Significance
The effectiveness of nucleic acid drugs is limited by inefficient delivery to target tissues and cells and by unwanted accumulation in off-target organs. Although thousands of chemically distinct nanoparticles can be synthesized, nanoparticles designed to deliver nucleic acids in vivo were first tested in cell culture, yielding poor predictions for delivery in vivo. To facilitate testing of many nanoparticles in vivo, we designed and optimized a high-throughput DNA barcoding system to simultaneously measure nucleic acid delivery mediated by dozens of distinct nanoparticles in a single mouse. This nano-barcoding system can be used to study hundreds, or even thousands, of nanoparticles directly in vivo and could dramatically accelerate the discovery and understanding of nanoparticle drug delivery systems.
Keywords: barcode, nanotechnology, nanoparticle, drug delivery, gene therapy
Abstract
Nucleic acid therapeutics are limited by inefficient delivery to target tissues and cells and by an incomplete understanding of how nanoparticle structure affects biodistribution to off-target organs. Although thousands of nanoparticle formulations have been designed to deliver nucleic acids, most nanoparticles have been tested in cell culture contexts that do not recapitulate systemic in vivo delivery. To increase the number of nanoparticles that could be tested in vivo, we developed a method to simultaneously measure the biodistribution of many chemically distinct nanoparticles. We formulated nanoparticles to carry specific nucleic acid barcodes, administered the pool of particles, and quantified particle biodistribution by deep sequencing the barcodes. This method distinguished previously characterized lung- and liver- targeting nanoparticles and accurately reported relative quantities of nucleic acid delivered to tissues. Barcode sequences did not affect delivery, and no evidence of particle mixing was observed for tested particles. By measuring the biodistribution of 30 nanoparticles to eight tissues simultaneously, we identified chemical properties promoting delivery to some tissues relative to others. Finally, particles that distributed to the liver also silenced gene expression in hepatocytes when formulated with siRNA. This system can facilitate discovery of nanoparticles targeting specific tissues and cells and accelerate the study of relationships between chemical structure and delivery in vivo.
The clinical and scientific potential of nucleic acid therapies is limited by inefficient drug delivery to target cells. Drug delivery vehicles must avoid clearance by the immune and reticuloendothelial systems, access the correct organ, and enter specific cells within a complex tissue microenvironment (1–3). At each of these steps, anatomical structures and biological molecules can actively engage the vehicles and influence their final destination. For example, fenestrations in endothelial cells lining the liver may improve access to hepatocytes, tight junctions in brain endothelial cells inhibit delivery across the blood–brain barrier, the basement membrane in renal tubules can disassemble cationic delivery vehicles, and serum proteins can bind nanoparticles in the blood and affect their interactions with target cells (4–7). It is not currently possible to recapitulate the totality of this complex process in cell culture.
Thousands of nanoparticles with distinct chemical structures and properties have been synthesized to overcome drug delivery obstacles and control nanoparticle biodistribution (8–15). Due to the expensive and laborious nature of in vivo experiments, the current practice is to characterize these diverse nanoparticle “libraries” in cell culture before selecting a small number to test in vivo (8–15). However, in vitro transfection can be a poor predictor of in vivo transfection, and in vitro screens cannot predict whole-body biodistribution, which influences off-target effects (16, 17).
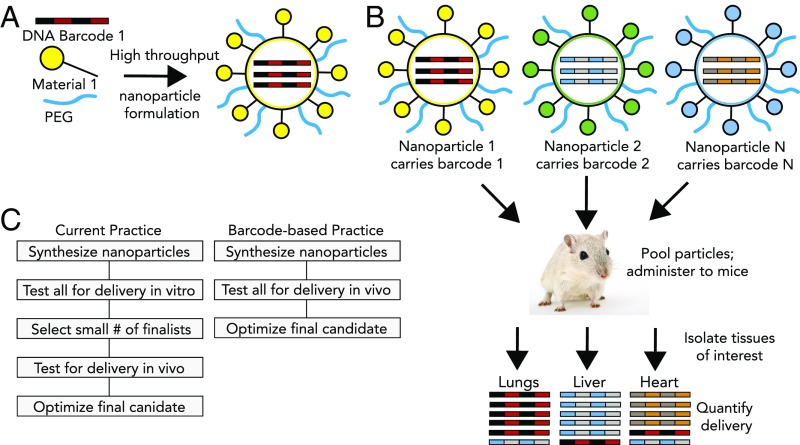
We sought to develop a system that increases the number of nanoparticles testable in vivo. To increase the throughput of in vivo studies, we used a rapid microfluidic mixing system to encapsulate nucleic acid barcodes inside nanoparticles and administered them as a single pool to mice (Fig. 1 A and B). We recovered the barcodes from tissues and cells and used deep sequencing to obtain counts for those barcodes in each sample of interest (18). Deep sequencing is a high throughput, cost-effective method to precisely quantitate nucleic acid species; it has led to the identification of molecules or peptides with specific biological activities and enabled pooled screening with shRNAs, cDNAs, and labeled pools of RNA (19–21). By associating specific nanoparticles with unique DNA barcode sequences, we can now reliably measure the biodistribution of many nanoparticles in a single animal (Fig. 1C).
Fig. 1.
DNA barcoded nanoparticles for high throughput in vivo nanoparticle delivery. (A) Using high-throughput fluidic mixing, nanoparticles are formulated to carry a DNA barcode. (B) Many nanoparticles can be formulated in a single day; each nanoparticle chemical structure carries a distinct barcode. Particles are then combined and administered simultaneously to mice. Tissues are then isolated, and delivery is quantified by sequencing the barcodes. In this example, nanoparticle 1 delivers to the lungs, nanoparticle 2 delivers to the liver, and nanoparticle N delivers to the heart. (C) This DNA barcode system enables multiplexed nanoparticle-targeting studies in vivo, improving upon the current practice, which relies on in vitro nanoparticle screening to identify lead candidates.
Results
We formulated chemically distinct lipid nanoparticles (LNPs) so they each carried a unique DNA barcode oligonucleotide (Fig. 1A). We pooled the different nanoparticle formulations together, and injected the pool i.v. into mice (Fig. 1B). At different time points, we isolated tissues or cells and recovered the oligonucleotides. We used PCR with indexed primers to amplify the oligonucleotides and label each tissue/animal (SI Appendix, Fig. S1) and performed deep sequencing. By counting the nanoparticle-associated barcode sequences obtained from each tissue, we measured the relative biodistribution of many nanoparticles simultaneously (Fig. 1C).
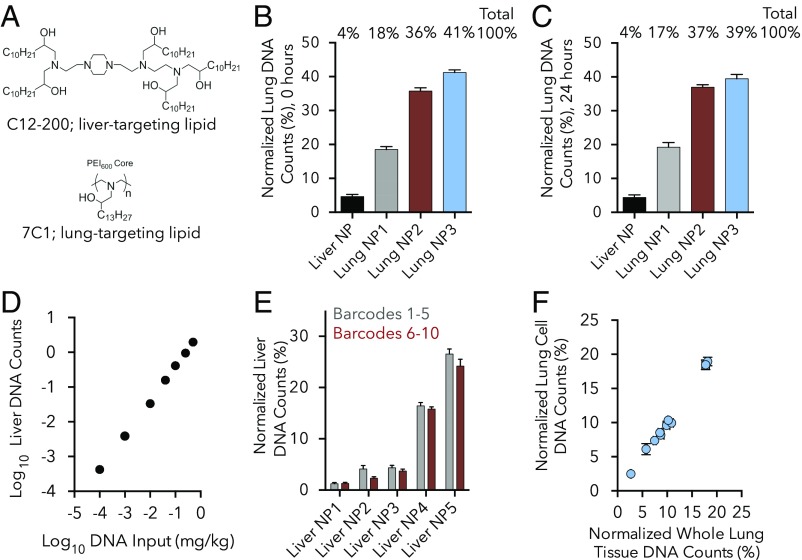
We first tested this approach using nanoparticles with known abilities to target nucleic acids to lung and liver (Fig. 2A) (13, 14, 22–24). For these experiments, we analyzed barcode distribution 4 h after injection, a length of time sufficient for these LNPs to be cleared by the bloodstream (14). We formulated four different nanoparticles with four different DNA barcodes. One barcode was formulated with LNPs made with the lipid C12-200; these “liver-targeting” LNPs deliver nucleic acids to hepatocytes at doses of nucleic acid as low as 0.01 mg/kg (13). Three other barcodes were formulated with LNPs made with the lipid 7C1; these “lung-targeting” LNPs deliver nucleic acids to pulmonary endothelial cells at doses as low as 0.01 mg/kg (14). As expected, barcodes delivered by 7C1 particles were enriched ∼4.5- to 10-fold in the lung relative to liver, compared with barcodes delivered by C12-200; these results were consistent across all four mice (Fig. 2B). Formulation details for all nanoparticles analyzed are listed in SI Appendix, Fig. S2.
Fig. 2.
DNA barcoded nanoparticle data are robust. (A) Nanoparticle (NP) biodistribution using 7C1 and C12-200, two well-validated NPs with known activity in the lung and liver, respectively. (B) Normalized DNA barcode counts in the lung 4 h after the administration of the liver-targeting C12-200 or three different formulations of the lung-targeting 7C1. Normalization technique used for all “normalized” figures is described in Materials and Methods. n = 4 mice/group. (C) Normalized DNA barcode counts in the lungs 4 h after administration of the same four-nanoparticle solution in B. In this case, the particles were administered the next day, after being allowed to mix for 24 h. No change in targeting was observed between the “freshly injected” and “24 h mixed” particles. n = 4 mice/group. (D) DNA barcode counts in the liver 4 h after administration of an “in vivo standard curve.” The same C12-200 nanoparticle formulation was made seven separate times with seven different barcodes. These solutions were mixed together at different doses (DNA inputs) to form an in vivo standard curve. DNA readouts align with this DNA input at doses between 0.0001 and 0.5 mg/kg DNA barcode. n = 5 mice/group. (E) Normalized DNA barcode counts in liver 4 h after administration of different DNA sequences. Five different C12-200 NPs were formulated twice, each with a different barcode. Delivery for each of the five NPs did not change with barcode sequence. n = 5 mice/group. (F) DNA barcode counts in the lung 4 h after 10 different 7C1 NPs were injected. Sequencing was performed on either whole-lung tissue or lung cells isolated isolated from the same lung by flow cytometry. The delivery of all 10 particles was the same for whole tissue and isolated cells. n = 4 mice/group. For all data presented in this figure, the detailed NP formulation parameters are listed in SI Appendix, Fig. S2, and the data are plotted as average ± SD.
Interpretation of results following injection of pooled nanoparticles requires that there is minimal reassortment of nanoparticles following formulation. To assess whether particle mixing occurred, we repeated the previous experiment, but allowed the particles to mix for 24 h before injection. We observed the same delivery efficiencies (Fig. 2C), suggesting that, for these particles and this timescale, appreciable mixing or “hybrid” nanoparticle formation does not occur and that nanoparticle barcode content is not differentially lost into the buffer over time.
Encouraged by these results, we assessed the sensitivity of our assay. We formulated seven identical C12-200 LNPs, so that each formulation carried a distinct barcode. We then mixed the formulations together so that the abundance of each barcode-containing particle varied, spanning a range of 0.0001 mg/kg to 0.5 mg/kg, forming an “in vivo standard curve.” Barcode counts in both lung and liver correlated linearly to the dose of injected LNP 4 h after injection (Fig. 2D and SI Appendix, Fig. S3A). These data demonstrate that DNA barcodes delivered by LNPs can be accurately measured at doses as low as 0.0001 mg/kg DNA barcode for each particle. Notably, sequencing at greater depth may allow for lower doses to be measured.
Barcodes were designed so that exonuclease-mediated degradation would not differentially impact the barcodes based on their sequence. We achieved this by including identical flanking nucleotides (21-bp adapters) on the 5′ and 3′ end of each barcode (SI Appendix, Fig. S1). To investigate whether using different barcode sequences would change results, we performed a barcode-swapping experiment in which we formulated five distinct C12-200–based LNPs with separate barcodes (barcodes 1–5). We then repeated the same five formulations, but used new barcodes (barcodes 6–10). We then administered all 10 nanoparticles together. The relative delivery efficiency of each of the five LNPs to lung or liver remained constant independently of the barcode sequence (Fig. 2E and SI Appendix, Fig. S3B). This suggests that the short barcode sequence does not change LNP behavior and does not appreciably influence the relative efficiency of PCR to amplify slightly different barcode sequences.
Nucleic acid therapeutics must reach affected tissues and access relevant cell types in those tissues. We assessed whether our platform could be used to measure delivery to cells, and not only whole tissue. We barcoded 10 distinct 7C1-based nanoparticles and assessed their ability to enter the lung; we selected the lung because we had previously established a protocol to isolate live lung cells from mouse tissue (14). Four hours after injection, half of the lung was processed as a whole tissue, and the other half was digested into a single-cell suspension (14). Live cells were selected by flow cytometry, and barcodes were recovered from cells and whole tissue. The relative delivery efficiency for each of these LNPs was similar in whole lung and flow-sorted lung cells (Fig. 2F). The ability to recover oligonucleotides from sorted cells suggests that this assay may be used to assess delivery to cell subtypes isolated from tissue.
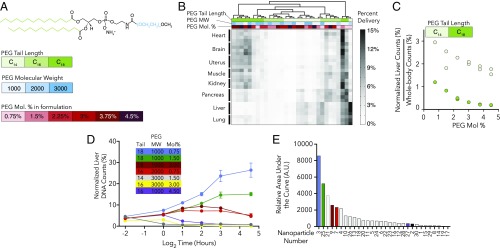
A high-throughput in vivo assay can enable systematic studies of how nanoparticle chemical properties impact particle biodistribution. Particle activity can vary with many factors, including nanoparticle size, shape, charge, the structure or molar ratio of hydrophilic polymers like polyethylene glycol (PEG), and the molar ratio of lipids including cholesterol (25). This complexity makes it difficult to systematically study the relationship between nanoparticle chemical structure and in vivo activity when a small number of nanoparticles are tested. For example, although PEG lipids are known to increase LNP circulation (26), the relationship between the PEG-lipid chemical structure and tissue delivery is less well understood.
To assess the feasibility of a systematic study, we generated a library of 30 distinct C12-200 LNPs. Among the 30 LNPs, we varied three PEG structural parameters: the PEG molecular weight (1, 2, or 3 kDa), the mole percentage of PEG in the LNP formulation (0.75–4.5%), and the length of the hydrophobic lipid attached to the PEG (C14, C16, C18) (Fig. 3A). Both PEG molecular weight and molar ratio affect particle shielding, whereas the lipid length can influence how securely PEG “anchors” into the LNP (27). We barcoded all 30 particles, pooled them together, and injected them into mice before isolating the brain, heart, kidney, liver, lung, skeletal muscle, uterus, and pancreas 4 h later. We observed a broad range in relative delivery efficiency to different tissues (Fig. 3B), which was reproducible across mouse replicates. Some tissues behaved similarly to other tissues in their ability to be targeted by certain particles. For example, particles that entered lung efficiently tended to enter the liver well and were distinct from particles that preferentially entered the heart and other organs (Fig. 3B).
Fig. 3.
DNA barcoded nanoparticles facilitate high-throughput in vivo analysis. (A) LNPs are often formulated with PEG lipids. The lipid length, PEG molecular weight, and the mole percentage of PEG used in the LNP can influence nanoparticle activity. We formulated 30 C12-200 LNPs with different PEGs, varying all three of these parameters. (B) Normalized DNA counts, measured 4 h after mice were injected with LNPs. Certain tissues “cluster”; the same tissues tend to be targeted by similar particles. (C) Normalized liver counts, divided by the normalized counts for the rest of the tested tissues, as a function of PEG mole percentage. This measure quantifies how delivery to the liver, compared with the rest of the body, changes. Increasing PEG decreased relative delivery to the liver. (D) Normalized liver DNA counts for seven nanoparticles 0.25, 1, 2, 4, 8, or 24 h after injection. The relative distribution of some nanoparticles increased over time, whereas others decreased. This kinetic analysis was performed with 30 nanoparticles; all data are plotted in SI Appendix, Fig. S5. (E) The AUC for 30 nanoparticles in the liver. In all cases, data are plotted as mean ± SD, and n = 2–4 mice/group. In B, each row is an individual mouse. All formulation details are listed in SI Appendix, Fig. S2.
Because many tissues can be analyzed at once using this method, we studied how nanoparticle delivery to the liver changed relative to the rest of the body. We found that liver delivery efficiency (relative to delivery averaged across all tissues) for each individual LNP increased as C14PEG or C18PEG was reduced (Fig. 3C). These data, gathered in a single multiplexed study, are consistent with previous studies of liver-targeting LNPs that tested one nanoparticle at a time (27). The relationship was not observed with C16PEG or nanoparticle diameter (SI Appendix, Fig. S4 A and B). Importantly, these experiments suggest that DNA barcoding can be used to systematically study how nanoparticle structure influences “whole-body” biodistribution, which could be important in studying on- and off-target effects.
Nanoparticle pharmacokinetics can affect efficacy and off-target effects. To this end, we performed a high-throughput pharmacokinetic experiment (Fig. 3D and SI Appendix, Fig. S5A). We chose to measure the relative area under the curve (AUC) because it is an important parameter that approximates how much nucleic acid accumulates in tissue over time. We formulated 30 distinct C12-200 LNPs and measured biodistribution 0.25, 1, 2, 4, 8, and 24 h after injection. Interestingly, nanoparticle distribution in the liver varied differentially over time; some nanoparticles became enriched with respect to the pool over time, whereas others decreased to near-zero values over time (Fig. 3D). Based on these values, we then calculated AUC for all 30 nanoparticles (Fig. 3E and SI Appendix, Fig. S6). We did not observe any simple statistically significant trends between nanoparticle PEG structure and AUC (SI Appendix, Fig. S5 B–D).
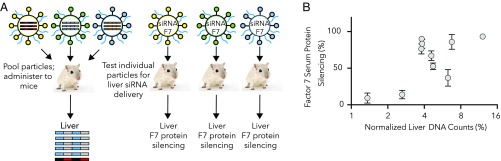
Many oligonucleotide therapeutics require intracellular delivery. To assess the ability of our screening approach to uncover particles that functionally deliver nucleic acids, and to eliminate nonfunctional nanoparticles from consideration, we compared liver biodistribution to functional hepatocyte gene silencing mediated by siRNA (Fig. 4A). We selected 10 of the 30 PEG particles that spanned a broad range of liver delivery as measured by barcode delivery and formulated them to carry siRNA against factor VII. Factor VII is an enzyme with a short half-life that is specifically secreted by hepatocytes; its silencing is used to assess functional siRNA delivery to hepatocytes (12, 13, 15). We administered each siRNA-carrying nanoparticle individually and measured factor VII levels 72 h later (12, 13, 15). Particles with low liver delivery were less effective at delivering factor VII siRNA than particles with higher liver delivery (Fig. 4B). These data suggest that our methodology may be useful as a “first-pass” screen to identify particles for further functional evaluation.
Fig. 4.
Comparing high-throughout analysis with individual analysis in vivo. (A) Workflow used to compare high-throughput nanoparticle analysis with traditional, individual analysis. Thirty nanoparticles with varying PEG characteristics were injected. Ten of the nanoparticles, with a range of liver biodistributions, were analyzed individually by formulating them with siRNA targeting factor 7 (F7), a gene expressed in hepatocytes. Mice were injected with one siRNA-containing nanoparticle at a siRNA dose of 0.10 mg/kg, and the resulting factor 7 protein knockdown was compared with the barcode liver data. (B) Factor 7 protein reduction, tested one nanoparticle at a time, plotted against normalized barcode delivery in the liver 15 min after i.v. injection, after 30 nanoparticles were tested simultaneously. Data are shown as average ± SD, and n = 3–4 mice/group.
Discussion
Genetic therapeutics, including aptamers, antisense oligonucleotides, RNAi, and gene-editing technologies, function through distinct biological mechanisms (28, 29). However, all genetic therapies are limited by the inability to predict delivery to on- and off-target tissues. Although syntheses of chemically distinct nanoparticles can be high throughput, characterization of nanoparticle behavior in vivo is still low throughput (8–15). Rapidly screening chemically distinct nanoparticles in vivo could accelerate preclinical screening and enable efforts to relate chemical structure to biological function. By incorporating deep sequencing, our approach dramatically increases the number of particles that can be simultaneously measured, as well as improves the sensitivity, specificity, and accuracy of those measurements. Our work, as well as DNA barcoded particles that were shown to target tumors, demonstrates the power of unbiased in vivo approaches (30). Notably, this platform is distinct from previous reports, which conjugate nucleic acids to the exterior of particles to fluorescently label them or use them to identify known pathogenic DNA sequences in bodily fluids (31–34).
We carefully tested our workflow to identify biases that may arise from particle mixing or differences in barcode sequence. DNA barcode readouts varied linearly with the input across four orders of magnitude encompassing typical doses of effective nucleic acid therapeutics (0.0001–0.5 mg/kg DNA) (25) (Fig. 2D); the ability to measure a single nanoparticle at a dose as low as 0.0001 mg/kg DNA suggests that dozens, or even hundreds, of nanoparticles can be multiplexed in a single experiment. DNA barcode amplification did not vary with barcode sequence (Fig. 2E). We did not observe any evidence of hybrid particle formation with 7C1 and C12-200 LNPs over 24 h (Fig. 2 B and C) or when 10 C12-200–based LNPs were tested simultaneously (Fig. 2D). However, because hybrid particle mixing may occur with other nanoparticles, especially if dozens or hundreds nanoparticles are tested simultaneously, it will be important to control for, and test, particle mixing. Nevertheless, these data demonstrate that the DNA barcoding approach can be used to rapidly study biodistribution and pharmacokinetics. For example, although we did not uncover any structure–function mechanisms or LNPs with new targeting functionality in this study, we did identify a LNP that performed well in many organs, as well as LNPs that distributed inefficiently in all organs (Fig. 3B).
We designed this system to be useful in many in vivo contexts. Because this approach can quantify delivery to cells isolated by flow cytometry (Fig. 2F), we anticipate that future studies will simultaneously study delivery to multiple cell types in a complex microenvironment. Similarly, we believe this system may be used to study how nanoparticle delivery changes with an animal disease state. Finally, although this system cannot directly differentiate between delivery to, and into, a cell, future work could use this approach to study nanoparticle delivery to intracellular and subcellular compartments using standard fractionation approaches, with the goal of identifying nanoparticles that evade lysosomes, remain in the cytoplasm, or, alternatively, enter the nucleus (35). By associating DNA barcodes with ligands, this high-throughput nanoparticle barcoding system may also be used to rapidly identify effective targeting sequences via a process that is akin to phage display.
It is unlikely that this system will work for every drug delivery vehicle; it will be most effective for well-tolerated nanoparticles that are stable in solution before injection. In future studies, it will be important to characterize nanoparticle stability before using this system to study the activity of different nanoparticles. Even with these constraints, we anticipate that this methodology will facilitate nanoparticle pharmacokinetic studies and will rapidly accelerate the discovery of nanoparticles with wide-ranging therapeutic and research applications.
Materials and Methods
Oligonucleotide Barcode Amplification and DNA Sequencing.
DNA barcodes were 61 nucleotides long, with three phosphorothioate bonds at each end (Integrated DNA Technologies) to increase barcode stability and decrease exonuclease degradation. Oligonucleotide sequences and primers are listed in SI Appendix, Fig. S1. The “barcode” portion comprised 10 nucleotides in the center of the oligonucleotide. Ten random bases were also included directly 3′ of the barcode. The random bases were incorporated to monitor excessive PCR amplification, which was never observed in any experiment (>90% of the randomized sequences were always unique). The 5′ and 3′ ends of each oligonucleotide contained priming sites for Illumina adapters. Tissues were lysed in 2-mL tubes using 1.4-mm ceramic beads, placed into a tissue-lyser machine that rapidly agitated the tubes. DNA oligonucleotides were isolated from this tissue lysate according to manufacturer instructions (Clarity OTX columns, Phenomenex). Crude oligonucleotide preparations were further purified on Zymo Oligo Clean and Concentrator columns. Each oligonucleotide pool was amplified by PCR using the following recipe: 5 μL 5× HF Phusion buffer, 0.5 μL 10 mM dNTPs (New England Biolabs), 1 μL oligonucleotide pool, 0.5 μL 5 μM Universal primer, 0.5 μL 5 μM Index primer, 0.5 μL 0.5 μM Index-base primer, 0.25 μL Phusion enzyme (New England Biolabs), 2 μL DMSO, and 14.75 μL H2O. DNA barcode and primer sequences are described in SI Appendix, Fig. S1. Cycling conditions were 98° for 15 s, 60° for 15 s, and 72° for 30 s, repeated 25–30 cycles. PCR products were run by gel electrophoresis on 1.4% Tris-acetate-EDTA agarose, and bands were excised, pooled, and purified by Zymo Gel Extraction columns. Agarose bands containing PCR products were pooled only if the Index primers were distinct. The purified products were kept frozen until deep sequencing.
Deep Sequencing.
All deep-sequencing runs were performed using multiplexed runs on Illumina Miseq machines. PCR product pools were quantitated using the KAPA Library Quantification Kit for next generation sequencing. PCR product pools were loaded onto flow cells at 4 nM concentration. Raw counts for all experiments can be found in SI Appendix, Fig. S7.
Nanoparticle Formulation.
All nanoparticle formulation details are listed in SI Appendix, Fig. S2. The 7C1 lipid was synthesized as previously described (14), and C12-200 lipid was purchased from Wuxi AppTec. LNPs were synthesized by mixing a lipid-containing ethanol phase with a nucleic acid-containing aqueous phase at a 1:3 volume ratio in a microfluidic chip as previously described (36). The ethanol phase was prepared by solubilizing a mixture including some of the following components: lipids 7C1 or C12-200, phospholipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and/or lipid-anchored PEG (e.g., C14 PEG 2000, C18 PEG 1000, and others) (Avanti Polar Lipids). The aqueous phase was prepared in 10 mM citrate buffer with DNA barcode or factor VII siRNA. The 7C1:nucleic acid and C12-200:nucleic acid weight ratios were 5:1 and 10:1, respectively. Immediately after mixing the ethanol and aqueous phases, the resultant LNPs were dialyzed against 1× PBS overnight at 4 °C. Formulation parameters for the 30-LNP screen were generated using statistical Design of Experiment software JMP (SAS Institute) using the Custom Design feature. In all cases (except for the in vivo standard curve in Fig. 2D), we administered 0.04 mg/kg DNA barcode per nanoparticle. This dose was selected because the “total dose” ranged between 0.16 mg/kg DNA barcode (e.g., Fig. 2B) and 1.2 mg/kg DNA (e.g., Fig. 3A), which are within commonly used doses for 7C1 and C12-200 (13, 14).
Nanoparticle Characterization.
LNP particle diameter and polydispersity were measured using dynamic light scattering (DLS, ZetaPALS, Brookhaven Instruments) as previously described (13, 14). Nanoparticles were diluted to ∼0.001 mg/mL nucleic acid in PBS and analyzed at room temperature. We quantified siRNA or DNA concentration and encapsulation as previously described and according to manufacturing instructions (Quant-iT RiboGreen RNA assay, Quant-iT OliGreen ssDNA, respectively; Invitrogen) (37). Doses for each nanoparticle and barcode, for each experiment, are listed in SI Appendix, Fig. S2.
Tissue/Cell Isolation.
For experiments in Fig. 2, as well as in Fig. 3 B and C, tissues were isolated 4 h after animals were injected. A 4-h time point was selected because C12-200– and 7C1-based nanoparticles have a half-life of ∼10 min; at 4 h, over 99.9% of the particles would be out of the circulation. In Fig. 3D, tissues were isolated 0.25, 1, 2, 4, 8, or 24 h after administration. In all cases, animals were killed, and tissues were snap-frozen in liquid nitrogen. To isolate live lung cells in Fig. 2F, we used a protocol that we previously developed (14, 38). Immediately after sacrificing the mouse, lungs were perfused with 37 °C 1× PBS. Lungs were cut into small slices, placed in buffer with Collagenase I, Collagenase XI, and Hyaluronidase, and digested for 30 min at 37 °C. Whole-tissue homogenates were then passed through a 100-μm filter to separate cells. Cells were stained to identify live cells (Biolegend Zombie Dyes) and sorted by flow cytometry.
Normalized DNA Counts.
For all figures with normalized DNA counts, the following calculations were made: The total number of sequencing reads from a given tissue were added together. The number of sequencing reads with a specific barcode was then calculated. As a simplified example, if a mouse lung generated a total of 10,000 barcode reads, and individual barcodes 1, 2, and 3 had 6,000, 3,000, and 1,000, of those reads, respectively, then the percentages delivered in that lung by nanoparticles 1, 2, and 3 were 60, 30, and 10%.
AUC Calculations.
The relative AUC for each LNP (units of mass lipid × time/mass liver) was calculated using the trapezoidal rule on a plot of LNP liver concentration vs. time. To translate normalized liver counts for each LNP to liver concentration, we used pharmacokinetic data reported from a previous study (12, 13, 27) in which liver-targeting siRNA-LNPs were formulated with 50% 14C-labeled ionizable lipid, 10% DSPC, 38.5% cholesterol, and 1.5 mol% C16PEG2000; following i.v. injection, the liver accumulation of 14C was monitored over time. In all cases, the percentage above refers to molar percentage (moles component/total moles of material) of the LNP formulation. We created an LNP (#6) with an identical ionizable lipid, DSPC, cholesterol, and C16PEG2000 composition to that of the radiolabeled LNP and assumed that the pharmacokinetic curve of the radiolabeled LNP would make an ideal approximation for LNP #6 in our study. A detailed description of the AUC calculation may be found in SI Appendix, Fig. S6.
Factor VII Analysis.
Factor VII siRNA was synthesized and modified to reduce off-target effects and immunostimulation as previously described (12, 13, 15, 39) (provided by Alnylam Pharmaceuticals). The sense sequence was 5′-GGAUfCfAUfCfUfCfAAGUfCfUfUfACfdTdT-3′, and the antisense sequence was 5′-GUfAAGACfUfUfGAGAUfGAUfCfCfdTdT-3′ (“Cf”denotes 2-fluoro modification to C base, and “dT” denotes DNA). siRNA was administered i.v. at a dose 0.10 mg/kg. After 72 h, blood was collected via the tail vein, and serum was isolated by spinning at 2000 × g for 10 min at 4 °C. Factor VII was quantified according to manufacturer instructions (Biophen FVII, Aniara Corporation), as described previously (12, 13, 15). Factor VII levels were compared with mice injected with PBS; factor VII expression in PBS-treated mice was treated as “100%” factor VII expression.
Animal Experiments.
The Massachusetts Institute of Technology (MIT) Institutional Animal Care and Use Committee approved all animal experiments. Seven- to 10-wk-old female C57BL/6 mice between 17 and 21 g were used in these experiments (Charles River Labs). Randomization of sample groups was not necessary because mouse replicates were administered the same pool of nanoparticles, except in Fig. 4B, where mice were randomly assigned different siRNA formulations. Results from experiments shown in Fig. 2 were very consistent among cohorts of four or five animals, and, as a result, later experiments shown in Figs. 3 and 4 were performed using cohorts of three to four animals.
Data Blinding.
In all experiments, preparation of deep-sequencing libraries was performed in a manner blinded to nanoparticle administration and tissue harvest.
Data Analysis.
Python scripts were written to count barcodes from Illumina fastq files. Additional python scripts were used to plot and analyze all data. Ordinary least-squares linear regression with two-sided P values was performed using scipy.linregress, as this test requires minimal assumptions about the data. Code is available upon request.
Supplementary Material
Acknowledgments
We thank the Animal Imaging and Preclinical Testing and Flow Cytometry Cores at the Koch Institute at MIT. J.E.D. was funded by an MIT Presidential Fellowship; National Defense Science and Engineering Graduate Fellowship; and National Science Foundation Graduate Research Fellowship Program Fellowship. K.J.K. was funded by the Marble Center for Cancer Nanomedicine and the Cancer Center Support (core) Grant P30-CA14051. Y.X., T.E.S., and C.C.D. were funded by the MIT Undergraduate Research Opportunities Program. Funding was provided by the Kathy and Curt Marble Cancer Research Fund/Koch Institute Frontier Grant (to E.T.W. and D.G.A.). Funding was also provided by the NIH Grant DP5-OD017865 (to E.T.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620874114/-/DCSupplemental.
References
- 1.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng CJ, Tietjen GT, Saucier-Sawyer JK, Saltzman WM. A holistic approach to targeting disease with polymeric nanoparticles. Nat Rev Drug Discov. 2015;14(4):239–247. doi: 10.1038/nrd4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol. 2012;7(12):779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 5.Zuckerman JE, Choi CHJ, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci USA. 2012;109(8):3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2(1):a006429. doi: 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Y, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci USA. 2014;111(11):3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porel M, Alabi CA. Sequence-defined polymers via orthogonal allyl acrylamide building blocks. J Am Chem Soc. 2014;136(38):13162–13165. doi: 10.1021/ja507262t. [DOI] [PubMed] [Google Scholar]
- 10.Hao J, et al. Rapid synthesis of a lipocationic polyester library via ring-opening polymerization of functional valerolactones for efficacious siRNA delivery. J Am Chem Soc. 2015;137(29):9206–9209. doi: 10.1021/jacs.5b03429. [DOI] [PubMed] [Google Scholar]
- 11.Siegwart DJ, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc Natl Acad Sci USA. 2011;108(32):12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 13.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlman JE, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol. 2014;9(8):648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehead KA, et al. In vitro-in vivo translation of lipid nanoparticles for hepatocellular siRNA delivery. ACS Nano. 2012;6(8):6922–6929. doi: 10.1021/nn301922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130(4):601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18(6):839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- 19.Mannocci L, et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc Natl Acad Sci USA. 2008;105(46):17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 21.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014;42(14):8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White K, et al. Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension. EMBO Mol Med. 2015;7(6):695–713. doi: 10.15252/emmm.201404511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herr KJ, et al. Loss of α-catenin elicits a cholestatic response and impairs liver regeneration. Sci Rep. 2014;4:6835. doi: 10.1038/srep06835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 26.Otsuka H, Nagasaki Y, Kataoka K. PEGylated nanoparticles for biological and pharmaceutical applications. Adv Drug Deliv Rev. 2003;55(3):403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 27.Mui BL, et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2:e139. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol. 2012;19(8):937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Yaari Z, et al. Theranostic barcoded nanoparticles for personalized cancer medicine. Nat Commun. 2016;7:13325. doi: 10.1038/ncomms13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eustaquio T, Leary JF. Nanobarcoding: Detecting nanoparticles in biological samples using in situ polymerase chain reaction. Int J Nanomedicine. 2012;7:5625–5639. doi: 10.2147/IJN.S37433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eustaquio T, Leary JF. Nanobarcoded superparamagnetic iron oxide nanoparticles for nanomedicine: Quantitative studies of cell-nanoparticle interactions by scanning image cytometry. Cytometry A. 2016;89(2):207–216. doi: 10.1002/cyto.a.22699. [DOI] [PubMed] [Google Scholar]
- 33.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc. 2006;128(26):8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 34.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem Int Ed Engl. 2006;45(20):3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 35.Rajendran L, Knölker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010;9(1):29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134(16):6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 37.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107(2):276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 38.Sager HB, et al. RNAi targeting multiple cell adhesion molecules reduces immune cell recruitment and vascular inflammation after myocardial infarction. Sci Transl Med. 2016;8(342):342ra80–342ra80. doi: 10.1126/scitranslmed.aaf1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Silencing or stimulation? siRNA delivery and the immune system. Annu Rev Chem Biomol Eng. 2011;2:77–96. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.