Significance
S-acylation, the attachment of different fatty acids onto cysteine residues, regulates the activity of a diverse array of cellular proteins. This reversible posttranslational modification is essential for normal physiology, and defects are linked to human disease. S-acylation is catalyzed by a large family of zDHHC S-acyltransferases that use a cellular pool of diverse fatty acyl-CoAs as substrates. Using chemically synthesized probes, we show that individual zDHHC enzymes have marked differences in fatty acid selectivity and identify the underlying molecular basis for this property. This study identifies how acyl chain heterogeneity of S-acylated proteins is generated and is significant because the chemical nature of the attached S-acyl chain can fundamentally impact protein behavior.
Keywords: S-acylation, palmitoylation, zDHHC enzyme, click chemistry, acyl CoA
Abstract
S-acylation is a major posttranslational modification, catalyzed by the zinc finger DHHC domain containing (zDHHC) enzyme family. S-acylated proteins can be modified by different fatty acids; however, very little is known about how zDHHC enzymes contribute to acyl chain heterogeneity. Here, we used fatty acid-azide/alkyne labeling of mammalian cells, showing their transformation into acyl-CoAs and subsequent click chemistry-based detection, to demonstrate that zDHHC enzymes have marked differences in their fatty acid selectivity. This difference in selectivity was apparent even for highly related enzymes, such as zDHHC3 and zDHHC7, which displayed a marked difference in their ability to use C18:0 acyl-CoA as a substrate. Furthermore, we identified isoleucine-182 in transmembrane domain 3 of zDHHC3 as a key determinant in limiting the use of longer chain acyl-CoAs by this enzyme. This study uncovered differences in the fatty acid selectivity profiles of cellular zDHHC enzymes and mapped molecular determinants governing this selectivity.
S-acylation is a reversible posttranslational modification (PTM) involving the attachment of fatty acids onto cysteines (1, 2). This PTM occurs on both soluble and transmembrane (TM) proteins and exerts a number of important effects, including mediating membrane binding (of soluble proteins or soluble loops of TM proteins), regulating protein trafficking and targeting to cholesterol-rich membrane microdomains, and modulating protein stability (3, 4). These actions of S-acylation on a diverse array of cellular proteins affect many important physiological pathways, and defects in this process are linked to a number of major diseases and disorders (2, 5).
S-acylation is mediated by the opposing actions of acyltransferases and thioesterases. S-acyltransferase enzymes belong to the zinc finger DHHC domain containing (zDHHC) protein family, which are encoded by 24 distinct genes (6–8). zDHHC enzymes are thought to share the same overall membrane topology, with four to six transmembrane domains and the N and C termini present in the cytosol (9). The catalytic DHHC cysteine-rich domain (CRD) of the enzymes lies in a cytosolic loop (9), allowing zDHHC enzymes to modify substrate cysteines present at the cytosol–membrane interface. The S-acylation reaction is thought to proceed through an enzyme-acyl intermediate, where the acyl chain is attached to the cysteine of the DHHC motif via a thioester linkage (often referred to as enzyme “autoacylation”) (10, 11). The S-acyl chain is then transferred to a cysteine residue of a substrate protein (10, 11). This overall process is referred to as a “ping-pong” reaction mechanism. There has been progress in identifying the zDHHC enzymes that are active against many substrate proteins (2), although we lack a detailed understanding of the protein substrate profiles of individual enzymes and how enzyme–substrate interaction specificity is achieved. Coexpression experiments have suggested that individual zDHHC enzymes might exhibit a level of overlap in their protein substrate profiles, suggesting some possible redundancy within the zDHHC family (2). Nevertheless, individual enzymes have been linked with many disorders, including intellectual disability, diabetes, and cancer, suggesting that any functional redundancy is limited (2).
In contrast to the steadily increasing knowledge about the specific interactions of zDHHC enzymes with their protein substrates, we know relatively little about the fatty acid selectivity of these enzymes. The term “palmitoylation” is often used as a synonym for S-acylation; however, this does not reflect the potential diversity of S-acyl chains added to substrate proteins. Indeed, an analysis of the acyl groups added to S-acylated proteins in platelets revealed that 74% were from palmitate (C16:0), 22% were from stearate (C18:0), and 4% were from oleate (C18:1) (12). Other studies have shown that S-acylated proteins can be modified by acyl chains from myristic acid (C14:0), palmitoleic acid (C16:1), linoleic acid (C18:2), and arachidonic acid (C20:4) (12–15). Furthermore, mass spectrometry analysis of the S-acyl chains attached to influenza hemagglutinin proteins has revealed that C16:0 and C18:0 fatty acids are attached in a site-specific manner (16). There is also potential for cell type-specific differences in the fatty acid profiles of S-acylated proteins; for example, one study reported that in RAW26.7 cells, <10% of the acyl-CoAs were greater than C20, whereas in MCF7 cells, C24:0 and C26:0 acyl-CoAs were present at similar amounts as C16:0 and C18:0 acyl-CoAs (17). The identity of the added acyl chain is central to the regulatory effects of S-acylation, because different acyl chains vary in their affinity for membranes and for cholesterol-rich membrane microdomains (18).
To date, only one study has explored the potential role of zDHHC enzymes in differential protein S-acylation using purified recombinant zDHHC enzymes in detergent micelles (10). This study reported that zDHHC2 catalyzed the S-acylation of substrate proteins with similar efficiency using palmitate (C16:0), stearate (C18:0), or C20 (C20:0, C20:4) fatty acids. In contrast, zDHHC3 displayed a marked preference for shorter-chain-length acyl-CoAs and incorporated C16:0 much more efficiently than either C18:0 or C20 fatty acids (10). This elegant study highlighted the potential for discrete patterns of fatty acid selectivity in the zDHHC family, but addressing these questions has been challenging, particularly in a cellular context. Recent developments at the chemistry–biology interface have identified new approaches to investigating this poorly defined area of the S-acylation field. Specifically, azide and alkyne fatty acid probes have provided novel and highly sensitive chemical tools for interrogating S-acylation by click chemistry (19–24). In this study, we report the use of chemically synthesized azide and alkyne fatty acid probes to investigate fatty acid selectivity in the zDHHC enzyme family and determine the molecular mechanisms governing this selectivity.
Results
Fatty Acid Azides Are Good Mimics of Endogenous Fatty Acids.
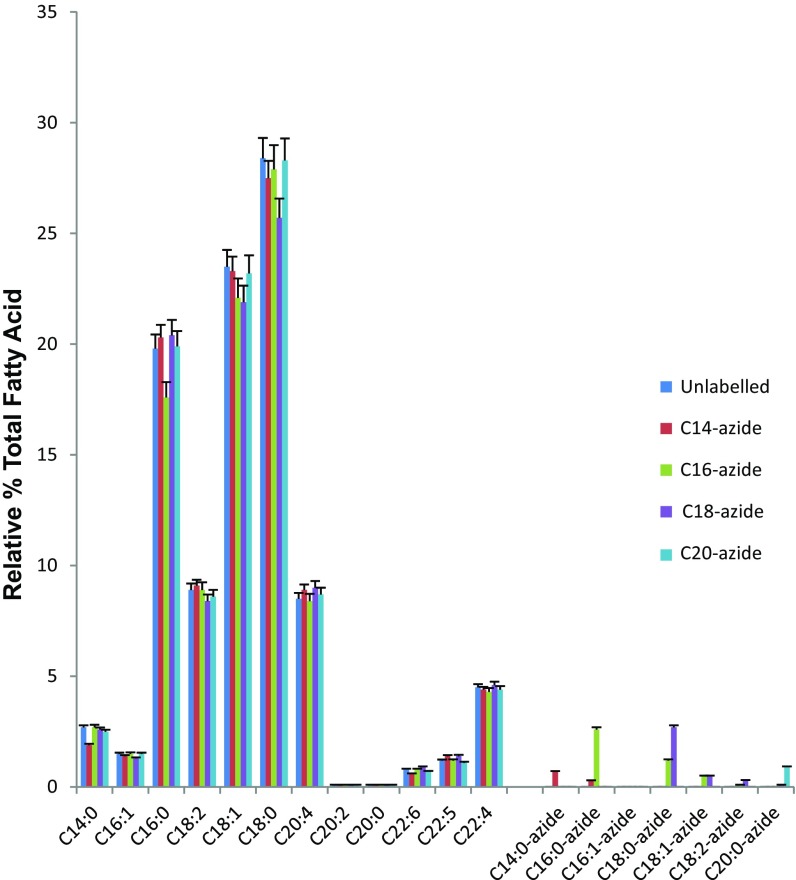
Fatty acid azide probes of different acyl chain lengths were synthesized as described in SI Appendix. To assess these probes’ ability to be taken up by cells, we determined the total fatty acid content of HEK293T cells treated with these synthesized fatty acid azides (Fig. 1). Untreated cells showed the expected range of saturated and unsaturated fatty acids, with C18:0 and C18:1 the most abundant. On the addition of the various fatty acid azides to the cells, it can clearly be seen that these were not only accumulated within the cells (with C16:0 and C18:0 azides taken up significantly better than C14:0 and C20:0 azides), but also metabolized by the cells. For example, the C16:0 azide was elongated to C18:0, which was subsequently desaturated to C18:1, whereas the added C18:0 azide was also desaturated to C18:1 and, to a very minor extent, C18:2; however, C18:0 azide was not significantly elongated to C20:0.
Fig. 1.
Relative percentage of the total fatty acid content of cells with and without treatments of fatty acid azides. HEK293T cells were incubated with or without fatty acid azides. Fatty acids released from cellular lipids and protein by acid were then converted to methyl esters and analyzed by GC-MS as described in Materials and Methods. Values shown are mean ± SEM (n = 3).
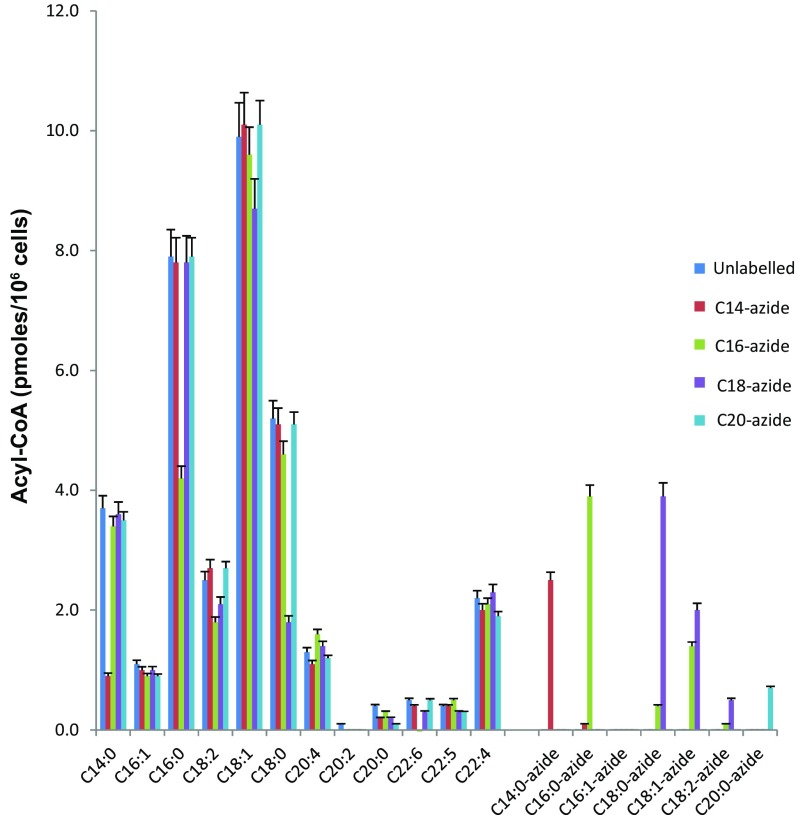
Given that S-acylation of proteins involves acyl-CoA donors, we deemed it important to investigate the impact on the cellular acyl-CoA pools. We did so using a multiple reaction monitoring mass spectrometry method in conjunction with an internal nonnatural (C17:0) acyl-CoA standard. This analysis revealed that in the untreated HEK293T cells, the C18:1-CoA and C16:0-CoA were the most abundant, accounting for almost one-half of the total acyl-CoA pool (Fig. 2). On addition of the various azide fatty acids, a significant amount of the corresponding acyl-CoA azides were formed, including some of the corresponding metabolized (elongated and desaturated) acyl-CoA azides, reflecting the total fatty acid content. It is interesting to note that there were decreases in the corresponding endogenous acyl-CoA pools, suggesting clear competition for the acyl-CoA synthases that have a preference for a particular acyl chain length.
Fig. 2.
Quantification of the acyl-CoA content of HEK293T cells with and without treatments of azide fatty acids. HEK293T cells were incubated with or without fatty acid azides, and acyl-CoAs were then quantified as described in Materials and Methods. Values shown are mean ± SEM (n = 3).
Collectively, these data strongly suggest that the synthesized fatty acid azides are good mimics of the natural saturated fatty acids and can be used for downstream metabolic processes involving fatty acids, such as lipid metabolism and protein lipidation.
zDHHC Enzymes Display Marked Differences in Fatty Acid Azide Selectivity in Cell-Based Substrate S-Acylation Assays.
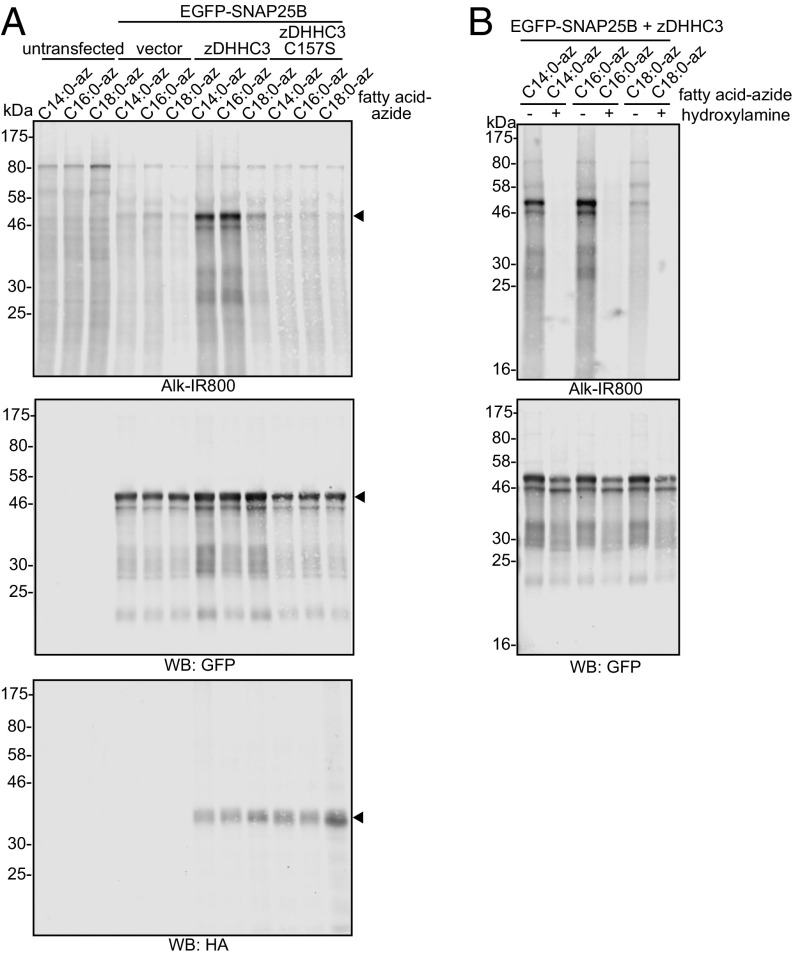
To test the ability of the synthesized fatty acid azide probes to reliably measure protein S-acylation, HEK293T cells were cotransfected with EGFP-tagged synaptosomal-associated protein of 25 kDa (SNAP25) and hemagglutinin (HA)-tagged zDHHC3 or an inactive mutant of this enzyme (C157S). Fig. 3A shows that zDHHC3 promoted a marked increase in incorporation of the azide probes into SNAP25, whereas in the absence of active enzyme (either empty vector or the C157S mutant), only a low signal was detected, presumably representing SNAP25 that is modified by endogenously expressed zDHHC enzymes. To test whether the incorporated azide probes were attached to SNAP25 via a thioester linkage, transfected cells were treated overnight with 1 M hydroxylamine (pH 7), which led to a marked loss of labeling, whereas incubation with 1 M Tris (pH 7) as a control had no effect (Fig. 3B).
Fig. 3.
S-acylation of EGFP-SNAP25B by zDHHC3. HEK293T cells were transfected with EGFP-SNAP25B and pEF-BOS-HA (vector), HA-zDHHC3, or HA-zDHHC3(C157S), or were left untransfected. Cells were then incubated with C14:0, C16:0, or C18:0 fatty acid azides for 4 h at 37 °C. Incorporated fatty acid azides were detected by click chemistry using an alkyne-800 infrared dye. Isolated proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes. Representative images are shown. (A) Click chemistry signal (Top), anti-GFP immunoblot (Middle), and anti-HA immunoblot (Bottom). Arrowheads denote the position of the EGFP-SNAP25 band (Top and Middle) and HA-zDHHC3 band (Bottom). (B) Following click chemistry, samples were incubated in hydroxylamine (+) or Tris (−) at a final concentration of 1 M overnight before SDS/PAGE. (Top) Click chemistry signal. (Bottom) Anti-GFP immunoblot. The positions of molecular weight markers are shown.
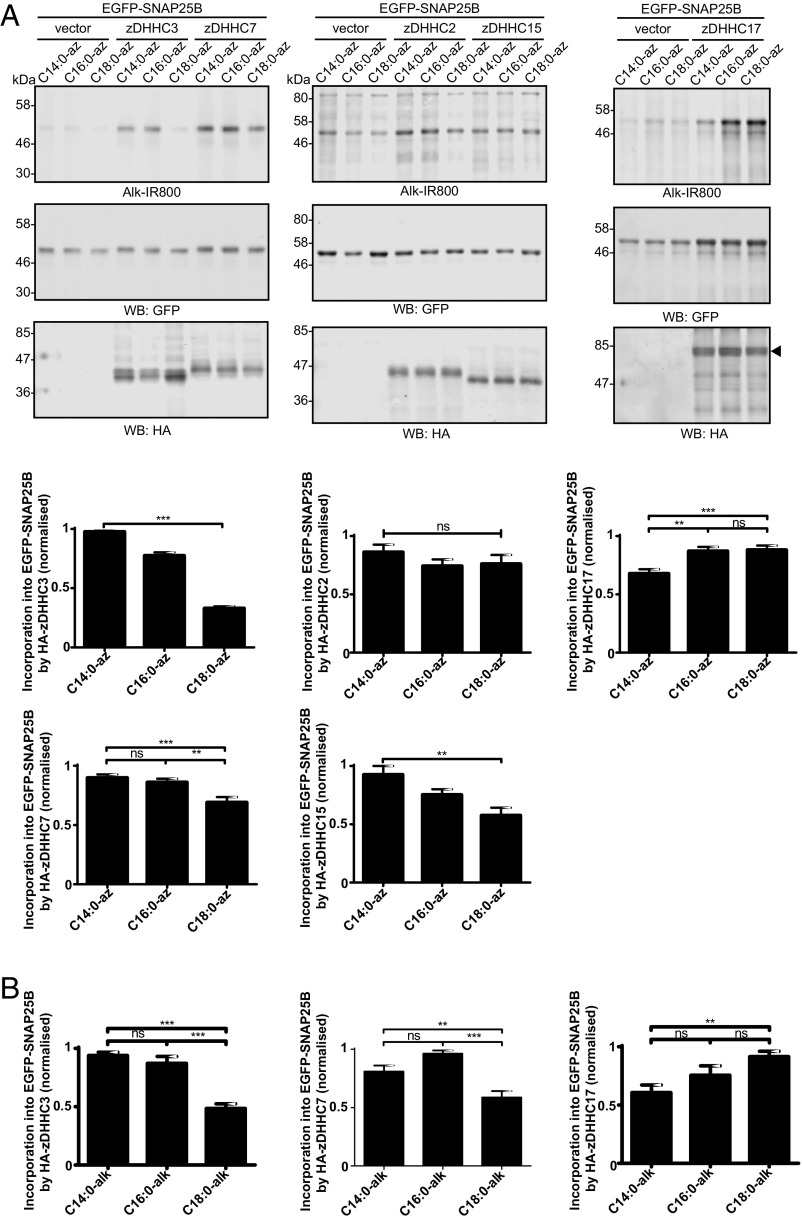
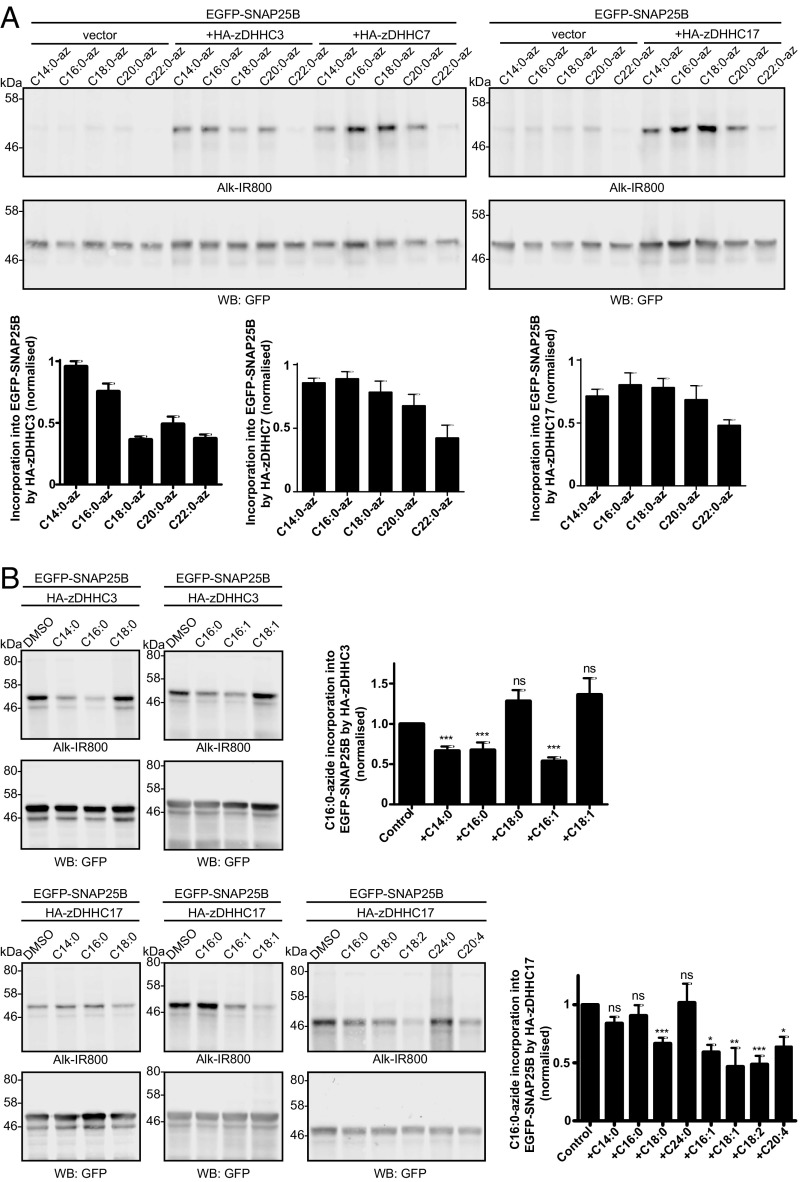
The results presented in Fig. 3 suggest that the level of C18:0 incorporation into SNAP25 by zDHHC3 is markedly lower than that of C14:0 and C16:0. Our previous work and that of others has shown that five zDHHC enzymes are active against SNAP25 in similar coexpression assays: zDHHC2, zDHHC3, zDHHC7, zDHHC15, and zDHHC17 (25). Thus, we undertook a quantitative comparison of SNAP25 S-acylation by these five zDHHC enzymes to examine whether zDHHC enzymes exhibit any differences in fatty acid selectivity when expressed in cells. The results, shown in Fig. 4A (Left), confirm that zDHHC3 has a marked preference for C14:0/C16:0 over C18:0 azide. Interestingly, zDHHC7, which is highly related to zDHHC3 at the sequence level (67.8% identical for mouse), incorporated C18:0-azide into SNAP25 more efficiently than zDHHC3, although there was still a significant reduction in C18:0 azide incorporation relative to C14:0/C16:0 azides with this enzyme (Fig. 4A, Left). zDHHC2 and zDHHC15 also displayed interesting differences in fatty acid azide selectivity despite being highly related at the amino acid level (65.4% identical). zDHHC2 exhibited no significant preference, whereas C18:0 azide incorporation into SNAP25 by zDHHC15 was markedly lower than C14:0/C16:0 azide incorporation (Fig. 4A, Middle). Finally, zDHHC17 showed a preference for longer-chain fatty acids and incorporated C16:0 and C18:0 with higher efficiency than C14:0 (Fig. 4A, Right).
Fig. 4.
S-acylation of EGFP-SNAP25B by different zDHHC enzymes. HEK293T cells were transfected with EGFP-SNAP25B and pEF-BOS-HA (vector), HA-zDHHC-2, -3, -7, -15, or -17. Cells were then incubated with C14:0, C16:0, or C18:0 fatty acid azides or alkynes as indicated for 4 h at 37 °C. Fatty acid azides/alkynes were labeled by click chemistry using an alkyne- or azide-800 infrared dye. Isolated proteins were then resolved by SDS/PAGE and transferred to nitrocellulose membranes. (A) S-acylation analysis of EGFP-SNAP25B using fatty acid azides. (Top) Representative images with positions of molecular mass standards indicated. (Bottom) Graphs showing mean ± SEM. zDHHC3, n = 46; zDHHC7, n = 26; zDHHC2, n = 10; zDHHC15, n = 9; zDHHC17, n = 22. **P < 0.01; ***P < 0.001. (B) S-acylation analysis of EGFP-SNAP25B in HEK293T cells labeled with fatty acid alkynes. Graphs show mean ± SEM (n = 10). ns, not significant; **P < 0.01; ***P < 0.001.
We also synthesized the corresponding alkyne probes and assayed their incorporation into SNAP25 by zDHHC-3, -7, and -17, which revealed the same distinct profiles as seen with the azide probes (Fig. 4B). To avoid confusion, the number of carbon groups given in the name for the alkyne probes reflects those of the fatty acid minus the alkyne group. Thus, C14:0 alkyne is a C14 fatty acid chain plus an alkyne group (16 carbon atoms total). Synthesis of these alkyne probes is described and their correct IUPAC nomenclature provided in SI Appendix.
To generate a more comprehensive understanding of the limits of zDHHC fatty acid selectivity, we also examined C20:0 and C22:0 azide probes. zDHHC3 showed no difference in ability to transfer C18:0, C20:0, or C22:0 (low incorporation for each), whereas zDHHC7 and zDHHC17 displayed a gradual decline in azide incorporation as chain length was increased (Fig. 5A). We also used competition assays to test the ability of unlabeled fatty acids to block C16:0 azide incorporation into SNAP25 by zDHHC3 and zDHHC17. Consistent with the observation that zDHHC3 did not incorporate the C18:0 azide probe with high efficiency, neither stearic acid (C18:0) nor oleic acid (C18:1) could block incorporation of C16:0 azide into SNAP25 catalyzed by this enzyme, whereas myristic acid (C14:0), palmitic acid (C16:0), and palmitoleic acid (C16:1) were effective inhibitors (Fig. 5B). In contrast to zDHHC3, stearic acid effectively blocked C16:0 azide incorporation into SNAP25 by zDHHC17, whereas myristic acid and palmitic acid had no significant inhibitory effect (Fig. 5B). Furthermore, oleic acid and linoleic acid (C18:2) blocked incorporation of C16:0 azide, and indeed, even arachidonic acid (C20:4) effectively blocked C16:0 azide incorporation by zDHHC17 (Fig. 5B). The inhibitory effects of these longer-chain unsaturated fatty acids are interesting, given that significant amounts of the respective acyl-CoAs are present in HEK293T cells (Fig. 2). In contrast, lignoceric acid (C24:0) did not inhibit C16:0 azide incorporation, consistent with the results presented in Fig. 5A showing that C22:0 azide displayed a marked reduction in incorporation by zDHHC17 compared with shorter-chain azide probes.
Fig. 5.
S-acylation of EGFP-SNAP25B by longer-chain saturated and unsaturated fatty acids. (A) S-acylation analysis of EGFP-SNAP25B by HA-zDHHC3, HA-zDHHC7, and HA-zDHHC17 in HEK293T cells with C14:0, C16:0, C18:0, C20:0, and C22:0 fatty acid azides. Representative images are shown, and the positions of molecular mass standards are indicated. (Top) Click chemistry signal. (Middle) Anti-GFP immunoblot. (Bottom) Quantified data. n ≥3; mean ± SEM. (B) Competition analysis of EGFP-SNAP25B S-acylation by HA-zDHHC3 (Top) or HA-zDHHC17 (Bottom) in HEK293T cells labeled with C16:0-azide in the presence of threefold excess of the indicated unlabeled fatty acids. Positions of molecular mass standards are shown. Graphs show mean values ± SEM (n ≥ 3). ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Different Fatty Acid Selectivities of zDHHC Enzymes Correspond to Autoacylation Status.
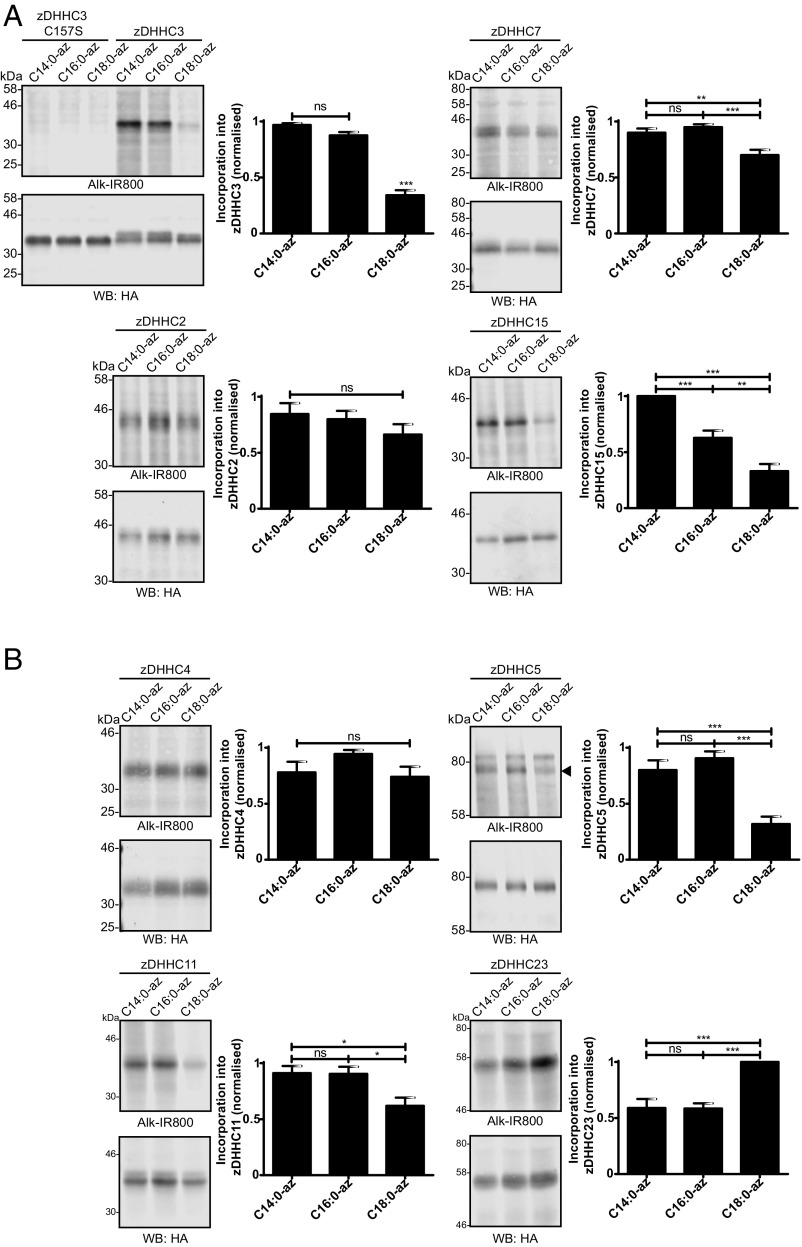
The S-acylation reaction proceeds via an enzyme-acyl intermediate (referred to as “autoacylation”) and is followed by transfer of the acyl chain to a substrate protein (10). Thus, we tested whether the different substrate S-acylation profiles observed in Figs. 4 and 5 correspond to the autoacylation status of zDHHC enzymes. Fig. 6A shows that the autoacylation profiles of zDHHC-2, -3, -7, and -15 were essentially identical to the profiles observed for these enzymes with SNAP25 S-acylation (Fig. 4). Despite repeated attempts, we were unable to reliably detect autoacylation of zDHHC17, and thus could not compare the autoacylation profile of this enzyme with its substrate S-acylation profile. Because enzyme autoacylation reliably reports on the fatty acid selectivity of zDHHC enzymes, we extended this analysis to provide a more comprehensive study of the zDHHC family. Further distinct fatty acid azide selectivity profiles were identified for a set of zDHHC enzymes for which autoacylation was readily detectable. zDHHC5 and zDHHC11 exhibited a preference for C14:0/C16:0 azides, zDHHC4 showed no preference for C14:0/C16:0/C18:0 azides, and zDHHC23 displayed a marked preference for C18:0 azide over C14:0 and C16:0 azides (Fig. 6B). Autoacylation of all zDHHC enzymes tested was dependent on an intact DHHC motif (SI Appendix, Fig. S1).
Fig. 6.
Autoacylation of zDHHC enzymes by fatty acid azides. HEK293T cells transfected with HA-tagged zDHHC constructs were incubated with C14:0, C16:0, or C18:0 fatty acid azides for 4 h at 37 °C. Fatty acid azides were then labeled by click chemistry using an alkyne-800 infrared dye. Isolated proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes. Representative click signals and Western blots with positions of molecular mass standards indicated, together with quantified data (mean ± SEM) are shown for each zDHHC enzyme. (A) Autoacylation of zDHHC enzymes active against SNAP25B. zDHHC2, n = 6; zDHHC3, n = 14; zDHHC7, n = 11; zDHHC15, n = 6. (B) Autoacylation of additional zDHHC enzymes. n ≥ 6. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001. The arrowhead on the zDHHC5 blot indicates the zDHHC5 band.
Differences in Fatty Acid Selectivity of zDHHC3 and zDHHC7 Are Linked to Residues in Transmembrane Domain 3.
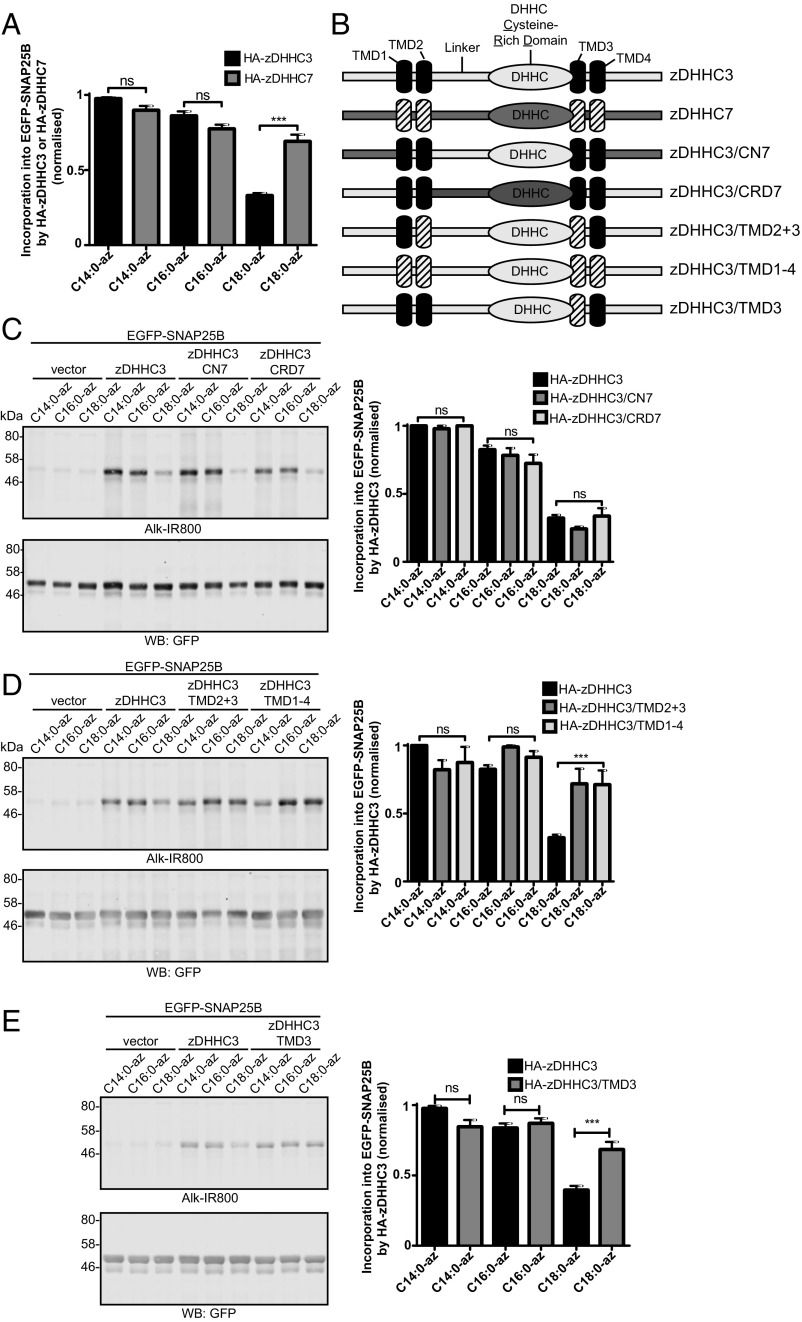
To understand how differences in fatty acid selectivity might be encoded at the molecular level, we undertook a systematic domain swapping analysis of zDHHC3 and zDHHC7. These enzymes are highly conserved, and thus we reasoned that domain swapping between these two isoforms should be less likely than swapping between other zDHHC enzymes to produce deleterious effects on protein folding. zDHHC3 and zDHHC7 exhibited a marked difference in the ability to incorporate the C18:0 azide probe into both SNAP25 (Fig. 7A), and a different substrate protein (cysteine-string protein; SI Appendix, Fig. S2). Thus, we focused on this difference in fatty selectivity of these two enzymes. zDHHC3 and zDHHC7 consist of four predicted transmembrane domains with cytosolic N and C termini and a central intracellular loop containing the 51-aa catalytic DHHC-CRD (Fig. 7B). Interestingly, replacing either the N- or C-terminal domain or the DHHC-CRD of zDHHC3 with the same domains from zDHHC7 had no detectable effect on the fatty acid selectivity profile of zDHHC3 (Fig. 7C). In contrast, replacing all four transmembrane domains or only transmembrane domains 2 and 3 of zDHHC3 with those from zDHHC7 resulted in a significant increase in C18:0 azide incorporation into SNAP25 by these mutant enzymes (Fig. 7D), mirroring the fatty acid profile of zDHHC7 (Fig. 7A). Further analysis revealed that substituting only transmembrane domain 3 of zDHHC3 with the same domain from zDHHC7 was sufficient to change the C18:0 selectivity profile of zDHHC3 (Fig. 7E).
Fig. 7.
S-acylation of EGFP-SNAP25B by zDHHC3/zDHHC7 chimeras. HEK293T cells were transfected with EGFP-SNAP25B and pEF-BOS-HA (vector) or the indicated WT or mutant zDHHC constructs. Cells were then incubated with C14:0, C16:0, or C18:0 fatty acid azides for 4 h at 37 °C. Fatty acid azides were labeled by click chemistry using an alkyne-800 infrared dye. Isolated proteins were then resolved by SDS/PAGE and transferred to nitrocellulose membranes for analysis. (A) Quantification of the relative levels of C14:0, C16:0, or C18:0 azide incorporation into EGFP-SNAP25B by zDHHC3 or zDHHC7 (mean ± SEM). n ≥ 26. (B) Schematic illustration detailing the zDHHC3/zDHHC7 chimeras that were constructed. (C–E) Analysis of EGFP-SNAP25B S-acylation by zDHHC3 chimeras with fatty acid azides. (Left) Representative images. (Right) Graphs showing mean ± SEM. (C) n ≥ 4. (D) n = 6. (E) n ≥12. ns, not significant; ***P < 0.001.
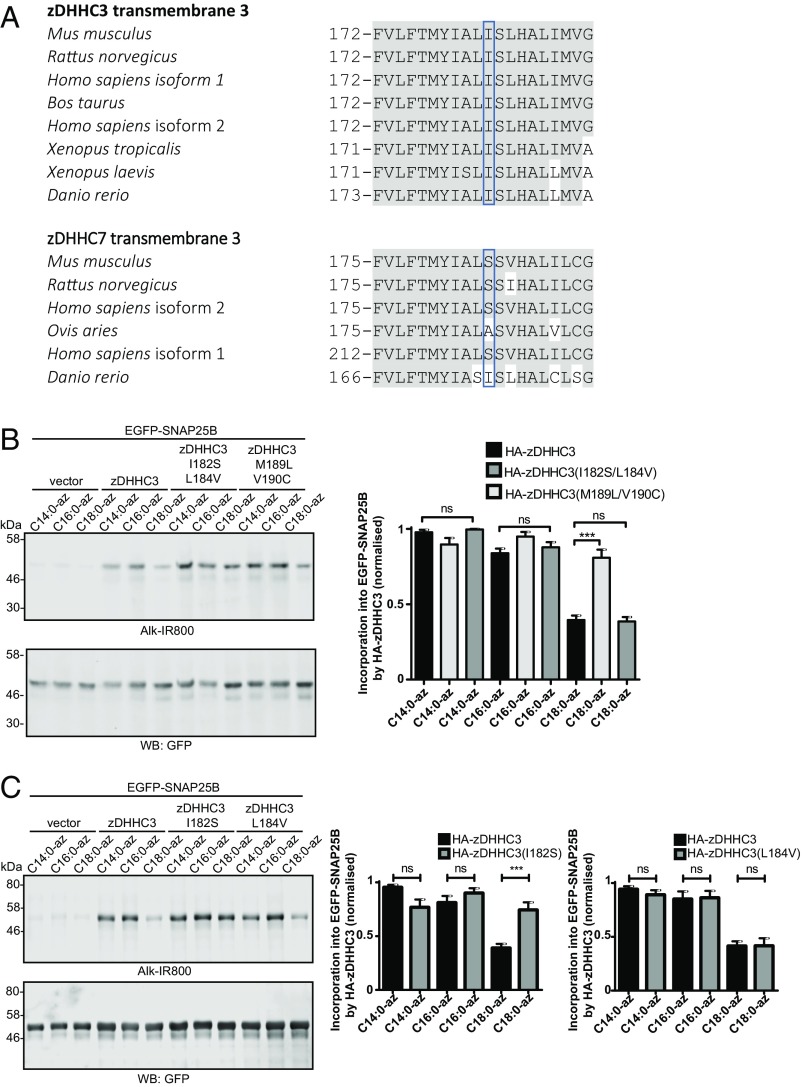
To pinpoint the specific features of TMD3 that are important for dictating the fatty acid selectivity profile of zDHHC3, we compared the amino acid sequences of this domain from zDHHC3 and zDHHC7 (Fig. 8A). There are four differences in the amino acid sequence between mammalian zDHHC3 and zDHHC7 isoforms in this region, and thus we generated two mutants containing either I182S/L184V or M189L/V190C mutations. Fig. 8B shows that the I182S/L184V mutation led to a marked increase in C18:0 azide incorporation into SNAP25 by zDHHC3, whereas the M189L/V190C mutations had no effect. Thus, we subsequently generated single I182S and L184V mutations, which clearly showed that the I182S mutation in zDHHC3 significantly enhanced the level of C18:0 incorporation by zDHHC3, whereas the L184V mutation had no effect (Fig. 8C). It is interesting to note that zDHHC7 in Danio rerio has an isoleucine rather than a serine residue at this position (Fig. 8A). Examination of autoacylation and substrate S-acylation profiles of zDHHC3 and zDHHC7 from this species revealed no significant difference, with both enzymes weakly incorporating the C18:0 azide (SI Appendix, Fig. S3). This finding further supports the key role played by these residues in determining the fatty acid selectivity profiles of zDHHC enzymes.
Fig. 8.
S-acylation of EGFP-SNAP25B by zDHHC3 TMD3 mutants. HEK293T cells were transfected with EGFP-SNAP25B and pEF-BOS-HA (vector) or the indicated WT or mutant zDHHC constructs. Cells were then incubated with C14:0, C16:0, or C18:0 fatty acid azides for 4 h at 37 °C. Fatty acid azides were labeled by click chemistry using an alkyne-800 infrared dye. Isolated proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes. (A) Sequence alignment of amino acids in transmembrane domain 3 of zDHHC3 and zDHHC7. Grey shading highlights amino acids in each zDHHC enzyme that are conserved between all species shown. The blue boxes highlight isoleucine-182 in zDHHC3 and serine-185 in zDHHC7. (B and C) Analysis of EGFP-SNAP25B S-acylation by zDHHC3 TMD3 mutants with fatty acid azides. (Left) Representative images. (Right) Graphs showing mean ± SEM. (B) n ≥ 6. (C) n ≥ 8. ns, not significant, ***P < 0.001.
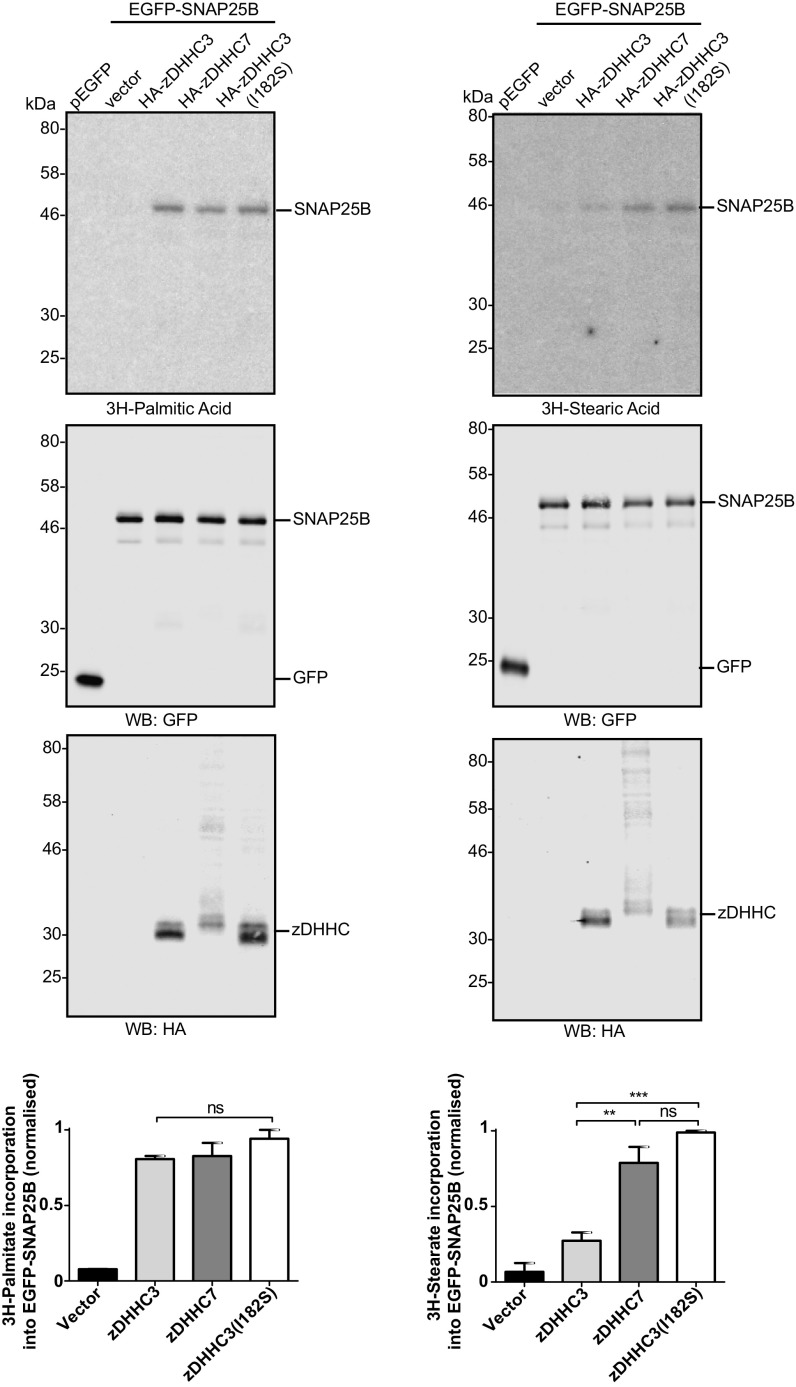
Finally, to exclude the possibility that any findings with the fatty acid azide probes were related to the presence of the azide/alkyne group, we examined the incorporation of [3H]-palmitic acid and [3H]-stearic acid into SNAP25 by zDHHC3, zDHHC7, and the zDHHC3 (I182S) mutant. Fig. 9 shows that the level of incorporation of [3H]palmitic acid catalyzed by these zDHHC enzymes was similar. In contrast, zDHHC7 more effectively incorporated [3H]stearic acid than zDHHC3, and the zDHHC3 (I182S) mutant exhibited a significant increase in [3H]stearic acid incorporation compared with wild-type (WT) zDHHC3 and to a similar level as seen with zDHHC7. Thus, the results of these experiments fully support the results obtained with the fatty acid azides.
Fig. 9.
Incorporation of [3H]palmitic and [3H]stearic acid into EGFP-SNAP25B by zDHHC3 and zDHHC7. HEK293T cells were transfected with pEGFPC2 or EGFP-SNAP25B together with pEF-BOS-HA (vector), HA-zDHHC3, HA-zDHHC7, or HA-zDHHC3(I182S). Cells were labeled with either [3H]palmitic acid (Left) or [3H]stearic acid (Right), lysed, and resolved by SDS/PAGE. (Top) [3H] fatty acid incorporation. (Top, Middle): expression levels of EGFP-SNAP25B. (Bottom, Middle) zDHHC protein expression. Molecular weight markers are shown on the left. (Bottom) Quantification of [3H] fatty acid incorporation normalized to EGFP-SNAP25B protein levels, expressed as mean ± SEM. n = 3. ns, not significant; **P < 0.01; ***P < 0.001.
Discussion
This study provides insight into zDHHC enzyme fatty acid selectivity in cells. The only other published study examining how zDHHC enzymes handle acyl-CoA substrates explored S-acylation by purified zDHHC2 and zDHHC3 in detergent micelles (10). Jennings and Linder demonstrated that zDHHC3 has a strong preference for C16:0 over C18:0 acyl-CoA, whereas zDHHC2 displays no overt preference (10). Importantly, in the present study, these differences were also seen when zDHHC2 and zDHHC3 were expressed in mammalian cells. zDHHC3 is Golgi-localized (26), whereas zDHHC2 associates with the plasma membrane and endosomes (27); therefore, the acyl-CoA selectivity profiles of zDHHC2/3 are preserved irrespective of whether they are in detergent micelles or native membranes and regardless of localization to distinct cellular compartments.
zDHHC autoacylation provides a robust measure of fatty acid selectivity, as demonstrated by the close match between the autoacylation profiles of zDHHC2, -3, -7, and -15 and their respective substrate S-acylation profiles. This property allowed us to investigate the acyl-CoA selectivity of an additional set of enzymes by measuring their autoacylation. The results of this analysis show that zDHHC-3, -5, -7, -11, and -15 prefer C14/C16 over C18; zDHHC-2 and -4 display no clear fatty acid preference; zDHHC17 prefers C16/C18 over C14; and zDHHC23 exhibits a strong preference for C18. These observations reemphasize that acyl-CoA specificities show no correlation with intracellular localization; for example, zDHHC-3, -7, -15, -17, and -23 all localize to the Golgi (26, 28), whereas zDHHC-2 and -5 are localized to the plasma membrane (25), and zDHHC4 is ER-localized (29).
zDHHC-3 and -7 are highly related at the sequence level, localize to Golgi membranes, and share many common protein substrates. However, these enzymes have significant differences in acyl-CoA selectivity, with zDHHC7 having a greater ability to incorporate C18:0 chains compared with zDHHC3. The high sequence conservation of zDHHC-3 and -7 provided an opportunity to undertake a comprehensive domain swapping analysis to pinpoint features that underpin acyl-CoA selectivity. This analysis identified a single amino acid in transmembrane domain 3 of zDHHC3 as a critical determinant limiting the use of longer-chain fatty acids. When Ile-182 was replaced by a serine, which is present at the same position in zDHHC7, a significant increase in the ability of the mutant protein to incorporate C18:0 was noted. We confirmed the importance of this residue using both C16:0/C18:0 azides with click chemistry detection and [3H]palmitic acid/stearic acid labeling. It is interesting that neither the azide group or the alkyne group present on the probes appeared to influence acyl chain selectivity, perhaps suggesting that these chemical groups have a flexible character that does not impede association with zDHHC enzymes. Consistent with this, we also found that the fatty acid azides selectively competed with endogenous fatty acids of the same carbon chain length to form acyl-CoAs when added to HEK293T cells. This shows that the azide probes compete with and are very good mimics of endogenous fatty acids, and thus are excellent tools for interrogating aspects of fatty acid biology, such as S-acylation.
Although we performed a comprehensive domain-swapping analysis between zDHHC3 and zDHHC7, the close similarity of these two enzymes means that other factors important for acyl chain selectivity could have been missed. Nevertheless, the approach taken here provides an important first step toward understanding the basis for acyl-CoA selectivity in the zDHHC family. Interestingly, a lysine has been identified in the transmembrane domain of an elongase component of the yeast very-long-chain fatty acid synthase complex that is also a key determinant of the final length of fatty acid acyl-CoA chain synthesized by this enzyme complex (30). In addition, a subsequent mutational analysis of the related rat elongases Elovl2 and Elovl5 highlighted the importance of the transmembrane domains of these enzymes in setting the substrate specificity profiles. Elovl2 is required for synthesis of omega-3 docosahexaenoic acid (DHA; 22:6n-3), because this elongase (but not Elovl5) can elongate docosapentaenoic acid (22:5n-3) to 24:5n-3, a precursor of DHA. This difference in substrate specificity between Elovl2 and Elovl5 was shown to involve a region encompassing transmembrane domains 6 and 7, with a cysteine-to-tryptophan switch in transmembrane domain 7 proving particularly important in setting specificity (31).
It is tempting to speculate that the transmembrane domains of zDHHC enzymes form “channels” that accommodate specific acyl-CoA molecules. This might involve different transmembrane domains in the same zDHHC molecule, or may require dimerization and multimerization of zDHHC enzymes (32). Because isoleucine occupies more space than serine, it is possible that this amino acid in TMD3 of zDHHC3 limits the length of the acyl chain that can be accommodated by blocking the acyl-CoA channel. This idea is consistent with the position of the isoleucine residue in zDHHC3, which is present in the middle of TMD3. Indeed, the catalytic DHHC-CRD of zDHHC enzymes is present immediately preceding TMD3, suggesting that the cysteine in the DHHC active site could be positioned close to the channel opening.
Mass spectrometry analysis of HA from influenza has shown that C18:0 is added specifically to a cysteine present in the transmembrane domain, whereas C16:0 is attached to cysteines in a membrane proximal region (33). How this highly selective process is achieved is not clear, but our results suggest that the modified cysteines in HA might be targets of different zDHHC enzymes with distinct acyl-CoA specificities. It is also possible that a contribution is sometimes made by the substrate in determining which fatty acids are attached. Thus, for transmembrane proteins, the sequence of the transmembrane domain could affect which fatty acids are added to membrane-proximal cysteines in a similar way as seen here for zDHHC enzymes.
There are two major properties of S-acylation considered central to its various effects on modified proteins: hydrophobicity and affinity for cholesterol-rich membranes (3, 34). Hydrophobicity is fundamental to the effects of S-acylation on the reversible membrane binding of many soluble proteins and in regulating the membrane association of soluble loops of transmembrane proteins (3). Association with cholesterol-rich membranes is thought to influence aspects of protein trafficking and membrane compartmentalization. It is well established from in vitro studies that saturated phospholipids cluster together with cholesterol, whereas unsaturated phospholipids are excluded from these domains (35). Although the effects of saturated vs. unsaturated S-acyl chains have not been studied in such detail, S-acylation clearly is a major signal for association with cholesterol-rich domains, at least in vitro (36). It is likely that the addition of saturated vs. unsaturated acyl chains onto proteins has a distinct influence on membrane partitioning and subsequent trafficking and function in the cell. Furthermore, the acyl chain added to S-acylated proteins is also likely to affect the strength of membrane association. Although two tandem lipid modifications (e.g., myristoylation/palmitoylation, prenylation/palmitoylation) provide a strong membrane anchor, a single lipid group (e.g., myristoyl, prenyl) is not sufficient for membrane association (37). However, single myristate, palmitate, or stearate groups added to an S-acylated protein could have different effects on membrane association. Indeed, stearate has a significantly stronger interaction with phospholipid membranes than palmitate (12).
This study identified clear differences in the acylation profiles of different zDHHC enzymes and their substrates expressed in HEK293T cells. In future work, it will be important to further advance the understanding of this area and the functional significance of fatty acid heterogeneity by investigating the fatty acid selectivity of endogenous proteins and how this is impacted by a dynamic and heterogeneous acyl-CoA pool. The distribution of different pools of acyl-CoAs and their availability for use in S-acylation reactions is unclear, and this is another important area for future investigations.
Materials and Methods
Materials.
Mouse GFP antibody was obtained from Clontech, and rat HA antibody was obtained from Roche. D. rerio zDHHC3 and zDHHC7 coding sequences were synthesized by GeneArt (Thermo Fisher Scientific). IR dye conjugated secondary antibodies and alkyne/azide probes were purchased from LI-COR. [9,10-3H] palmitic acid and [9,10-3H] stearic acid (specific activity for each, 1.11–2.22 TBq/mmol) were obtained from Hartmann Analytic.
Cell Transfection.
For substrate S-acylation assays, HEK293T cells plated on 24-well plates were transfected using Lipofectamine 2000 (Invitrogen) with 0.8 μg of EGFP-SNAP25 and 1.6 μg of zDHHC plasmid (in a pEF-BOS-HA vector backbone). Autoacylation assays were performed using 3 μg of zDHHC plasmid. Cells were labeled and processed the day after transfection.
Quantification of Fatty Acids and Acyl-CoAs.
Cells were collected by centrifugation and washed with serum-free DMEM, before incubation at 37 °C for 15 min with DMEM only, followed by the addition of defatted BSA coupled to the appropriate fatty acid azide (100 µM final concentration) and incubation for 4 h at 37 °C. Cells were then harvested by centrifugation and either washed in ice-cold PBS and freeze-dried for total fatty acid determination or processed for acyl-CoA extraction (see below).
For total fatty acid determination, the freeze-dried cells were subjected to acid hydrolysis using constant boiling HCl (6 M, 200 μL) vortexing/sonication, followed by incubation for 16 h at 110 °C. After cooling, the samples were spiked with 100 pmoles of C17:0 fatty acid (as an internal control) and processed and derivatised to fatty acid methyl esters (FAMEs), before analysis by gas chromatography-mass spectrometry (GC-MS) as described previously (38). The individual azide fatty acids were also converted to their corresponding FAMEs and analyzed by GC-MS to determine retention time and fragmentation patterns.
For acyl-CoA extraction and quantification, the absolute number of cells was determined (typically 2–4 × 107 cells). Cells were harvested by centrifugation at 15,000 × g for 1 min at room temperature and the supernatant media removed. The pellet was washed with 200 μL of ice-cold PBS and completely lysed with ice-cold TCA (100 μL, 1 M) and vortexing, and stored on ice to prevent sample hydrolysis.
The internal standard C17:0-CoA (150 pmol) was added to the lysate, and the sample was centrifuged at 15,000 × g for 10 min at 4 °C. The resulting supernatant was transferred to a fresh precooled tube. EDTA (25 μL, 10 mM, pH 7.0) was added, followed by chloroform (50 μL) triethylammonium acetate (50 μL), and the mixture was vortexed and centrifuged at 15,000 × g for 10 min at 4 °C. The upper phase was carefully removed to a fresh Eppendorf tube, flash-frozen in liquid nitrogen, and freeze-dried. The dried sample was kept at −80 °C before analysis by electrospray-mass spectrometry using multiple reaction monitoring (MRM), similar to the method described by Haynes et al. (17).
Samples were suspended in 15 μL of a 1:2 (vol/vol) chloroform/methanol and 5 μL of acetonitrile/isopropanol/water (6:7:2) and delivered using a NanoMate electrospray ionization system (Advion) to a AB Sciex 4000 QTRAP triple-quadrupole mass spectrometer with a nanoelectrospray source, using nitrogen as the collision gas. An MRM approach was used to quantify acyl-CoA. MRM mass transition for the acyl-CoAs was determined in positive ion mode, (entrance potential, 8 eV; collision cell exit potential, 12 eV; interface temperature, 30 °C; gas pressure, 0.5 psi; tip voltage, 1.25–1.5 kV; dwell time, 500 ms), and spectra were acquired for 2 min (SI Appendix, Table S1). All MRM data were normalized relative to the internal standard before the generation of standard curves (0.1–500 pmol) for the acyl-CoAs (C14:0, C16:0, C17:0, C18:0, C20, and C20:4) which were obtained from either Sigma-Aldrich or Avanti Polar Lipids, allowing determination of their own response factor. Samples were analyzed in the same manner, allowing quantification of the extracted acyl-CoAs.
Cell Labeling with Fatty Acid Azide and Alkyne Probes for Analysis of S-Acylation.
HEK293T cells were incubated with 100 μM of the fatty acid azide probes (in DMEM with 1 mg/mL defatted BSA) for 4 h at 37 °C. For competition experiments, transfected cells were labeled with 100 µM of the C16:0 azide probe in the presence of a threefold excess (300 µM) of the relevant fatty acid. For cell labeling with [3H]palmitic acid and [3H]stearic acid (Hartmann Analytic), transfected cells were incubated in DMEM/BSA containing 0.5 mCi/mL of the tritiated fatty acid probes for 4 h at 37 °C.
Detection of Fatty Acid Azide and Alkyne Probes in S-Acylated Proteins.
Cells were washed twice in PBS, then lysed on ice in 100 μL of 50 mM Tris pH 8.0 containing 0.5% SDS and protease inhibitors (Roche). Conjugation of azide or alkyne IR-800 Dye (LI-COR) to fatty acid azide or alkyne probes was carried out for 1 h at room temperature with end-over-end rotation by adding an equal volume (100 μL) of freshly mixed click chemistry reaction mixture containing 10 μM IRDye 800CW azide or alkyne infrared dye, 4 mM CuSO4, 400 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, and 8 mM ascorbic acid in dH20. Proteins were isolated by acetone precipitation and resuspended in 100 μL of SDS sample buffer containing 25 mM DTT. Samples were incubated at 95 °C for 5 min, and 10–15 μL was resolved by SDS/PAGE.
For detection of [3H] fatty acid probes, cell lysates were resolved by SDS/PAGE and transferred to duplicate nitrocellulose membranes. One membrane was used for immunoblotting, and the other was exposed to light-sensitive film in the presence of a Kodak Biomax Transcreen LE intensifier screen for detection of [3H].
Generation of Mutant zDHHC Constructs.
The zDHHC3 chimeras containing the N- and C-terminal domains (zDHHC3/CN7) or intracellular domain, including the zDHHC cysteine-rich domain of zDHHC7 (zDHHC3/CRD7) were generated within the HA-tagged constructs by inserting NheI and SalI restriction sites at the boundaries of the domains that were swapped (upstream of C47 and F235 in zDHHC3 and C50 and F238 in zDHHC7 for zDHHC3/CN7, and S93 and T176 in zDHHC3 and S96 and T179 in zDHHC7 for zDHHC3/CRD7) by site-directed mutagenesis. The regions were then swapped by restriction/ligation and the NheI/SalI restriction sites removed using site-directed mutagenesis. The zDHHC3 chimeras containing the transmembrane domains of zDHHC7 were constructed by GeneArt and subcloned into pEF-BOS-HA vector using BamHI restriction sites. The transmembrane domains predicted by UniProt were defined as follows: zDHHC3 A48-V68 (TMD1), Y73-S93 (TMD2), F172-F192 (TMD3), I215-F235 (TMD4), zDHHC7 A51-L71 (TMD1), F76-S96 (TMD2), F175-G194 (TMD3), and I218-F238 (TMD4). Site-directed mutants were generated using PCR. The validity of all clones was confirmed by sequencing.
Data Quantification and Statistical Analysis.
Quantification of all click chemistry experiments was performed by expressing the click signal relative to the corresponding protein signal (immunoblot). For substrate S-acylation assays, this was then normalized to empty vector control. Statistical analysis was performed by one-way ANOVA with a Tukey posttest, using Graphpad Prism software.
Supplementary Material
Acknowledgments
We thank Mike Shipston and Heather McClafferty for the zDHHC23 cysteine mutant. We also thank the EPSRC Mass Spectrometry Service, Swansea, for high-resolution spectra. This work was funded by Biotechnology and Biological Sciences Research Council Grant BB/L022087/1 (to T.K.S., N.C.O.T., and L.H.C.) and Wellcome Trust Grant 093228 (to T.K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.C.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612254114/-/DCSupplemental.
References
- 1.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu Rev Biochem. 2004;73(1):559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. 2015;95(2):341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. 2010;191(7):1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 5.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36(5):245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44(6):987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277(43):41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 8.Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159(1):23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Politis EG, Roth AF, Davis NG. Transmembrane topology of the protein palmitoyl transferase Akr1. J Biol Chem. 2005;280(11):10156–10163. doi: 10.1074/jbc.M411946200. [DOI] [PubMed] [Google Scholar]
- 10.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287(10):7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285(49):38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muszbek L, Haramura G, Cluette-Brown JE, Van Cott EM, Laposata M. The pool of fatty acids covalently bound to platelet proteins by thioester linkages can be altered by exogenously supplied fatty acids. Lipids. 1999;34(0) Suppl:S331–S337. doi: 10.1007/BF02562334. [DOI] [PubMed] [Google Scholar]
- 13.Hallak H, et al. Covalent binding of arachidonate to G protein alpha subunits of human platelets. J Biol Chem. 1994;269(7):4713–4716. [PubMed] [Google Scholar]
- 14.O’Brien PJ, Zatz M. Acylation of bovine rhodopsin by [3H]palmitic acid. J Biol Chem. 1984;259(8):5054–5057. [PubMed] [Google Scholar]
- 15.Veit M, Reverey H, Schmidt MF. Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem J. 1996;318(Pt 1):163–172. doi: 10.1042/bj3180163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brett K, et al. Site-specific S-acylation of influenza virus hemagglutinin: The location of the acylation site relative to the membrane border is the decisive factor for attachment of stearate. J Biol Chem. 2014;289(50):34978–34989. doi: 10.1074/jbc.M114.586180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haynes CA, et al. Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49(5):1113–1125. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang X, et al. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276(33):30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 19.Hang HC, et al. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J Am Chem Soc. 2007;129(10):2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 20.Charron G, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131(13):4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 21.Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: Omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol. 2009;4(7):581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 22.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6(7):498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 23.Yap MC, et al. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res. 2010;51(6):1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111(10):6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaves J, Gorleku OA, Salaun C, Chamberlain LH. Palmitoylation of the SNAP25 protein family: Specificity and regulation by DHHC palmitoyl transferases. J Biol Chem. 2010;285(32):24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves J, Salaun C, Fukata Y, Fukata M, Chamberlain LH. Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J Biol Chem. 2008;283(36):25014–25026. doi: 10.1074/jbc.M802140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greaves J, Carmichael JA, Chamberlain LH. The palmitoyl transferase DHHC2 targets a dynamic membrane cycling pathway: Regulation by a C-terminal domain. Mol Biol Cell. 2011;22(11):1887–1895. doi: 10.1091/mbc.E10-11-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian L, McClafferty H, Knaus H-G, Ruth P, Shipston MJ. Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels. J Biol Chem. 2012;287(18):14718–14725. doi: 10.1074/jbc.M111.335547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorleku OA, Barns A-M, Prescott GR, Greaves J, Chamberlain LH. Endoplasmic reticulum localization of DHHC palmitoyltransferases mediated by lysine-based sorting signals. J Biol Chem. 2011;286(45):39573–39584. doi: 10.1074/jbc.M111.272369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denic V, Weissman JS. A molecular caliper mechanism for determining very-long-chain fatty acid length. Cell. 2007;130(4):663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 31.Gregory MK, Cleland LG, James MJ. Molecular basis for differential elongation of omega-3 docosapentaenoic acid by the rat Elovl5 and Elovl2. J Lipid Res. 2013;54(10):2851–2857. doi: 10.1194/jlr.M041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai J, Linder ME. Oligomerization of DHHC protein S-acyltransferases. J Biol Chem. 2013;288(31):22862–22870. doi: 10.1074/jbc.M113.458794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordyukova LV, Serebryakova MV, Baratova LA, Veit M. Site-specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology. 2010;398(1):49–56. doi: 10.1016/j.virol.2009.11.039. [DOI] [PubMed] [Google Scholar]
- 34.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2(11):584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 35.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275(23):17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 36.Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts: Many raft proteins are acylated, while few are prenylated. J Biol Chem. 1999;274(6):3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 37.Shahinian S, Silvius JR. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34(11):3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 38.Trindade S, et al. Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host Microbe. 2016;19(6):837–848. doi: 10.1016/j.chom.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.