings confirm that the IRE1-XBP1 signaling branch of the UPR maintains the expression of key proatherogenic cytokines and chemokines in macrophages.
Induction of IL-1β and CCL2 Depends on IRE1 During Lipotoxicity.
Our finding that IRE1 maintains the expression of several important proatherogenic genes in macrophages suggests that, when induced by metabolic stress, heightened IRE1 activity could drive the atherosclerotic process. One important activating signal for the UPR in macrophages is exposure to excessive amounts of lipids, which elicits toxicity (14, 28, 41). This lipotoxicity results in increased production of reactive oxygen species (ROS), ER stress, and inflammation, and it can result in apoptosis (41).
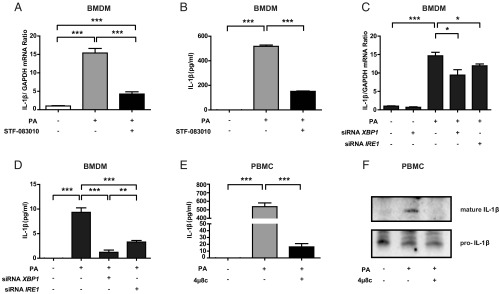
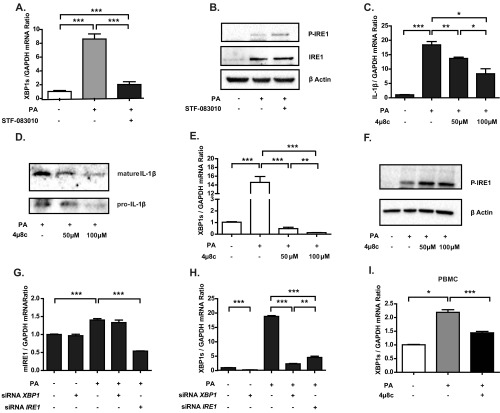
Based on these observations, we next investigated if lipid-induced IRE1 activation plays a role in IL-1β induction. Inhibition of IRE1 with STF-083010 led to a significant block in lipid-induced Il-1β mRNA production and secretion from BMDMs (Fig. 2 A and B and Fig. S2 A and B). As expected, inhibition of IRE1 activity with 4µ8c showed the same effects (Fig. S2 C–F). To further delineate the role of IRE1 and XBP1s in the regulation of IL-1β, we transfected BMDMs with a specific siRNA. We found that transfection of siRNAs against Ire1α (the ubiquitously expressed IRE1 homolog) or Xbp1 resulted in a significant reduction in lipid-induced Il-1β mRNA production and IL-1β secretion in BMDMs [Fig. 2 C and D (P < 0.05, P < 0.05, P < 0.001, and P < 0.001) and Fig. S2 G and H]. Moreover, treatment of human peripheral blood monocytes (PBMCs) with lipids induced IL-1β secretion, which was blocked by preincubation with 4µ8c (Fig. 2 E and F and Fig. S2I). These findings show that IRE1-XBP1 signaling plays an important role in Il-1β mRNA up-regulation and secretion from both mouse and human macrophages.
Fig. 2.
IRE1 regulates lipid-induced IL-1β secretion in mouse and human macrophages. Il-1β (A) mRNA and (B) protein levels measured from LPS-primed and PA-stimulated mouse BMDMs treated with 150 μM STF-083010 or DMSO (control) by qRT-PCR and ELISA, respectively. Il-1β (C) mRNA and (D) protein levels measured from LPS-primed and PA-stimulated mouse BMDMs transfected with siRNAs against Ire1α or Xbp1 by qRT-PCR and ELISA, respectively. (E) Secreted IL-1β from LPS-primed, PA-stimulated human PBMCs treated or not treated with 100 μM 4μ8c measured by ELISA. (F) Same as E but protein levels were measured by immunoblotting to show the immature and processed forms of the cytokine (representative image of three independent blots). Statistics are the same as in Fig. 1. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Fig. S2.
Inhibition of IRE1 by various methods in lipid-stressed macrophages: validation of its impact on IL-1β. (A and B) Validation of results in Fig. 2 A and B. (A) qRT-PCR analysis of spliced Xbp1 mRNA levels in LPS-primed, PA-stimulated mouse BMDMs treated with 150 μM STF-083010 or DMSO (control). (B) Western Blot analysis of p-IRE in LPS-primed, PA-stimulated mouse BMDMs treated with 150 μM STF-083010 or DMSO (control). (C–F) Inhibition of IRE1 RNase activity by 4µ8c and its impact on IL-1β. (C) qRT-PCR analysis of Il-1β mRNA levels in LPS-primed, PA-stimulated mouse BMDMs treated with 4µ8c (at the indicated doses). (D) Western blot analysis of secreted mature IL-1β from LPS-primed, PA-stimulated BMDMs treated with 4µ8c (at the indicated doses) or DMSO (control). (E) qRT-PCR analysis of spliced Xbp1 mRNA levels in LPS-primed, PA-stimulated mouse BMDMs treated with 4µ8c (at the indicated doses) or DMSO (control). (F) Western blot analysis of IRE1 phosphorylation in LPS-primed, PA-stimulated mouse BMDMs treated with 4µ8c (at the indicated doses) or DMSO (control). (G and H) Validation of Ire1α and Xbp1 knockdown in Figs. 2 C and D and 3B. qRT-PCR analysis of (G) Ire1α or (H) spliced Xbp1 mRNA levels in LPS-primed, PA-stimulated mouse BMDMs transfected with siRNAs against Ire1α or Xbp1. (I) Validation of results in Fig. 2 E and F. qRT-PCR analysis of spliced Xbp1 mRNA levels in LPS-primed human PBMCs treated with PA or a combination of PA and 4µ8c. Statistics are the same as in Fig. 1. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
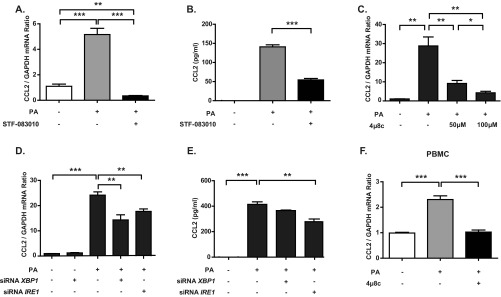
We also investigated if the proatherogenic chemokine CCL2 is regulated by IRE1 under lipotoxic ER stress conditions. Both STF-083010 and 4µ8c treatments of BMDMs resulted in complete inhibition of lipid-induced Ccl2 mRNA production and protein secretion (Fig. S3 A–C). Consistent with this observation, siRNAs against Ire1α or Xbp1 also suppressed lipid-induced CCL2 production and secretion in BMDMs (Fig. S3 D and E). Moreover, 4μ8c blocked lipid-induced CCL2 production in human PBMCs (Fig. S3F). Collectively, these data show that IRE1 is important for the inflammatory response to lipids and the production of proatherogenic cytokines in both mouse and human macrophages.
Fig. S3.
IRE1 regulates lipid-induced CCL2 production in macrophages. (A) qRT-PCR analysis of Ccl2 mRNA levels in LPS-primed, PA-stimulated BMDMs that were treated with 150 μM STF-083010 or DMSO (control). (B) ELISA measurements of secreted CCL2 from the conditioned medium used to culture the cells in A. (C and D) qRT-PCR analysis of Ccl2 mRNA levels in LPS-primed, PA-stimulated BMDMs that were (C) treated with 4µ8c (at the indicated doses) or DMSO (control) or (D) transfected with siRNAs against Ire1α or Xbp1. (E) ELISA measurements of secreted CCL2 from the cells in D. (F) qRT-PCR analysis of Ccl2 mRNA levels in LPS-primed, PA-stimulated human PBMCs treated with 4µ8c (at the indicated doses) or DMSO (control). Statistics are the same as in Fig. 1. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Inflammasome Activation Depends on IRE1 During Lipotoxicity.
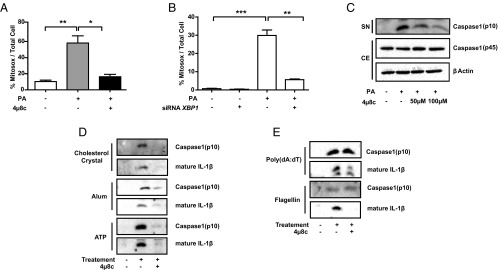
Because IRE1 inhibition leads to a strong suppression of IL-1β secretion, we reasoned that IRE1 may contribute to the lipid-induced activation of the Nod-like receptor family, pyrin domain-containing protein-3 (NLRP3) inflammasome, a multicomponent platform that contains caspase-1 and induces the caspase-1–dependent secretion of the proinflammatory cytokines IL-1β and IL-18 (42, 43). Previous studies showed that ER stress induces inflammasome activation through several mechanisms, including calcium mobilization and the release of reactive oxygen species from damaged mitochondria (mtROS) (44). Because earlier studies from our laboratory and the laboratories of others showed that treatment of macrophages with saturated fatty acids activates IRE1 and because these lipids specifically activated the NLRP3 inflammasome through inducing mtROS production, we sought to investigate this connection further (42, 43, 45). To this end, we first measured mtROS production in cells exposed to lipotoxic stress in the presence of IRE1 inhibitors. We observed that lipid-induced ER stress in BMDMs resulted in a dramatic elevation of mtROS, which was completely blocked by 4μ8c treatment as well as XBP1 knockdown (Fig. 3 A and B and Fig. S4A).
Fig. 3.
IRE1 inhibitors block lipid-induced mtROS release and inflammasome activation. (A and B) mtROS production was measured in LPS-primed, PA-stimulated mouse BMDMs after (A) 100 μM 4µ8c or DMSO (control) treatment or (B) transfection with scrambled or Xbp1-specific siRNAs. (C–E) Immunoblots of the levels of the zymogen (p45) and mature (p10) forms of caspase-1 in LPS-primed mouse BMDMs pretreated with 4μ8c (at the indicated doses) or DMSO (control) and stimulated with (C) PA, (D) other NLRP3 agonists (5 nM ATP, 200 μg/mL alum, or 400 μg/mL cholesterol crystals), or (E) specific activators of other inflammasome complexes [5 μg/mL poly(dA:dT) and 1 μg/mL flagellin] according to previously published protocols (described in detail in Experimental Procedures). Blots shown are representative of three independent experiments. Statistics are the same as in Fig. 1. CE, cell extract; SN, supernatant. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Fig. S4.
IRE1 perpetuates mtROS production and inflammasome activation. (A) Representative confocal microscopy images of mtROS production in BMDMs (Fig. 3A). MitoSOX fluorescence indicator (red) and Mitotracker (green) are shown. mtROS (yellow) levels are quantified and depicted in the graph in Fig. 3A. (Scale bar: 20 µm.) (B and C) PA-stimulated BMDMs treated with the indicated doses of 4μ8c (at the indicated doses) or DMSO (control) were analyzed with (B) qRT-PCR for Txnip mRNA and (C) calcium uptake. (D and E) Western blot analysis of caspase-1 activation in LPS-primed, PA-stimulated mouse BMDMs (D) treated with 150 μM STF-083010 or DMSO (control) or (E) transfected with siRNAs against Ire1α or Xbp1 Procaspase-1 (p45) and cleaved caspase-1 (p10) levels are indicated. Statistics are the same as in Fig. 1. CE, cell extract. SN, supernatant. *P ≤ 0.05; ***P ≤ 0.001.
The impact of IRE1 signaling on inflammasome activation has been postulated to be mediated by the IRE1-dependent accumulation of the thioredoxin-interacting protein (TXNIP), a thioredoxin inhibitor with increased levels that promote activation of the NLRP3 inflammasome (35). In stark contrast to these earlier findings, which used cells treated with canonical ER poisons, lipid-induced ER stress led to a profound suppression of TXNIP in macrophages, which was partially blocked by inhibiting IRE1 (Fig. S4B). Another mechanism by which ER stress can induce mitochondrial oxidative stress and activation of the NLRP3 inflammasome complex is through calcium mobilization (46). Indeed, we observed that palmitic acid (PA) leads to a significant increase (threefold) of calcium levels in the mitochondria, but this increase could not be prevented by IRE1 inhibition (Fig. S4C). Moreover, lipid-induced ER stress also induced procaspase-1 maturation (indicated by the appearance of the p10 fragment), an effect that was reduced by treatment with IRE1 inhibitors or siRNA-mediated silencing of Ire1α and Xbp1 (Fig. 3C and Fig. S4 D and E). Taken together, these results indicate that IRE1 plays a crucial role in perpetuating mtROS production and NLRP3 inflammasome activation in cells experiencing lipotoxic ER stress but that this effect is independent of TXNIP induction and calcium mobilization.
We further investigated if IRE1 affects the activation of the NLRP3 inflammasome by other stimuli or the activation of different inflammasome complexes by specific inducers. Although IRE1 inhibition blocked NLRP3 inflammasome activation, cleavage of caspase-1, and secretion of IL-1β by various different NLRP3 inflammasome inducers (including ATP, Alum, and cholesterol crystals) (Fig. 3D), it did not alter the activation of other inflammasome complexes, such as NLR family CARD domain-containing protein 4 by flagellin or absent in melanoma 2 by poly(deoxyadenylic-deoxythymidylic acid) [poly(dA:dT)] as evident by no changes in caspase-1 cleavage (Fig. 3E). Because IRE1-XBP1 controls Il-1β mRNA production, we observed reduction in secreted IL-1β protein levels in all of the treatments (Fig. 3 D and E). Finally, together with our results showing that IRE1 induces IL-1β, our data show that IRE1 plays a major role in controlling IL-1β signaling both transcriptionally and by promotion of its maturation via the activation of the NLRP3 inflammasome.
Pharmacological Inhibition of IRE1 Combats Atherosclerosis.
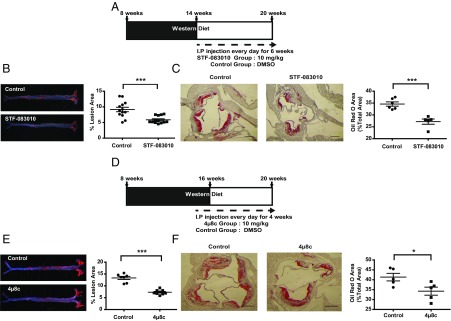
The evidence presented above in conjunction with previous results showing that restoring or improving ER function alleviates atherosclerosis (14, 17, 47) suggest that inhibiting IRE1 may impair atherosclerosis progression. Therefore, we postulated that administration of IRE1 modulators might have beneficial effects by limiting the inflammatory signaling associated with elevated ER stress in a mouse model of atherosclerosis. To test this notion, we challenged ApoE−/− mice with a Western diet (12 weeks) and then treated them daily with STF-083010 by intraperitoneal (i.p.) injection (6 weeks) (Fig. 4A). Although limited pharmacodynamic data are available for the IRE1 inhibitors that we used, target engagement has been shown in cells and tissues (21, 39). We based our in vivo drug dosage and delivery on earlier in vivo studies that successfully administered the IRE1 modulators without toxicity (21, 24, 31). Furthermore, we confirmed target engagement in tissues by assessing drug-induced suppression of Xbp1 mRNA splicing and RIDD activities by observing a significant reduction in the spliced Xbp1 mRNA (P < 0.05) and a modest increase in canonical RIDD target mRNAs (P < 0.05) (Fig. S5 A–C). Importantly, the IRE inhibitor that we used did not alter IRE1 phosphorylation in vivo (Fig. S5D). Furthermore, we detected no differences in body weights, blood glucose levels, liver morphology, plasma alanine amino transferase (ALT) activity, and cholesterol profiles between the STF-083010–treated and control groups (Figs. S5 E and F and S6 and Table S3). However, the analysis of en face aorta preparations showed that chronic administration of STF-083010 led to a significant decrease (35.8%; P < 0.001) in atherosclerotic lesions compared with the control group (Fig. 4B). Furthermore, when we evaluated the impact of STF-0803010 on plaque development in the aortic root, we observed a significant reduction (21.4%; P < 0.001) in the foam cell area (visualized by Oil Red O staining) in the inhibitor-treated group compared with control mice (Fig. 4C).
Fig. 4.
IRE1 inhibitors reduce plaque area in a mouse model of atherosclerosis. (A) Experimental design in ApoE−/− mice using STF-083010 (10 mg/kg per day). (B) En face aorta analysis of atherosclerotic plaques in ApoE−/− mice. (Left) Sudan IV staining of atherosclerotic plaques. (Right) Quantification of plaque area (n = 13–14). (C) Analysis of aortic sinus plaque area in the animals in A and B. (Left and Center) Oil Red O staining of aorta cross-sections. (Right) Quantification of plaque area (n = 5–6). (D–F) Experimental design and data for analogous experiments in ApoE−/− mice using 4μ8c (10 mg/kg per day; E, n = 7–9; F, n = 5). Statistics are the same as in Fig. 1. (Scale bar: 350 µm.) *P ≤ 0.05; ***P ≤ 0.001.
Fig. S5.
Treatment of ApoE−/− mice with IRE1 inhibitors suppresses Xbp1 mRNA splicing and degradation of canonical RIDD targets. (A) qRT-PCR analysis of spliced Xbp1 mRNA levels from spleen tissue of ApoE−/− mice that were fed a Western diet for 12 weeks and treated with STF-083010 (10 mg/kg per day) or vehicle (control) for 6 weeks. (B and C) qRT-PCR analysis of the mRNA levels of reported RIDD targets in the same tissues as in A. (D) Liver protein lysates from the same mice as in A were analyzed for the phosphorylated form of IRE1 by Western blotting. (E) H&E staining of the livers from the same mice in A. (Scale bar: 50 µm.) (F) Plasma ALT activity measurements from the same mice in A. Statistics are the same as in Fig. 1. ns, Not significant. *P ≤ 0.05.
Fig. S6.
Lipid and lipoprotein profiles do not change in animals treated with IRE1 inhibitors. (A) Total plasma triglyceride and lipoprotein [very LDL (VLDL), LDL, and HDL] triglyceride levels in STF-083010– and vehicle-treated groups. (B) Total plasma cholesterol and lipoprotein (VLDL, LDL, and HDL) cholesterol levels in STF-083010– and vehicle-treated groups. (C and D) Lipoprotein profiles in control (blue) and STF-083010–treated (red) ApoE−/− mice. Statistics are the same as in Fig. 1. ns, not significant.
Table S3.
Physical and biochemical characterization of ApoE−/− mice in STF-083010–treated and control groups
| Variables and treatment | Control | STF-083010 |
| n | 6 | 6 |
| Body weight (g) | ||
| Before | 30.3 ± 3.5 | 28.3 ± 2.2 |
| After | 33.3 ± 5.1 | 29.8 ± 2.5 |
| Glucose (mg/dL) | ||
| Before | 120.5 ± 18.5 | 134 ± 13.3 |
| After | 79.0 ± 11.8 | 72.6 ± 15.1 |
Analogous experiments using 4μ8c in the same animal model [using previously published doses that showed no toxicity (33, 37, 40, 48)] produced similar results (Fig. 4 D–F): 4μ8c treatment led to a significant reduction (45.2%; P < 0.001) in atherosclerotic lesion area in en face aorta preparations (Fig. 4E) and a significant reduction in the spliced Xbp1 mRNA (Fig. S7A) (P < 0.05) but no change in IRE1 phosphorylation in the spleens (Fig. S7B). Furthermore, 4μ8c treatment led to a reduced foam cell area (Fig. 4F) without overt differences in body weight, blood glucose levels (Table S4), liver morphology, and plasma ALT activity between the inhibitor-treated and control mice (Fig. S7 C and D). These in vivo findings show that pharmacological inhibition of IRE1 can effectively mitigate plaque development in mice.
Fig. S7.
Suppression of Xbp1 mRNA splicing by IRE1 inhibitors in vivo. (A) qRT-PCR analysis of spliced Xbp1 mRNA levels in spleen of ApoE−/− mice that were fed a Western diet for 12 weeks and treated with 4µ8c (10 mg/kg per day) or vehicle (control) for 4 weeks. (B) Liver protein lysates from the same mice as in A were analyzed for the phosphorylated form of IRE1 by Western blotting. (C) H&E staining of the livers from the same mice in A. (Scale bar: 50 µm.) (D) Plasma ALT activity measurements from the same mice in A. Statistics are the same as in Fig. 1. ns, not significant. *P ≤ 0.05.
Table S4.
Physical and biochemical characterization of ApoE−/− mice in 4µ8c-treated and control groups
| Variables and treatment | Control | 4µ8c |
| n | 7 | 9 |
| Body weight (g) | ||
| Before | 28.8 ± 2.7 | 26.6 ± 1.9 |
| After | 31.1 ± 2.5 | 28.6 ± 1.2 |
| Glucose (mg/dL) | ||
| Before | 93.9 ± 30.8 | 79.2 ± 19.6 |
| After | 91.3 ± 17.0 | 91.9 ± 20.0 |
Pharmacological Inhibition of IRE1 Alters Plaque Composition.
Endothelial cells, vascular smooth muscle cells (VSMCs), and immune cells, such as lymphocytes, dendritic cells, neutrophils, and macrophages, play important roles in the development of atherosclerotic plaques in the arterial wall. The UPR is activated in many of these cell types and at all stages of atherosclerotic plaque development. This increase in ER stress is also associated with plaque progression, vulnerability to rupture, and acute coronary syndrome in humans (7). Given that IRE1 inhibitors alleviated atherosclerosis in ApoE−/− mice, we next analyzed the impact of these inhibitors on the cellular composition of the lesions. STF-083010 treatment led to a significant reduction (35%; P < 0.01) in macrophages [as visualized by monocyte/macrophage marker-2 (MOMA-2) staining] infiltrating the aortic root plaques (Fig. 5A). This reduction in macrophage numbers was not the product of increased apoptosis as determined by TUNEL (in situ cell death detection) assays in macrophage-enriched areas of the plaques (Fig. 5B). Furthermore, there were no differences in the necrotic core area between the treatment and control groups (Fig. S8A). During plaque formation, VSMCs migrate from adventitia to intima, secreting collagen and sealing the fibrous cap of the plaque. The analysis of the lesions in STF-083010–treated mice (with Masson’s Trichrome staining) showed that there is a significant increase in collagen content that is responsible for tensile strength and elasticity of the plaques (22%; P < 0.05) without changes in the numbers of the VSMCs infiltrating the lesions (Fig. S8 B and D). However, we did not observe significant differences in the fibrous cap thickness between the treatment and control groups (Fig. S8C) Finally, STF-083010 treatment did not alter CD3+ T-cell numbers in the adventia/lesions (Fig. S8E). Taken together, these results indicate that the major consequences of IRE1 inhibition include a reduction in macrophages and an increase in collagen deposition in atherosclerotic plaques.
Fig. 5.
IRE1 inhibitors alter plaque composition and inflammation. Immunohistochemical and TUNEL assay analyses of proximal aorta cryosections from ApoE−/− mice (Fig. 4) treated with an IRE1 inhibitor. In each case, a representative image is shown in Left and Center, and the quantification of the data appears on in Right. (A) MOMA-2. (Scale bar: 100 µm.) (B) TUNEL assay (apoptotic cells are shown with arrowheads). (Scale bar: 50 µm.) (C) IL-1β. (Scale bar: 100 μm.) Statistics are the same as in Fig. 1. ns, not significant. *P ≤ 0.05; **P ≤ 0.01.
Fig. S8.
IRE1 inhibitors alter plaque composition and inflammation. Immunohistochemical and immunofluorescence analyses of proximal aorta sections obtained from ApoE−/− mice treated with STF-083010 or controls. In each case, representative images are shown in Left and Center, and the quantification of the data appears in Right. (A) Necrotic area was calculated (from indicated area) in the H&E-stained plaque sections. (Scale bar: 150 µm.) (B) Masson’s Trichrome collagen stain (collagen, blue; cytoplasm and muscle fibers, red; n = 5). (Scale bar: 200 µm.) (C) Fibrous cap thickness calculation from samples in A. (Scale bar: 100 µm.) (D) Anti–α-smooth muscle actin (VSMCs marker). (Scale bar: 200 µm.) (E) Anti-CD3 (T-cell marker). Arrowheads show CD3-positive cells (n = 5). Statistics are the same as in Fig. 1. (Scale bar: 200 µm.) ns, not significant. *P ≤ 0.05.
Last, we sought in vivo evidence for the observed inhibition of IL-1β by IRE1 inhibition in macrophages (Figs. 1 and 2). We observed that STF-083010 treatment reduced the expression of IL-1β in the aortic root lesions stained with a specific antibody against IL-1β (Fig. 5C). Together, these results validate our earlier in vitro findings and show that the antiatherogenic effect of IRE1 inhibitors involves a blockage of inflammation in the lesions.
IRE1 Inhibitors Suppress Hyperlipidemia-Induced Th-1 Immune Responses.
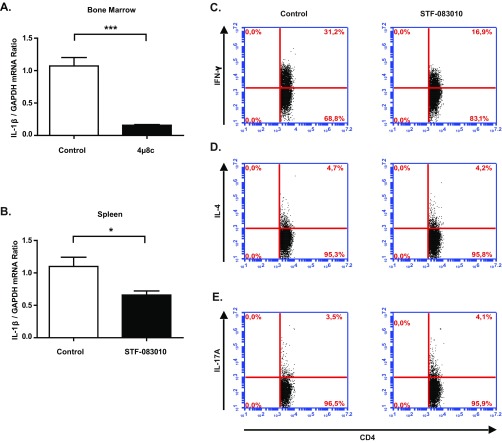
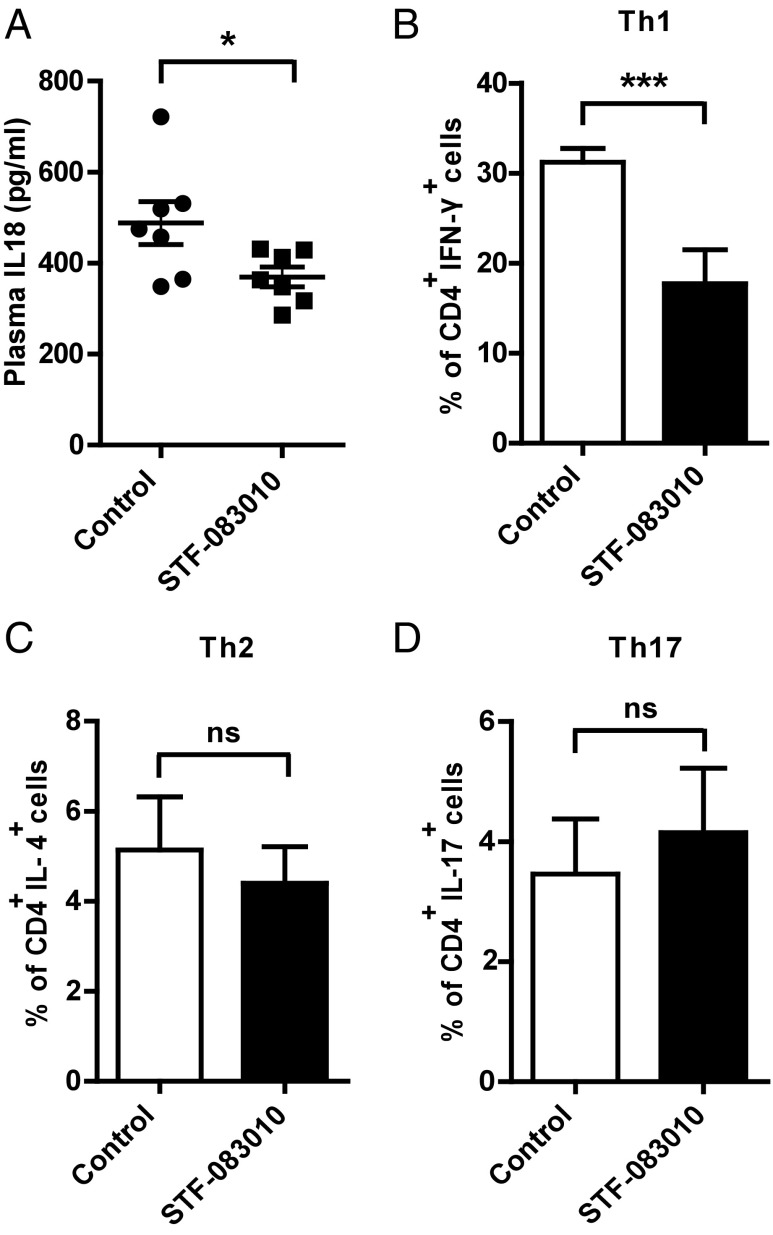
Atherosclerosis initiation and progression depend on both innate and adaptive immunity pathways. T cells orchestrate adaptive immunity, whereas macrophages bridge innate and adaptive immune processes that contribute to lesion development. Th cells form the majority of lymphocytes in the atherosclerotic plaques. Th-1 cells are proinflammatory, produce high amounts of IFN-γ, and contribute to the progression of atherosclerosis. Two other types of lymphocytes implicated in atherosclerosis progression include Th-2 cells, which produce IL-4, and Th-17 cells, which produce IL-17 (49–51). The inflammasome-induced cytokines IL-18 and IL-1β play an important role in the polarization of Th-1 and Th-17 responses (52). Because inhibition of IRE1 suppressed inflammasome activation (Fig. 3C and Fig. S4 D and E) and IL-1β production in lipid-challenged macrophages (Fig. 2) as well as lesions and tissues (Fig. 5C and Fig. S9 A and B), we next assessed the impact of IRE1 inhibition on systemic IL-18 levels and Th cell differentiation in hyperlipidemic mice. ApoE−/− mice (on Western diet) that were treated with STF-083010 displayed a significant decrease in plasma IL-18 levels (Fig. 6A) (P < 0.05) and a marked reduction in the secretion of IFN-γ—but not IL-4 or IL-17—from splenocytes (P < 0.001) (Fig. 6 B–D and Fig. S9 C–E). We did not observe changes in the overall T-cell counts in atherosclerotic lesions after STF-083010 treatment (Fig. S8E), indicating that decreased lymphokine production is intrinsic to intracellular signaling and does not result from a decline in the infiltrating immune cells that produce them. In conclusion, the reduced inflammasome activity in these mice (as measured by IL-1β and IL-18 levels in Figs. 5C and 6A) after STF-083010 treatment correlates with the suppression of the Th-1 inflammatory response that is known to promote atherosclerosis development.
Fig. S9.
IRE1 inhibitors reduce IL-1β levels in tissues and suppress Th1 immune responses. qRT-PCR analysis of Il-1β mRNA levels in (A) bone marrow or (B) spleen tissue of ApoE−/− mice that were fed a Western diet and treated with IRE1 inhibitors or vehicle (control). (C–E) Flow cytometry analysis of (C) IFN-γ, (D) IL-4, and (E) IL-17 levels in splenocytes activated with PMA/ionomycin (n = 5). Statistics are the same as in Fig. 1. *P ≤ 0.05; ***P ≤ 0.001.
Fig. 6.
IRE1 inhibitors suppress hyperlipidemia-induced Th-1 immune responses and IL-18 cytokine levels. (A) Plasma IL-18 in ApoE−/− mice (Fig. 4) treated with an IRE1 inhibitor was measured by ELISA (n = 7). (B–D) Flow cytometry analysis of IFN-γ, IL-4, and IL-17 in splenocytes from ApoE−/− mice treated with IRE1 inhibitor and activated with PMA/ionomycin (n = 5). Statistics are the same as in Fig. 1. ns, not significant. *P ≤ 0.05; ***P ≤ 0.001.
Discussion
Studies in mice and humans suggest that chronic ER stress plays an important role in atherosclerosis progression. Therefore, pharmacological manipulation of the UPR—the network of signaling pathways that respond to ER stress—represents a promising therapeutic approach to manage atherosclerosis (7, 14, 53). The recent discovery of highly selective UPR modulators provides unique opportunities to investigate the contribution of individual UPR branches to the pathogenesis of this disease. Using small molecules that target IRE1, we showed that modulating IRE1 signaling counteracts atherosclerotic plaque formation in mouse models.
First, IRE1 inhibition altered plaque cellular composition mainly by reducing the numbers of macrophages in the atherosclerotic lesions without altering apoptosis. We infer that this effect is likely to stem from reduction in CCL2, a strong macrophage chemoattractant, consistent with our observations in macrophages treated with IRE1 inhibitors. Alternatively, IRE1 modulators could impact macrophage clearance from lesions by phagocytosis of dying cells. We observed no change in the apoptotic cell counts in lesions, arguing against this possibility. Nevertheless, more detailed future studies are required to discriminate between these two possibilities.
Second, IRE1 inhibitor-treated mice displayed an increased collagen content in atherosclerotic lesions, which imparts tensile strength and elasticity to the plaques (54). However, we did not observe an increase in fibrous cap thickness on IRE1 inhibitor treatment. Because we observed no differences in the number of VSMCs in the lesions, the increased collagen deposition may be related to increased collagen folding and secretion, which is consequential to enhanced ER function coupled to reduced cleavage by MMPs. In fact, early in our study, we observed that MMP9 is regulated by IRE1, whereas another study reported RIDD-dependent collagen degradation during ER stress (55). Both observations lend support to our findings and substantiate our hypotheses.
Third, the results from our RNA-seq analyses in macrophages that were treated with IRE1 inhibitors strongly hinted at IRE1’s involvement in the production of several proatherogenic cytokines, chemokines, and their receptors, including IL-1β, CCL2, and chemokine receptor 2. Indeed, IRE1, through XBP1s, regulates Il-1β and Ccl2 mRNA induction in lipid-stressed macrophages. Moreover, oxidative stress can activate NF-κB and production of CCL2, resulting in the recruitment of monocytes to a growing plaque (7).
Fourth, cholesterol crystals, saturated fatty acids, and ROS accumulate in plaque areas and provide activation signals for activation of the NLRP3 inflammasome and subsequent secretion of IL-1β and IL-18 (44). These cytokines generate Th-1–type immune responses that promote plaque progression and unstable lesions (52). Treatment of macrophages with the IRE1 inhibitors suppressed lipid-induced mtROS production, activation of the NLRP3 inflammasome, and subsequent IL-1β secretion. Our results, therefore, implicate IRE1 activation in the perpetuation of lipid-induced mitochondrial oxidative stress upstream of NLRP3 inflammasome activation, but our data show that this effect is independent of TXNIP induction or calcium mobilization (35, 44). Macrophage mitochondrial oxidative stress plays an important role in atherogenesis by amplifying inflammation (56). Our results reveal that IRE1 is crucial for the regulation of mtROS generation during lipid-induced ER stress; however, future detailed studies will be needed to uncover how this is achieved by IRE1. In addition to our findings in cells, we confirmed the inhibitory effect of IRE1 inhibitors on hypercholesterolemia-induced IL-1β and IL-18 production in vivo in plaques and plasma, respectively. Consistent with these observations, treatment with IRE1 inhibitors led to a marked suppression of hyperlipidemia-induced Th-1 immune responses in these mice. We observed no differences in T-cell numbers in adventia/lesions between the IRE1 inhibitor treatment and control groups, and previous studies have shown the IRE1 inhibitor used in this study does not affect Treg cells (33, 37). Collectively, these findings show that prevention of inflammasome-associated cytokine production by IRE1 inhibitors in vivo has dramatic effects on counteracting atherosclerotic disease progression. Our findings correlating suppression of IL-1β with atheroprotection are timely in view of a major ongoing clinical trial on the effect of an anti–IL-1β trial in coronary artery disease (by Cantos). Although our data show a clear impact of IRE1 inhibition on macrophage inflammatory functions, activation of the UPR also occurs in many other lesion-resident cell types. Thus, our results do not exclude the possibility that the antiatherogenic effects of IRE1 inhibition could also involve other lesion-resident cell types that contribute to atherogenesis.
Fifth, the reduction in plaque inflammation and size occurred independent of a correction of elevated plasma lipid levels in IRE1 inhibitor-treated ApoE−/− mice. This notion contrasts with results of a previous study, in which mice bearing a liver-specific deletion of Xbp1 displayed a profound reduction in plasma cholesterol and triglyceride levels (57). However, it is important to note that this apparent disparity may exclusively result from feedback activation of IRE1 on genetic loss of Xbp1 (57). Notably, additional siRNA-mediated IRE1 depletion partially reverted the hypolipidemic phenotype in vivo, hinting at an RIDD-dependent function that controls lipid metabolism (30). Furthermore, we did not observe feedback activation of IRE1 or any signs of liver toxicity with the IRE1 modulators. Because XBP1s promotes the transcriptional up-regulation of a lipogenic gene program, these findings could be interpreted to mean that the splicing and RIDD outputs of IRE1 have opposing effects on lipid metabolism (30). Although most of the direct targets of XBP1 participating in triglyceride and sterol lipogenic programs were also identified as RIDD substrates in this study (ref. 30, figure 3), IRE1 (through its RIDD modality) seems to be able to reduce the steady-state expression of a larger number of lipogenic genes independent of XBP1, hinting at a complex lipogenic regulatory program that depends on the interplay between XBP1 and IRE1 signaling (30, 57). Thus, the reduction in plaque size that we observed in our study is likely to result from IRE1 inhibitor-mediated antiinflammatory changes and not to result from the product of changes in lipid metabolism.
Although we have seen important gains in mitigating atherosclerosis by pharmacologically targeting IRE1 in our experimental models, it is important to note that the other UPR branches, particularly the PERK-CHOP branch, are also induced as atherosclerosis progresses and seem to be instrumental for macrophage apoptosis (5, 11). Previous studies focusing on the engagement of apoptotic pathways initiated by the UPR (such as those mediated by CHOP and JNK) showed that mice deficient for these apoptotic effectors are protected from atherosclerosis (5, 11–13). Independent of CHOP or JNK engagement, here, we found that modulating IRE1 signaling in vivo with small molecule IRE1 inhibitors modifies a different branch of UPR signaling that impinges on metaflammation and alters the course of atherosclerosis. These results support the notion that it may be possible to uncouple metabolically induced ER stress from inflammation by calibrating UPR signaling, thereby improving the clinical outcome of atherosclerosis. With the advent of specific inhibitors that can target different UPR branches, exploring the efficacy of combinatorial UPR calibration in this chronic disease setting becomes a promising endeavor.
Experimental Procedures
A list of primers used in the study can be found in Dataset S1.
Reagents and Plasmids.
IRE1−/− MEFs were provided by Gokhan Hotamişligil, Harvard School of Public Health, Cambridge, MA. BMDMs were isolated from C57BL/6 mice. Plasmids encoding XBP1s (21833) and WT IRE1α (13009) were purchased from Addgene. l-Glutamine, DMEM, PBS, HBSS, penicillin/streptomycin (P/S), FBS, and Roswell Park Memorial Institute (RPMI)-1640 medium were obtained from Thermo Scientific or HyClone (GE Healthcare Life Sciences). Trypsin, ampicillin, kanamycin, Bradford assay reagents, ultrapure LPS, PA (palmitic acid), phosphatase inhibitor mixture, and STF-083010 were purchased from Sigma-Aldrich; 4μ8c was purchased from Calbiochem. Primary antibodies used for immunoblotting were purchased from the following suppliers: anti-IRE1 phospho-S724 antibody (ab48187; Abcam), IL-1β (ab9722; Abcam), IRE1α rabbit mAb (3294; Cell Signaling), β-Actin (sc-47778; Santa Cruz Biotechnology), β-actin (sc-9104; Santa Cruz Biotechnology), Caspase-1 (M20; sc-514; Santa Cruz Biotechnology), and IL-1β (AF-401-NA; R&D Systems). ECL Prime Western Blot Detection Kit was purchased from Amersham Pharmacia. ALT assay (MAK052) was from Sigma-Aldrich.
Preparation of PA-BSA Complex.
PA was dissolved in absolute ethanol to yield a stock concentration of 500 mM and stored at −80 °C. Stock PA was diluted to the working concentration and suspended with 1% fatty acid-free BSA in RPMI-1640 medium (without serum) by mixing at 50 °C for 30 min.
Preparation of Cholesterol Crystal.
The 10-mg/mL cholesterol solution was prepared in ethanol, heated to 60 °C, brought to room temperature to allow for crystallization, washed with PBS, and resuspended in RPMI-1640 medium at 5 mg/mL; 400-μg/mL cholesterol crystals were used for treating the cells (for 20 hours) as described (58).
Cell Culture and Treatments.
Isolation of BMDMs.
Bone marrows were collected from the tibia and femurs of mice into RPMI-1640 medium containing 1% P/S. After filtering through a cell strainer (352350; BD Biosciences), the cells were centrifuged at 400 × g for 5 min and resuspended in RPMI-1640 medium enriched with 15% (vol/vol) L929 conditioned medium and 1% P/S followed by seeding for growth and differentiation into macrophages on Petri dishes for 7 d.
Inflammasome activation.
BMDMs were pretreated (1 h) with 150 µM STF-083010 or the indicated concentrations of 4μ8c followed by stimulation with ultrapure LPS (200 ng/mL) for 3 hours and then, followed by treatment with PA-BSA (1,000 µM) for 20 hours. Human PBMCs were purchased from Zenbio and grown in lymphocyte medium (RPMI-1640, 2 mM l-glutamine, 10% (vol/vol) FBS, 1% P/S) according to the instructions provided by the manufacturer. Cells were pretreated with 100 µM 4µ8c before stimulation with ultrapure LPS (200 ng/mL) for 3 hours. These incubations were followed by treatment with ethanol-BSA (control) or PA-BSA (500 µM). LPS-primed BMDMs were treated with various inflammasome inducers, including 5 nM ATP (Sigma-Aldrich), 200 μg/mL Imject Alum (Thermo Scientific), and 1 μg/mL Flagellin (Invivogen) either alone or in combination with 100 µM 4µ8c. Lipofectamine 3000 was used for poly(dA:dT) DNA (5 μg/mL) transfection as described (43).
Induction of mtROS.
BMDMs were pretreated (1 h) with 100 µM 4µ8c followed by sequential stimulation with 200 ng/mL LPS (3 hours) and then, PA-BSA (1,000 µM) or vehicle (ethanol)-BSA (control) for 20 hours.
Western Blot Analysis.
Cells were lysed in lysis buffer [50 mM Hepes, pH 7.9, 100 mM sodium chloride, 10 mM EDTA, 10 mM sodium fluoride, 4 mM tetrasodium diphosphate, 1% Triton X-100, 2 mM sodium orthovanadate, 1 mM PMSF (phenylmethylsulfonyl), 1× phosphatase inhibitor mixture (Sigma-Aldrich), 1× protease inhibitor mixture (Sigma-Aldrich)]. Lysates were cleared by brief centrifugation followed by the addition of 5× SDS loading dye. For the detection of cleaved caspase-1 (active p10 form) in the cell medium, cell culture supernatants were collected, mixed with 5× SDS loading dye, and heated at 95 °C for 5 min before loading on SDS/PAGE gels. Proteins were subjected to SDS/PAGE separation and transferred onto PVDF membranes. Blocking and antibody incubation were carried out in TBS (Tris-buffered saline) with 0.1% (vol/vol) Tween-20 and 5% (wt/vol) dry milk or BSA and visualized by ECL in a BioRad Imager.
Transfection.
IRE1−/− MEFs that reached 60–80% confluency were transfected with the indicated plasmids (2 µg DNA for every 4.5 × 105 cells) and polyethylenimine (Sigma-Aldrich), and BMDMs were electroporated using a Neon Electroporator (Invitrogen) according to the protocols provided by the manufacturer.
RNAi.
BMDMs were transfected with 50 nM siRNA against ire1 (SI0099588; Qiagen), 70 nM siRNA against Xbp1 (SI01473227; Qiagen), or scrambled siRNAs (1027281; Qiagen); 24 hours after transfection, the cells were treated with indicated reagents.
RNA Isolation and qRT-PCR.
Trisure (Bioline) was used to isolate total RNA from cells and reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (K1691; Thermo Scientific) according to the manufacturer’s protocols. cDNAs were amplified using specific primers on a Rotor Gene (Qiagen) real-time PCR instrument. Roche SYBR Green was used for qRT-PCR. Quantifications were performed using the ΔΔCt (threshold cycle) method, and gene expression levels were normalized to Gapdh transcript levels using the following expression: (primer efficiency)−ΔΔCt, where ΔΔCt means ΔCt (target gene) − ΔCt (reference gene). We analyzed the results from three or more independent experiments using the Student’s t test.
RNA-Seq Library Preparation and Sequencing.
Total RNA was isolated from control and IRE1-inhibited (with STF-083010) BMDM samples using Trisure (Bioline). To remove genomic DNA contamination, the RNA samples were treated with 20 U DNase I (New England Biolabs). rRNA was depleted from 5 µg total RNA using Ribo-Zero Gold rRNA Removal Kit (Epicentre Biotechnologies) following the manufacturer’s recommendations. Sequencing libraries for whole-transcriptome analysis were prepared using the ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre Biotechnologies) following the manufacturer’s recommendations. After 3′-terminal tagging, the di-tagged cDNA was purified using column concentrators (DNA clean-up and concentrators; Zymo Research). The cDNA libraries were bar-coded to allow sample multiplexing using ScriptSeqTM Index PCR Primers (Epicentre Biotechnologies). The libraries were amplified by 12 cycles of PCR, and the amplified libraries were size-selected and purified using 8% TBE (Tris/borate/EDTA) acrylamide gels. The libraries were quantified using Agilent Technologies 2100 Bioanalyzer. Up to four RNA-seq libraries were then multiplexed in a single lane of an Illumina HiSeq2500 Deep-Sequencer Flow Cell (University of California, San Francisco Center for Advanced Technologies) and sequenced using 50-bp single-end sequencing chemistry.
RNA Sequencing Data Processing.
The 3′ adapter sequences (AGATCGGAAGAGCACACGTCTGAAC) were removed from the sequenced libraries using the FastQ/A clipper found in the FastX Toolkit (hannonlab.cshl.edu/fastx_toolkit/) after d-multiplexing, and only reads longer than 20 nt were kept for alignment. The adapter-stripped reads were then aligned with the Bowtie indices for the mouse genome reference version 10 of the University of California, Santa Cruz Genome Browser (mm10) using the splice junction mapper Tophat2 v2.0.9 and the sequence aligner Bowtie 2 V2.2.3.0 with default parameters. The transcript assembler Cufflinks V2.1.1 was then used on the list of mapped reads to assemble and quantify transcripts using an mm10 reference annotation and masking mitochondrial, rRNA, and tRNA sequences. To estimate the changes in gene expression levels, the number of sequenced reads that align to a gene of interest was then compared among biological samples using Cuffdiff (default parameters). Changes in the levels of expression of normalized Cufflinks-quantified transcripts are expressed as fragments per kilobase of transcript per million mapped reads (FPKM).
Flow Cytometric Analysis of Intracellular Cytokine Staining.
Fresh splenocytes were prepared from mice spleen, and erythrocytes were removed using RBC (red blood cell) lysis buffer as described earlier (43). Cells were stimulated for 4 hours with phorbol-myristate-acetate (PMA; 50 ng/mL; Abcam) and ionomycin (1 µg/mL; Abcam) in the presence of Golgi stop (BD Biosciences). Live cells were discriminated from dead ones by using Zombie Green (Bio Legend). Cell surfaces were stained with PerCP-Cy5.5–conjugated anti-CD4 antibody (BD Biosciences) followed by incubation in Cytofix/Cytoperm Solution (BD Biosciences) at room temperature for 15 min. Then, intracellular cytokines were stained with allophycocyanin (APC)-conjugated IFN-γ, phycoerythrin (PE)-conjugated IL-17A, and PE-conjugated IL-4 antibodies (all from BD Biosciences). Data were analyzed on BD Accuri C6 software.
Measurement of Secreted IL-1β and IL-18 and CCL2 Cytokines.
An IL-1β Elisa Kit (Abcam) was used for detecting IL-1β; a Mouse IL-18 ELISA Kit (Medical & Biological Laboratories) and a Mouse CCL2 Elisa Kit (Abcam) were used for detecting the cytokines in mouse plasma or conditioned medium, respectively, as indicated according to the manufacturer’s instructions.
Plasma Measurements.
FPLC was used for analyzing the size distribution of lipoproteins. All measurements were carried out at the Mouse Metabolic Phenotyping Center at the University of Cincinnati. For the resolution of major lipoprotein classes from plasma; the columns were equilibrated in 50 mM PBS. Using a microtiter plate enzyme-based assay, the major lipoprotein classes were measured in cholesterol or triglyceride assays from collected fractions.
Staining of Cryosections.
Cryosections (7-μm thick) were cut from the aortic root of the frozen heart tissue with a cryostat (Leica CM1850) and stained with Oil Red O, anti–MOMA-2 (monocyte/macrophage marker) (ab33451; Abcam), anti–CD3-Alexa488 (1:400; Biolegend), TUNEL Kit (11684795910; Roche), anti–α-SMA (ab5694; Abcam), and IL-1β antibody (ab9722; Abcam). Immunofluorescent staining was mounted with an antifade reagent including DAPI (GR211467-2; Abcam). Representative images were taken with a Zeiss Fluorescent Microscope. Collagen content of the lesions and fibrous cap thickness were determined in Masson’s Trichrome-stained lesions (as per the manufacturer’s protocol; Bio-Optica). Fibrous cap thickness was calculated according to the published protocol (59). Heart tissue sections were stained with Oil Red O stain for plaque area quantifications and H&E for necrotic core quantification in accord with previously published protocols (43). All quantifications were performed using ImageJ (NIH). Percentage of average cross-sectional stained area per leaflet was calculated from all three valves.
Atherosclerotic Lesion Analysis.
Aortas were pinned on a black wax surface, and atherosclerotic lesions were analyzed in the aortic arch and descending aorta by Sudan IV staining as described earlier (43). Areas were quantified using ImageJ and expressed as the percentages of the total aorta area.
Quantification of mtROS.
mtROS production was measured with MitoSOX Red Mitochondrial Superoxide Indicator (M36008; Life Technologies) according to the protocol provided by the company. Representative images were acquired with a confocal microscope (LSM 510; Zeiss) and analyzed with ImageJ.
Mitochondrial Calcium Measurement.
RHOD-2 AM (cell-permeable fluorescent calcium indicator) (Thermo Scientific) was used for mitochondria-specific calcium measurement according to previously published protocols (60).
Mice and Treatments.
ApoE−/− mice in a C57BL/6 background (Charles River WIGA GmbH) were used in atherosclerosis experiments. Starting from 8 weeks of age, male mice were fed a Western diet (TD88137 mod. containing 21% fat and 0.2% cholesterol; Ssniff) for 6 weeks. Then, the mice were injected with STF-083010 (10 mg/kg) or DMSO, both given in 16% (vol/vol) Cremophor EL (Sigma-Aldrich) saline solution via i.p. injections as described previously, for 6 more weeks while mice were continued on the Western diet (21). The other ApoE−/− mice that were used in atherosclerosis experiments were fed a Western diet for 8 weeks. Then, they were injected with 4µ8c (10 mg/kg) or DMSO, both given in 16% (vol/vol) Cremophor EL saline solution via i.p. injections as described previously (33), for 4 more weeks while mice were continued on Western diet. Weights were measured every other day, whereas blood glucose concentrations were measured before and after treatments. At the end of the experiment, mice were anesthetized, and blood was collected by cardiac puncture. Bone marrow, spleen, and liver tissues were collected, frozen immediately into liquid nitrogen, and stored at −80 °C. Perfusion was performed with ice-cold PBS and heparin (1,000 U/mL) followed by 10% formalin solution (Sigma). After fixation, the aorta was dissected intact, immersed immediately in 10% formalin, and stored at 4 °C until analysis. The heart was removed at the proximal aorta, placed into a tissue mold, covered with OCT (optimal cutting temperature compound) (Tissue-Tek), frozen in cold isobutene solution, and stored at −80 °C. All animal experiments were performed according to protocols approved by the Experimental Animal Care Committee at Bilkent University.
Statistical Analysis.
Values are expressed as mean ± SEM. No samples were treated as outliers and left out of analysis. Statistical significance was evaluated using the Student’s t test or the Mann–Whitney test (for in vivo analysis as indicated in the figures). P < 0.05 was considered as significant.
Supplementary Material
Acknowledgments
We thank Gokhan S. Hotamisligil (Harvard University) for providing IRE1−/− MEFs, Esther Lutgens and Dirk Lievens for technical advice on bone marrow-derived macrophages (BMDMs), Buket Gültekin for administrative help, and Mojtaba Beyramzadeh for technical assistance with atherosclerosis studies. This work was funded, in part, by Scientific and Technological Research Council of Turkey (TUBITAK)-German Federal Ministry of Education and Research (BMBF) Intensified Cooperation Grant 110S293 (to C.W. and E.E.), TUBITAK Grant 113Z023 (to E.E.), and European Research Council (ERC) Starting Grant 336643 (to E.E.). D.A.-A. was supported by an Irvington Postdoctoral Fellowship from the Cancer Research Institute. P.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621188114/-/DCSupplemental.
References
- 1.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 4.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186(3):323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou AX, Tabas I. The UPR in atherosclerosis. Semin Immunopathol. 2013;35(3):321–332. doi: 10.1007/s00281-013-0372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol. 1995;15(5):551–561. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabas I. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Circ Res. 2010;107(7):839–850. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myoishi M, et al. Increased endoplasmic reticulum stress in atherosclerotic plaques associated with acute coronary syndrome. Circulation. 2007;116(11):1226–1233. doi: 10.1161/CIRCULATIONAHA.106.682054. [DOI] [PubMed] [Google Scholar]
- 9.McAlpine CS, Werstuck GH. The development and progression of atherosclerosis: Evidence supporting a role for endoplasmic reticulum (ER) stress signaling. Cardiovasc Hematol Disord Drug Targets. 2013;13(2):158–164. doi: 10.2174/1871529x11313020009. [DOI] [PubMed] [Google Scholar]
- 10.Zeng L, et al. Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc Natl Acad Sci USA. 2009;106(20):8326–8331. doi: 10.1073/pnas.0903197106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorp E, et al. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe-/- and Ldlr-/- mice lacking CHOP. Cell Metab. 2009;9(5):474–481. doi: 10.1016/j.cmet.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci R, et al. Requirement of JNK2 for scavenger receptor A-mediated foam cell formation in atherogenesis. Science. 2004;306(5701):1558–1561. doi: 10.1126/science.1101909. [DOI] [PubMed] [Google Scholar]
- 13.Babaev VR, et al. Jnk1 deficiency in hematopoietic cells suppresses macrophage apoptosis and increases atherosclerosis in low-density lipoprotein receptor null mice. Arterioscler Thromb Vasc Biol. 2016;36(6):1122–1131. doi: 10.1161/ATVBAHA.116.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erbay E, et al. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat Med. 2009;15(12):1383–1391. doi: 10.1038/nm.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimura S, et al. Reduction of endoplasmic reticulum stress inhibits neointima formation after vascular injury. Sci Rep. 2014;4:6943. doi: 10.1038/srep06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, et al. The modulation of endoplasmic reticulum stress by chemical chaperone upregulates immune negative cytokine IL-35 in apolipoprotein E-deficient mice. PLoS One. 2014;9(1):e87787. doi: 10.1371/journal.pone.0087787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engin F, Hotamisligil GS. Restoring endoplasmic reticulum function by chemical chaperones: An emerging therapeutic approach for metabolic diseases. Diabetes Obes Metab. 2010;12(Suppl 2):108–115. doi: 10.1111/j.1463-1326.2010.01282.x. [DOI] [PubMed] [Google Scholar]
- 18.Ong DS, Kelly JW. Chemical and/or biological therapeutic strategies to ameliorate protein misfolding diseases. Curr Opin Cell Biol. 2011;23(2):231–238. doi: 10.1016/j.ceb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12(9):703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 20.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73(6):1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 21.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das I, et al. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 2015;348(6231):239–242. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mimura N, et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119(24):5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gargalovic PS, et al. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26(11):2490–2496. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- 26.Gora S, et al. Phospholipolyzed LDL induces an inflammatory response in endothelial cells through endoplasmic reticulum stress signaling. FASEB J. 2010;24(9):3284–3297. doi: 10.1096/fj.09-146852. [DOI] [PubMed] [Google Scholar]
- 27.Outinen PA, et al. Homocysteine-induced endoplasmic reticulum stress and growth arrest leads to specific changes in gene expression in human vascular endothelial cells. Blood. 1999;94(3):959–967. [PubMed] [Google Scholar]
- 28.Feng B, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5(9):781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Lhoták S, Hilditch BA, Austin RC. Activation of the unfolded protein response occurs at all stages of atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation. 2005;111(14):1814–1821. doi: 10.1161/01.CIR.0000160864.31351.C1. [DOI] [PubMed] [Google Scholar]
- 30.So JS, et al. Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16(4):487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkmann K, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286(14):12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cross BC, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109(15):E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu Q, et al. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32(18):2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upton JP, et al. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338(6108):818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerner AG, et al. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16(2):250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemp KL, et al. The serine-threonine kinase inositol-requiring enzyme 1α (IRE1α) promotes IL-4 production in T helper cells. J Biol Chem. 2013;288(46):33272–33282. doi: 10.1074/jbc.M113.493171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bright MD, Itzhak DN, Wardell CP, Morgan GJ, Davies FE. Cleavage of BLOC1S1 mRNA by IRE1 is sequence specific, temporally separate from XBP1 splicing, and dispensable for cell viability under acute endoplasmic reticulum stress. Mol Cell Biol. 2015;35(12):2186–2202. doi: 10.1128/MCB.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross BC, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109(15):E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Nosak C, Sollazzo P, Odisho T, Volchuk A. IRE1 inhibition perturbs the unfolded protein response in a pancreatic β-cell line expressing mutant proinsulin, but does not sensitize the cells to apoptosis. BMC Cell Biol. 2014;15:29. doi: 10.1186/1471-2121-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaffer JE. Lipotoxicity: When tissues overeat. Curr Opin Lipidol. 2003;14(3):281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 42.Wen H, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12(5):408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Çimen I, et al. Prevention of atherosclerosis by bioactive palmitoleate through suppression of organelle stress and inflammasome activation. Sci Transl Med. 2016;8(358):358ra126. doi: 10.1126/scitranslmed.aaf9087. [DOI] [PubMed] [Google Scholar]
- 44.Guo H, Callaway JB, Ting JP. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robblee MM, et al. Saturated fatty acids engage an IRE1α-dependent pathway to activate the NLRP3 inflammasome in myeloid cells. Cell Reports. 2016;14(11):2611–2623. doi: 10.1016/j.celrep.2016.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami T, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci USA. 2012;109(28):11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol Sci. 2012;33(2):53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Wu T, et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28(7):708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Paulsson G, Stemme S, Hansson GK. Hypercholesterolemia is associated with a T helper (Th) 1/Th2 switch of the autoimmune response in atherosclerotic apo E-knockout mice. J Clin Invest. 1998;101(8):1717–1725. doi: 10.1172/JCI1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitman SC, Ravisankar P, Daugherty A. IFN-gamma deficiency exerts gender-specific effects on atherogenesis in apolipoprotein E-/- mice. J Interferon Cytokine Res. 2002;22(6):661–670. doi: 10.1089/10799900260100141. [DOI] [PubMed] [Google Scholar]
- 51.Huber SA, Sakkinen P, David C, Newell MK, Tracy RP. T helper-cell phenotype regulates atherosclerosis in mice under conditions of mild hypercholesterolemia. Circulation. 2001;103(21):2610–2616. doi: 10.1161/01.cir.103.21.2610. [DOI] [PubMed] [Google Scholar]
- 52.Ait-Oufella H, Taleb S, Mallat Z, Tedgui A. Recent advances on the role of cytokines in atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31(5):969–979. doi: 10.1161/ATVBAHA.110.207415. [DOI] [PubMed] [Google Scholar]
- 53.Austin RC. The unfolded protein response in health and disease. Antioxid Redox Signal. 2009;11(9):2279–2287. doi: 10.1089/ars.2009.2686. [DOI] [PubMed] [Google Scholar]
- 54.Adiguzel E, Ahmad PJ, Franco C, Bendeck MP. Collagens in the progression and complications of atherosclerosis. Vasc Med. 2009;14(1):73–89. doi: 10.1177/1358863X08094801. [DOI] [PubMed] [Google Scholar]
- 55.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313(5783):104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Wang GZ, Rabinovitch PS, Tabas I. Macrophage mitochondrial oxidative stress promotes atherosclerosis and nuclear factor-κB-mediated inflammation in macrophages. Circ Res. 2014;114(3):421–433. doi: 10.1161/CIRCRESAHA.114.302153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320(5882):1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freigang S, et al. Nrf2 is essential for cholesterol crystal-induced inflammasome activation and exacerbation of atherosclerosis. Eur J Immunol. 2011;41(7):2040–2051. doi: 10.1002/eji.201041316. [DOI] [PubMed] [Google Scholar]
- 59.Fredman G, et al. An imbalance between specialized pro-resolving lipid mediators and pro-inflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat Commun. 2016;7:12859. doi: 10.1038/ncomms12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, Armstrong JS. Bax and the mitochondrial permeability transition cooperate in the release of cytochrome c during endoplasmic reticulum-stress-induced apoptosis. Cell Death Differ. 2007;14(4):703–715. doi: 10.1038/sj.cdd.4402072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.