Significance
Cardiac myosin binding protein C (cMyBP-C) is an important regulator of myocardial contraction, but its mechanism of action is unclear. In this study, we examined the structure of thick filaments from the hearts of mice in which the three serine residues that are phosphorylated by protein kinase A in the m-domain of cMyBP-C were replaced by either alanine or aspartic acid to mimic either the nonphosphorylated or phosphorylated state of cMyBP-C. In contrast to earlier work on rat cardiac filaments, the results support a model in which nonphosphorylated cMyBP-C stabilizes the relaxed/superrelaxed ordered “off-state” conformation of the heads while phosphorylation weakens the binding of the heads to the thick filament surface, increasing the probability of interaction with actin.
Keywords: cMyBP-C, cardiac myosin binding protein C, cardiac thick filament, cardiac thick filament structure, cardiac myosin binding protein C function
Abstract
Cardiac myosin binding protein C (cMyBP-C) has a key regulatory role in cardiac contraction, but the mechanism by which changes in phosphorylation of cMyBP-C accelerate cross-bridge kinetics remains unknown. In this study, we isolated thick filaments from the hearts of mice in which the three serine residues (Ser273, Ser282, and Ser302) that are phosphorylated by protein kinase A in the m-domain of cMyBP-C were replaced by either alanine or aspartic acid, mimicking the fully nonphosphorylated and the fully phosphorylated state of cMyBP-C, respectively. We found that thick filaments from the cMyBP-C phospho-deficient hearts had highly ordered cross-bridge arrays, whereas the filaments from the cMyBP-C phospho-mimetic hearts showed a strong tendency toward disorder. Our results support the hypothesis that dephosphorylation of cMyBP-C promotes or stabilizes the relaxed/superrelaxed quasi-helical ordering of the myosin heads on the filament surface, whereas phosphorylation weakens this stabilization and binding of the heads to the backbone. Such structural changes would modulate the probability of myosin binding to actin and could help explain the acceleration of cross-bridge interactions with actin when cMyBP-C is phosphorylated because of, for example, activation of β1-adrenergic receptors in myocardium.
Myosin binding protein C is a major accessory protein associated with myosin in the vertebrate thick filament (1). The importance of the cardiac isoform of MyBP-C (cMyBP-C) in myocardial contraction is indicated by evidence that mutations in its encoding gene (MYBPC3) are a major cause of familial hypertrophic cardiomyopathy (2, 3). Further evidence from studies using biochemical extraction, genetic ablation, and phosphorylation of cMyBP-C has demonstrated that it is a major regulator of cross-bridge kinetics and myocardial contraction (4–12).
cMyBP-C is located at nine sites 42.9-nm apart along a portion of the cross-bridge region in each half of the cardiac thick filament (13): the C-zone (14, 15). The protein consists of 11 repeating fibronectin-like and Ig-like domains, labeled C0 to C10, from the N terminus to the C terminus (10, 16, 17), with binding sites for both myosin S2 and F-actin at its N terminus (18–23) and sites for binding to light meromyosin and titin at its C terminus (24, 25). Its regulatory role is mediated by an m-domain or m-motif that lies between the C1 and C2 domains. This region has four serines that are phosphorylated in response to β-adrenergic agonist stimulation of living myocardium (10, 19, 26–31). Three of these sites are principally phosphorylated by protein kinase A (PKA) (10), whereas the fourth is principally phosphorylated by Ca2+-calmodulin–dependent kinase (CAMK) (10, 32). When nonphosphorylated, binding of this region and associated N-terminal domains to both the S2-region of myosin (10, 19, 21, 22, 33, 34) and F-actin (35–45) have been demonstrated. Phosphorylation of this region, in contrast, abolishes binding to myosin S2 and weakens the binding to F-actin (10, 20, 38, 46, 47).
These data are consistent with a mechanism in which cMyBP-C in the nonphosphorylated state normally depresses cross-bridge kinetics and its interaction with the thin filament, whereas phosphorylation of cMyBP-C accelerates cross-bridge kinetics and increases the interactions of myosin with the thin filament (reviewed in ref. 48). However, the precise molecular mechanism by which this occurs remains an enigma. Because cMyBP-C has binding sites for both myosin S2 and F-actin, regulation of contraction by cMyBP-C could involve its binding to myosin S2, F-actin, or both.
Although much of the recent work on cMyBP-C has focused on its ability to bind to F-actin (35–45) as a possible regulatory mechanism, reversible phosphorylation-dependent binding of the N-terminal region of cMyBP-C to the S2-region of myosin (10, 20–23, 33, 34, 49) or to other regions of myosin (50, 51) also remains a strong possibility as a mechanism by which cMyBP-C could regulate contraction.
Consistent with such a mechanism, several studies have implicated phosphorylation-regulated binding of the N-terminal region of cMyBP-C to the S2 region of myosin, with correlated changes in thick filament structure, and its binding to the thin filament, as either an additional or alternative mechanism for regulation of cross-bridge kinetics and contraction by cMyBP-C (10, 19–22, 52–54). Support for regulation involving binding of cMyBP-C to the S2-region of myosin is provided by studies showing that mutations in myosin S2 which produce familial hypertrophic cardiomyopathy (HCM) may abolish the interaction with the regulatory domain of myosin-binding protein-C (19). Similarly, it has been demonstrated that the E258K HCM-causing mutation in cMyBP-C abolishes interaction between the N terminal of cMyBP-C and myosin-S2 by directly disrupting the cMyBP-C–S2 interface independent of cMyBP-C phosphorylation (49). This disruption accelerates contractile kinetics (49), similar to the effects of ablation of cMyBP-C or phosphorylation. In addition, the results of Calaghan et al. (33), showing that exogenous S2 when added to myofibrils affects their contractile properties, also appear to support a role for the binding of cMyBP-C to myosin S2 as part of the regulatory mechanism. This binding could act as a tether of the myosin head to the filament backbone or stabilize the relaxed cross-bridge array, thus constraining the cross-bridge kinetics. For example, the interaction of cMyBP-C with the S2 region of myosin is abolished upon phosphorylation (20), which may free the tether or change the properties of the myosin head to allow faster cross-bridge kinetics (6, 8, 49). Similarly, recent studies showing that the C0 domain of cMyBP-C may bind to the regulatory light chain (50), or possibly to the mesa region of the myosin head (51), are consistent with a regulatory mechanism in which binding of cMyBP-C to myosin could affect the spatial arrangement of myosin heads on the thick filament and their interactions with the thin filament.
Although earlier studies have examined the effect of phosphorylation of cMyBP-C on the cross-bridge arrangement in isolated rat cardiac thick filaments (52–54), the extent of phosphorylation of cMyBP-C for the individual filaments examined in the electron microscope was unknown in these studies. This finding, along with our more recent results from cMyBP-C knockout cardiac thick filaments (55, 56), which appear to differ from the earlier results for the isolated rat cardiac thick filaments, has left open questions about the effect of phosphorylation of cMyBP-C on cross-bridge arrangement.
Because the levels of phosphorylation of cMyBP-C in the wild-type filaments can be variable, to determine directly the impact of cMyBP-C phosphorylation on the arrangement of cross-bridges on the thick filament, we have compared the structure of cardiac thick filaments from previously generated transgenic mouse lines cMyBP-C(t3SA) and cMyBP-C(t3SD), in which the three m-domain serine residues (Ser273, Ser282, and Ser302) that are phosphorylated by PKA were replaced by either alanine or aspartic acid, respectively, to mimic the nonphosphorylated or the phosphorylated states of cMyBP-C (9, 29–31). For brevity, we will refer to these as the CT-SA and CT-SD mouse lines, respectively.
These transgenic mice have been well characterized as models for the nonphosphorylated and phosphorylated states of cMyBP-C (9, 29–31) and the effects of phosphorylation on the kinetics of cardiac contraction (reviewed in ref. 48). The modified proteins have been demonstrated by immunofluorescence to properly incorporate into the myofibril (9, 31), with expression levels of 74% (CT-SA) and 84% (CT-SD) (31) relative to true wild-type. Because the mutations were produced in mice with a null-background, 100% of the cMyBP-C is mutated (9, 29–31) and endogenous cMyBP-C is absent. Western blots have verified that the serine residues (Ser273, Ser282, and Ser302) in the cMyBP-C of the CT-SA myocardium, unlike wild-type myocardium, are nonphosphorylated under both basal conditions and in the presence of dobutamine (29), thus establishing CT-SA myocardium as a good model for studying the functional and structural role of cMyBP-C in the nonphosphorylated state. Similarly, the CT-SD myocardium has comparable contractile properties to those produced by PKA phosphorylation or ablation of cMyBP-C (31, 48).
Thus, these transgenic mice provide a unique, well-characterized model system in which cMyBP-C is present on all of the isolated filaments and the phospho-mimetic state of cMyBP-C on each thick filament is known. Our results show that thick filament structural effects caused by mimicked phosphorylation in the CT-SD mouse filaments are virtually identical to those we previously obtained from the cMyBP-C knockout mouse hearts (55, 56), where we demonstrated that the absence of cMyBP-C results in a tendency toward disorder of the myosin heads. The filaments from the CT-SA mouse hearts appeared well ordered and similar to well-ordered wild-type filaments isolated by the same procedure. These results provide a compelling structural basis for recent results and models (48, 57, 58), suggesting that nonphosphorylated cMyBP-C stabilizes the relaxed highly ordered off-state configuration of the thick filament. Moreover, it is now even more evident that models of the regulation of contraction via phosphorylation of cMyBP-C must involve modulation of its interactions with myosin.
Results
Appearance of the Filaments.
Thick filaments were isolated from both the CT-SA and CT-SD mouse hearts and their structures compared. Filaments from wild-type mouse hearts isolated in parallel using the same procedure appeared similar in structure to those we have previously described (55, 56). As shown in the low-magnification electron micrographs in Figs. 1A and 2A, the thick filaments from both the CT-SA and CT-SD mouse hearts appear normal, with a length of ∼1.6 µm and a bipolar structure with a central bare zone (asterisks in Figs. 1A and 2A) similar to that of the wild-type mouse cardiac thick filaments and to the structure previously shown for vertebrate cardiac thick filaments (55, 56, 59–63). The measured diameters for both the CT-SA and CT-SD filaments were very similar to each other: 31.1 ± 1.6 nm (mean ± SD, n = 597 measurements from 60 filaments) for the CT-SA filaments and 31.0 ± 1.8 nm (n = 581 measurements from 60 filaments) for the CT-SD filaments. This finding is very similar to the diameters of 31.0 ± 1.7 nm and 31.1 ± 1.4 nm, which we reported previously for wild-type and cMyBP-C knockout cardiac thick filaments (55). Histograms of the distributions of diameters of both the CT-SA and CT-SD filaments were unimodal (Fig. S1), suggesting single populations of filaments for each preparation.
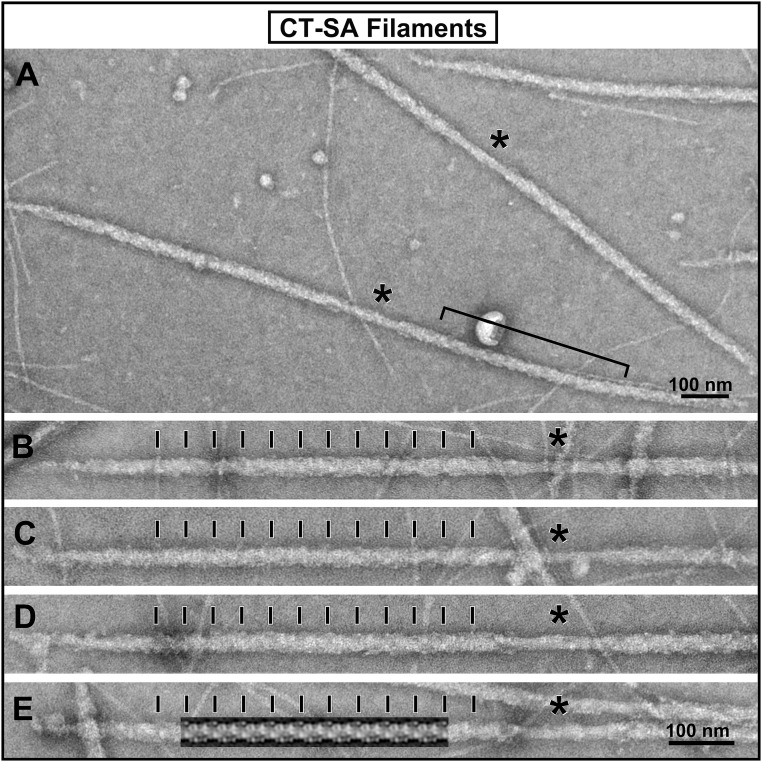
Fig. 1.
Electron micrograph images of the isolated and negatively stained thick filaments from the CT-SA mouse hearts. (A) Lower-magnification image of a field of the isolated thick filaments from the CT-SA hearts. The bipolar structure of the filaments with a central bare zone (asterisks) is apparent and consistent with previous images of isolated mouse cardiac thick filaments. Even at this lower magnification, the well-ordered periodic appearance of the thick filaments is apparent. The bracket marks a region of a filament in which the distinctive saw-tooth staining pattern of the myosin heads on the filament shown in the Fourier-filtered image of a filament in E is apparent. (B–D) A gallery of higher-magnification images of the individual CT-SA filaments is shown. The individual images were straightened using the straightening function in ImageJ for better comparison. The cross-bridge regions of the filaments are well ordered with the 42.9-nm axial periodicity of the myosin head array indicated by the black tick marks. (E) The Fourier transform filtered image of the filament obtained using the first six layer lines of the transform is shown overlaid on the micrograph of the filament. It illustrates the distinctive saw- tooth pattern of staining of the myosin cross-bridge array, which we have previously described, and which can also be seen in the aligned filament images in B–D. The black bars at the bottom right hand corner of the micrographs indicate the scale.
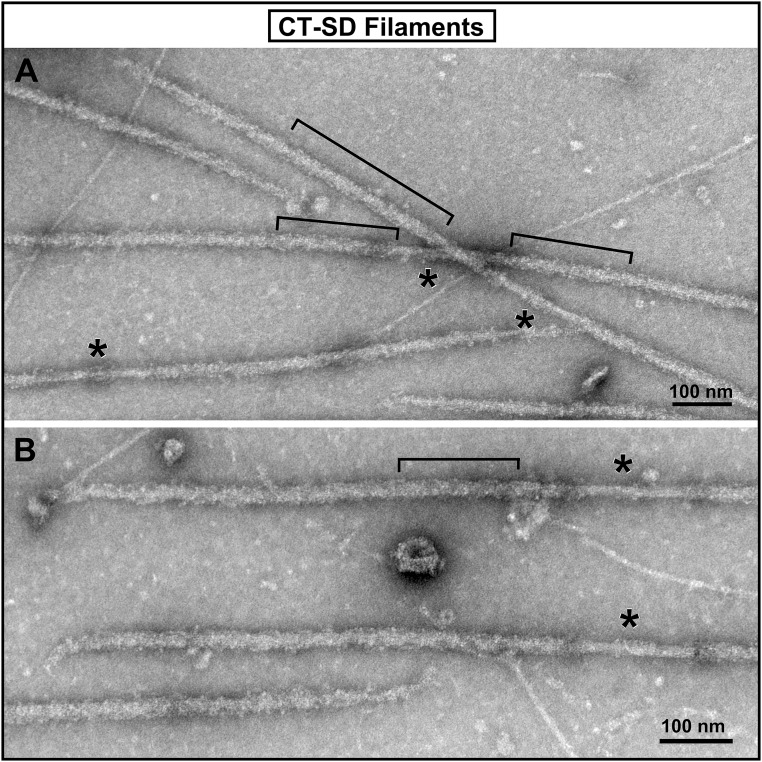
Fig. 2.
Electron micrograph images of the isolated and negatively stained thick filaments from the CT-SD mouse hearts. (A and B) Lower-magnification and higher-magnification images of fields of the isolated filaments from the hearts of the CT-SD mice. Compared with the filaments from the CT-SA hearts, the CT-SD filaments typically appear much more disordered, as seen in both panels. Although the 42.9-nm axial periodicity can still be recognized, particularly in regions, such as those indicated by the black brackets, many areas of the filaments appear to have at least partial disordering of the myosin head array.
Fig. S1.
Histograms and normal curves of the distribution of diameters measured for the CT-SA and CT-SD filaments. The distributions are unimodal suggesting a single population of filaments for both the CT-SA and CT-SD filaments. The calculated means and SDs for the distributions were 31.1 ± SD 1.6 nm (n = 597 measurements from 60 filaments) for the CT-SA filaments and 31.0 ± SD 1.8 nm (n = 581 measurements from 60 filaments) for the CT-SD filaments.
Despite the similarity in their length, diameter, and bipolar structure to control wild-type filaments, differences between the CT-SA and CT-SD cardiac filaments were apparent in the extent of order versus disorder of their cross-bridge arrangements.
As shown in Fig. 1, the thick filaments from CT-SA mouse hearts, in which the nonphosphorylated state of cMyBP-C is mimicked by replacement of serines with nonphosphorylatable alanines, typically appear very periodic with well-ordered arrangements of the myosin heads. This is apparent both in the low-magnification electron micrograph of a field of the filaments (Fig. 1A) and in the higher-magnification gallery of images of straightened individual filaments (Fig. 1 B–E). Both the quasi-helical paths of the myosin heads and the 42.9-nm axial periodicity (indicated by the black bars in Fig. 1 B–E) are clearly evident along the cross-bridge regions of the filaments in the form of the “saw-tooth” staining pattern of the cross-bridge array, which we have previously shown for other vertebrate striated muscle thick filaments (55, 59, 62). This saw-tooth staining pattern is illustrated in Fig. 1E, in which the periodic Fourier layer-line filtered image of the cross bridge arrangement of that filament was overlaid on the micrograph image. This periodic staining pattern, as we have previously shown (62), results from the well-ordered three-stranded quasi-helical arrangement of the myosin heads on the filament. It can be clearly seen in the bracketed region of the filament at low magnification in Fig. 1A and in all of the other filaments (Fig. 1 A–D) as well. The ordered CT-SA filaments thus appear very similar to the examples of well-ordered wild-type mouse cardiac thick filaments previously described (55, 56), and to control wild-type filaments observed in the current study (Fig. S2). In contrast, as shown in Fig. 2, the filaments from the CT-SD hearts, with the mimicked fully phosphorylated state of cMyBP-C were much more variable in their periodicity. Although we were able to find some filaments that appeared relatively well-ordered and maintained the expected 42.9-nm quasi-helical periodicity, most of the CT-SD filaments appeared much more disordered in their cross-bridge arrangement compared with the CT-SA filaments. Typically, most of the filaments retained some evidence of the 42.9-nm quasi-helical arrangement of the cross-bridges along their cross-bridge regions, but looked “ragged” and disordered compared with the filaments from the CT-SA hearts. Often the CT-SD filaments had short regions of partial order (brackets in Fig. 2) with the saw-tooth pattern of staining of the myosin heads discernible, whereas the rest of the filament was much more disordered.
Fig. S2.
A gallery comparing isolated filaments from wild-type, CT-SA, and CT-SD hearts. As shown the CT-SA filaments (D–F) display, a well-ordered cross-bridge array similar to that of the wild-type filaments (A–C), whereas the CT-SD filaments (G–I) display a less well-ordered cross-bridge array. In A from the wild-type filaments and D from the CT-SA filaments, a Fourier-filtered image of that filament obtained using the first six layer lines of the filament’s Fourier transform has been overlaid on the filament image. As shown, both the wild-type filament and the CT-SA filament give similar filtered images, which display what we term the saw-tooth pattern of the cross-bridges that we have previously described for ordered cardiac thick filaments. The tick marks along the side of the filaments in A–F indicate the 42.9-nm periodicity along the filaments. In G–I of the CT-SD filaments, the brackets indicate regions in which the ordered saw-tooth motif corresponding to the 42.9-nm periodicity can still be recognized; the arrows point to regions in which the filament appears to be disordered. All of the images were straightened using the straightening function in ImageJ to facilitate comparison. The bars at the bottom of the figures indicate the magnification.
Thus, the CT-SD filaments, although they can be well-ordered, appear to show a much stronger tendency toward disorder of the cross-bridges compared with the filaments from the CT-SA hearts (compare Figs. 1 and 2, and Fig. S2). This result is similar to our previous observations of isolated filaments from cMyBP-C knockout mouse cardiac thick filaments (55, 56), which also showed a greater tendency toward disorder of the heads compared with the wild-type filaments with cMyBP-C present.
There were slight differences in the number of well-ordered filaments present in the multiple filament isolations we carried out for both the CT-SA and CT-SD hearts; however, this distinction between the CT-SA cardiac thick filaments and the CT-SD filaments was quite reproducible.
Assessment of Filament Periodicity with Fourier Transforms.
Fourier transform analysis of the C-zone region of images of the filaments isolated from the CT-SA and CT-SD mouse hearts confirmed the visual assessment of the degree of order of the myosin head array in each of the filament groups (Figs. 3 and 4).
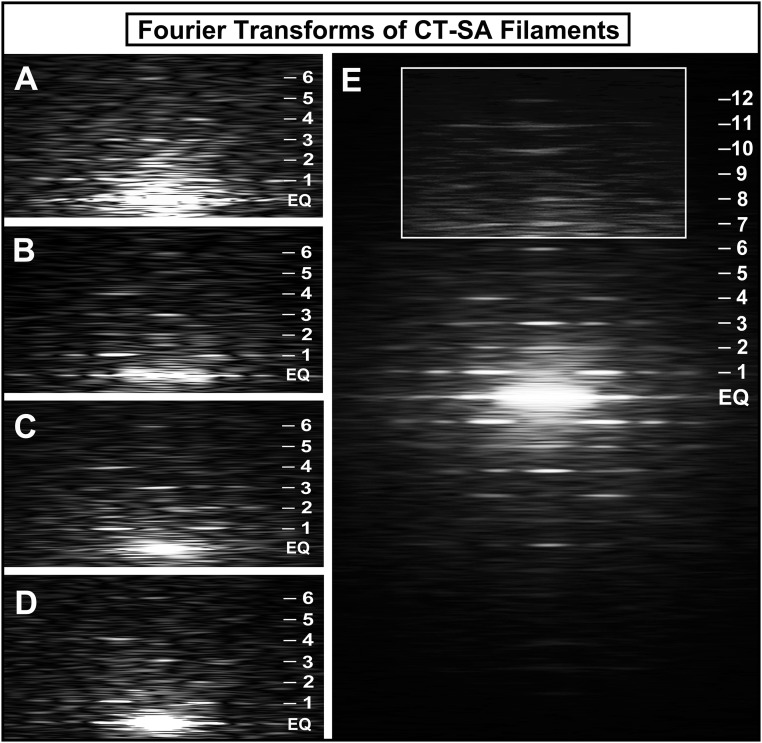
Fig. 3.
Fourier transforms of the thick filaments isolated from the CT-SA mouse hearts. (A–D) A gallery of Fourier transforms computed from images of regions along the arms of individual thick filaments isolated and negatively stained from the CT-SA mouse hearts. The transforms show a strong set of layer lines corresponding to the 42.9-nm axial repeat of the myosin heads out to at least the sixth layer line (indicated by numbered tick marks on the side; EQ is the equator of the pattern). The transforms confirm the highly ordered arrangement of the myosin heads on the CT-SA filaments with cMyBP-C unable to be phosphorylated. (E) Fourier transform obtained by averaging 10 of the Fourier transforms obtained from the individual CT-SA thick filaments. The transform shows layer lines extending to at least the 12th order of the 42.9-nm axial repeat of the pseudohelical arrangement of the myosin heads on the filament. The boxed and lightened region shows the meridional reflections on the layer lines beyond the strong inner six layer lines. This transform clearly shows that the filaments from the CT-SA mouse hearts with nonphosphorylated cMyBP-C are very well ordered.
Fig. 4.
Fourier transforms of the thick filaments isolated from the CT-SD mouse hearts. (A–D) A gallery of Fourier transforms computed from images of regions along the arms of individual thick filaments isolated and negatively stained from the CT-SD mouse hearts. Although the transforms showed that most of the CT-SD filaments retained some of the pseudohelical arrangement of the heads, only a few filaments gave strong transforms (A and B) with a sharp set of off-meridional layer lines extending to at least the sixth layer line (indicated by numbered tick marks on the side; EQ is the equator of the pattern) similar to that seen in the CT-SA filaments. Many of the CT-SD filaments gave transforms (C and D) in which the inner layer lines—particularly the first and second—were often blurred or absent, suggesting disorder of regions of the myosin heads on the filaments. (E) An averaged Fourier transform obtained by averaging 10 of the Fourier transforms obtained from the individual CT-SD thick filaments. As in the case of the averaged transform obtained from the CT-SA filaments, layer lines can be recognized out to approximately the 12th layer line. However, the layer lines were found to be weaker, blurred, and poorly defined compared with the averaged transform from the CT-SA filaments, again consistent with increased disorder of the myosin heads. The lower radial extent of the layer lines is also consistent with a lower resolution of the periodic information. The boxed and lightened region shows the meridional reflections on the layer lines beyond the strong inner six layer lines.
Large numbers of the filaments from the CT-SA mouse hearts gave Fourier transforms with a strong set of sharp layer lines extending to at least the 11th or 12th meridional reflections of the thick filament quasi-helical 42.9-nm periodicity of the myosin heads (Fig. 3). The transforms from the individual CT-SA filaments (Fig. 3 A–D) were consistent with those we have previously shown for well-ordered wild-type mouse cardiac thick filaments (55, 56). A strong set of off-meridional layer lines corresponding to the quasi-helical arrangement of the myosin heads was present up to the sixth layer line. Strong meridional reflections corresponding to the 14.3-nm average axial spacing of the cross-bridge levels were generally present on the 3rd and 6th layer lines, and occasionally on the 9th and 12th layer lines as well. The “forbidden” meridional reflections (reflections not expected from helical symmetry) previously shown both in X-ray diffraction patterns of living skeletal (64) and heart (65) muscle and in transforms of other isolated vertebrate striated muscle thick filaments, including mouse cardiac thick filaments (55, 56, 61–63), were frequently present on the 1st, 2nd, 4th, 5th, 8th, 10th, and 11th layer lines. These features of the transforms, indicating the highly ordered structure of the filaments, were particularly evident in an average of 10 transforms from individual CT-SA filaments (Fig. 3E). This averaged transform can extend to at least the 12th layer line of the 42.9-nm axial periodicity, suggesting that the periodicity in the CT-SA filaments extends to at least 3.58 nm.
The individual Fourier transforms from the CT-SD filaments in contrast were much more variable (Figs. 4 A–D). Although some of the filaments yielded transforms with sets of off-meridional layer lines and meridional reflections similar to those seen in the well-ordered CT-SA filaments (Fig. 4 A and B), many of the filaments gave transforms with only weak or diffuse off-meridional layer lines, even on the first and second layer lines (Fig. 4 C and D), which are the strongest in the CT-SA filaments. The presence of the meridional reflections on both the third and sixth layer lines was consistent with preservation of the average 14.3-nm spacing of the cross-bridge density on the filaments. Thus, the weakness and the diffuse nature of the reflections on the off-meridional layer lines in many of the transforms from the CT-SD filaments most likely results from disorder of the myosin heads along the quasi-helical paths on the filament. This finding is consistent with the filament images showing a tendency toward disorder of the myosin heads in the CT-SD filaments. Overall, a much smaller percentage of the filaments from the CT-SD mouse hearts gave strong transforms indicative of a well-ordered array of the myosin heads, compared with the filaments from the CT-SA mouse hearts.
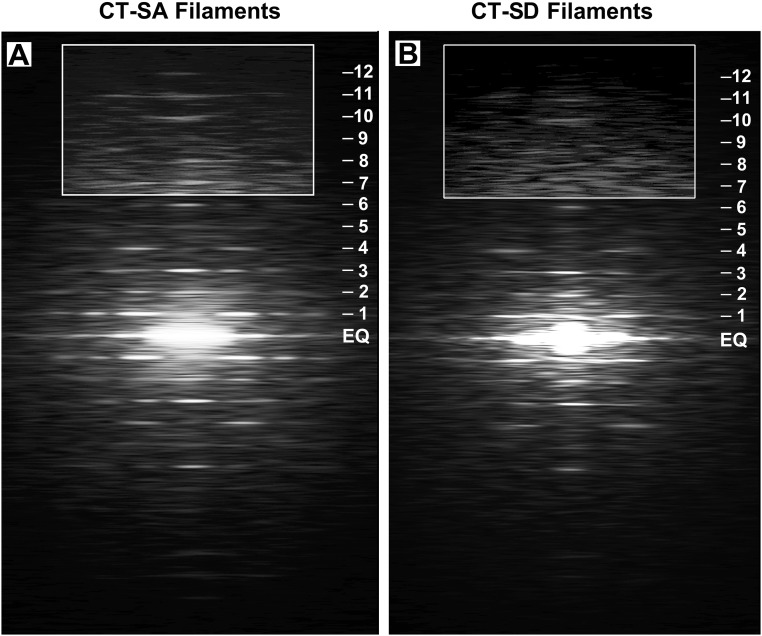
The general weakness in the layer line pattern of the transforms from the individual filaments from the CT-SD hearts was also apparent in the transform obtained by averaging 10 of the best transforms from the CT-SD cardiac filaments (Fig. 4E). Although this averaged transform showed a similar set of layer lines to that seen in the CT-SA transforms, the pattern of the off-meridional layer lines was weaker and the layer line reflections more diffuse than seen for the averaged transform of the CT-SA filaments (Fig. 3E). The layer lines of the averaged CT-SD transform appeared to extend to a lower radius from the meridian with weaker subsidiary maxima and a more rapid fall-off in intensity of the maxima compared with the transform from the CT-SA filaments (Figs. 3 and 4 and Fig. S3). This finding was confirmed in measurements of individual transforms of the CT-SD and CT-SA filaments where periodic information on the first layer line extended to only 8.4 ± 1.6 nm−1 (mean ± SD, n = 50 transforms) resolution for the CT-SD transforms compared with 6.5 ± 0.9 nm−1 (n = 50 transforms) resolution for the CT-SA transforms, the larger SD for the CT-SD transforms reflecting the greater variation in these transforms.
Fig. S3.
A direct comparison of the averaged transforms of the CT-SA (A) and CT-SD filaments (B). The averaged transform from the CT-SA filaments is stronger with sharper reflections and subsidiary reflections extending to a higher radius along the layer lines compared with the averaged transform (B) of the CT-SD filaments. The weaker transform from the CT-SD filaments is consistent with the greater variation in the individual transforms from the CT-SD filaments and the greater variation in cross-bridge order of the CT-SD filaments.
These results are consistent with our observation that although the CT-SD filaments retained the 42.9-nm pseudohelical periodicity in some filaments and along short regions of other filaments, overall the filaments appeared much more prone to disordering of the arrangement of the myosin heads compared with the filaments from the CT-SA mouse hearts.
One interesting feature of the transforms from the CT-SD filaments was the relatively strong intensity of the “forbidden” meridional reflections, particularly those on the first and second layer lines, compared with the CT-SA filaments (Fig. 3) and to transforms of the cMyBP-C knockout cardiac thick filaments, in which these reflections were weak or absent (55). A possible explanation for this finding is that the disorder of the heads is exposing periodic structure along the backbone, such as the density of the C8-C10 domains of cMyBP-C which are present axially every 42.9 nm, or that cMyBP-C has changed to a conformation more perpendicular to the thick filament axis, strengthening its contribution to the 42.9-nm meridional reflection. In the earlier case of the cMyBP-C knockout filaments, cMyBP-C was absent and could not contribute intensity to these reflections.
Discussion
In this study, we examined the structure of isolated thick filaments from mice in which the nonphosphorylated and phosphorylated states of cMyBP-C are mimicked by amino acid substitutions of either alanine or aspartic acid for the PKA-phosphorylatable serines in the m-domain (9, 29–31). As described at the beginning of this report, both the phospho-deficient CT-SA [cMyBP-C(t3SA)] and phospho-mimetic CT-SD [cMyBP-C(t3SD)] transgenic mouse heart models studied here have been well characterized in their physiological properties and differences, including protein expression, proper insertion into the myofibril, and inability of the PKA-phosphorylatable serines in the m-domain to be phosphorylated in the CT-SA mouse hearts (9, 29–31). Basal levels of phosphorylation of the regulatory light chain (RLC), which has been shown to affect the cross-bridge order in skeletal muscle (66) and the radius of the cross-bridge in cardiac muscle (67), has been demonstrated to be similar (∼40%) for both the CT-SA and CT-SD mouse hearts (29, 31). This finding, together with the report by Chang et al. (68) that the phosphorylation levels of mouse cardiac RLC are not maintained, but rapidly decrease in homogenates and isolated filament preparations, means that RLC phosphorylation levels should be similar and very low (∼10%) in our filament preparations. Thus, differences in phosphorylation levels of RLC cannot explain differences seen between the filaments from the CT-SA and CT-SD hearts. These cMyBP-C mutant mice thus provide a unique model system for the study of the effects of dephosphorylation and phosphorylation of cMyBP-C, both for physiological studies of contractile kinetics (reviewed in ref. 48) and for structural studies such as reported here.
We found that the thick filaments from the hearts of the CT-SA mice, which mimic the nonphosphorylated state of cMyBP-C, typically maintained the well-ordered pseudohelical relaxed arrangement of the myosin heads previously shown for relaxed vertebrate cardiac thick filaments (55, 56, 59–63). The thick filaments from the CT-SD mouse hearts, which mimic the phosphorylated state of cMyBP-C, showed a much stronger tendency toward disorder of the myosin heads.
Comparison with Earlier Studies.
These results, like those of our earlier studies of the cMyBP-C knockout mouse cardiac thick filaments (55, 56), were surprising because they differed from previous results reported by Weisberg and Winegrad (52, 53) and Levine et al. (54) for the effects of phosphorylation of cMyBP-C on the cross-bridge arrangement in isolated rat cardiac thick filaments.
The studies of Weisberg and Winegrad (52, 53) reported that phosphorylation of cMyBP-C produced an increase both in the cross-bridge order of the filament and in the diameter of isolated rat cardiac thick filaments. Consistent with this, Levine et al. (54) presented data supporting a model in which the cross-bridge array on the filament was disordered when cMyBP-C was nonphosphorylated and that upon phosphorylation of cMyBP-C, the myosin array on the thick filament changed from disordered to a highly ordered array with expansion of the filament diameter, extension of the myosin heads, and loosening of the backbone structure. It was further proposed that upon phosphorylation of cMyBP-C, the cross-bridges were less flexible and that the loosening of the backbone would be associated with an increase in diameter of a collar-like structure formed by the three cMyBP-C molecules present at every third cross-bridge level (69, 70).
Although the Levine et al. (54) model is widely cited as a mechanism by which cMyBP-C phosphorylation regulates contraction, one major ambiguity in these studies is that the actual state of phosphorylation of cMyBP-C for any individual filament examined in the electron microscope was unknown. The degree of phosphorylation was measured for the whole population of filaments and not the individual filaments that were examined. Thus, a direct correlation of the degree of cross-bridge order for a filament and the state of phosphorylation of its cMyBP-C was not possible.
Our subsequent studies comparing the structure of thick filaments isolated from cMyBP-C knockout mouse hearts to those isolated from wild-type hearts (55, 56) raised some questions about this model. The absence of cMyBP-C on the thick filaments in the knockout mouse could be argued to be analogous to the phosphorylated state of cMyBP-C, because in both cases the interaction of cMyBP-C with myosin S2 would be weakened or abolished (20). Interestingly, the cardiac filaments from the cMyBP-C knockout mouse hearts showed no difference in diameter from the wild-type filaments and showed a strong tendency to have a disordered cross-bridge array, not the increased diameter and increased cross-bridge order predicted by the studies of Levine et al. (54). These results were found both in a study by Kensler and Harris (55) and a separate study by Zoghbi et al. (56). One caveat to these results, however, was the question whether the genetic ablation of cMyBP-C and phosphorylation of cMyBP-C are entirely analogous, although both have similar physiological effects (reviewed in ref. 48).
In contrast to the earlier studies by Weisberg and Winegrad (52, 53) and Levine et al. (54), in the present study of filaments from the cMyBP-C phospho-deficient CT-SA and the cMyBP-C phospho-mimetic CT-SD mutant mouse hearts, the state of pseudophosphorylation of cMyBP-C is known for each examined filament (9, 29–31). With the change from serine to alanine for each of the three PKA-phosphorylatable serines in the m-domain of cMyBP-C in the CT-SA mouse hearts, the cMyBP-C molecules cannot be phosphorylated at these sites and should be analogous to the completely nonphosphorylated state of cMyBP-C (9, 29, 31). Similarly, the substitution of aspartic acid for the serines in the cMyBP-C in CT-SD hearts should mimic the fully phosphorylated state of the cMyBP-C (30, 31). This idea is supported by evidence that the cMyBP-C phosphomimetic mice exhibit physiological effects on contractile kinetics very similar to those reported in earlier studies using ablation of cMyBP-C or PKA-induced phosphorylation of cMyBP-C (reviewed in ref. 48). In addition, unlike the cMyBP-C knockout mouse hearts, cMyBP-C is still present on the filaments of both the CT-SA mouse hearts and the CT-SD mouse hearts.
The results for the filaments isolated from the CT-SA mouse hearts are particularly striking because the three serines in the m-domain of cMyBP-C, which can normally be phosphorylated by PKA, are completely nonphosphorylatable. In contrast to the Levine et al. (54) model, which would predict that these filaments with completely nonphosphorylated cMyBP-C should be highly disordered in their cross-bridge arrangement, these filaments tend to be very well ordered. Similarly, the filaments from the hearts of the CT-SD mice, which should mimic the fully phosphorylated state of the cMyBP-C, would have been predicted by the Levine et al. (54) model to be very well ordered, but were found instead to have a strong tendency toward disorder of the myosin heads. This disorder of the heads in the filaments of both the CT-SD mouse hearts and the filaments from the cMyBP-C knockout mouse hearts suggests that the myosin heads are likely less tightly bound to the backbone and perhaps more mobile, not less flexible or less free to move, as suggested by Levine et al. (54).
Interestingly, in contrast to the results of Weisberg and Winegrad (52, 53) and Levine et al. (54), we did not see a significant change in diameter of either the filaments from the CT-SD (31.0 ± 1.8 nm) or CT-SA filaments (31.1 ± 1.6 nm) from the wild-type diameter of 31.0 ± 1.7 nm we reported previously (56). Histograms of the filament diameter distributions for the CT-SA and CT-SD filaments (Fig. S1) show similar unimodal distributions of the diameters. In the earlier results by Weisberg and Winegrad (52, 53) and Levine et al. (54), it was reported that the filaments with phosphorylated cMyBP-C were highly ordered and had a diameter of 36–37 nm, greater than the 30–32 nm found for the filaments with nonphosphorylated cMyBP-C. We did not observe this increase in either diameter or cross-bridge order for the CT-SD filaments, which should have corresponded to the filaments with phosphorylated cMyBP-C observed by Weisberg and Winegrad (52, 53) and Levine et al. (54). Instead, the results complement our previous observations demonstrating that cMyBP-C knockout heart filaments did not differ significantly in diameter from the wild-type filaments (55), but showed a greater tendency toward disorder.
The reason for the difference between the earlier studies by Weisberg and Winegrad (52, 53) and Levine et al. (54) for isolated rat cardiac thick filaments and our studies of the isolated thick filaments from cMyBP-C knockout mouse hearts (55, 56) and the phosphomimetic mouse hearts is not certain. However, there are several differences between our studies and theirs. In addition to knowing the mimicked state of phosphorylation of cMyBP-C for each filament in our studies of the mutant mice, we also used the 42.9-nm axial periodicity of the thick filaments as an internal ruler for the measurements of filament diameter, in both our earlier measurements of the diameter of the wild-type and cMyBP-C mouse heart filaments (55) and in the filaments from the phosphomimetic mice reported here. This avoided possible differences in magnification in the electron microscope from day to day, which can affect measurements of the apparent diameter of the filaments. In addition, the use of Fourier transforms in our studies, rather than the optical diffraction patterns used in their studies (52–54), has allowed us to more easily assess the extent of order of the filaments. The averaging of the transforms, in both our previous cMyBP-C knockout cardiac filament studies (55) and the studies here of the phosphomimetic mouse heart filaments, provides a particularly useful tool for assessing the order of a population of filaments that is not available with optical diffraction. Furthermore, many of the optical diffraction patterns used in the earlier studies of the isolated rat cardiac thick filaments (52–54) were relatively weak, with only one or two strong off-meridional layer lines, thus making it difficult to assess the extent of order of the filaments in those studies.
Interpretation of Results for the CT-SA and CT-SD Thick Filaments.
Our previous results from studies of cMyBP-C knockout mice (55, 56) provided evidence that, although cMyBP-C is not a requirement for maintenance of the highly ordered relaxed state of the myosin heads, it may help to stabilize that order. The results from the current study of cMyBP-C phosphomimetic mouse cardiac thick filaments add to this conclusion by showing that when nonphosphorylated, cMyBP-C stabilizes and promotes the relaxed quasi-helical arrangement of the myosin heads, whereas when phosphorylated, it does not. Phosphorylation or ablation of cMyBP-C thus have similar effects, abolishing or reducing cMyBP-C’s stabilizing effect.
This interpretation is supported by the observation that both the filaments from the CT-SD hearts and the filaments from the cMyBP-C knockout mouse hearts (55, 56) retain significant evidence of the 42.9-nm relaxed pseudohelical arrangement of the myosin heads in some regions, and are variable in the extent of disorder. The myosin heads are not completely disordered, as might be expected if phosphorylation or ablation of cMyBP-C directly caused an order-to-disorder transition in their arrangement. Instead, the disorder is likely to be a result of the weakening of the binding of the heads to the filament surface predisposing them to become disordered by factors, such as interactions with actin or other elements during the isolation procedure or their binding to the carbon substrate during the negative-staining procedure (71).
This interpretation is also consistent with our finding that the diameters of the CT-SD filaments are similar to those of the CT-SA filaments, and also to the diameters previously measured for the filaments from the cMyBP-C knockout mouse and wild-type hearts (55). Although the similarity in diameters was initially surprising, it is consistent with the possibility that many of the myosin heads in the isolated CT-SD filaments and the cMyBP-C knockout mouse filaments, although partially disordered in their orientation, are still weakly bound to the filament backbone. Within the lattice of the thick and thin filaments in the intact muscle, however, this loss or weakening of binding of the myosin heads to the thick filament would likely increase the potential interaction of the myosin heads with the adjacent thin filaments. This finding is suggested by X-ray diffraction evidence indicating that when cMyBP-C is ablated or phosphorylated in intact muscle, the myosin heads can extend out to a higher radius (30, 72, 73), and by the acceleration of cardiac contractile kinetics observed when cMyBP-C is ablated or phosphorylated (reviewed in ref. 48).
Relationship to Recent Studies of cMyBP-C Phosphorylation and Ablation.
Our results complement several recent studies using techniques other than electron microscopy that also suggest that cMyBP-C, when nonphosphorylated, helps to stabilize the relaxed, ordered arrangement of the myosin heads (57, 58).
Kampourakis et al. (57), using fluorescent probes on both troponin C and the myosin regulatory light chain, provided evidence suggesting that in addition to activating the actin thin filament, the nonphosphorylated N-terminal region of cMyBP-C may help to maintain the thick filament in the relaxed “off-state.” They demonstrated that the addition of the C1mC2 N-terminal fragment of cMyBP-C to cardiac fibers in the presence of blebbistatin (74, 75) induced a change in the orientation of the regulatory light chain toward the off-state structure beyond that induced by blebbistatin alone. This change in the orientation of the light chain clearly is consistent with the idea that the N-terminal region of cMyBP-C, when nonphosphorylated, helps to stabilize the relaxed off-state of the myosin heads and their quasi-helical arrangement on the vertebrate cardiac thick filament.
Also in support of this idea, McNamara et al. (58) examined the role of cMyBP-C in regulating the population of heads in the superrelaxed state (SRX) of cardiac myosin by comparing the rate of single ATP turnover of myosin in cMyBP-C knockout mouse hearts to that in wild-type hearts. In the SRX state (75; for review, see ref. 76), the myosin heads are thought to be in the off-state relaxed configuration suggested by the models of Wendt et al. (77) and 3D reconstructions of both tarantula (78) and vertebrate cardiac muscle thick filaments (56, 59, 60), and have highly inhibited ATPase activity. Approximately 20% of cardiac myosin in the wild-type mouse is in the SRX state and may represent a reserve population of myosin heads that could modulate cardiac contraction in response to factors such as stress, changes in preload, and increased arterial pressures (76). A significant reduction was reported in the proportion of heads in the SRX state in homozygous cMyBP-C knockout mice compared with that in the wild-type mouse hearts (58). Because in the SRX state the myosin heads are thought to be bound in a highly ordered arrangement to the filament backbone, the results were interpreted as supporting the proposal that cMyBP-C stabilizes this arrangement and that the loss of cMyBP-C results in an untethering of the heads.
The results of these two studies appear to be consistent with our earlier results by electron microscopy showing that the loss of cMyBP-C is associated with an increased tendency for the relaxed ordered myosin head array to become disordered (55, 56). These results also complement our structural results here for the cMyBP-C phosphomimetic mouse hearts. Because ablation or PKA phosphorylation of cMyBP-C appear to have similar effects on contractility (reviewed in ref. 48), our finding that the fully phosphorylated state of cMyBP-C in the CT-SD mouse hearts results in an increased tendency toward disorder of the myosin heads complements the results of McNamara et al. (58) in demonstrating the similarity of mechanisms by which ablation and phosphorylation of cMyBP-C confer enhanced cardiac contractility.
Our studies, as well as these other recent studies, do not in any way negate evidence that binding of cMyBP-C to the thin filament contributes to the mechanism for regulation by cMyBP-C, but instead support the increasing consensus that regulation of myocardial contractility involves a mechanism in which both the binding of cMyBP-C to the myosin head/S2 and to F-actin in the thin filament are modulated by the phosphorylation state of cMyBP-C.
Conclusions.
Comparison by electron microscopy of the structure of thick filaments isolated from cMyBP-C phospho-deficient (CT-SA) and phospho-mimetic (CT-SD) mouse hearts supports the conclusion that cMyBP-C, when nonphosphorylated, acts to stabilize the highly ordered off-state conformation of the myosin heads on the thick filament, whereas phosphorylation of cMyBP-C weakens this stabilization, predisposing the myosin heads to disorder and increasing the probability of their interacting with the thin filament.
Materials and Methods
Transgenic CT-SA and CT-SD Mouse Hearts and Wild-Type Hearts.
The transgenic CT-SA [cMyBP-C(t3SA)] mice and CT-SD mice [cMyBP-C(t3SD)] (29) were bred and maintained at the University of Wisconsin (UW) School of Medicine and Public Health, whereas the wild-type BALB/c mice were maintained at the University of Puerto Rico. As previously reported (9, 29, 31), the CT-SA mice have substitution of alanine residues for serine residues Ser273, Ser282, and Ser302 in the m-domain of the cMyBP-C expressed in their hearts. The nonphosphorylatable alanine replacements in the cMyBP-C should mimic the nonphosphorylated state of the cMyBP-C. In the CT-SD mice, these serines in the m-domain were similarly replaced with aspartic acid to mimic the fully phosphorylated state of the cMyBP-C in the hearts of the mice (30, 31). Three-month-old male and female mice were used in the studies. Following Institutional Animal Care and Use Committee-approved killing procedures at the UW School of Medicine and Public Health, hearts from both the CT-SA and the CT-SD mice were dissected from the animals, sliced in half, and placed in an EGTA-Mincing solution (100 mM NaCl, 2.5 mM EGTA, 5 mM MgAc, 1 mM DTT in 7 mM phosphate buffer, pH 7.0) and flash-frozen in liquid nitrogen. The flash-frozen hearts on dry ice were shipped by overnight express to the University of Puerto Rico and either immediately used to prepare thick filament suspensions or stored at −80 °C until used. No differences were seen between the immediately processed hearts and those stored at −80 °C. The wild-type BALB/c mice were killed following Institutional Animal Care and Use Committee-approved procedures at the University of Puerto Rico Medical School. Both male and female wild-type mice were used. Typically, two hearts from the CT-SA, CT-SD, and wild-type mice were used for each filament preparation. Four separate replica isolations of filaments from the CT-SA and CT-SD hearts were performed, thus a total of 16 transgenic mice were used.
Isolation of Thick Filament Suspensions.
Suspensions of thick filaments from the CT-SA and CT-SD mouse hearts were isolated, as previously described for mouse (55) and zebrafish cardiac thick filaments (59). In initial experiments, thick filaments from wild-type hearts were isolated in parallel by the same procedure for comparison and to ensure that the isolation procedure was working as previously described (55, 59). In brief, small fiber bundles of the hearts, rinsed with three to four changes of the mincing solution (1 h each), were left overnight at 4 °C, and the following morning incubated for 30 min at room temperature (∼25 °C) with slow stirring in a relaxing solution composed of EGTA-mincing solution containing 2.5 mM ATP, 10 mM creatine phosphate, and a proteolytic inhibitor mixture comprised of five components: N-p-tosyl-l-phenylalanine chloromethyl ketone, leupeptin, aprotinin, pepstatin A, and trypsin inhibitor, each at a final concentration of 0.004 mg/mL, at pH 6.8. Separation of the thick filaments was performed using the previously described elastase procedure for cardiac tissue (59, 60), with elastase and trypsin inhibitor each at a final concentration of 0.44 mg/mL. The supernatant containing the thick filaments was used to prepare electron microscopy grids. As explained in the Discussion, inhibitors of regulatory light chain were not used because the CT-SA and CT-SD hearts have similar in vivo basal levels of phosphorylation (29, 31) and are expected to be unphosphorylated or minimally phosphorylated with the long incubation times and low calcium levels involved in the isolation procedure (68).
Negative Staining and Electron Microscopy.
Negative staining was performed with 1% uranyl acetate, as previously described (55, 59, 60). All solutions were at 25 °C to avoid the disordering of the filaments which can occur at temperatures less than 15 °C (55, 59, 60). Electron microscopy was performed as previously described (55) at 80 kV with a JEOL 1200EXII electron microscope with an AMT HR60 high-resolution digital CCD camera (2K × 2K). Magnification was calibrated using the 42.9-nm axial periodicity of the filaments as a standard.
Assessment of Filament Helical Order.
The thick filament periodicity was assessed both visually and by calculation of Fourier transforms in 2,048 × 2,048 pixel arrays as previously described (55, 61–63). The images were straightened, rescaled, and rotated as necessary to ensure that the helical layer lines fell on the sampled lines by sizing the filament images to be equivalent to thirty 42.9-nm axial repeats per 2,048 pixels (55, 61–63), which is equivalent to a sampling spacing of ∼0.63 nm per pixel. Assessment of the filament helical order was based upon the number, sharpness, and strength of the layer lines in the transform. The radial extent of periodic information along the first layer line and other periodicities in the transforms were calculated relative to the spacings of the 14.29-nm−1 and 7.15-nm−1 meridional reflections on the third and sixth layer lines.
Measurement of Filament Diameters.
The diameters of both the CT-SA and CT-SD filaments were measured in ImageJ. Because the actual magnification can vary by up to 5% or more from that indicated by the electron microscope, the 42.9-nm axial periodicity of the filaments was used as an internal standard of magnification. For each micrograph, a filament that appeared to retain at least some evidence of the 42.9-nm axial repeat was selected, straightened, and a Fourier transform performed. The layer line spacing was measured based upon the meridional reflections on the third and sixth layer lines. This value was used to calculate a scaling factor that was applied to the micrograph to ensure that the spacing of the layer lines in the transforms from the filaments was exactly 30 pixels. The straightened images of the filaments from the micrograph were displayed horizontally, and the maximum diameter of the filament was measured using a rectangular box whose longitudinal edges ran along the outer tips of the myosin heads in regions ∼one repeat (42.9 nm) in length along the filament. The diameter in nanometers of each of the measured regions was calculated by simple proportion relative to the spacing in pixels of the 42.9-nm axial repeat, and a mean and SD of all of the measured values for the CT-SA and CT-SD filaments calculated.
Acknowledgments
We thank Dr. Dan Fitzsimmons in R.L.M.’s laboratory for his technical support. This work was supported by NIH Grants SC1 HL096017 (to R.W.K.), R01 AR067279 (to R.C. and David Warshaw), and R37 HL82900 and P01 HL094291 (to R.L.M.). R.C. also received funding through Program Project Grant P01 HL059408 (to David Warshaw).
Footnotes
Conflict of interest statement: R.L.M. is Chief scientific officer for Wisconsin Innovation Initiative (Wi2), a not-for-profit company affiliated with the University of Wisconsin, but is prohibited from realizing any personal benefit, financial or otherwise, from Wi2.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614020114/-/DCSupplemental.
References
- 1.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973;74(4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 2.Marian AJ, Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2001;33(4):655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108(6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann PA, Hartzell HC, Moss RL. Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. J Gen Physiol. 1991;97(6):1141–1163. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93(8):752–758. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 6.Stelzer JE, Dunning SB, Moss RL. Ablation of cardiac myosin-binding protein-C accelerates stretch activation in murine skinned myocardium. Circ Res. 2006;98(9):1212–1218. doi: 10.1161/01.RES.0000219863.94390.ce. [DOI] [PubMed] [Google Scholar]
- 7.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90(5):594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 8.Palmer BM, et al. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Mol Cell Biochem. 2004;263(1):73–80. doi: 10.1023/B:MCBI.0000041849.60591.45. [DOI] [PubMed] [Google Scholar]
- 9.Tong CW, Stelzer JE, Greaser ML, Powers PA, Moss RL. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ Res. 2008;103(9):974–982. doi: 10.1161/CIRCRESAHA.108.177683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao L, et al. PKCepsilon increases phosphorylation of the cardiac myosin binding protein C at serine 302 both in vitro and in vivo. Biochemistry. 2007;46(23):7054–7061. doi: 10.1021/bi700467k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardswell SC, et al. Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. J Biol Chem. 2010;285(8):5674–5682. doi: 10.1074/jbc.M109.066456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luther PK, et al. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol. 2008;384(1):60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1976;192(1109):451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 15.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell Motil. 1986;7(6):550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 16.Yasuda M, Koshida S, Sato N, Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J Mol Cell Cardiol. 1995;27(10):2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- 17.Kasahara H, et al. Autoimmune myocarditis induced in mice by cardiac C-protein. Cloning of complementary DNA encoding murine cardiac C-protein and partial characterization of the antigenic peptides. J Clin Invest. 1994;94(3):1026–1036. doi: 10.1172/JCI117416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem J. 1978;171(3):813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruen M, Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999a;286(3):933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 20.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999b;453(3):254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 21.Ababou A, Gautel M, Pfuhl M. Dissecting the N-terminal myosin binding site of human cardiac myosin-binding protein C. Structure and myosin binding of domain C2. J Biol Chem. 2007;282(12):9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
- 22.Ababou A, et al. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1. J Mol Biol. 2008;384(3):615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhuiyan MS, Gulick J, Osinska H, Gupta M, Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C’s interactions. J Mol Cell Cardiol. 2012;53(6):838–847. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okagaki T, et al. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol. 1993;123(3):619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235(1-2):317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 26.Hartzell HC, Titus L. Effects of cholinergic and adrenergic agonists on phosphorylation of a 165,000-dalton myofibrillar protein in intact cardiac muscle. J Biol Chem. 1982;257(4):2111–2120. [PubMed] [Google Scholar]
- 27.Hartzell HC, Glass DB. Phosphorylation of purified cardiac muscle C-protein by purified cAMP-dependent and endogenous Ca2+-calmodulin-dependent protein kinases. J Biol Chem. 1984;259(24):15587–15596. [PubMed] [Google Scholar]
- 28.Schlender KK, Hegazy MG, Thysseril TJ. Dephosphorylation of cardiac myofibril C-protein by protein phosphatase 1 and protein phosphatase 2A. Biochim Biophys Acta. 1987;928(3):312–319. doi: 10.1016/0167-4889(87)90191-1. [DOI] [PubMed] [Google Scholar]
- 29.Tong CW, et al. Phosphoregulation of cardiac inotropy via myosin binding protein-C during increased pacing frequency or β1-adrenergic stimulation. Circ Heart Fail. 2015;8(3):595–604. doi: 10.1161/CIRCHEARTFAILURE.114.001585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colson BA, et al. Myosin binding protein-C phosphorylation is the principal mediator of protein kinase A effects on thick filament structure in myocardium. J Mol Cell Cardiol. 2012;53(5):609–616. doi: 10.1016/j.yjmcc.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosas PC, et al. Phosphorylation of cardiac myosin-binding protein-C is a critical mediator of diastolic function. Circ Heart Fail. 2015;8(3):582–594. doi: 10.1161/CIRCHEARTFAILURE.114.001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bardswell SC, Cuello F, Kentish JC, Avkiran M. cMyBP-C as a promiscuous substrate: Phosphorylation by non-PKA kinases and its potential significance. J Muscle Res Cell Motil. 2012;33(1):53–60. doi: 10.1007/s10974-011-9276-3. [DOI] [PubMed] [Google Scholar]
- 33.Calaghan SC, Trinick J, Knight PJ, White E. A role for C-protein in the regulation of contraction and intracellular Ca2+ in intact rat ventricular myocytes. J Physiol. 2000;528(Pt 1):151–156. doi: 10.1111/j.1469-7793.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunst G, et al. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86(1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 35.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122(6):761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razumova MV, et al. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J Biol Chem. 2006;281(47):35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 37.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: Implications for cardiac function. Proc Natl Acad Sci USA. 2008;105(47):18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2011;174(1):44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mun JY, et al. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410(2):214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luther PK, et al. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci USA. 2011;108(28):11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mun JY, et al. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci USA. 2014;111(6):2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mun JY, Kensler RW, Harris SP, Craig R. The cMyBP-C HCM variant L348P enhances thin filament activation through an increased shift in tropomyosin position. J Mol Cell Cardiol. 2016;91:141–147. doi: 10.1016/j.yjmcc.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orlova A, Galkin VE, Jeffries CM, Egelman EH, Trewhella J. The N-terminal domains of myosin binding protein C can bind polymorphically to F-actin. J Mol Biol. 2011;412(3):379–386. doi: 10.1016/j.jmb.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris SP, Belknap B, Van Sciver RE, White HD, Galkin VE. C0 and C1 N-terminal Ig domains of myosin binding protein C exert different effects on thin filament activation. Proc Natl Acad Sci USA. 2016;113(6):1558–1563. doi: 10.1073/pnas.1518891113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): Who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33(1):83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 47.Weith A, et al. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52(1):219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ Res. 2015;116(1):183–192. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Lange WJ, et al. E258K HCM-causing mutation in cardiac MyBP-C reduces contractile force and accelerates twitch kinetics by disrupting the cMyBP-C and myosin S2 interaction. J Gen Physiol. 2013;142(3):241–255. doi: 10.1085/jgp.201311018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: Cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286(14):12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spudich JA. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem Soc Trans. 2015;43(1):64–72. doi: 10.1042/BST20140324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weisberg A, Winegrad S. Alteration of myosin cross bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc Natl Acad Sci USA. 1996;93(17):8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weisberg A, Winegrad S. Relation between crossbridge structure and actomyosin ATPase activity in rat heart. Circ Res. 1998;83(1):60–72. doi: 10.1161/01.res.83.1.60. [DOI] [PubMed] [Google Scholar]
- 54.Levine R, Weisberg A, Kulikovskaya I, McClellan G, Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys J. 2001;81(2):1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kensler RW, Harris SP. The structure of isolated cardiac Myosin thick filaments from cardiac myosin binding protein-C knockout mice. Biophys J. 2008;94(5):1707–1718. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kampourakis T, Yan Z, Gautel M, Sun YB, Irving M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc Natl Acad Sci USA. 2014;111(52):18763–18768. doi: 10.1073/pnas.1413922112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McNamara JW, et al. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J Mol Cell Cardiol. 2016;94:65–71. doi: 10.1016/j.yjmcc.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González-Solá M, Al-Khayat HA, Behra M, Kensler RW. Zebrafish cardiac muscle thick filaments: Isolation technique and three-dimensional structure. Biophys J. 2014;106(8):1671–1680. doi: 10.1016/j.bpj.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci USA. 2013;110(1):318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kensler RW. Mammalian cardiac muscle thick filaments: Their periodicity and interactions with actin. Biophys J. 2002;82(3):1497–1508. doi: 10.1016/S0006-3495(02)75503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kensler RW. The mammalian cardiac muscle thick filament: Crossbridge arrangement. J Struct Biol. 2005a;149(3):303–312. doi: 10.1016/j.jsb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Kensler RW. The mammalian cardiac muscle thick filament: Backbone contributions to meridional reflections. J Struct Biol. 2005b;149(3):313–324. doi: 10.1016/j.jsb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 64.Huxley HE, Brown W. The low-angle X-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30(2):383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 65.Matsubara I, Millman BM. X-ray diffraction patterns from mammalian heart muscle. J Mol Biol. 1974;82(4):527–536. doi: 10.1016/0022-2836(74)90246-0. [DOI] [PubMed] [Google Scholar]
- 66.Levine RJ, Kensler RW, Yang Z, Stull JT, Sweeney HL. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys J. 1996;71(2):898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colson BA, et al. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. J Physiol. 2010;588(Pt 6):981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang AN, et al. Constitutive phosphorylation of cardiac myosin regulatory light chain in vivo. J Biol Chem. 2015;290(17):10703–10716. doi: 10.1074/jbc.M115.642165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winegrad S. Cardiac myosin binding protein C. Circ Res. 1999;84(10):1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- 70.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: Its role in physiology and disease. Circ Res. 2004;94(10):1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 71.Knight P, Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J Mol Biol. 1984;177(3):461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- 72.Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J Mol Biol. 2007;367(1):36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colson BA, et al. Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ Res. 2008;103(3):244–251. doi: 10.1161/CIRCRESAHA.108.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao FQ, Padrón R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95(7):3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107(7):1637–1646. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNamara JW, Li A, dos Remedios CG, Cooke R. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys Rev. 2015;7:5–14. doi: 10.1007/s12551-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98(8):4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436(7054):1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]