Significance
Our results demonstrate that SUPPRESSOR OF THYLAKOID FORMATION 1 (SOT1), an Arabidopsis pentatricopeptide repeat (PPR) protein with a small MutS-related (SMR) domain, has endonuclease activity. The SMR moiety of SOT1 performs the endonucleolytic maturation of 23S and 4.5S rRNA through the PPR domain specifically recognizing a 13-nucleotide RNA sequence in the 5′ end of the chloroplast 23S–4.5S rRNA precursor. Our results also show that SOT1 can be engineered to recognize and cleave a predicted RNA substrate. Our findings suggest that SOT1 could be used as a tool for RNA manipulation in the future.
Keywords: photosynthesis, PPR-SMR protein, RNA endonuclease, rRNA biogenesis
Abstract
Numerous attempts have been made to identify and engineer sequence-specific RNA endonucleases, as these would allow for efficient RNA manipulation. However, no natural RNA endonuclease that recognizes RNA in a sequence-specific manner has been described to date. Here, we report that SUPPRESSOR OF THYLAKOID FORMATION 1 (SOT1), an Arabidopsis pentatricopeptide repeat (PPR) protein with a small MutS-related (SMR) domain, has RNA endonuclease activity. We show that the SMR moiety of SOT1 performs the endonucleolytic maturation of 23S and 4.5S rRNA through the PPR domain, specifically recognizing a 13-nucleotide RNA sequence in the 5′ end of the chloroplast 23S–4.5S rRNA precursor. In addition, we successfully engineered the SOT1 protein with altered PPR motifs to recognize and cleave a predicted RNA substrate. Our findings point to SOT1 as an exciting tool for RNA manipulation.
Sequence-specific RNA endonucleases are crucial to establishing RNA manipulation technology (1). Compared with DNA editing, RNA manipulation could be more useful and reversible because it does not result in permanent changes to the genome. In addition, sequence-specific RNA endonucleases could potentially be used as an RNA silencing tool to complement RNAi, because RNAi is sometimes ineffective in certain organisms and RNAi machinery is not present in cellular compartments such as chloroplasts and mitochondria. Despite extensive investigations, a natural RNA endonuclease that recognizes RNA in an intrinsic sequence-specific manner has not yet been identified.
Pentatricopeptide repeat (PPR) proteins exist in eukaryotes, have greatly expanded in terrestrial plants, and take part in most RNA metabolic processes in organelles (2–5). The PPR domain can specifically recognize RNAs in an intrinsic sequence-specific manner (5–8). The 2nd, 5th, and 35th (or 1st, 4th, and 34th or 3rd, 6th, and 1st in other numbering systems) residues at each repeat are considered to be RNA selection “codes” (9–11). Based on these codes, several PPR proteins have been successfully modified to recognize predictable RNA targets (9, 12–17).
The small MutS-related (SMR) domain was originally identified at the C terminus of MutS2 in the cyanobacterium Synechocystis (18). SMR proteins are widely distributed in almost all organisms (19). Recent studies demonstrated the SMR domain exhibits DNA nicking nuclease activity in vitro (20–25). Furthermore, a C-terminal SMR domain in Leishmania donovani S-phase mRNA cycling sequence binding protein (CSBP) has RNA cleavage activity in vitro (26). These findings suggest that the SMR domain has nuclease activity.
Interestingly, a small protein family containing both PPR and SMR domains was recently described (27). PPR-SMR proteins are found mainly in land plants. Arabidopsis thaliana contains eight PPR-SMR proteins localized to the organelles, including mitochondria and chloroplasts (27). Currently, four PPR-SMR proteins have been characterized. They play an essential role in plastid retrograde signaling, plastid transcription, and RNA biogenesis (28–34). Thus, PPR-SMR proteins clearly play important roles in organelle biogenesis. However, the molecular mechanisms underlying the functions of PPR-SMR proteins are largely unclear. In particular, the functions of the enigmatic SMR domain of these PPR-SMRs are unknown.
Considering the sequence-specific RNA binding capacity of the PPR domain and the potential nuclease activity of the SMR domain, it has been suggested that PPR-SMR proteins may represent natural sequence-specific RNA endonucleases (27). If the endonuclease activity of the SMR domain of PPR-SMRs can be confirmed, the PPR-SMR proteins may serve as sequence-specific RNA endonucleases in nature and could be potentially used as tools for RNA manipulation (27).
Here, we show that SUPPRESSOR OF THYLAKOID FORMATION 1 (SOT1) has endonuclease activity and performs the endonucleolytic maturation of 23S and 4.5S rRNA through the PPR domain, specifically recognizing a 13-nucleotide RNA sequence in the 5′ end of the chloroplast 23S–4.5S rRNA precursor. We also show that SOT1 can be modified to recognize and cleave a predicted RNA substrate. Our findings suggest that SOT1 could be used as a tool for RNA manipulation in the future.
Results
Disruption of SOT1 Impairs Translation in Chloroplasts.
To identify factors required for chloroplast development, we screened an Arabidopsis mutant library and isolated a mutant line (ultimately named sot1-3, as described below) with a high chlorophyll fluorescence phenotype (SI Appendix, Fig. S1A). The mutant displayed retarded growth and a virescent-leaf phenotype compared with the wild type (WT) (SI Appendix, Figs. S1 A and B and S2A). Chloroplasts in the mutant displayed a vesicular shape and few thylakoids in contrast to the crescent-shaped chloroplasts and well-formed thylakoid structure of WT (SI Appendix, Fig. S1C). Analyses of chlorophyll fluorescence induction curves and P700 redox kinetics revealed a defect in the functions of photosystem II (PSII) and photosystem I (PSI) in the mutant (SI Appendix, Fig. S3).
Because photosynthetic function was clearly defective in this mutant, we investigated changes in the core subunits of key photosynthetic complexes, including PSII, PSI, the cytochrome b6f complex, ATP synthase, the NADH dehydrogenase-like complex (NDH), and Rubisco. Whereas the levels of nuclear-encoded proteins (PsbO, Fd, and FNR) were largely unchanged, the levels of chloroplast-encoded proteins (D1, D2, PsaA, Cyt f, CF1β, RbcL, and ndhI) were considerably decreased in the mutant (SI Appendix, Fig. S2B). Despite the changes in protein levels, the accumulation of transcripts corresponding to D1, PsaA, Cyt f, CF1β, and PsbO were not reduced in the mutant (SI Appendix, Fig. S4). These results suggest that the mutant harbors a defect in chloroplast translation. In vivo 35S pulse-labeling experiments showed that the overall protein biosynthesis rate was dramatically lower in the mutant than in WT (SI Appendix, Fig. S2C), supporting the idea that chloroplast translation was impaired in the mutant. The reduced biosynthesis of chloroplast-encoded proteins is likely responsible for the defects in chloroplast development, photosynthetic function, and plant growth observed in the mutant.
Map-based cloning identified a PPR-SMR gene (AT5G46580) in which a 31-base pair (bp) deletion resulted in a premature stop codon in the eighth PPR domain (SI Appendix, Fig. S2D). AT5G46580 was previously assigned the name SOT1 because the corresponding mutants sot1-1 and sot1-2 (suppressor of thf1 1/2) were identified in a suppressor screen for the leaf variegation phenotype of thylakoid formation 1 (thf1) (34). Accordingly, we designated the mutant identified in our screen as sot1-3. sot1-3 is a knockout mutant (SI Appendix, Fig. S5) and genetic complementation confirmed that disruption of SOT1 is responsible for the phenotypes observed in sot1-3 (SI Appendix, Figs. S1–S3).
SOT1 Functions with Miniribonuclease III in the Maturation of 23S and 4.5S rRNA.
SOT1 and its maize ortholog PPR53 encode proteins with 11 PPR motifs in the N-terminal region and a SMR domain in the C-terminal region and both are involved in the maturation of 23S and 4.5S rRNA (33, 34). Our results with sot1-3 show that the loss of SOT1 resulted in less mature 23S and 4.5S rRNA as well as staggered 23S rRNA 5′ ends (SI Appendix, Fig. S6), which is similar to results observed in sot1-1 and sot1-2 (34).
Chloroplast miniribonuclease III proteins RNC3 and RNC4 are known to cleave the 5′ and 3′ regions of 23S–4.5S rRNA precursor simultaneously and the loss of miniribonuclease III results in staggered 23S rRNA 5′ ends and less mature 4.5S rRNA (35). To investigate how SOT1 might function in the maturation of 23S and 4.5S rRNA, we compared the 5′ end of 23S rRNA and 3′ end of 4.5S rRNA in sot1-3 and miniribonuclease III mutant rnc3/4 using RACE assays. The staggered 23S rRNA 5′ ends and levels of mature 4.5S rRNA in sot1-3 were strikingly similar to those in rnc3/4 (SI Appendix, Fig. S7). These results indicate that miniribonuclease III processing is disrupted in sot1-3 and suggest that miniribonuclease III processing is mediated by SOT1 during maturation of 23S and 4.5S rRNA.
The SMR Domain of SOT1 Is Required for the Maturation of 23S and 4.5S rRNA.
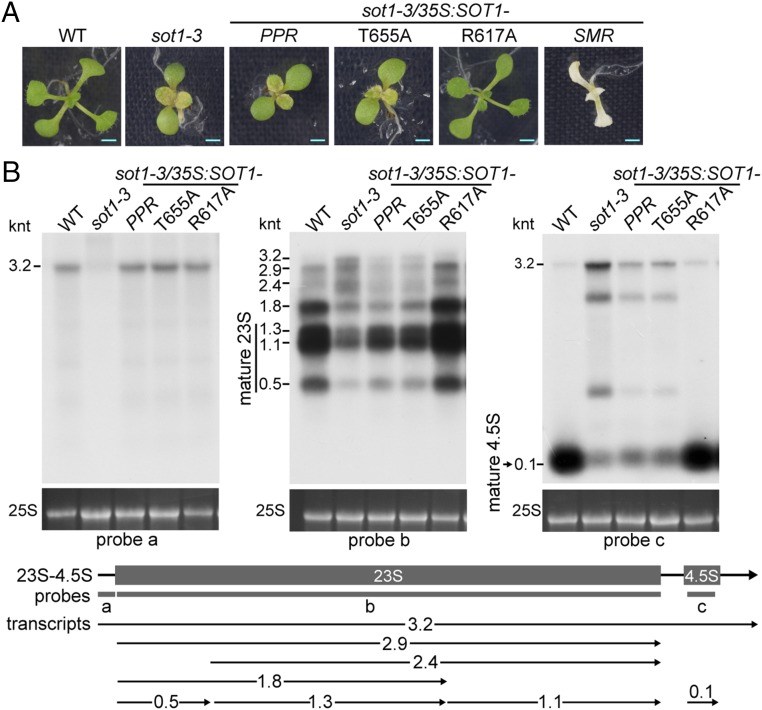
It has been suggested that SOT1/PPR53 acts in the maturation of 23S and 4.5S rRNA through directly binding to the 5′ end of the 23S–4.5S rRNA precursor via the PPR domain and hence blocking the attacks of 5′ exonuclease (33, 34). To address whether the SMR domain of SOT1 functions in the maturation of 23S and 4.5S rRNA, we attempted to complement sot1-3 plants with the PPR domain alone (sot1-3/35S:SOT1PPR-HA). The expression of the PPR domain in sot1-3/35S:SOT1PPR-HA plants was confirmed by RT-PCR and immunoblot assays (SI Appendix, Fig. S8). The sot1-3/35S:SOT1PPR-HA plants displayed a virescent-leaf phenotype and had considerably reduced mature 23S and 4.5S rRNA, similar to sot1-3 (Fig. 1 A and B). The lack of phenotype recovery with the PPR domain alone suggests that the SMR domain of SOT1 plays an important role in the maturation of 23S and 4.5S rRNA.
Fig. 1.
Genetic evidence for the role of the SMR domain of SOT1 in the maturation of 23S and 4.5S rRNA. (A) Phenotypes of sot1-3 and its genetic complementation lines grown for 12 d. The sot1-3 mutant was complemented with the PPR domain of SOT1 (sot1-3/35S:SOT1PPR-HA), SOT1 T655A (sot1-3/35S:SOT1T655A-HA), SOT1 R617A (sot1-3/35S:SOT1R617A-HA), and the SMR domain of SOT1 (sot1-3/35S:SOT1SMR-HA). (Scale bars, 0.2 cm.) (B) RNA gel blot analysis of sot1-3 and its genetic complementation lines. Mature transcripts and precursors of 23S and 4.5S rRNA were detected by RNA gel blot using probes a–c shown below the gene model. The transcript size is shown in kilonucleotides (knt) to the left of each panel. The positions of transcripts detected by RNA gel blot are marked with horizontal arrows below the gene model. The 25S rRNA (loading control) was stained with ethidium bromide.
The SMR Domain of SOT1 Has Nuclease Activity.
Because SMR domains typically have DNA nicking activity (20–26), we investigated whether the SMR domain of SOT1 retained the ability to nick supercoiled DNA. We found that recombinant SMR protein SOT1SMR (amino acids 603–710 of SOT1) cleaved supercoiled pUC19 to open circular and linear conformations and that the metal ions Mn2+ and Mg2+ increased the DNA endonuclease activity of SOT1SMR (Fig. 2 A–C), indicating that the SMR domain of SOT1 indeed has DNA endonuclease activity.
Fig. 2.
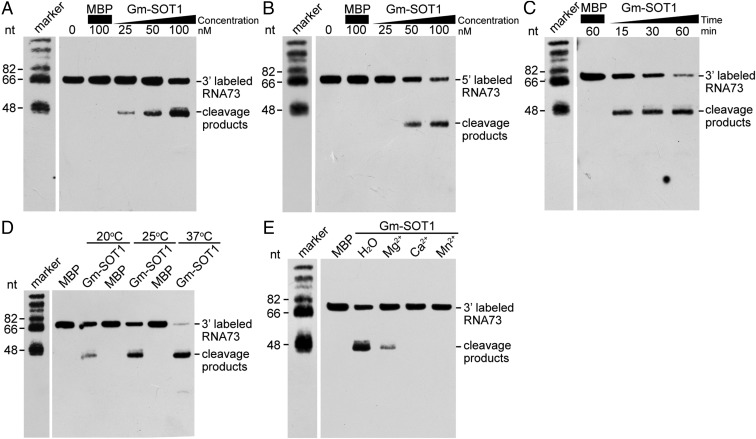
The SMR domain of SOT1 has nuclease activity. (A) Purified MBP, SOT1SMR, and Gm-SOT1SMR used for nuclease activity were resolved by SDS/PAGE with Coomassie Brilliant Blue (CBB) staining or with the analyses of immunoblot using the MBP antibody. The marker sizes are shown to the Left. (B) DNA endonuclease activities of the SMR domains of SOT1 and Gm-SOT1SMR. MBP, SOT1SMR, and Gm-SOT1SMR at different concentrations were incubated with 5 ng pUC19 plasmid DNA at 25 °C for 60 min in the presence of 3 mM MgCl2. The reactions were stopped by loading buffer and were electrophoresed on 1.2% (wt/vol) agarose gels. Parallel experiments were carried out with EcoRI to linearize pUC19 or with H2O as a control. OC, open circular; CCC, covalently closed-circular forms of pUC19. (C) Effects of Mg2+, Ca2+, and Mn2+ on DNA nicking activities of the SMR domains of SOT1 and Gm-SOT1SMR. A total of 5 ng pUC19 was incubated with H2O, MgCl2, CaCl2, or MnCl2 and 100 nM MBP, SOT1SMR, and Gm-SOT1SMR. The concentration of each cation was 3 mM. (D) Analyses of DNA nicking activity of Gm-SOT1. MBP and Gm-SOT1 were incubated with 5 ng pUC19 plasmid DNA at 25 °C for 60 min in the presence of 3 mM MgCl2. (E) RNA nuclease activities of the SMR domains of SOT1 and Gm-SOT1SMR. MBP, SOT1SMR, and Gm-SOT1SMR with different concentrations were incubated with total wild-type Arabidopsis RNA at 25 °C for 30 min. The reaction products were separated in agarose/formaldehyde gels and observed by ethidium bromide staining. (F) Effects of Mg2+, Ca2+, and Mn2+ on RNA nuclease activities of the SMR domains of SOT1 and Gm-SOT1SMR. A total of 100 nM MBP, SOT1SMR, and Gm-SOT1SMR were incubated with total wild-type Arabidopsis RNA in the presence of MgCl2, CaCl2, or MnCl2. The concentration of each cation was 3 mM. (G) Accumulation of total RNAs in WT, sot1-3/35S:SOT1SMR-HA, and sot1-3 plants. A total of 3 μg total RNAs from 12-d-old WT, sot1-3/35S:SOT1SMR-HA, and sot1-3 seedlings were separated in agarose/formaldehyde gels and observed by ethidium bromide staining. (H) Analyses of RNA nuclease activity of Gm-SOT1. MBP and Gm-SOT1 with different concentrations were incubated with total wild-type Arabidopsis RNA at 25 °C for 30 min. The reaction products were detected by electrophoretic separation in agarose/formaldehyde gels with ethidium bromide staining.
Given the fact that SOT1 is able to bind the 5′ region of the 23S–4.5S rRNA precursor (34), we then investigated whether the SMR domain of SOT1 has RNA nuclease activity. SOT1SMR efficiently degraded total rRNA isolated from wild-type Arabidopsis, suggesting that the SMR domain of SOT1 has RNA nuclease activity in vitro (Fig. 2E). However, the metal ions Mn2+, Ca2+, and Mg2+ inhibited the RNA nuclease activity of SOT1SMR under these conditions (Fig. 2F). To investigate the potential nuclease activity of the SMR domain in vivo, we complemented sot1-3 with a chloroplast transit peptide-SMR domain of SOT1 fusion protein (sot1-3/35S:SOT1SMR-HA) (SI Appendix, Fig. S8). This overexpression of the SMR domain in sot1-3 caused a lethal phenotype at the early development of seedlings (Fig. 1A). Smeared rRNA bands on the RNA gel (Fig. 2G) indicated that the SMR domain of SOT1 has in vivo nuclease activity. Taken together, these results establish that the SMR domain of SOT1 possesses both DNA and RNA nuclease activities.
SOT1 Has RNA Endonuclease Activity and Cleaves the 5′ Region of the 23S–4.5S rRNA Precursor.
SOT1 specifically binds the 5′ region of 23S–4.5S rRNA precursor with a 73-nt segment (denoted RNA73 hereafter) (34–36); thus, we reasoned that RNA73 should be an in vivo substrate for SOT1. To start, we aimed to investigate whether SOT1 could specifically cleave RNA73 using the recombinant SOT1 in vitro. Unfortunately, it was quite difficult to express intact SOT1 in Escherichia coli. Therefore, we tried to express the homologous proteins of SOT1 from other species such as Arabidopsis lyrata, Glycine max, Zea mays, and Oryza sativa. After numerous unsuccessful attempts, we ultimately obtained Gm-SOT1 from G. max (SI Appendix, Fig. S9). The sequence identity of SOT1 and Gm-SOT1 is ∼70%, and they share key residues for RNA recognition (residues 5 and 35 in each PPR motif, marked as red frames in SI Appendix, Fig. S10A). The 5′ region of 23S–4.5S rRNA precursor is highly conserved between A. thaliana and G. max (SI Appendix, Fig. S10B). In addition, similar to the SMR domain of SOT1, the SMR domain of Gm-SOT1 had both DNA and RNA nuclease activities (Fig. 2 A, C, E, and F). Thus, we used the recombinant protein Gm-SOT1 as an appropriate substitute for SOT1.
We used 3′-end and 5′-end biotin-labeled RNA73 to examine the catalytic activity of Gm-SOT1. With increasing Gm-SOT1 concentration, more cleavage products could be detected (Fig. 3 A and B). These results suggest that Gm-SOT1 can efficiently cleave the 5′ end of 23S–4.5S rRNA precursor and that the cleavage products are released due to the RNA endonuclease activity of Gm-SOT1. In addition, increasing amounts of the cleavage products were detected with increasing incubation time (Fig. 3C). Compared with 20 °C incubation, incubation at higher temperatures (25 °C and 37 °C) led to the production of more obvious cleavage products (Fig. 3D). The metal ions Mn2+, Ca2+, and Mg2+ inhibited the RNA nuclease activity of Gm-SOT1 (Fig. 3E). The cleavage products of RNA73 arose specifically due to the activity of Gm-SOT1 and not a contaminating ribonuclease, as the incubation of RNA73 with MBP protein that was purified from E. coli in parallel did not show any RNA cleavage. Together, these results demonstrate that Gm-SOT1 has RNA endonuclease activity.
Fig. 3.
Gm-SOT1 has RNA endonuclease activity. (A and B) Analyses of RNA endonuclease activity using Gm-SOT1. A total of 10 nM 3′-end biotin-labeled RNA73 (A) and 5′-end biotin-labeled RNA73 (B) were incubated with different concentrations of Gm-SOT1 at 25 °C for 30 min, followed by separation of RNA on 10% (wt/vol) denaturing polyacrylamide gels. φX174 DNA/Hinf I dephosphorylated markers (Promega) were biotinylated, and the marker sizes are shown in the Left margins. (C) Effects of different reaction times on Gm-SOT1 RNA endonuclease activity. The reactions were performed with 100 nM Gm-SOT1 at 25 °C for 15, 30, or 60 min. (D) Effects of different temperatures on Gm-SOT1 RNA endonuclease activity. The reactions were performed with 100 nM Gm-SOT1 for 30 min at 20 °C, 25 °C, or 37 °C. (E) Effects of different metal ions on Gm-SOT1 RNA endonuclease activity. The reactions were performed using 100 nM Gm-SOT1 and 3 mM solutions of various metal ions at 25 °C for 30 min.
Our above results show that the SMR domain of SOT1 possesses DNA and RNA nuclease activities, cleaving pUC19 plasmid and Arabidopsis total RNA, respectively (Fig. 2 B and E). Thus, we investigated whether the full-length Gm-SOT1 can cleave pUC19 plasmid and Arabidopsis total RNA. The full-length Gm-SOT1 did not cleave pUC19 plasmid and total rRNA efficiently (Fig. 2 D and H).
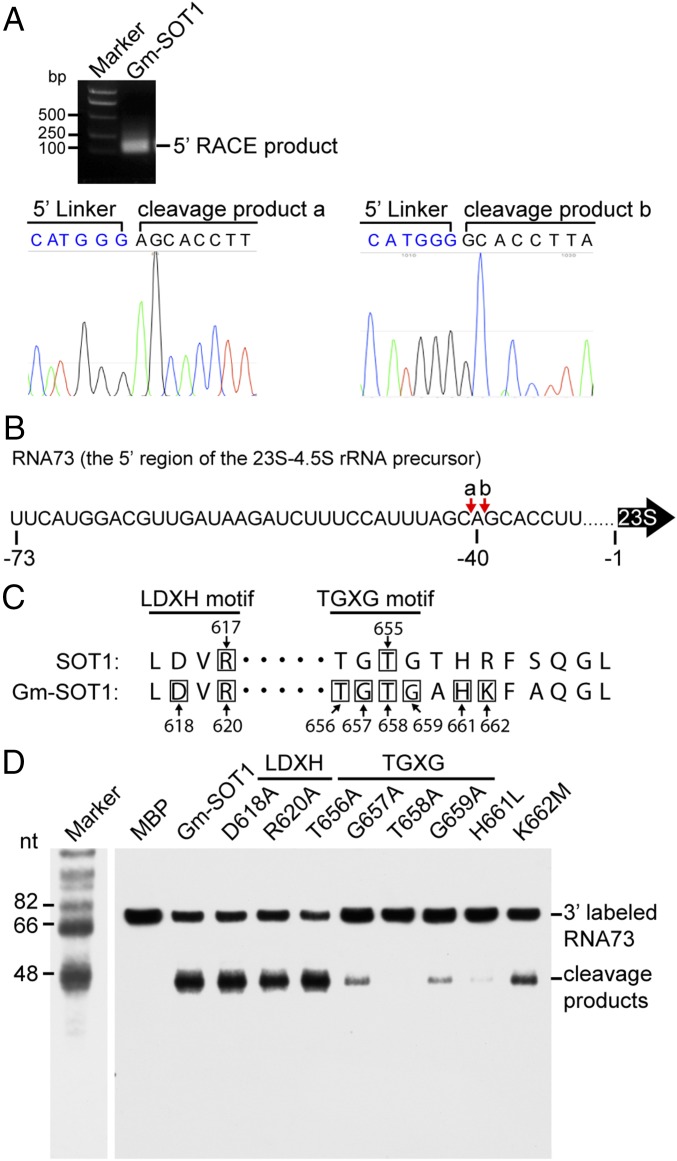
We observed clear cleavage products of RNA73 produced by Gm-SOT1 (Fig. 3). To identify the cleavage sites of SOT1, the cleavage products were recovered and analyzed by 5′-rapid amplification of cDNA ends (5′ RACE), followed by sequencing (Fig. 4A). Most of the clones derived from 5′-RACE revealed two cleavage sites by Gm-SOT1 that were separated by only one base. These two cleavage sites were located at around −40 relative to the 5′ end of mature 23S (Fig. 4B).
Fig. 4.
Cleavage sites in RNA73 by Gm-SOT1 and critical amino acids in the SMR domain of SOT1 for RNA endonuclease activity. (A) Mapping of the sites in RNA73 cleaved by Gm-SOT1. The cleavage products in RNA73 produced by Gm-SOT1 were recovered from the SDS/PAGE gel and reverse transcribed to cDNA by 5′ RACE, followed by sequencing. The sequencing chromatograms were constructed using four peaks (nucleobytes.com/4peaks/). (B) Schematic illustration of the cleavage sites in RNA73. The cleavage sites are indicated by red arrows and shown above the sequence of RNA73. The numbers below the sequence indicate the positions from the 5′ end of mature 23S rRNA. (C) Alignment of conserved LDXH and TGXG motifs of the SMR domain of SOT1 and Gm-SOT1. The numbers indicate the amino acid positions with respect to the translation initiation site. Black boxes indicate the amino acids in site-directed mutagenesis. (D) The TGXG motif is critical for the RNA endonuclease activity of Gm-SOT1. Site-directed mutagenesis was performed to investigate the effects of amino acids in the LDXH motif and TGXG motifs on the RNA endonuclease activity of Gm-SOT1. A total of 10 nM 3′-end biotin-labeled RNA73 was incubated with 100 nM MBP, Gm-SOT1, and Gm-SOT1 variants at 25 °C for 30 min, followed by separation of RNA on 10% (wt/vol) denaturing polyacrylamide gels. Marker sizes are shown at Left.
Critical Amino Acids of the SMR Domain of SOT1 for RNA Cleavage.
The SMR domain in eukaryotes contains two conserved motifs, LDXH and TGXG (27). Because it has been proposed that amino acids D and R in the LDVR motif, and the TGXG motif might function in nucleic acid binding or nuclease activity (19), we investigated the possible roles of these amino acids in RNA endonuclease activity. These two conserved motifs of the SMR domains in SOT1 and Gm-SOT1 correspond to residues LDVR and TGTG, respectively (Fig. 4C). We used site-directed mutagenesis to test the effects of these amino acids on RNA endonuclease activity in Gm-SOT1 (SI Appendix, Fig. S11, Left). The G657A, T658A, and G659A substitutions resulted in dramatic decreases in the catalytic activity of Gm-SOT1, but D618A, R620A, and T656A substitutions had no effects on the catalytic activity of Gm-SOT1 (Fig. 4D). It is most likely that G657A and G659A substitutions affect the SMR structure, thereby influencing the catalytic pocket. According to the “catalytic triad model” (37), T658 should function as the nucleophile in this region and H661 and K662 nearby the TGTG motif may function as two other catalytic sites. We thus investigated the roles of H661 and K662 in endonuclease activity. A H661L mutation almost completely blocked catalytic activity and K662M led to moderate inhibition of catalytic activity (Fig. 4D). Based on these results, we conclude that T658, H661, and K662 as critical amino acids of the SMR domain may form a catalytic triad and play a critical role in RNA endonuclease activity of Gm-SOT1.
Based on our above results, Gm-SOT1 T658 is important in endonuclease activity but Gm-SOT1 R620 apparently has no effect on endonuclease activity. We selected these two amino acids for analysis of their potential roles in the maturation of 23S and 4.5S rRNA using genetic approaches. SOT1 T655 (corresponding to Gm-SOT1 T658) and SOT1 R617 (corresponding to Gm-SOT1 R620) were mutated to SOT1 T655A and SOT1 R617A, respectively, and tested for their ability to complement the sot1-3 mutant. The sot1-3/35S:SOT1T655A-HA plants had virescent leaves and considerably decreased mature 23S and 4.5S rRNA similar to sot1-3, whereas the phenotypes were rescued in sot1-3/35S:SOT1R617A-HA plants in terms of the leaf phenotype and the maturation of 23S and 4.5S rRNA (Fig. 1). These results suggest that the critical amino acids identified above play an important role in the maturation of 23S and 4.5S rRNA.
The above results suggest that the TGXG motif of the SMR domain is critical for RNA cleavage. However, our results showed that for the SMR domain, the DNA nicking activity was mainly dependent on Mg2+, Mn2+, but not Ca2+, and RNA cleavage was inhibited by all three divalent ions (Fig. 2 C and F). These results indicate that DNA nicking might catalyze at a different active site as RNA cleavage. To investigate this possibility, we mutated the critical amino acid T of the TGXG motif in SOT1SMR and Gm-SOT1SMR and obtained two variants of the SMR domain, SOT1SMR T655A and Gm-SOT1SMR T658A (SI Appendix, Fig. S12 A and B). SOT1SMR T655A and Gm-SOT1SMR T658A had DNA nicking activity but not RNA cleavage activity (SI Appendix, Fig. S12 C and D), suggesting that the TGXG motif is not critical for DNA nicking. We further mutated the two conserved amino acids D and R in the LDXH motif to examine whether the LDXH motif is involved in DNA nicking. The D615A and R617A substitutions in SOT1SMR and the D618A and R620A substitutions in Gm-SOT1SMR showed a considerable decrease in DNA nicking activity but had no effects on RNA cleavage activity (SI Appendix, Fig. S12 C and D). Taken together, these results indeed indicate that for the SMR domain, DNA nicking catalyzes at a different active site as RNA cleavage.
Programmable RNA Recognition and Cleavage by Gm-SOT1.
Site-specific recognition and cleavage of RNA is crucial for in vitro RNA manipulation and in vivo gene silencing (1). Therefore, we investigated whether SOT1 can be engineered to specifically recognize and cleave new substrates. To engineer SOT1 successfully, it is crucial to identify the minimal binding sequence by SOT1. A 20-nt/24-nt segment in the 5′ region of the 23S–4.5S rRNA precursor was identified as the binding sequence by SOT1/PPR53 (33, 34) (illustrated in SI Appendix, Fig. S13A). Our results showed that the 13-nt sequence 5′-AUGGACGUUGAUA-3′ (RNA13) spanning −70 to −58 (relative to the 5′ end) of mature 23S rRNA contributed to the minimal binding sequence of SOT1 (SI Appendix, Fig. S13).
According to the “PPR code,” a simple combination of amino acids at position 5 and 35 in a PPR motif determines its nucleotide recognition (9–11) (Fig. 5A). Accordingly, we mutated the fifth amino acids in PPR motifs 1–4 in Gm-SOT1. Thus, the fifth amino acid N, S, S, and S at the PPR motif 1, 2, 3, and 4 were mutated as T, N, N, and N, respectively. This mutated Gm-SOT1 was termed as “Gm-SOT1m” (SI Appendix, Fig. S11, Right). According to the PPR code, the predicted RNA targets in the PPR motifs 1–4 in Gm-SOT1 were U, G, G, and A, respectively and in Gm-SOT1m, they were G, U, U, and C, respectively (Fig. 5 B and C).
Fig. 5.
Programmable RNA recognition and cleavage by SOT1. (A) The PPR code (9–11). (B) Alignment between Gm-SOT1 and its RNA targets. Individual boxes indicate PPR motifs. The amino acids at positions 5 and 35 of each PPR motif are shown in the box. RNA13 is the minimal binding sequence of SOT1. RNA73 is the sequence of the 5′ region of the 23S–4.5S precursor, including the binding and cleavage sites of SOT1. (C) Alignment between the mutated Gm-SOT1m and its mutated RNA targets. Gm-SOT1m is a mutated Gm-SOT1 protein in which the fifth amino acids in PPR motifs 1–4 (red boxes) were mutated. Mutated ribonucleotides in RNA13m and RNA73m are indicated in red. (D) Programmable RNA recognition by Gm-SOT1. EMSA shows the binding of Gm-SOT1 and the variant Gm-SOT1m to Cy5-labeled RNA13 and Cy3-labeled RNA13m. Increasing concentrations of protein (100, 200, and 400 nM) were incubated with 40 nM probes. (E) Programmable RNA cleavage by Gm-SOT1. RNA cleavage analysis shows the cleavage of FAM-labeled RNA73 and Cy3-labeled RNA73m by Gm-SOT1 and the variant Gm-SOT1m, respectively. Increasing concentrations of protein (100, 200, and 400 nM) were incubated with 40-nM probes.
We determined whether Gm-SOT1m could recognize its predicted targets. Because RNA13 was the minimal binding sequence of SOT1 (SI Appendix, Fig. S13), we used this sequence as the substrate for binding analysis. The RNA13 was modified into RNA13m (Fig. 5 B and C). We used a system with multiple fluorescent probes in a single assay (15). RNA13 and RNA13m were 5′ labeled with Cy5 and Cy3, respectively, to produce Cy5 RNA13 and Cy3 RNA13m. As expected, the wild-type Gm-SOT1 recognized Cy5 RNA13 but not Cy3; the mutated form Gm-SOT1m recognized Cy3 RNA13m but not Cy5 RNA13 (Fig. 5D).
The RNA13 used in this study likely has little residual structure. Thus, programmable RNA binding by Gm-SOT1 may be interfered by RNA structure that can render the intended binding site less accessible. To address this issue, we expanded the EMSA assay by varying the structural content of RNA13 by adding short, unlabeled oligos (oligo1–5) that progressively base pair with the RNA13 (SI Appendix, Fig. S14A). Our results showed that Gm-SOT1 was still capable of binding the RNA13 in the presence of oligo1–5, respectively (SI Appendix, Fig. S14B).
Then, we determined whether Gm-SOT1m could cleave its predicted targets. Because RNA73 contained the binding and cleavage sites for Gm-SOT1, RNA73 was used as the substrate for cleavage analysis. The sequence modification in RNA73 was the same as that in RNA13, and the modified sequence of RNA73 was designated RNA73m (Fig. 5 B and C). RNA73 and RNA73m were 5′ labeled with FAM and Cy3, respectively, producing FAM RNA73 and Cy3 RNA73m. Importantly, Gm-SOT1 cleaved FAM RNA73 but not Cy3 RNA73m, and Gm-SOT1m cleaved Cy3 RNA73m but not FAM RNA73 (Fig. 5E).
To further confirm Gm-SOT1 could be engineered to recognize and cleave RNA targets in a predictable manner, we generated three additional Gm-SOT1 variants in which the fifth amino acids were mutated in PPR motifs either 5–6, or 5–7, or 1–6. The cognate RNA target for each Gm-SOT1 variant was predicted according to the PPR code (SI Appendix, Fig. S15 A–C). As expected, the Gm-SOT1 variants solely recognized and cleaved their cognate RNA targets (SI Appendix, Fig. S15 D and E). Together, these results demonstrate that both RNA recognition and cleavage by SOT1 can be altered in a predictable manner.
Discussion
Because sequence-specific RNA endonucleases would be potentially powerful tools for RNA manipulation (1, 27), many efforts have been invested to identify a natural RNA endonuclease that recognizes RNA in an intrinsic sequence-specific manner. Here, we report that SOT1 has sequence-specific RNA endonuclease activity. Even more importantly, we successfully engineered SOT1 protein with an altered PPR motif to recognize and cleave a predicted RNA substrate, suggesting that SOT1 could be used for RNA manipulation in the future.
The PPR-SMR SOT1 Protein Has RNA Endonuclease Activity.
PPR-SMR proteins represent a small subset of the large PPR protein family in higher plants (27). Despite being few in number, they play critical roles in chloroplast biogenesis and retrograde signaling (28–34). The PPR domain can specifically recognize RNAs in an intrinsic sequence-specific manner (5). Previous studies of SMR-containing proteins in other organisms have provided evidence for endonuclease activity of the SMR domain (20–23, 26). However, the function of the enigmatic SMR domain in PPR-SMR proteins has not yet been comprehensively investigated.
In this study, we demonstrated that the SMR domain of SOT1 has metal-dependent DNA endonuclease activity (Fig. 2 B and C), which is consistent with the DNA endonuclease activity observed in other SMR-containing proteins (20–26). In addition, our results showed that the SMR domain of SOT1 has RNA endonuclease activity (Fig. 2E). In this regard, SOT1 is similar to the conserved endoribonuclease YbeY, which is able to cleave Arabidopsis total rRNA (38). The expression of the SMR domain of SOT1 in sot1-3 plants resulted in a lethal phenotype (Fig. 1A), similar to the PilT N-terminal (PIN) ribonuclease domain (39). Overall, our results clearly indicate that the SMR domain of SOT1 has both DNA and RNA endonuclease activities.
Given that the SMR domain has these activities, does the full-length SOT1 exhibit DNA and RNA endonuclease activities? Our results show that the full-length Gm-SOT1 could not cleave the plasmid DNA (Fig. 2D), indicating that the full-length SOT1 likely has no DNA endonuclease activity. Similarly, the full-length Gm-SOT1 could not cleave Arabidopsis total rRNA (Fig. 2H). These results suggest that the endonuclease activity of the full-length SOT1 may be dependent on the specific RNA sequence. Previous study has shown that SOT1 specifically binds a 73-nt segment (RNA73) of the 5′ region of 23S–4.5S rRNA precursor (34). Our results showed that the full-length Gm-SOT1 could cleave RNA73 at two sites that were located at around −40 relative to the 5′ end of mature 23S (Fig. 4 A and B). Thus, the full-length SOT1 has sequence-specific RNA endonuclease activity.
It would be invaluable to identify catalytic residues in the SMR domain of SOT1 for its RNA endonuclease activity (27). It has been suggested that two motifs, LDXH and TGXG, are the potential catalytic residues of the SMR domain (19, 22, 27). Our site-directed mutagenesis and genetic complementation analyses indicated that the motif TGXG, but not the motif LDXH, is critical for RNA endonuclease activity of SOT1 (Figs. 1 and 4 C and D).
How is SOT1 involved in the maturation of 23S and 4.5S rRNA precursor? We detected effective cleavage at the 5′ end of the 23S–4.5S rRNA by SOT1 in vitro (Fig. 3). Further analyses of the SOT1 cleavage products demonstrated that the SOT1 cleavage site is located at approximately −40 relative to the 5′ end of mature 23S (Fig. 4 A and B). The miniribonuclease III cleavage site is thought to be located in an RNA duplex of ∼20 bp formed by complementary sequences in the 5′ proximal region of 23S and the 3′ proximal region of 4.5S. Miniribonuclease III a_ppears to cleave the 23S–4.5S precursor to simultaneously produce the mature 5′ end of 23S and the 3′ end of 4.5S (35). The SOT1 cleavage site is located ∼40 nucleotides upstream of the miniribonuclease III cleavage site. The comparison of the 5′ end of 23S rRNA and 3′ end of 4.5S rRNA in sot1-3 and the miniribonuclease III mutant rnc3/4 show that there was the disruption of the miniribonuclease III processing in sot1-3 (SI Appendix, Fig. S7). Based on our results, we propose that the endonucleolytic cleavage performed by SOT1 is required for the processing by miniribonuclease III during maturation of 23S and 4.5S rRNA.
In bacteria, it is thought that the final maturation of 23S rRNA is completed by miniribonuclease III through a one-step cleavage (40, 41). However, in chloroplasts, one or two small RNAs adjacently to the upstream of the miniribonuclease III cleavage site were identified (34, 35), indicating that additional, unknown processing events exist before miniribonuclease III cleavage in the maturation of 23S rRNA in chloroplasts. Ribonucleases that perform these additional unknown processing events remain elusive. Our results showed that SOT1 performed an endonucleolytic cleavage adjacently to the upstream of the miniribonuclease III cleavage during maturation of 23S rRNA in chloroplasts. Such a finding provides us deep understanding of the molecular mechanism for maturation of 23S rRNA in chloroplasts and suggests that new ribonucleases have emerged as being required for the 23S rRNA processing during evolution of chloroplasts. Our finding also expands the understanding of the biological functions of PPR proteins, because it is generally believed that PPR proteins stabilize or remodel their RNA targets (5). However, the exact molecular mechanism of SOT1 in maturation of 23S rRNA in chloroplasts remains to be investigated further.
SOT1 Could Be a Powerful Tool for RNA Manipulation.
DNA restriction enzymes were first described ∼40 y ago. However, no analogous RNA endonuclease that recognizes RNA in a sequence-specific manner was forthcoming. Despite their affinity for many types of RNA, RNA endonucleases exhibit limited, imprecise recognition of RNA sequences, which limits their value for RNA manipulation. Until now, many RNA binding proteins, such as RNA-recognition motif (RRM), K homology (KH), zinc finger (ZF), and Pumilio/FBF homology protein (PUF), were found to have modular structures to recognize RNA sequences and/or structures (42). Among these RNA binding proteins, PUF proteins may represent a candidate for engineering highly sequence-specific RNA binding (42). The classical PUF proteins contain eight repeat domains and each repeat can recognize a single RNA nucleotide (43). The RNA specificity of the PUF domain has been well decoded (44–46). Based on the properties of the PUF proteins, the programmable RNA binding by the engineered PUF protein has been realized (47–50). Moreover, the PUF domains fusing with a general RNA cleavage domain have been generated to the artificial site-specific RNA endonucleases that specifically recognize and cleave RNA targets (51, 52).
Similar to PUF proteins, several PPR proteins have been successfully modified to recognize corresponding RNA targets through simply modifying the critical amino acids of each PPR motif (9, 12–17). Considering the PPR motifs have been designed to target a specific transcript, if RNA endonuclease activity can be confirmed for the SMR domain of PPR-SMR proteins, the PPR-SMR protein will enable “engineering” of sequence-specific RNA endonucleases, which will have exciting applications for RNA manipulation (27). Our current results demonstrated that the PPR-SMR protein SOT1 has RNA endonuclease activity and can be engineered to recognize and cleave RNA with customizable sequence specificity. We altered the critical amino acids for RNA recognition in the SOT1 PPR motifs, finding that the modified protein could recognize and cleave a new RNA substrate in the expected manner (Fig. 5 and SI Appendix, Fig. S15).
Recently, the CRISPR/Cas9 system has been widely used in the field of genome editing (53, 54). This system can also be used for programmable RNA recognition and cleavage (55). In addition, it was found recently that C2c2 is a programmable RNA-guided RNA-targeting CRISPR effector (56). These findings suggest that the CRISPR/Cas system can be used to develop RNA-targeting tools (55–57). Compared with the CRISPR/Cas system that uses RNA-mediated base pairing to recognize RNA targets, SOT1 relies upon PPR–RNA interactions to recognize RNA targets. The results in this study reveal that SOT1 can achieve programmable RNA recognition and cleavage, suggesting that SOT1, similar to the CRISPR/Cas system, can be used as a tool for RNA manipulation. Currently, chloroplast and mitochondrial transformation is technically very difficult, labor intensive, and time consuming and achieved only in very few organisms (58, 59). It is unclear whether CRISPR/Cas9 could cleave or edit the mitochondrial genome because it is challenging to import the guide RNA component into mitochondria (60). Thus, developing chloroplast and mitochondrial RNA-targeting tools will be significant for chloroplast and mitochondrial biology. SOT1 is a chloroplast-localized protein and could be targeted into mitochondria by fusing a mitochondrial signaling peptide. Therefore, an advantage for SOT1 is that it can be used for programmable chloroplast/mitochondrial genes-encoded RNA recognition and cleavage, which enables SOT1 to be a potential RNA-targeting tool in chloroplasts and mitochondria. Just as DNA restriction enzymes have revolutionized the field of molecular biology, we look forward to the potential application of SOT1 to the broad field of RNA manipulation, especially in chloroplasts and mitochondria. Nevertheless, further elucidation of the detailed catalytic mechanism used by SOT1 is essential to make use of this exciting tool.
Materials and Methods
Plant Materials and Growth Conditions.
The A. thaliana wild-type line (ecotype Columbia-0) was obtained from the Arabidopsis Biological Resource Center (abrc.osu.edu/). The mutant sot1-3 line was isolated from a pSKI15 T-DNA-mutagenized A. thaliana library (ecotype Columbia-0) based on its high chlorophyll fluorescence phenotype. The sot1-3 plants were backcrossed three times to wild-type plants. The mutant rnc3/4 line was kindly provided by D. B. Stern, Boyce Thompson Institute for Plant Research, Ithaca, NY.
The Arabidopsis seeds were surface sterilized and plated on Murashige and Skoog (MS) medium containing 2% (wt/vol) sucrose and 0.8% agar. After incubation in darkness for 2 d at 4 °C, the plants were grown under 100 μmol m−2⋅s−1 14-h light/10-h-dark cycles at 23 °C and 50% relative humidity.
Expression of Recombinant Proteins.
The pASK-IBA44 expression system (IBA GmbH) was used to express recombinant proteins in E. coli. The gene encoding the MBP tag was inserted into pASK-IBA44 to facilitate the protein purification. The coding sequences for the SOT1 SMR domain (amino acids 603–710), the Gm-SOT1 SMR domain (amino acids 606–711), and Gm-SOT1 (amino acids 182–706) were subcloned into the pASK-IBA44 vector. Expression of MBP, SOT1SMR, and Gm-SOT1 was induced upon addition of 200 μg anhydrotetracycline per liter E. coli JM83 in a shaking culture (A550 = 0.4). After induction at 17 °C for 3 h, E. coli cultures were harvested and homogenized in buffer containing 50 mM phosphate, pH 7.5, 400 mM NaCl, 100 mM KCl, and 10% (vol/vol) glycerol. After sonication and centrifugation, the supernatant was applied to Ni-NTI (Qiagen) and subsequently loaded onto an amylose resin column (NEB). The purified proteins were further fractionated by size exclusion chromatography (GE Superdex 200 10/300 GL).
To generate the constructs of Gm-SOT1 variants, SOT1SMR variants, and Gm-SOT1SMR variants, site-directed mutagenesis and multisite-directed mutagenesis were performed using a Fast Mutagenesis System (Transgen FM111) and a Fast MultiSite Mutagenesis System (Transgen FM201), respectively, according to the manufacturer’s instructions (for primers, see SI Appendix, Table S1). Expression and purification of the Gm-SOT1 variants, SOT1SMR variants and Gm-SOT1SMR variants were performed as described for Gm-SOT1.
Nuclease Activity Analysis of the SMR Domain of SOT1.
DNA nicking nuclease activity analysis was performed as described previously (21). The pUC19 (Takara) plasmid DNA (5 ng μL−1) was incubated with MBP, SOT1SMR, SOT1SMR variants, Gm-SOT1SMR, Gm-SOT1SMR variants, or Gm-SOT1 in buffer [20 mM phosphate, pH 7.5, 160 mM NaCl, 40 mM KCl, and 4% (vol/vol) glycerol] at 25 °C for 60 min. The reaction products for pUC19 plasmid DNA were detected by electrophoretic separation in 1% agarose gels with ethidium bromide.
RNA nuclease activity was performed as described previously (38). The total RNA (3 μg) extracted from wild-type plants was incubated with MBP, SOT1SMR, SOT1SMR variants, Gm-SOT1SMR, Gm-SOT1SMR variants, or Gm-SOT1 in buffer [20 mM phosphate, pH 7.5, 160 mM NaCl, 40 mM KCl, and 4% (vol/vol) glycerol] at 25 °C for 30 min. The reaction products for total RNAs were separated in 1.2% (wt/vol) agarose/formaldehyde gels and visualized by ethidium bromide staining.
RNA Endonuclease Activity Analysis of SOT1.
The 5′-end biotin-labeled RNA73, 3′-end biotin-labeled RNA73, 5′-end FAM-labeled RNA73, 5′-end Cy3-labeled RNA73m, 3′-end biotin-labeled RNA73PPR5-6, 3′-end biotin-labeled RNA73PPR5-7, and 3′-end biotin-labeled RNA73PPR1-6 were synthesized and labeled by Takara. The reaction buffer contained 20 mM phosphate, pH 7.5, 160 mM NaCl, 40 mM KCl, and 4% (vol/vol) glycerol. The reaction products for the 5′-and 3′-end biotin-labeled RNA73, 3′-end biotin-labeled RNA73PPR5-6, 3′-end biotin-labeled RNA73PPR5-7, and 3′-end biotin-labeled RNA73PPR1-6 were separated in 10% (wt/vol) polyacrylamide gels, transferred into nylon membranes, and subsequently detected using a chemiluminescent detection kit (Thermo, 89880). The reaction products for the 5′-end FAM-labeled RNA73, and 5′-end Cy3-labeled RNA73m were resolved on 10% (wt/vol) polyacrylamide gels and detected using a Typhoon Trio imager (GE Healthcare).
Electrophoretic Mobility Shift Assays.
Electrophoretic mobility shift assays (EMSA) were carried out with a LightShift Chemiluminescent RNA EMSA Kit (Thermo 20158) following the manufacturer’s instructions (61). The 5′-end Cy3-labeled RNA14/RNA13/RNA12a/RNA12b, 5′-end Cy5-labeled RNA13, 5′-end Cy3-labeled RNA13m, 5′-end biotin-labeled RNA13PPR5-6, 5′-end biotin-labeled RNA13PPR5-7, 5′-end biotin-labeled RNA13PPR1-6, and oligo1–5 were synthesized and labeled by Takara. For the RNAs labeled with different fluorescent dyes (Cy5 and Cy3), the binding reaction mixture consisted of 10 mM Hepes (pH 7.3), 20 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% (vol/vol) glycerol, and 40 nM RNA. The protein sample was incubated with the binding reaction mixture at 20 °C for 30 min. The reaction products for the 5′-end Cy3-labeled RNA14/RNA13/RNA12a/RNA12b, 5′-end Cy5-labeled RNA13, and 5′-end Cy3-labeled RNA13m were resolved on 6% (wt/vol) native polyacrylamide gels and were detected using a Typhoon Trio imager (GE Healthcare). For the RNAs labeled with biotin, the binding reaction mixture consisted of 10 mM Hepes (pH 7.3), 20 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% (vol/vol) glycerol, and 10 nM RNA. The protein sample was incubated with the binding reaction mixture at 20 °C for 30 min. The reaction products for the 5′-end biotin-labeled RNA13, 5′-end biotin-labeled RNA13PPR5-6, 5′-end biotin-labeled RNA13PPR5-7, and 5′-end biotin-labeled RNA13PPR1-6 were resolved on 6% (wt/vol) native polyacrylamide gels and transferred into nylon membranes and were subsequently detected according to standard protocol of the chemiluminescent detection kit (Thermo, 89880).
Supplementary Material
Acknowledgments
We are grateful to Professor D. B. Stern for providing the mutant rnc3/4 line and to the Arabidopsis Biological Resource Center for the seed stocks. This work was supported by the Key Research Plan of Frontier Sciences of the Chinese Academy of Sciences (Grant QYZDJ-SSW-SMC003), the State Key Basic Research and Development Plan of China (Grant 2015CB150105), and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB17030100).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612460114/-/DCSupplemental.
References
- 1.Choudhury R, Wang Z. Manipulation of RNA using engineered proteins with customized specificity. Adv Exp Med Biol. 2014;825(6):199–225. doi: 10.1007/978-1-4939-1221-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Aubourg S, Boudet N, Kreis M, Lecharny A. In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol Biol. 2000;42(4):603–613. doi: 10.1023/a:1006352315928. [DOI] [PubMed] [Google Scholar]
- 3.Small ID, Peeters N. The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25(2):46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 4.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16(8):2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65(1):415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 6.Ke J, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20(12):1377–1382. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 7.Yin P, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504(7478):168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 8.Gully BS, et al. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 2015;43(3):1918–1926. doi: 10.1093/nar/gkv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkan A, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8(8):e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takenaka M, Zehrmann A, Brennicke A, Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8(6):e65343. doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8(3):e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coquille S, et al. An artificial PPR scaffold for programmable RNA recognition. Nat Commun. 2014;5:5729. doi: 10.1038/ncomms6729. [DOI] [PubMed] [Google Scholar]
- 13.Okuda K, et al. Quantitative analysis of motifs contributing to the interaction between PLS-subfamily members and their target RNA sequences in plastid RNA editing. Plant J. 2014;80(5):870–882. doi: 10.1111/tpj.12687. [DOI] [PubMed] [Google Scholar]
- 14.Gully BS, et al. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 2):196–208. doi: 10.1107/S1399004714024869. [DOI] [PubMed] [Google Scholar]
- 15.Kindgren P, Yap A, Bond CS, Small I. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell. 2015;27(2):403–416. doi: 10.1105/tpc.114.134189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen C, et al. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol Plant. 2015;8(4):667–670. doi: 10.1016/j.molp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Shen C, et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat Commun. 2016;7:11285. doi: 10.1038/ncomms11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira D, Philippe H. Smr: A bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem Sci. 1999;24(8):298–300. doi: 10.1016/s0968-0004(99)01419-x. [DOI] [PubMed] [Google Scholar]
- 19.Fukui K, Kuramitsu S. Structure and function of the small MutS-related domain. Mol Biol Int. 2011;2011:691735. doi: 10.4061/2011/691735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukui K, Kosaka H, Kuramitsu S, Masui R. Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain. Nucleic Acids Res. 2007;35(3):850–860. doi: 10.1093/nar/gkl735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukui K, et al. Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression. J Biol Chem. 2008;283(48):33417–33427. doi: 10.1074/jbc.M806755200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diercks T, et al. Solution structure and characterization of the DNA-binding activity of the B3BP-Smr domain. J Mol Biol. 2008;383(5):1156–1170. doi: 10.1016/j.jmb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Gui WJ, et al. Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli. J Struct Biol. 2011;174(2):282–289. doi: 10.1016/j.jsb.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, et al. Structural and functional studies of MutS2 from Deinococcus radiodurans. DNA Repair (Amst) 2014;21:111–119. doi: 10.1016/j.dnarep.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Damke PP, Dhanaraju R, Marsin S, Radicella JP, Rao DN. The nuclease activities of both the Smr domain and an additional LDLK motif are required for an efficient anti-recombination function of Helicobacter pylori MutS2. Mol Microbiol. 2015;96(6):1240–1256. doi: 10.1111/mmi.13003. [DOI] [PubMed] [Google Scholar]
- 26.Bhandari D, Guha K, Bhaduri N, Saha P. Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Lett. 2011;585(5):809–813. doi: 10.1016/j.febslet.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Melonek J, Boykin LM, Small I, Howell KA. PPR-SMRs: Ancient proteins with enigmatic functions. RNA Biol. 2013;10(9):1501–1510. doi: 10.4161/rna.26172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18(1):176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316(5825):715–719. [PubMed] [Google Scholar]
- 30.Liu X, Yu F, Rodermel S. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 2010;154(4):1588–1601. doi: 10.1104/pp.110.164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoschke R, et al. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72(4):547–558. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- 32.Zoschke R, Qu Y, Zubo YO, Börner T, Schmitz-Linneweber C. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J Plant Res. 2013;126(3):403–414. doi: 10.1007/s10265-012-0527-1. [DOI] [PubMed] [Google Scholar]
- 33.Zoschke R, Watkins KP, Miranda RG, Barkan A. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016;85(5):594–606. doi: 10.1111/tpj.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu W, et al. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016;85(5):607–621. doi: 10.1111/tpj.13126. [DOI] [PubMed] [Google Scholar]
- 35.Hotto AM, et al. Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell. 2015;27(3):724–740. doi: 10.1105/tpc.114.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollenbach TJ, et al. RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005;33(8):2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23(9):347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, et al. The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol. 2015;168(1):205–221. doi: 10.1104/pp.114.255000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arcus VL, McKenzie JL, Robson J, Cook GM. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng Des Sel. 2011;24(1–2):33–40. doi: 10.1093/protein/gzq081. [DOI] [PubMed] [Google Scholar]
- 40.Redko Y, Bechhofer DH, Condon C. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol Microbiol. 2008;68(5):1096–1106. doi: 10.1111/j.1365-2958.2008.06207.x. [DOI] [PubMed] [Google Scholar]
- 41.Redko Y, Condon C. Ribosomal protein L3 bound to 23S precursor rRNA stimulates its maturation by Mini-III ribonuclease. Mol Microbiol. 2009;71(5):1145–1154. doi: 10.1111/j.1365-2958.2008.06591.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013;280(16):3734–3754. doi: 10.1111/febs.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 44.Filipovska A, Razif MFM, Nygård KKA, Rackham O. A universal code for RNA recognition by PUF proteins. Nat Chem Biol. 2011;7(7):425–427. doi: 10.1038/nchembio.577. [DOI] [PubMed] [Google Scholar]
- 45.Campbell ZT, Valley CT, Wickens M. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat Struct Mol Biol. 2014;21(8):732–738. doi: 10.1038/nsmb.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall TMT. Expanding the RNA-recognition code of PUF proteins. Nat Struct Mol Biol. 2014;21(8):653–655. doi: 10.1038/nsmb.2863. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Cheong C-G, Hall TMT, Wang Z. Engineering splicing factors with designed specificities. Nat Methods. 2009;6(11):825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooke A, Prigge A, Opperman L, Wickens M. Targeted translational regulation using the PUF protein family scaffold. Proc Natl Acad Sci USA. 2011;108(38):15870–15875. doi: 10.1073/pnas.1105151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J, Arha M, Sudrik C, Schaffer DV, Kane RS. Bidirectional regulation of mRNA translation in mammalian cells by using PUF domains. Angew Chem Int Ed Engl. 2014;53(19):4900–4904. doi: 10.1002/anie.201402095. [DOI] [PubMed] [Google Scholar]
- 50.Adamala KP, Martin-Alarcon DA, Boyden ES. Programmable RNA-binding protein composed of repeats of a single modular unit. Proc Natl Acad Sci USA. 2016;113(19):E2579–E2588. doi: 10.1073/pnas.1519368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, et al. Treatment of type 1 myotonic dystrophy by engineering site-specific RNA endonucleases that target (CUG)(n) repeats. Mol Ther. 2014;22(2):312–320. doi: 10.1038/mt.2013.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 54.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164(1–2):29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 55.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.East-Seletsky A, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larosa V, Remacle C. Transformation of the mitochondrial genome. Int J Dev Biol. 2013;57(6–8):659–665. doi: 10.1387/ijdb.130230cr. [DOI] [PubMed] [Google Scholar]
- 59.Bock R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu Rev Plant Biol. 2015;66:211–241. doi: 10.1146/annurev-arplant-050213-040212. [DOI] [PubMed] [Google Scholar]
- 60.Patananan AN, Wu TH, Chiou PY, Teitell MA. Modifying the mitochondrial genome. Cell Metab. 2016;23(5):785–796. doi: 10.1016/j.cmet.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA. 2011;108(1):415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.