Significance
Methylation of genomic DNA is an epigenetic modification at the interface between genetic information and environmental stimuli underlying many phenotypic variations in the human population as well as the pathogenesis of complex diseases. Accordingly, mutations in the de novo DNA methyltransferase enzyme DNMT3A have been identified in a number of diseases, including mast cell-related disorders. However, the role of DNA methylation and DNMT3A in regulating mast cell physiology still needs to be elucidated. Here, we found that Dnmt3a plays a critical role in modulating mast cell responsiveness to acute and chronic stimulation, potentially implicating DNA methylation-mediated processes in all types of mast cell-related diseases.
Keywords: DNA methylation, epigenetics, inflammation, mast cells
Abstract
DNA methylation and specifically the DNA methyltransferase enzyme DNMT3A are involved in the pathogenesis of a variety of hematological diseases and in regulating the function of immune cells. Although altered DNA methylation patterns and mutations in DNMT3A correlate with mast cell proliferative disorders in humans, the role of DNA methylation in mast cell biology is not understood. By using mast cells lacking Dnmt3a, we found that this enzyme is involved in restraining mast cell responses to acute and chronic stimuli, both in vitro and in vivo. The exacerbated mast cell responses observed in the absence of Dnmt3a were recapitulated or enhanced by treatment with the demethylating agent 5-aza-2′-deoxycytidine as well as by down-modulation of Dnmt1 expression, further supporting the role of DNA methylation in regulating mast cell activation. Mechanistically, these effects were in part mediated by the dysregulated expression of the scaffold protein IQGAP2, which is characterized by the ability to regulate a wide variety of biological processes. Altogether, our data demonstrate that DNMT3A and DNA methylation are key modulators of mast cell responsiveness to acute and chronic stimulation.
DNA methylation is an epigenetic process in which a methyl group is covalently linked to a cytosine base in the genomic DNA, predominantly at CpG dinucleotides, yielding 5-methylcytosine (5mC). Such a process is carried out by three DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B), and has a critical role in the control of gene expression (1, 2). In general, high levels of DNA methylation are associated with transcriptional silencing (3), especially when present at promoter regions and at repetitive elements (4), although the function of DNA methylation at other genomic features and its correlation with gene expression are more uncertain (3–5). Whereas DNMT1 is thought to be primarily responsible for copying the preexisting methylation to the newly synthesized DNA strand during replication, DNMT3A and DNMT3B display significant affinity also for unmethylated DNA, and are therefore considered de novo methyltransferases (6, 7). DNA methylation is essential during development: Various mouse models have shown that the absence of Dnmt1 or Dnmt3b is embryonically lethal, and mice lacking Dnmt3a die within 4 wk after birth because of their failure to thrive (6, 8); in humans, mutations in the DNMT3A gene are associated with an overgrowth syndrome with intellectual disability (9). More specific to the hematopoietic compartment, loss of Dnmt1 in hematopoietic stem cells (HSCs) led to defects in self-renewal, niche retention, as well as altered cell differentiation, especially toward the myeloid lineage (10), whereas loss of both Dnmt3a and Dnmt3b impaired HSC self-renewal capabilities (11). Importantly, aberrant DNA methylation is a hallmark of many diseases, including autoimmune diseases and especially various types of cancer (4, 5). Mutations in DNMT3A have been found in a variety of hematological malignancies (4, 12, 13), including systemic mastocytosis, a clonal proliferative disorder of mast cells (14), pointing toward a role for DNMT3A in modulating mast cell biology. Further correlating DNA methylation with the biology of mast cells (which are key effector cells in asthmatic and allergic responses), a recent survey compared atopic and asthmatic patients with healthy controls and identified 81 differentially methylated regions (15); the hypomethylated regions included genes such as IL13, which is not only crucial in asthma pathogenesis but is also expressed at high levels by mast cells (16). Finally, highlighting the potential relevance of understanding the role of DNA methylation in mast cell biology, altered DNA methylation patterns were identified in patients with mast cell activation disease, a complex disorder characterized by aberrant release of mast cell-derived mediators (17). We therefore set out to investigate the role of DNA methylation in general and DNMT3A in particular in regulating mast cell differentiation and function.
We found that mast cells lacking Dnmt3a appeared to be more responsive to stimuli compared with their wild-type counterparts. Among other phenotypes, stimulation with IgE and antigen complexes triggered a significantly stronger acute response in mast cells lacking Dnmt3a, including higher cytokine production and increased degranulation capacity. Such phenotypes were recapitulated or exacerbated by treatment of the cells with the demethylating agent 5-aza-2′-deoxycytidine as well as by reducing Dnmt1 expression, further supporting the notion that DNA methylation-regulated processes are important modulators of mast cell activation. Mechanistically, these effects were likely to be mediated, at least in part, by the dysregulated expression of the scaffold protein IQGAP2 (IQ motif-containing GTPase-activating protein 2), and led to exacerbated in vivo responses in both acute and chronic models of mast cell activation, namely passive cutaneous anaphylaxis and oxazolone-induced dermatitis. Our results indicate that appropriate regulation of DNMT3A-mediated processes modulates mast cell responses to environmental stimuli, both in vitro and in vivo, and may be relevant in all types of mast cell activation diseases.
Results
Increased Susceptibility to IgE Stimulation of Mast Cells Lacking Dnmt3a.
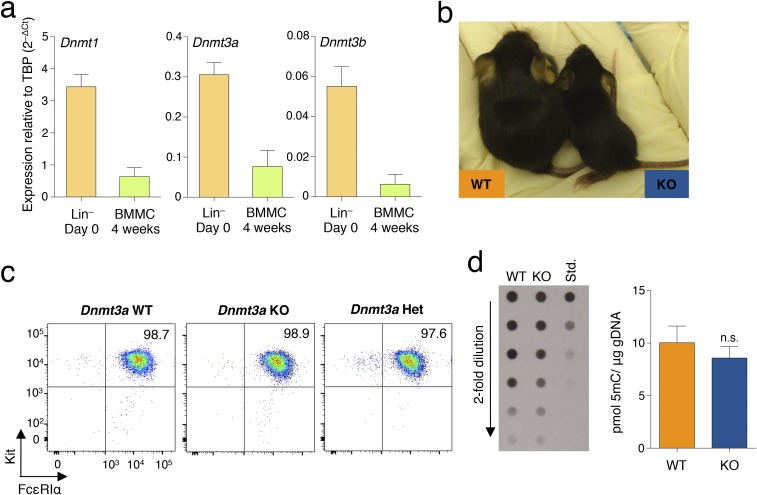
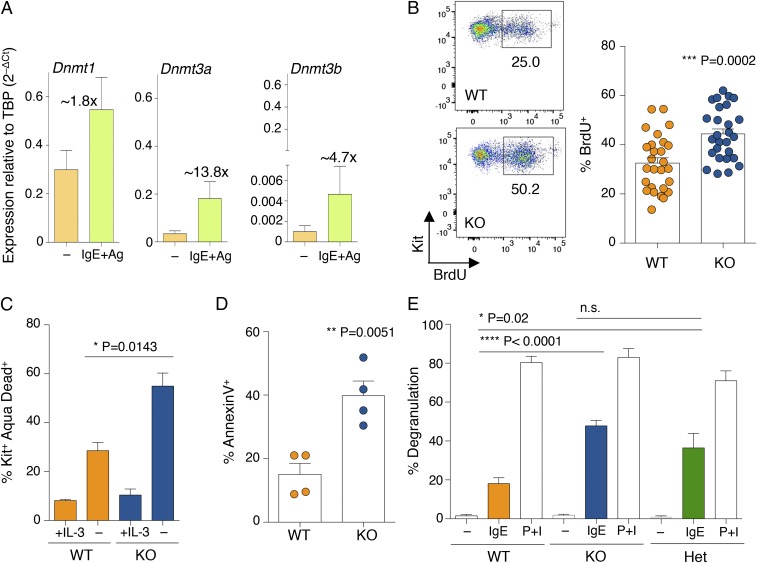
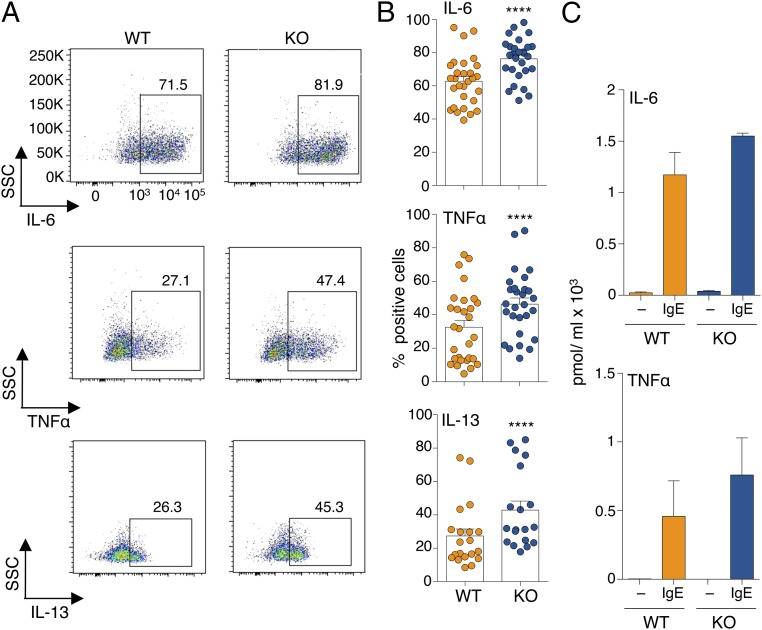
The mRNAs of the three DNA methyltransferases were expressed at relatively high levels in HSCs, and their levels decreased upon differentiation to mast cells (Fig. S1A). However, all of them remained expressed at detectable levels in differentiated mast cells, with Dnmt1 displaying the highest relative expression, Dnmt3b the lowest, and Dnmt3a expressed at intermediate levels (Fig. 1A and Fig. S1A). Expression of all Dnmts was induced upon acute stimulation of mast cells with IgE and antigen complexes (Fig. 1A), with Dnmt3a being the most inducible (about 13.8-fold after 6 h of stimulation), suggesting a potential role for this enzyme in modulating mast cell responses following activation. Mice genetically deleted for Dnmt3a are runt (Fig. S1B), and die perinatally within 4 wk after birth (6). We found that differentiation to mast cells was unchanged in the absence of Dnmt3a, and the resulting gross phenotype was undistinguishable regardless of the genotype (Fig. S1C). However, Dnmt3a knockout (KO) mast cells showed a significant increase in proliferation compared with their wild-type (WT) counterparts (Fig. 1B). This is in accordance with the fact that DNMT3A is frequently mutated in myeloid neoplasms, including systemic mastocytosis (14), and deletion of this enzyme predisposes the cells to myeloid transformation (18). To assess whether such increased proliferation was also associated with increased survival in vitro, mast cells were transiently deprived of the essential survival factor IL-3. Removal of IL-3 from the culture medium led to an overall increase in cell death, which was, however, much more pronounced in the absence of Dnmt3a (Fig. 1C), potentially pointing toward an increased susceptibility of these cells to perturbations in their microenvironment. We therefore assessed mast cell responses to physiological stimulation with IgE and antigen. Interestingly, upon acute stimulation, mast cells lacking Dnmt3a showed significantly increased ability to release the content of their cytoplasmic granules, as assessed both by annexin V binding [which occurs at sites of secretory granule fusion with the plasma membrane (19–21)] and release of β-hexosaminidase (an enzyme normally stored in cytoplasmic granules) in the culture supernatant (Fig. 1 D and E). Similarly, production of IL-6, TNF-α, and IL-13 (cytokines secreted by mast cells at high levels in response to IgE stimulation) was also increased in Dnmt3a-deleted cells, as assessed both by intracellular staining (Fig. 2 A and B) and ELISA (Fig. 2C).
Fig. S1.
(A) Reduced expression of Dnmts upon differentiation of hematopoietic precursors into mast cells. Lin− hematopoietic precursors were isolated from murine bone marrow using a Lineage Cell Depletion Kit (Miltenyi Biotec). Cells were either immediately lysed in TRIzol for RNA extraction or differentiated into mast cells for 4 wk, after which expression of Dnmt1, Dnmt3a, and Dnmt3b was assessed by qRT-PCR. n = 3 independent experiments. Mean ± SEM. (B) Dnmt3a KO mice are runt and fail to thrive. Shown is one example of a Dnmt3a KO pup at 2.5 wk of age, together with an age-matched littermate. (C) Dnmt3a KO mast cells are indistinguishable from their WT counterparts. Bone marrow cells were obtained from age- and sex-matched Dnmt3a WT, KO, and heterozygous mice and were in vitro differentiated into mast cells. Surface staining for the mast cell markers Kit and FcεRIα revealed no gross phenotypic differences among the three genotypes. (D) Deletion of Dnmt3a does not lead to a significant reduction in overall levels of genomic 5mC. Genomic DNA (gDNA) was extracted from WT and Dnmt3a KO mast cells, and overall levels of 5mC were measured by dot blot assay using an anti-5mC antibody. Data in the histogram are from n = 18 measurements for WT cells and n = 17 measurements for Dnmt3a KO mast cells. Mean ± SEM; unpaired t test, two-tailed.
Fig. 1.
Mast cells lacking Dnmt3a display a complex phenotype. (A) WT differentiated mast cells were either stimulated with IgE and antigen (Ag) for 6 h or left untreated. After RNA extraction, expression of the indicated Dnmt mRNA was assessed by qRT-PCR. PCR primers used in this study are listed in Table S1. n = 5 independent experiments. Mean ± SEM. TBP, TATA-box-binding protein. (B) Mast cell proliferation was assessed between weeks 4 and 7 of differentiation by BrdU incorporation. One representative experiment is shown (Left); each dot represents one experiment. Mean ± SEM; unpaired t test, two-tailed. (C) Mast cells were deprived of IL-3 for 48 h prior to staining with an Aqua Dead dye to determine cell survival. Shown is the percentage of dying mast cells (Kit+ Aqua Dead+) in the absence of IL-3 in n = 3 independent experiments. Mean ± SEM; unpaired t test, two-tailed. (D) Degranulation of mast cells was measured by annexin V staining upon stimulation with IgE and antigen complexes for 30 min. Each dot represents one experiment. Mean ± SEM; unpaired t test, two-tailed. (E) Mast cell degranulation upon stimulation with either IgE and antigen complexes or phorbol 12-myristate 13-acetate (PMA) and ionomycin (P+I) was measured by β-hexosaminidase release. P+I stimulation was used as a positive control to induce maximum degranulation. n = 4 independent experiments. Mean ± SEM; unpaired t test, two-tailed. n.s., not significant.
Fig. 2.
Increased cytokine production in the absence of Dnmt3a. (A) Intracellular staining for the indicated cytokines in mast cells stimulated with IgE and antigen for 3 h. One representative experiment is shown. SSC, side-scatter. (B) Same as A; each dot represents one experiment. Mean ± SEM; paired t test, two-tailed (****P < 0.0001). (C) Release of IL-6 and TNF-α in the supernatant was measured by ELISA after 12 h of stimulation. n = 4 independent experiments. Mean ± SEM.
Dysregulated DNA Methylation Activity Leads to Increased Mast Cell Responses.
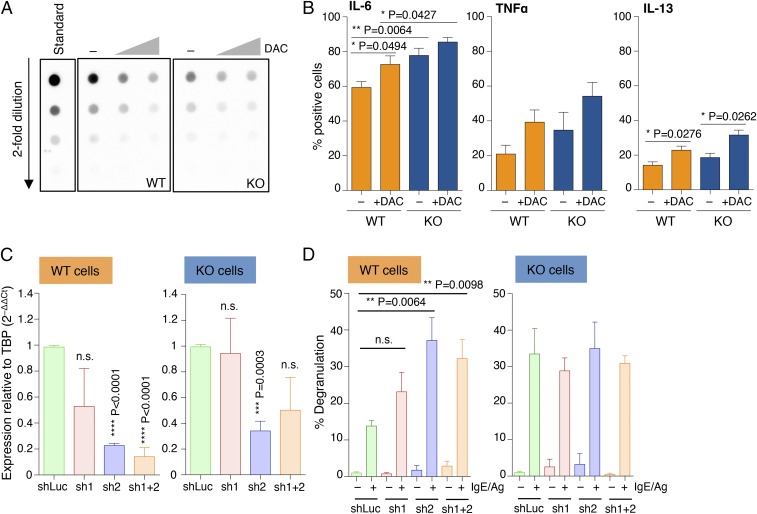
To further evaluate the impact of DNA methylation on the ability of mast cells to respond to external stimuli, we treated WT and KO cells for 48 h with 5-aza-2′-deoxycytidine (decitabine; DAC), a drug commonly used for the treatment of myelodysplastic syndromes and known to inhibit DNA methyltransferase enzymes (22). Such treatment led to global genomic hypomethylation in mast cells (Fig. 3A), which not only correlated with increased capacities of WT mast cells to produce cytokines but also further exacerbated the phenotype displayed by KO mast cells (Fig. 3B). To further investigate the role of DNA methylation in mast cell responses, we used shRNAs to down-modulate the expression of Dnmt1 (Dnmt3b was expressed by mast cells only at extremely low levels; Fig. 1A). Reducing Dnmt1 expression provided WT mast cells with increased degranulation capabilities (Fig. 3 C and D). Such an increase in degranulation was very prominent especially for one of the two shRNAs tested (sh2), which also showed the highest reduction in Dnmt1 expression (Fig. 3C). Interestingly, reduction of Dnmt1 expression did not further increase the ability of Dnmt3a KO cells to degranulate in response to IgE stimulation (Fig. 3D), suggesting that Dnmt3a KO cells may have already reached their maximum degranulation capacity in response to IgE and antigen stimulation.
Fig. 3.
Increased mast cell responses upon disruption of DNA methylation activities. (A) Differentiated mast cells were treated with 0.5 and 5 μM 5-aza-2′-deoxycytidine for 48 h, after which genomic DNA was extracted and overall levels of 5mC were quantified by dot blot. (B) Mast cells were treated with 0.5 μM DAC for 3 d before stimulation with IgE and antigen and measurement of cytokine production by intracellular cytokine staining. Mean ± SEM; unpaired t test, two-tailed. (C) Mast cells were transduced with lentiviral vectors (sh1 and sh2) expressing shRNAs to knock down expression of Dnmt1. A vector expressing an irrelevant hairpin (shLuc) was used as control. After puromycin selection of the transduced cells, total RNA was extracted and the extent of Dnmt1 down-regulation was measured by qRT-PCR. Shown are the compiled results of three independent experiments. Mean ± SEM; unpaired t test (relative to the shLuc sample), two-tailed. (D) Same as C, except that cells were stimulated with IgE and antigen prior to measurement of the extent of degranulation by β-hexosaminidase release. Shown are the compiled results of three independent experiments. Mean ± SEM; unpaired t test, two-tailed.
The DNA methylation status of different cell types, including cells of the hematopoietic lineage, with or without Dnmt3a, has been previously studied (23–25): First, HSCs lacking Dnmt3a showed global 5mC levels that were comparable to those of control cells, with a very modest number of differentially methylated regions (24). In agreement with these findings, we found that overall levels of genomic 5mC were not significantly different in WT and KO mast cells, despite a trend toward slightly diminished levels in KO cells (Fig. S1D). Second, the differentially methylated regions identified in Dnmt3a-null cells showed substantial hyper- as well as hypomethylation at a variety of genomic regions, with very little correlation between changes in methylation and differential gene expression in the absence of Dnmt3a in both mouse and human cells (4, 24, 25). These observations suggest the existence of both direct and indirect effects due to the loss of Dnmt3a, and potentially also the existence of DNMT3A activities that are independent of its DNA catalytic activity (26), similar to what has been described for the catalytically inactive DNMT3L (27), some of the DNMT3B isoforms (28), and even for the TET family of DNA-modifying enzymes (29, 30). To gain more insight into the mechanisms that may underlie increased mast cell responses in the absence of Dnmt3a, we therefore investigated global gene expression in WT and KO mast cells.
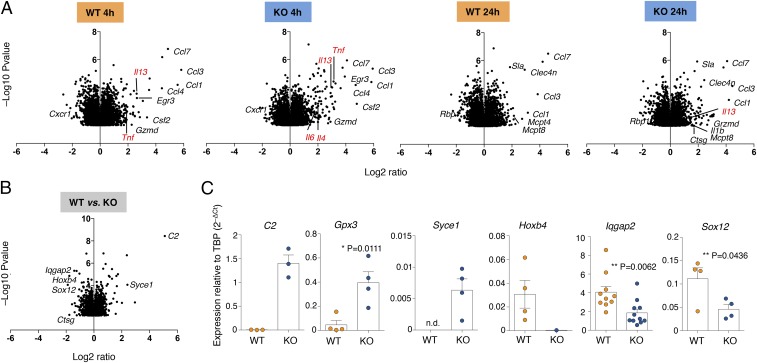
Altered Gene Expression and Chromatin Accessibility in the Absence of Dnmt3a.
Using Illumina microarrays, we performed gene expression analyses of WT and KO mast cells, either resting or stimulated with IgE and antigen, for 4 and 24 h. Triplicate biological samples were used and data were quantile-normalized. Differentially expressed transcripts were identified by analysis of variance (ANOVA) (P value < 0.05). We found that upon stimulation of KO cells, most transcriptional changes were of limited magnitude compared with WT cells, and these especially included changes in the levels of expression of many cytokine and chemokine genes (Fig. 4A). For instance, the Tnf gene was induced 8.7-fold in KO cells and 3.6-fold in WT cells after 4 h (Fig. 4A). Similarly, the Il13 gene was induced 7.9-fold in KO cells and 6.2-fold in WT cells. These findings were also in agreement with the increased cytokine production observed in KO cells by intracellular staining and ELISA (Fig. 2). We did not detect any compensatory increase in Dnmt1 and/or Dnmt3b expression in the absence of Dnmt3a. We therefore focused our attention on genes that were differentially expressed between WT and KO cells, regardless of stimulation (Fig. 4B). Overall, 2,654 genes were significantly differentially expressed in mast cells in the absence of Dnmt3a; of these, only 76 genes were up-regulated at least 1.5-fold and 100 genes were down-regulated at least 1.5-fold, indicating that the extent of transcriptional effects was overall rather subtle. We validated some selected genes by quantitative (q)RT-PCR using independent biological samples (Fig. 4C). Genes that were up-regulated in KO cells included C2, encoding for the complement component C2 of the classical C3 convertase, Gpx3 (glutathione peroxidase 3), and Syce1 (synaptonemal complex central element protein 1), encoding for a protein involved in chromosome segregation (31). Genes that were down-regulated in KO cells included Hoxb4, encoding for a transcription factor (TF) involved in development and a positive regulator of HSC self-renewal (32); the TF Sox12 (33); and Iqgap2, encoding for a scaffold protein with a variety of different functions (34).
Fig. 4.
Altered gene expression in the absence of Dnmt3a. (A) Mast cells (three independent biological samples) were either left untreated or stimulated with IgE and antigen for 4 or 24 h; RNA was extracted and gene expression was analyzed by microarray. Shown is the effect of stimulation on gene expression in either WT or KO cells. (B) Same as A, except that genes differentially expressed between WT and KO cells (regardless of stimulation) are shown. (C) Selected genes from B were validated in independent biological samples by qRT-PCR. Each dot represents one experiment. Mean ± SEM; unpaired t test, two-tailed.
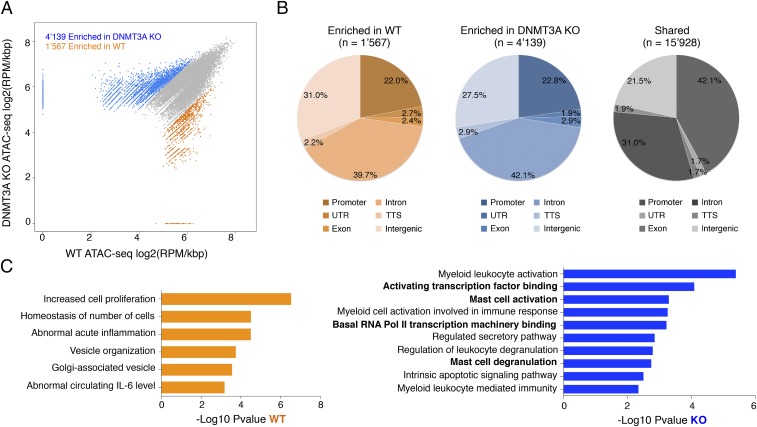
To gain a better understanding of the mechanisms that could lead to dysregulated gene expression in the absence of Dnmt3a, we performed ATAC-seq (assay for transposase-accessible chromatin sequencing) (35) to assess the overall accessibility of genomic regions in WT and Dnmt3a KO mast cells. We found that many regions were more accessible in Dnmt3a KO mast cells compared with WT (4,139 regions enriched in KO cells vs. 1,567 regions enriched in WT cells) (Fig. 5A). Such regions were similarly distributed in WT and KO cells between promoter, intronic, and intergenic regions (Fig. 5B). Most importantly, a GREAT (Genomic Regions Enrichment of Annotations Tool) analysis (36) of the regions specifically enriched in KO cells revealed gene ontology categories that were highly correlated with the phenotype observed in the absence of Dnmt3a (Fig. 5C), most notably “mast cell degranulation” and “mast cell activation.” Moreover, pointing toward dysregulated TF binding and RNA polymerase II (Pol II) recruitment, other significant categories for Dnmt3a KO cells included “activating transcription factor binding” and “basal RNA Pol II transcription machinery binding.” Overall, these data indicate that the absence of Dnmt3a results in increased accessibility of genomic regions that may be involved in regulation of mast cell activation.
Fig. 5.
Chromatin accessibility in WT and KO cells. (A) The scatter plot shows ATAC-seq signals as reads per million (RPM) per kilobase at chromatin accessible regions in WT and Dnmt3a KO cells. Regions enriched in WT cells are depicted in orange; regions enriched in KO cells are in blue; shared regions are in gray. (B) The diagram illustrates the overall distribution of enriched and shared peaks into promoter, untranslated region (UTR), exon, intron, transcription termination site (TTS), and intergenic regions. (C) Functional enrichment analysis of the enriched regions was performed using GREAT. The full list of categories is provided in Dataset S1.
Reducing Iqgap2 Expression Is Sufficient to Increase Mast Cell Degranulation.
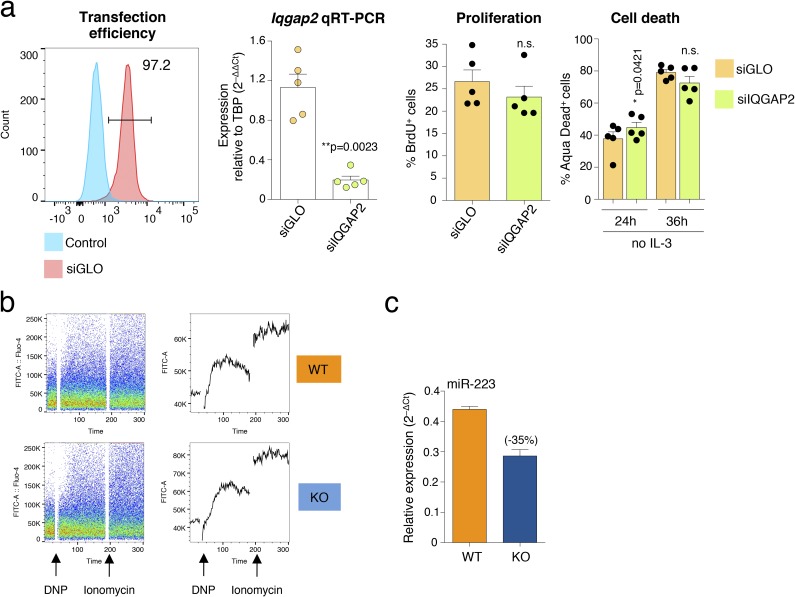
Among the differentially expressed genes in the absence of Dnmt3a, Iqgap2 showed a large decrease in expression in KO cells (about −54.5%; Fig. 4C). IQGAP proteins (IQGAP1, 2, and 3) are widely expressed proteins with the ability to regulate very diverse biological processes, including cytoskeleton dynamics, vesicle trafficking, cell proliferation, intracellular signaling, and TF activity (34). Most importantly, T lymphocytes from IQGAP1-deficient mice displayed increased production of cytokines, and both IQGAP1 and IQGAP2 were shown to interact with the long noncoding RNA NRON (noncoding RNA repressor of NFAT), known to interfere with nuclear import of the TF NFAT (37, 38). We therefore hypothesized that altered expression of Iqgap2 could explain, at least in part, the various phenotypes observed in activated mast cells lacking Dnmt3a, including increased cytokine production and degranulation. First, we measured the expression of the other two family members, Iqgap1 and Iqgap3, and found that, contrary to Iqgap2, their expression was slightly increased in KO cells, although in a nonsignificant manner (Fig. 6A). IQGAP proteins are, however, known to act as a scaffold for many interacting partners: We therefore assessed the effect of depleting Iqgap2 itself by transient transfection of siRNAs in WT differentiated mast cells. Two days after transfection, the average reduction in Iqgap2 expression compared with a control oligonucleotide was ∼68% (Fig. 6B), indicating effective knockdown to levels comparable to those observed in KO cells (Fig. 4C). Importantly, such down-regulation of Iqgap2 expression corresponded to a significant increase in mast cell degranulation (Fig. 6C), whereas cytokine expression showed just a very modest increase, which was statistically significant only for IL-13 (Fig. 6D). Notably, mast cell proliferation and survival were not significantly altered in WT mast cells upon knockdown of Iqgap2 (Fig. S2A), despite an initial modest increase in cell death, which, however, normalized over time. These data point toward different mechanisms of regulation of acute (degranulation, cytokine production) and homeostatic (proliferation, survival) responses in mast cells.
Fig. 6.
Reduced expression of Iqgap2 is sufficient to increase mast cell degranulation. (A) Expression of IQGAP family members Iqgap1 and Iqgap3 was measured by qRT-PCR in WT and KO mast cells. Each dot represents one experiment. Mean ± SEM; unpaired t test, two-tailed. (B) Mast cells were transiently transfected with a pool of siRNAs to knock down Iqgap2 expression. Forty-eight hours after transfection, total RNA was extracted and the extent of Iqgap2 down-modulation was assessed by qRT-PCR. Each dot represents one experiment. Mean ± SEM; paired t test, two-tailed (****P < 0.0001). (C) Same as B, except that transfected cells were stimulated with IgE and antigen, and their ability to degranulate was measured by the release of β-hexosaminidase enzyme. n = 4 independent experiments. Mean ± SEM; unpaired t test, two-tailed. (D) Cells were treated as in C, except that cytokine production was measured by intracellular staining. Each dot represents one experiment. Mean ± SEM; paired t test, two-tailed. (E) WT and KO mast cells were transduced with a lentiviral vector to stably express HA-NFAT1(4–460)-GFP. After selection, cells were stimulated with IgE and antigen for the indicated times, and nuclear translocation of HA-NFAT1(4–460)-GFP was followed over time using a fluorescence microscope. Representative of n = 8 independent experiments. (F) Same as B, except that protein extracts were prepared from transduced cells and the extent of HA-NFAT1(4–460)-GFP dephosphorylation upon stimulation was assessed by Western blot using an anti-HA antibody. (G) Nuclear translocation of endogenous NFAT1 upon stimulation of WT and KO cells with 0.5 μM ionomycin was visualized by immunofluorescence using an anti-NFAT1 antibody. Nuclei were counterstained with DAPI. (H) Calcium flux was measured by flow cytometry in WT and KO cells loaded with Fluo-4 AM and stimulated with IgE anti-DNP antibody and HSA-DNP antigen as well as ionomycin. Shown are the compiled data of n = 5 independent experiments. Mean ± SEM; two-way ANOVA. MFI, mean fluorescence intensity.
Fig. S2.
(A) Analysis of mast cell proliferation and survival upon knockdown of Iqgap2. WT mast cells were transfected with Amaxa with the indicated siRNAs; siGLO is a fluorescent conjugated siRNA used to assess efficiency of transfection, which was reproducibly close to 100%. Efficient knockdown of Iqgap2 was assessed by qRT-PCR, cell proliferation was assessed by BrdU incorporation assay, and cell death upon withdrawal of the essential cytokine IL-3 was assessed by Aqua Dead staining as described in SI Materials and Methods. Each dot represents one independent experiment. Mean ± SEM; paired t test, two-tailed. (B) Example of calcium measurement upon stimulation of WT and KO mast cells. (C) Reduced miR-223 expression in Dnmt3a KO mast cells. Total RNA was extracted from WT and KO mast cells. miR-223 expression was measured by qRT-PCR using Exiqon microRNA LNA PCR primer sets. n = 2 independent experiments. Mean ± SEM.
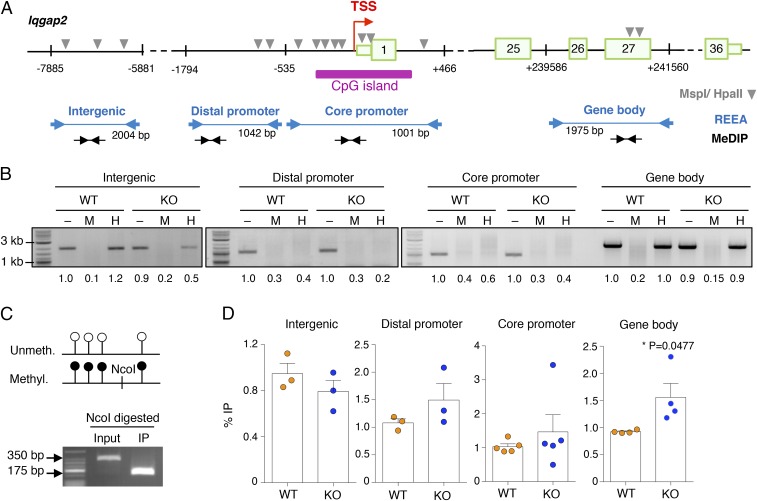
Because IQGAP1 and IQGAP2 were shown to interfere with nuclear translocation of NFAT (37, 38), and NFAT is required for efficient transactivation of cytokine genes in mast cells (39, 40), we assessed whether nuclear translocation of NFAT was somehow affected in KO cells, due to the altered expression of Iqgap family members. First, we transduced WT and KO cells to stably express comparable levels of a reporter NFAT-GFP fusion protein (41), and then we assessed nuclear translocation of the reporter protein with a fluorescence microscope (Fig. 6E). Upon stimulation with IgE and antigen, we observed comparable NFAT nuclear translocation in both WT and KO cells, with comparable kinetics. Moreover, NFAT-GFP nuclear export at later time points of stimulation also appeared to be comparable in WT and KO cells (Fig. 6E). Similarly, we found that the extent and kinetics of NFAT-GFP dephosphorylation and rephosphorylation upon stimulation with IgE and antigen were also comparable between WT and KO cells by Western blot (Fig. 6F). A similar trend was observed also for endogenously expressed NFAT1 (Fig. 6G), ruling out the possibility that increased NFAT translocation in KO cells could somehow contribute to the increased responsiveness of Dnmt3a KO cells. Accordingly, flow cytometry measurements of changes in calcium concentration upon WT and KO mast cell stimulation revealed only a modest trend toward slightly increased Ca2+ concentration in KO cells, which was, however, not statistically significant and of insufficient magnitude to alter the kinetics of NFAT translocation (Fig. 6H and Fig. S2B). We also assessed the levels of DNA methylation specifically at the Iqgap2 locus (Fig. 7A) by restriction enzyme accessibility assay (REAA) and methylated DNA immunoprecipitation (MeDIP). First, by performing REAA analysis, we observed that the gene body of Iqgap2 was overall methylated in both WT and KO cells, whereas both the core and distal promoter regions were mostly hypomethylated (Fig. 7B). Consistent with the published observation that differentially methylated regions in Dnmt3a-null cells showed substantial hyper- as well as hypomethylation at a variety of genomic regions (4, 24, 25), we found that an intergenic region in the proximity of the Iqgap2 locus was substantially hypomethylated in KO cells (Fig. 7B). Because REAA is not sensitive enough to measure small differences in methylation levels, we performed a MeDIP analysis on these same regions. First, we optimized the IP method using as spike-in a mixture of two plasmids, either unmethylated or in vitro methylated, at a ratio of 4:1. As shown in Fig. 7C, before the IP, a PCR specific for the spike-in plasmids amplified primarily the more abundant unmethylated plasmid, whereas after the IP, we recovered only the methylated form of the plasmid (containing an extra NcoI restriction site that allowed for size discrimination of the PCR product), confirming the specificity of the process. By performing MeDIP on WT and KO mast cells, we found that the distal promoter, core promoter, and gene body regions of Iqgap2 showed a modest increase in DNA methylation in the absence of Dnmt3a (Fig. 7D). At the core promoter region, results were more variable and differences were smaller, most likely due to the fact that this region coincided with a CpG island and was for the most part hypomethylated. The intergenic region appeared to be slightly demethylated in KO cells, concordant with the results of the REAA. Whether such differences in DNA methylation between WT and KO cells are sufficient to justify the observed altered expression of the Iqgap2 gene remains to be understood. Other mechanisms, direct or indirect, may also lead to altered Iqgap2 expression in Dnmt3a KO cells. Overall, our data suggest that diminished Iqgap2 expression may contribute, at least in part, to the increased degranulation and cytokine production observed in mast cells lacking Dnmt3a, that such phenotypes appear to be NFAT-independent, and that it only partially correlates with changes in DNA methylation at the Iqgap2 locus.
Fig. 7.
Methylation analysis of the Iqgap2 locus. (A) Schematic representation (not to scale) of the Iqgap2 locus, with the location of the primers used for REAA and MeDIP-PCR indicated, as well as the location of the restriction sites (MspI/HpaII) required for REAA. Numbered boxes represent exons. (B) REAA of the Iqgap2 locus: Purified genomic DNA of WT and KO cells was either left untreated (–) or digested with the methylation-insensitive enzyme MspI (M) or its CpG methylation-sensitive isoschizomer HpaII (H) before PCR analysis. Representative of n = 2 or 3 independent experiments, depending on the region. Numbers below the bands represent the relative quantification (to input WT) of band intensity. The sizes of the PCR products are indicated in the scheme in A. (C) To check for the efficiency of the IP reaction in all MeDIP experiments, control DNA, containing in vitro methylated and nonmethylated pUC19 plasmid at a ratio of 1:4, was added to every sample before the IP. The combination of plasmid-specific PCR primers with NcoI restriction digestion (site present only in the methylated plasmid) confirmed the specific enrichment of methylated DNA in all MeDIP experiments. Shown is one representative experiment out of at least four. (D) MeDIP-PCR of the Iqgap2 locus: Sonicated genomic DNA from WT and KO cells was immunoprecipitated with an anti-5mC antibody. Shown is the percentage of immunoprecipitated methylated DNA relative to the input for each region. Each dot represents one independent experiment. Mean ± SEM; unpaired t test, two-tailed.
Exacerbated in Vivo Responses in the Absence of Dnmt3a on Acute and Chronic Mast Cell Activation.
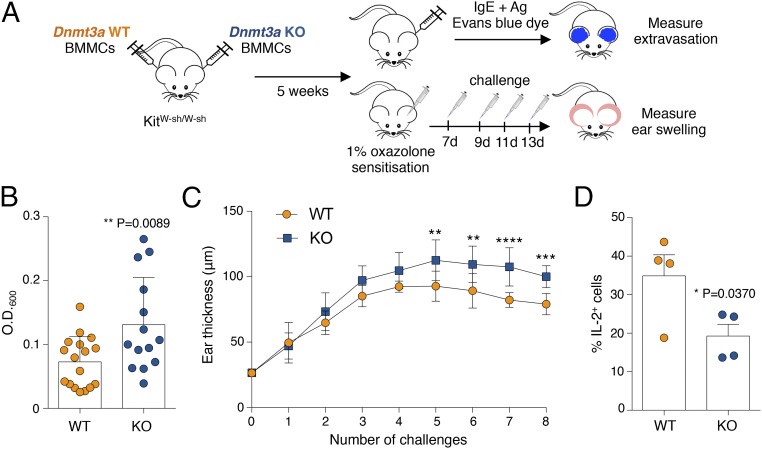
Because mast cells lacking Dnmt3a appeared phenotypically normal but with an increased propensity to respond to stimulation with enhanced cytokine production and degranulation activity, we assessed acute responses (which are primarily dependent on the release of mast cell granules) in an in vivo model of anaphylaxis-associated vascular hyperpermeability (passive cutaneous anaphylaxis) as well as chronic responses in a model of oxazolone-induced dermatitis (Fig. 8A). For passive cutaneous anaphylaxis, we performed adoptive transfer of WT and KO cells in KitW-sh/W-sh mice (42) by intradermal injection. Following reconstitution, mast cells were sensitized with IgE–anti-DNP antibody followed by systemic injection of antigen together with Evans blue dye (Fig. 8 A and B). As a measure of mast cell activation, we quantified the cutaneous extravasation of the dye after extraction from the tissue (Fig. 8B). In agreement with the increased responses observed in KO mast cells in vitro, we observed a significantly enhanced response of KO mast cells also during acute responses in vivo (Fig. 8B).
Fig. 8.
Increased acute and chronic inflammatory responses in vivo in the absence of Dnmt3a. (A) Schematic representation of the experimental design for passive cutaneous anaphylaxis and oxazolone-induced contact sensitivity experiments. BMMCs, bone marrow-derived mast cells. (B) For passive cutaneous anaphylaxis experiments, the ear pinnae of KitW-sh/W-sh mice were reconstituted with WT or KO mast cells. After ∼5 wk, ears were sensitized intradermally with IgE and then challenged 24 h later by i.v. injection of antigen in the presence of Evans blue dye. The dye was then extracted and measured. Each dot represents one mouse (WT, n = 17; KO, n = 14). Mean ± SD; unpaired t test, two-tailed. (C) Mice were reconstituted as in B, except that they were then challenged with 1% oxazolone, followed, 7 d later, by repeated challenges with 0.5% oxazolone. The extent of inflammation was assessed by measuring swelling of the ears. Data include measurements of eight mice (mean ± SD) and are representative of n = 2 independent experiments. Comparisons between the groups were performed by two-way ANOVA. **P = 0.0061, ***P < 0.0001, ****P < 0.0001. (D) WT and KO cells were stimulated for 5 h with PMA and ionomycin before intracellular cytokine staining for IL-2 expression. Each dot represents one experiment. Mean ± SEM; paired t test, two-tailed.
Allergic diseases are usually associated with repeated exposures to the allergen, which over time lead to chronic inflammation and tissue remodeling. Chronic responses in tissues are the result of a complex interplay between stromal cells and various immune cells, including mast cells, which can secrete several mediators with the potential to influence tissue remodeling. Oxazolone induces a T cell-dependent allergic contact hypersensitivity, which also involves mast cells and evolves over time into a chronic allergic inflammatory response that resembles atopic dermatitis (43). We therefore investigated whether the increased inflammatory response of Dnmt3a KO mast cells could also play a role during chronic responses. After adoptive transfer of WT and KO mast cells into KitW-sh/W-sh mice, the skin was first sensitized with 1% oxazolone, followed by repeated challenges with 0.5% oxazolone (Fig. 8A). We found that also in this chronic model, Dnmt3a KO mast cells acted in a more proinflammatory manner compared with their WT counterparts, leading over time to significantly increased ear thickness (Fig. 8C). Because the extent of inflammation in this model was shown to be dependent on mast cell-derived IL-2 production (43), which has been implicated in the control of allergic diseases, we also measured mast cell-derived IL-2 by intracellular cytokine staining. We found a significant decrease in the ability of Dnmt3a KO cells to produce IL-2 (Fig. 8D), strongly suggesting that the capacity of KO cells to produce increased levels of inflammatory cytokines, coupled with the reduced production of IL-2, determines overall exacerbated responses to both acute and chronic stimulation. Overall, our data uncover a role for Dnmt3a in restraining mast cell proinflammatory responses.
Discussion
DNMT3A is one of the most frequently mutated genes in hematological malignancies and, due to extensive research, its role in cancer is becoming clearer (44, 45). Although we found a relative increase in the ability of mast cells to proliferate in the absence of Dnmt3a, most of the major phenotypes we observed in these cells were actually linked to acute and chronic cell activation, both in vitro and in vivo. Although the role of DNA methylation in general and DNMT3A in particular in allergy, asthma, and mast cell activation disorders is far from being completely understood, several studies have investigated the role of Dnmt3a in regulating cytokine production in T lymphocytes (reviewed in ref. 5). For example, the Il2, Il4, and Ifng cytokine genes are regulated by DNA methylation in T cells (46–48) and, in the absence of Dnmt1 or Dnmt3a (but not Dnmt3b), T lymphocytes were unable to appropriately silence the expression of Ifng or Il4, resulting in unrestrained cytokine expression (49–51). T cells lacking Dnmt3a also showed increased Il13 transcription (51), similar to the phenotype we observed in mast cells and, accordingly, these lymphocytes led to increased lung inflammation in a murine model of asthma, further highlighting the importance of DNA methylation in modulating the activity and pathogenesis of complex diseases such as allergy and asthma. Here, we found that not only did loss of Dnmt3a lead to enhanced cytokine production in mast cells in response to IgE and antigen complexes but it also led to increased degranulation, suggesting that dysregulated DNA methylation may have a profound effect on mast cell-related diseases.
The complex phenotype resulting from the genetic deletion of Dnmt3a is likely the end result of a number of direct and indirect effects acting on the regulation of gene expression at multiple levels. Regulation of gene expression is not a linear one-to-one process but a network of interactions between multiple chromatin- and DNA-modifying enzymes, TFs, and cofactors, as well as posttranscriptional regulators such as RNA-binding proteins and microRNAs (miRNAs), all acting in concert to determine a final outcome (52). For instance, altered DNA methylation patterns can interfere directly with the recruitment of readers of DNA methylation and with the binding of TFs, potentially leading to a cascade of altered TF recruitment and gene expression, which eventually hinders normal cellular functions. Apart from TFs and cofactors able to directly or indirectly bind the chromatin and translate DNA methylation information into a transcriptional outcome, miRNAs represent some of the factors with a complex interplay with DNA methylation. Indeed, not only DNA methylation can alter miRNA expression and consequently expression of downstream genes; miRNAs are also known to fine-tune the expression of target DNA-modifying enzymes. For example, the miR-29 family was shown to target DNMT1, DNMT3A, and DNMT3B, leading to global DNA hypomethylation and reexpression of tumor suppressor genes in acute myeloid leukemia cells (53). Although our preliminary screening did not identify any major significant modification in the global miRnome of Dnmt3a KO mast cells, we identified miR-223 as a dysregulated miRNA in the absence of Dnmt3a (Fig. S2C). We found that expression of this miRNA was reproducibly reduced in Dnmt3a KO mast cells compared with their WT counterparts, and such reduction could also indirectly contribute to the increased ability of Dnmt3a KO mast cells to degranulate in response to acute stimulation. Indeed, down-regulation of miR-223 was recently shown to promote mast cell degranulation in response to IgE stimulation (54).
Although the role of DNA methylation in stabilizing gene expression during development and cell-fate decisions and in maintaining memory of cellular identity is well-established (55, 56), its role during acute stimulation of already-differentiated cells is more nuanced and less understood. In this respect, as mentioned above, T lymphocytes have been studied more extensively than other cell types, showing in general unrestrained expression of genes related to effector functions. Our data provide clear evidence about a role for DNA methylation in general and Dnmt3a in particular in restraining responses of differentiated mast cells, suggesting that a more general role of DNA methylation in at least some types of differentiated cells may be to buffer and restrict excessive responses to environmental stimuli.
Materials and Methods
Mice and Cell Cultures.
Dnmt3a knockout mice were purchased from The Jackson Laboratory, where they were generated as described (57). The Dnmt3a-deleted allele lacks exons 13 to 17 of the protein, roughly corresponding to part of the ATRX–DNMT3–DNMT3L domain and part of the catalytic domain, and leads to no detectable expression. Heterozygous intercrosses were used to generate age- and sex-matched KO and WT littermates for use in all experiments shown. Mast cells were differentiated in the presence of IL-3 as the essential survival and proliferation factor, as described (58). All animal studies were performed in accordance with Swiss Federal Veterinary Office guidelines and approved by the Cantonal animal experimentation committee, Dipartimento della Sanità e della Socialità Cantone Ticino (authorization nos. 02/2015 and 07/2015).
Gene Expression Omnibus Accession Numbers.
Gene expression profiling: Data are available for download at the Gene Expression Omnibus (GEO) database under accession no. GSE87483. ATAC-seq: Data are available in the GEO database under accession no. GSE91036.
Statistical Analysis.
Statistical analysis was performed with Prism software (GraphPad). Data are represented as mean ± SEM or SD, and significance was assessed by paired or unpaired Student’s t test, two-tailed, or two-way ANOVA.
Full materials and methods can be found in SI Materials and Methods.
SI Materials and Methods
Surface and Intracellular Staining.
For staining of surface receptors, the following antibodies were used (BioLegend): CD117 (Kit)-APC/Cy7 (or APC); and FcεRIα-PE. Intracellular cytokine staining was performed exactly as described (29, 59, 60). Briefly, cells were stimulated with 1 μg/mL IgE–anti-DNP and 0.2 μg/mL HSA-DNP (both from Sigma) for 3 h. To detect IL-2 production, 20 nM PMA and 2 µM ionomycin were used. Brefeldin A (10 μg/mL) was added in the last 2 h of stimulation, after which cells were fixed with 4% (wt/vol) paraformaldehyde, permeabilized with 0.5% saponin, and stained with the following antibodies: IL-6-PE and TNF-α-PE/Cy7 (BioLegend) and IL-13-PE (eBioscience).
ELISA and Immunofluorescence.
The measurement of cytokine release in the culture supernatant was performed using the Mouse TNFα (or IL-6) High Sensitivity ELISA (eBioscience) following the manufacturer’s instructions. Briefly, 3 × 105 cells were stimulated with 1 μg/mL IgE–anti-DNP and 0.2 μg/mL HSA-DNP (both from Sigma) for 12 h in a 48-well plate before collection of the supernatant and measurement of the cytokine release. Immunofluorescence experiments were performed as follows: Mast cells (1.5 × 105) were spun on cytospin slides (4 min at 15 × g) and subsequently fixed with 4% paraformaldehyde for 10 min. Cells were then permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature (RT) and blocked with 1% BSA, 0.05% Tween 20 in PBS at RT for 1 h. Endogenous levels of NFAT1 were detected using an anti-NFAT1 antibody (D43B1; Cell Signaling; diluted 1:1,000) and a 1:400 dilution of an anti-rabbit secondary antibody conjugated with Alexa Fluor 488. Nuclei were counterstained with DAPI. For microscope image acquisition, a Nikon Eclipse E800 upright microscope was used; images were acquired with an EM-CCD camera (Hamamatsu; C9100 digital camera), using the acquisition software VisiView (Visitron Systems) version 2.0.8.
Cell Proliferation and Cell Death.
Mast cell proliferation in response to IL-3 was measured by BrdU incorporation exactly as described previously (29, 59), using an APC BrdU Flow Kit from BD Biosciences. For analysis of cell survival, mast cells were deprived of IL-3 for 24 to 48 h prior to staining with an Aqua Dead dye (LIVE/DEAD Fixable Aqua Dead Cell Stain Kit; Life Technologies) to determine the extent of cell death.
Degranulation Assays.
Degranulation assays were performed as described (19). Briefly, 8 to 15 × 105 mast cells were preloaded with 0.5 to 1 μg/mL IgE–anti-DNP overnight at 37 °C. Cells were then resuspended in Tyrode’s buffer and stimulated in a 96-well U-bottom plate with 0.2 μg/mL HSA-DNP for 1 h at 37 °C. As positive control, cells were stimulated for 1 h with 20 nM PMA and 2 µM ionomycin. After stimulation, 10 µL of supernatant was collected in a flat-bottom 96-well plate. Cell pellets were then lysed in 0.5% Triton X-100 in Tyrode’s buffer, and 10 µL from the cell lysate was transferred to a second flat-bottom 96-well plate. Samples of supernatant and cell lysate were then incubated for 1 h with 50 μL β-hexosaminidase substrate (4-nitrophenyl N-acetyl-β-d-glucosaminide; 4 mM; Sigma). The reaction was stopped with 150 µL of 0.2 M glycine (pH 10.7), and the absorbance was read at 405 nm. The percentage of degranulation was calculated as the ratio between the absorbance of the supernatants and the total absorbance of the supernatants and cell lysates. Alternatively, degranulation was assessed using a PE Annexin V Kit (BD Pharmingen), because during the membrane fusion process of degranulation, annexin V binding occurs at sites of secretory granule exposure to the cell surface (19–21). Briefly, cells were stimulated for 30 min with 1 μg/mL IgE–anti-DNP and 0.2 μg/mL HSA-DNP, before staining using a PE Annexin V Staining Kit (BD Pharmingen) following the manufacturer’s instructions exactly.
Calcium Flux Assay.
To measure calcium mobilization upon receptor triggering, we preloaded mast cells (2 × 106 cells per mL) with 0.5 μg/mL IgE–anti-DNP antibody for 4 h in medium without IL-3 and then loaded with 3 μM Fluo-4 AM (Molecular Probes) calcium indicator for 30 min at 37 °C. Calcium flux was induced by adding HSA-DNP (30 ng/mL) and ionomycin (2 μM) and measured by flow cytometry. Analysis of the kinetics of changes in intracellular calcium concentration in different experiments was performed exactly as described (61).
qRT-PCR.
For quantitative RT-PCR, total RNA was extracted using TRI reagent (MRC), and cDNA was prepared with qScript cDNA SuperMix (Quanta Biosciences). Primer sequences are listed in Table S1. qPCR was performed with an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Data analysis was performed using the 2–ΔΔCt or 2–ΔCt method (62).
Table S1.
List of primers used in this study
| Primers | Forward, 5′-3′ | Reverse, 5′-3′ |
| qRT-PCR | ||
| Dnmt1 | TAAGGAGGACAAGGAGAATGC | GCCACCAAACTTCACCATATC |
| Dnmt3a | GTATGAGGTGCGCCAGAAGT | CATGCCTCCAATGAAGAGTG |
| Dnmt3b | GCTGGGTACAGTGGTTTGGT | GTGTGGTACATGGCCTTCCT |
| C2 | CTTGATGCTTCTCAGAGTGTGACAG | GGATCTCTCACTCAGGATCGAC |
| Gpx3 | CCGGGGACAAGAGAAGTCTA | TAGCTGGCTACGTTGACAAAGA |
| Syce1 | CAGCCGTTGGGTATGGAG | ACGGTTAATCAGGACCTCAATC |
| Hoxb4 | AGAGCCCGTCGTCTACCC | CGTCAGGTAGCGATTGTAGTGA |
| Sox12 | GAGGAGACGGTGGTATCTGG | CGCATCATCTCGGTAACCTC |
| Iqgap1 | AATAACAGCAAGTGGGTGAAGC | TATATGCAGCGGTTACTCCAGA |
| Iqgap2 | CAAGTACCAAGACATTCTCAACGAG | TACACAGGTCTTGATGTAGGTGTC |
| Iqgap3 | AATGTCGCCTATCAGTACCTCTG | TCCACGTCGTAGATCTTCTTCAG |
| REAA | ||
| Intergenic | GCCAGTGTGCACTAAGCTACTCCTCTC | CACATTGCTTCTATAGCACCAAATGCC |
| Iqgap2 distal promoter | GATAGGATGTTCTCCACCCAAC | GACAAGAAACGGTAAGGGTGAG |
| Iqgap2 core promoter | TGCTGTGGTTTTTCTTGCTCT | AGGAAAGTGCCTTCACCTCTC |
| Iqgap2 gene body | TGCTCACTTAGCCTTTTTCCA | AGGGAGCTAAGAGCATTGAGG |
| MeDIP | ||
| Intergenic | CTTGCTCTACATGCTGCTTCC | GCCTCATTTCTAGGCTCCACT |
| Iqgap2 distal promoter | CCTGTGGCCATAGCATTAGAA | GAAGTTGTGCCAGCCTATGAG |
| Iqgap2 core promoter | GAGGAGAAAGGAAACCTGCTG | CGGGGCAAAAGTGAACTATCT |
| Iqgap2 gene body | TTTTCCCTTGTGCAGATTGTT | TCCCAAGTTTCTCCTTTGGTT |
RNA Interference.
Oligonucleotide pools for RNAi against mouse Iqgap2 were purchased from Thermo Fisher Scientific (SMARTpool; ON-TARGETplus Iqgap2 siRNA). For transfection, 1 to 1.5 × 106 cells were transfected with 300 pmol of double-stranded oligonucleotides using the Amaxa Mouse T Cell Nucleofector Kit (program X-01). Efficiency of transfection was evaluated in each case by transfecting cells with a fluorescent oligonucleotide (siGLO; Dharmacon).
Plasmids and Lentiviral Transductions.
Lentiviral vectors were generated using standard cloning techniques. The shRNAs against Dnmt1 were cloned into the pAPM lentiviral vector (19, 58) and had the following sequences: sh1, 5′-GAGTGTGTGAGGGAGAAA-3′; sh2, 5′-GTGACGTCGAAGACTCCTG-3′. HA-NFAT1(4–460)-GFP was a gift from Anjana Rao, La Jolla Institute for Allergy and Immunology, La Jolla, CA (Addgene; plasmid 11107) (41), and was subcloned into the pScalps lentiviral vector (29, 59). Lentiviral transductions of mast cells were performed exactly as described (29, 58, 59). Briefly, HEK293T cells were transfected with the desired lentiviral vector together with packaging vectors psPAX2 and pMD2.G (Addgene; plasmids 12260 and 12259). After concentration as described (58), viral particles were added to the mast cell cultures. Successfully transduced cells were selected 2 to 3 d after transduction with 2 μg/mL puromycin for 24 to 48 h.
Gene Expression Profiling.
Total RNA was extracted from three independent cultures of bone marrow-derived mast cells from different animals, either KO or WT littermates, age and sex (females) matched. Gene expression was analyzed using the MouseRef-8 v2.0 Expression BeadChip (Illumina). Data were first extracted with Illumina GenomeStudio software and then imported into Genomics Suite 6.4 (Partek) and quantile-normalized. Differentially expressed transcripts were identified by ANOVA in KO mast cells (P < 0.05).
ATAC-Seq.
To profile chromatin accessibility, we used the original ATAC-seq protocol (35) with a few modifications (63). Briefly, 50,000 cells were lysed in 50 µL of cold lysis buffer (10 mM Tris⋅HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Igepal CA-630). The supernatant was discarded and nuclei were resuspended in 25 µL of reaction buffer (50 mM Tris⋅HCl, pH 8.4, 25 mM MgCl2) containing 1 µL Tn5 transposase (made in-house). The tagmentation reaction was incubated at 37 °C for 30 min, and then stopped with 5 µL of cleanup buffer (900 mM NaCl, 300 mM EDTA), 2 µL 5% (wt/vol) SDS, and 2 µL proteinase K (20 µg/µL) (New England Biolabs) and incubation for 30 min at 40 °C. Tagmented DNA was purified using 2× SPRI beads. Finally, 2 µL indexing primers and KAPA HiFi HotStart ReadyMix were used to PCR-amplify the obtained DNA library of tagmented DNA. Fragments smaller than 600 bp were isolated by size selection (using 0.65× SPRI beads) and then purified with 1.8× SPRI beads. DNA concentration was then measured with a Qubit fluorometer (Life Technologies). Before sequencing, ATAC-seq library size was assessed using a Bioanalyzer High Sensitivity DNA chip (Agilent Technologies). Libraries were sequenced on an Illumina HiSeq 2000 at 10M read depth (50-bp single reads). For ATAC-seq data analysis, short reads were quality-filtered according to the Illumina pipeline. Sequencing adaptors were removed using Trimmomatic (64), and trimmed reads were aligned to the GRCm38/mm10 assembly of the mouse genome using Bowtie 2 v2.2.6 (65) with the “–very-sensitive” preset of parameters. Reads that did not align to the nuclear genome or aligned to the mitochondrial genome were removed. Moreover, duplicate reads were marked and removed using SAMtools (66), and read positions were corrected for transposon insertion offset as described (35). Peaks with significantly enriched signal were called using MACS2 (version 2.1.1.20160309) (67) using the “–nomodel,” “–extsize 100,” and “–qvalue 0.01” flags and arguments. Peaks with a fold enrichment <5 (as determined by MACS2) and those blacklisted by the ENCODE Consortium analysis of artifactual signals in mouse cells (https://sites.google.com/site/anshulkundaje/projects/blacklists) were removed using BEDTools (68). To categorize ATAC-seq peaks into those with similar or differing levels of accessibility in WT versus Dnmt3a KO cells, all peaks were merged to provide one set of combined peaks. CoverageBed (68) was then used to compute coverage, which was transformed to reads per kilobase per million mapped reads and log2-transformed for each region. R software (https://www.r-project.org, version 3.2.1) was used to generate the scatter plot. Regions with absolute fold change >2 were classified as differentially enriched, and all others were considered shared regions. To annotate genomic locations and assign each region to the nearest transcription start site, the annotatePeaks script from the HOMER package (69) was used, based on the GENCODE mouse release M11 (70). For each list of enriched regions of interest, GREAT 3.0.0 (36) was used with default parameters and selecting the whole mm10 genome as background. The complete lists of functional categories are provided in Dataset S1. We applied RPM normalization to all datasets, and tracks for visualization in the UCSC Genome Browser (71) were generated using the bedGraphToBigWig tool.
DAC Treatment and 5mC Quantification.
For DAC treatment, cells were treated with 0.5 and 5 μM 5-aza-2′-deoxycytidine for 48 to 72 h. Genomic DNA was isolated with a QIAamp DNA Mini Kit (Qiagen), and twofold dilutions from a starting amount of 500 ng were spotted on a nitrocellulose membrane before incubation with an anti-5mC antibody (clone 33D3; EpiGentek), as previously described (72).
Restriction Enzyme Accessibility Assay and Methylated DNA Immunoprecipitation.
For REAA, the methylation status of CCGG restriction sites in selected genomic regions was analyzed using MspI (CpG methylation-insensitive) and HpaII (blocked by CpG methylation) restriction enzymes. Genomic DNA from WT and KO mast cells was purified with the DNeasy Blood & Tissue Kit (Qiagen); 1 μg of DNA was then digested for 18 h with either MspI or HpaII (5,000 units per mL; New England Biolabs) or left untreated. Digested DNA was further treated with 1 mg/mL proteinase K (Ambion) for 30 min at 40 °C, followed by heat inactivation at 95 °C for 10 min and PCR amplification. For MeDIP, 2.5 µg of genomic DNA was fragmented to a size of 300 to 500 bp using a Sonicator Bioruptor Plus System (Diagenode), and the IP was then performed using a MeDIP Kit (Zymo Research) following the manufacturer’s instructions. Briefly, 1 µg of sonicated DNA was diluted in DNA-denaturing buffer and denatured for 5 min at 98 °C; 500 ng was then saved as input and 500 ng underwent IP using 5 µg of anti-5mC antibody for 1 h at 37 °C, followed by DNA purification. qPCR was performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems), and the percentage of immunoprecipitated DNA was calculated using the following formula: % input = 2(Ct input − Ct MeDIP) × input dilution factor × 100. As a control for the efficiency of IP, control DNA containing both in vitro methylated and nonmethylated pUC19 plasmid was spiked into every sample prior to IP. PCR amplification using plasmid-specific primers, combined with digestion with NcoI (whose site was present only in the PCR from the methylated plasmid), allowed for the assessment of the efficiency of the IP.
Western Blots.
For protein extracts, 2 to 3 × 106 cells were lysed in 1% Triton-RIPA buffer (50 mM Tris⋅HCl, pH 7.4, 1% Nonidet P-40, 0.5% Na-deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA) supplemented with phosphatase (PhosSTOP EASYPack; Roche) and protease inhibitors (Sigma). Protein concentration was measured with a Pierce BCA Protein Assay Kit (Thermo Fisher). Samples (20 µg) were run on 10% SDS/polyacrylamide gels, and immunodetection was performed with 1:1,000 dilutions of either an anti-NFAT1 (D43B1; Cell Signaling) or anti-HA (072M4847; Sigma) antibody. An anti-GAPDH antibody (2606352; Millipore) was used for loading controls.
Passive Cutaneous Anaphylaxis.
In vivo anaphylaxis experiments were performed as previously described (59, 60). Briefly, the ear pinnae of mice lacking mast cells (KitW-sh/W-sh; The Jackson Laboratory) (42) were reconstituted with 2 × 106 WT or KO mast cells, injected intradermally. After 4 to 5 wk, ears were sensitized with an intradermal injection of 1 µg IgE–anti-DNP antibody before challenge with 250 µg HSA-DNP antigen, injected i.v. in the presence of Evans blue dye (10 mg/mL). The extravasated blue dye was then extracted from the tissues by incubation in formamide at 63 °C overnight, and its intensity (correlating with the extent of extravasation) was measured spectrophotometrically (OD600).
Oxazolone-Induced Dermatitis.
In vivo experiments of chronic mast cell stimulation were performed exactly as previously described (43, 59). Briefly, the ear pinnae of KitW-sh/W-sh mice were reconstituted with 2 × 106 WT or KO mast cells. After 4 to 5 wk, sensitization was performed with 20 μL of 2% oxazolone in acetone; after another week, mice were challenged every other day with 0.5% oxazolone for up to seven challenges. As readout of inflammation, ear swelling was measured over time with a dial thickness gauge (Mitutoyo).
Supplementary Material
Acknowledgments
A special thank you to Gioacchino Natoli for help with the ATAC-seq experiments and critical reading of the manuscript, Jessica Marcandalli for technical help with all in vivo experiments, and all laboratory members for their daily help and support. This work was supported by Swiss National Science Foundation Grant 156875 (to S. Monticelli), the Ceresio Foundation (S. Monticelli and C.L.), the Gelu Foundation (F.B.), and Marie Heim-Vögtlin Postdoctoral Fellowship 164489 (to S. Montagner).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The gene expression profiling data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE87483). The ATAC-seq data reported in this paper have been deposited in the GEO database (accession no. GSE91036).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616420114/-/DCSupplemental.
References
- 1.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leoni C, Vincenzetti L, Emming S, Monticelli S. Epigenetics of T lymphocytes in health and disease. Swiss Med Wkly. 2015;145:w14191. doi: 10.4414/smw.2015.14191. [DOI] [PubMed] [Google Scholar]
- 6.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 7.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Tatton-Brown K, et al. Childhood Overgrowth Consortium Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204(4):715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shlush LI, et al. HALT Pan-Leukemia Gene Panel Consortium Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traina F, et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS One. 2012;7(8):e43090. doi: 10.1371/journal.pone.0043090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang IV, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136(1):69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monticelli S, Lee DU, Nardone J, Bolton DL, Rao A. Chromatin-based regulation of cytokine transcription in Th2 cells and mast cells. Int Immunol. 2005;17(11):1513–1524. doi: 10.1093/intimm/dxh329. [DOI] [PubMed] [Google Scholar]
- 17.Haenisch B, Fröhlich H, Herms S, Molderings GJ. Evidence for contribution of epigenetic mechanisms in the pathogenesis of systemic mast cell activation disease. Immunogenetics. 2014;66(5):287–297. doi: 10.1007/s00251-014-0768-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48(9):1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayoral RJ, et al. MiR-221 influences effector functions and actin cytoskeleton in mast cells. PLoS One. 2011;6(10):e26133. doi: 10.1371/journal.pone.0026133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demo SD, et al. Quantitative measurement of mast cell degranulation using a novel flow cytometric annexin-V binding assay. Cytometry. 1999;36(4):340–348. doi: 10.1002/(sici)1097-0320(19990801)36:4<340::aid-cyto9>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Nugroho AE, Shudou M, Maeyama K. Regulation of mucosal mast cell activation by short interfering RNAs targeting syntaxin4. Immunol Cell Biol. 2012;90(3):337–345. doi: 10.1038/icb.2011.41. [DOI] [PubMed] [Google Scholar]
- 22.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 23.Bock C, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47(4):633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiedemann RL, et al. Acute depletion redefines the division of labor among DNA methyltransferases in methylating the human genome. Cell Reports. 2014;9(4):1554–1566. doi: 10.1016/j.celrep.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koya J, et al. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nat Commun. 2016;7:10924. doi: 10.1038/ncomms10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99(26):16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duymich CE, Charlet J, Yang X, Jones PA, Liang G. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat Commun. 2016;7:11453. doi: 10.1038/ncomms11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montagner S, et al. TET2 regulates mast cell differentiation and proliferation through catalytic and non-catalytic activities. Cell Reports. 2016;15(7):1566–1579. doi: 10.1016/j.celrep.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa Y, et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118(Pt 12):2755–2762. doi: 10.1242/jcs.02402. [DOI] [PubMed] [Google Scholar]
- 32.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109(1):39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 33.Hoser M, et al. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28(15):4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep. 2015;16(4):427–446. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci USA. 2011;108(28):11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 39.Monticelli S, Rao A. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur J Immunol. 2002;32(10):2971–2978. doi: 10.1002/1521-4141(2002010)32:10<2971::AID-IMMU2971>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 40.Monticelli S, Solymar DC, Rao A. Role of NFAT proteins in IL13 gene transcription in mast cells. J Biol Chem. 2004;279(35):36210–36218. doi: 10.1074/jbc.M406354200. [DOI] [PubMed] [Google Scholar]
- 41.Aramburu J, et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285(5436):2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 42.Grimbaldeston MA, et al. Mast cell-deficient W-sash c-kit mutant KitW-sh/W-sh mice as a model for investigating mast cell biology in vivo. Am J Pathol. 2005;167(3):835–848. doi: 10.1016/S0002-9440(10)62055-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hershko AY, et al. Mast cell interleukin-2 production contributes to suppression of chronic allergic dermatitis. Immunity. 2011;35(4):562–571. doi: 10.1016/j.immuni.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125(4):629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;29(6):922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kersh EN, et al. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J Immunol. 2006;176(7):4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 47.Makar KW, et al. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol. 2003;4(12):1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 48.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4(3):235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 49.Gamper CJ, Agoston AT, Nelson WG, Powell JD. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol. 2009;183(4):2267–2276. doi: 10.4049/jimmunol.0802960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 51.Yu Q, et al. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc Natl Acad Sci USA. 2012;109(2):541–546. doi: 10.1073/pnas.1103803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macneil LT, Walhout AJ. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011;21(5):645–657. doi: 10.1101/gr.097378.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q, et al. Down-regulation of microRNA-223 promotes degranulation via the PI3K/Akt pathway by targeting IGF-1R in mast cells. PLoS One. 2015;10(4):e0123575. doi: 10.1371/journal.pone.0123575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 56.Monticelli S, Natoli G. Short-term memory of danger signals and environmental stimuli in immune cells. Nat Immunol. 2013;14(8):777–784. doi: 10.1038/ni.2636. [DOI] [PubMed] [Google Scholar]
- 57.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayoral RJ, Monticelli S. Stable overexpression of miRNAs in bone marrow-derived murine mast cells using lentiviral expression vectors. Methods Mol Biol. 2010;667:205–214. doi: 10.1007/978-1-60761-811-9_14. [DOI] [PubMed] [Google Scholar]
- 59.Deho’ L, et al. Two functionally distinct subsets of mast cells discriminated by IL-2-independent CD25 activities. J Immunol. 2014;193(5):2196–2206. doi: 10.4049/jimmunol.1400516. [DOI] [PubMed] [Google Scholar]
- 60.Rusca N, et al. MiR-146a and NF-κB1 regulate mast cell survival and T lymphocyte differentiation. Mol Cell Biol. 2012;32(21):4432–4444. doi: 10.1128/MCB.00824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schepers E, Glorieux G, Dhondt A, Leybaert L, Vanholder R. Flow cytometric calcium flux assay: Evaluation of cytoplasmic calcium kinetics in whole blood leukocytes. J Immunol Methods. 2009;348(1–2):74–82. doi: 10.1016/j.jim.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujita PA, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leoni C, et al. Reduced DNA methylation and hydroxymethylation in patients with systemic mastocytosis. Eur J Haematol. 2015;95(6):566–575. doi: 10.1111/ejh.12537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.