Significance
Cancer is the leading cause of death worldwide, although studies revealed that dysregulation of the Hippo pathway contributes to tumorigenesis, whereas its roles in tumor invasion and cell migration remain paradoxical and largely elusive. Using Drosophila as a model, we herein find cross-talk between the Hippo and JNK pathways in regulating cell migration and invasion. Mechanistically, we identify bantam-Rox8 modules as essential components downstream of Yorkie in mediating JNK-dependent cell invasion. Our finding is particularly important as it offers a wake-up call for therapeutic interventions of Hippo-related cancers, because simply increasing Hippo signal activity may paradoxically accelerate cell invasion and metastasis.
Keywords: Hippo, JNK, Drosophila, migration, Rox8
Abstract
Overwhelming studies show that dysregulation of the Hippo pathway is positively correlated with cell proliferation, growth, and tumorigenesis. Paradoxically, the detailed molecular roles of the Hippo pathway in cell invasion remain debatable. Using a Drosophila invasion model in wing epithelium, we show herein that activated Hippo signaling promotes cell invasion and epithelial-mesenchymal transition through JNK, as inhibition of JNK signaling dramatically blocked Hippo pathway activation-induced matrix metalloproteinase 1 expression and cell invasion. Furthermore, we identify bantam-Rox8 modules as essential components downstream of Yorkie in mediating JNK-dependent cell invasion. Finally, we confirm that YAP (Yes-associated protein) expression negatively regulates TIA1 (Rox8 ortholog) expression and cell invasion in human cancer cells. Together, these findings provide molecular insights into Hippo pathway-mediated cell invasion and also raise a noteworthy concern in therapeutic interventions of Hippo-related cancers, as simply inhibiting Yorkie or YAP activity might paradoxically accelerate cell invasion and metastasis.
The Hippo pathway is a highly conserved tumor-suppressor pathway recently identified in Drosophila melanogaster via genetic screens for growth-regulating genes (1). In Drosophila, the core Hippo pathway acts through a serine-threonine kinase cascade, consisting of Hippo (Hpo) and Warts (Wts), to inactivate the transcriptional coactivator Yorkie (Yki) (2–7). Once the Hippo pathway is deactivated, Yki can translocate into the nucleus to interact with different DNA-binding transcription factors to initiate transcription of growth-regulating genes, including cyclin E (cycE), dmyc, bantam (ban), and Drosophila inhibitor of apoptosis protein 1 (Diap1) (1). However, despite the well-documented roles of the Hippo pathway in regulating various aspects of tumorigenesis, including cell growth, proliferation, and survival (1, 4–7), the role and underlying mechanism of Hippo signaling in tumor metastasis and cell invasion remains controversial. For one, overwhelming studies have shown hyperactivation of YAP (Yes-associated protein, Yki ortholog) in various human cancers (1, 7, 8), and YAP overexpression can promote cell invasion and the epithelial-mesenchymal transition (EMT) of cultured cells (9–13). Paradoxically, recent data suggest that YAP is silenced in a subset of highly aggressive human colorectal carcinomas (14), and acts as an inhibitor of cell invasion in some breast cancer cell lines (15). Furthermore, clinical data indicate that individuals affected by multiple myeloma with low YAP1 expression had a significant shorter survival than those with high YAP1 expression (16), suggesting that YAP also has a tumor-suppressor activity in some contexts.
To elucidate the Hippo pathway’s contradictory roles in regulating cell migration and invasion in vivo, we use Drosophila as a model to investigate the underlying mechanism. Here we show that Hippo pathway activation induces JNK-dependent cell invasion and EMT through ban miRNA, and identify Rox8 as an essential downstream mediator for Hippo activation-induced cell invasion.
Results and Discussion
Hippo Pathway Activation Promotes Cell Invasion.
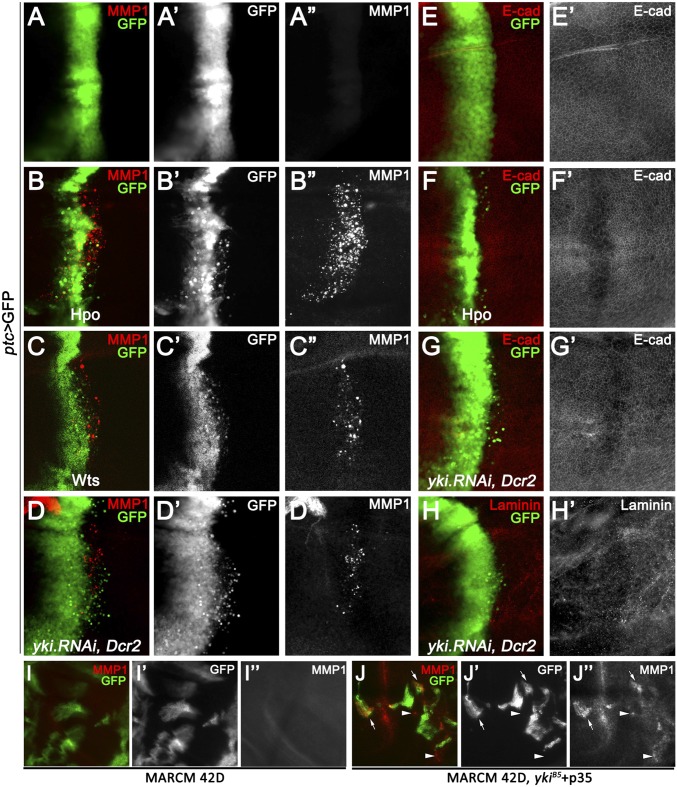
In the Drosophila wing epithelia, knocking down the cell polarity gene scribbled (scrib) along the anterior/posterior boundary using a patched-Gal4 (ptc-Gal4) driver produces an invasive migration phenotype (17–19), which has been used to model cell invasion in vivo. First, to investigate whether Hippo signaling activation could modulate cell invasion, we overexpressed Hpo or Wts, or knocked down yki, by ptc-Gal4. Compared with controls (Fig. 1A), activated Hpo signaling triggered invasive migration toward the posterior part of discs, a significant number of GFP+ cells detached and migrated away basally from the ptc expression domain (Fig. 1 B′–D′ and Fig. S1), along with up-regulated matrix metalloproteinase 1 (MMP1) expression (Fig. 1 B′′–D′′), a protein required for basement membrane degradation and cancer malignant transformation (20). Furthermore, activated Hippo signaling resulted in down-regulation of E-cadherin and Laminin (Fig. 1 E′–H′), two common molecular characteristics of EMT. Taken together, these results suggest that Hippo activation in epithelia cells promotes cell invasion and EMT.
Fig. 1.
Activation of Hpo signaling promotes cell invasion. (A–H) Fluorescence micrographs of wing discs are shown, anterior is to the Left, and cells are labeled with GFP expression. Compared with the control (A), ectopic expression of Hpo (B), or Wts (C), or loss of yki (D) induces cell invasion and MMP1 expression (red). (E–G) Compared with the control (E), cells expressing Hpo or knocking down yki show reduced E-cadherin expression (F and G). (H) yki-depleted cells show reduced Laminin B1 expression. (I–J) Compared with wild-type MARCM clones (I), yki mutant clones coexpressing p35 induces autonomous (arrow) and nonautonomous (arrowhead) MMP1 activation (J). [Magnification: (A–H) 20×; (I–J) 40×.] Genotypes: (A and E) ptc-Gal4 UAS-GFP/+; (B and F) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/+; (C) ptc-Gal4 UAS-GFP/+; UAS-Wts/tub-Gal80ts; (D, G, and H) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+; (I) UAS-GFP, hs-Flp; FRT42, tub-Gal80/FRT 42; tub-Gal4/+; and (J) UAS-GFP, hs-Flp; FRT42, tub-Gal80/FRT 42, ykiB5; tub-Gal4/+.
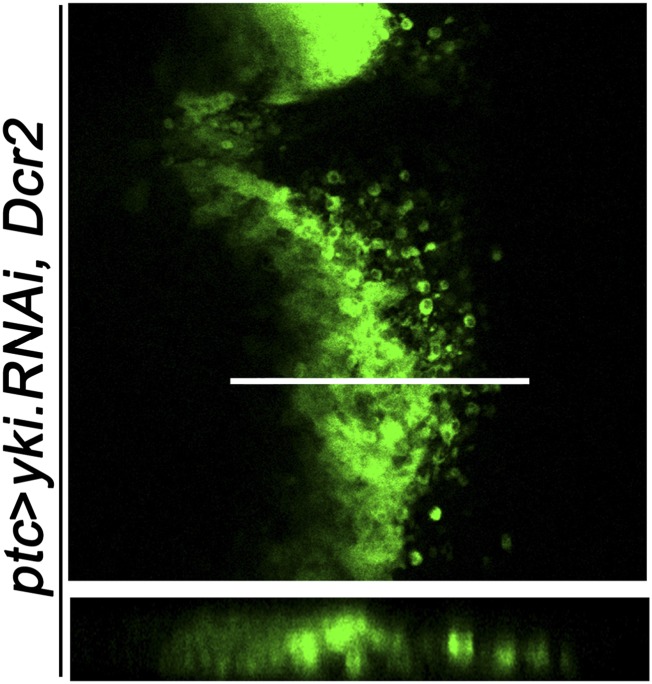
Fig. S1.
Loss of Yki induces basal cell invasion. Genotypes: ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+. The white line indicates the site of Z stack in bottom panel. (Magnification: 20×.)
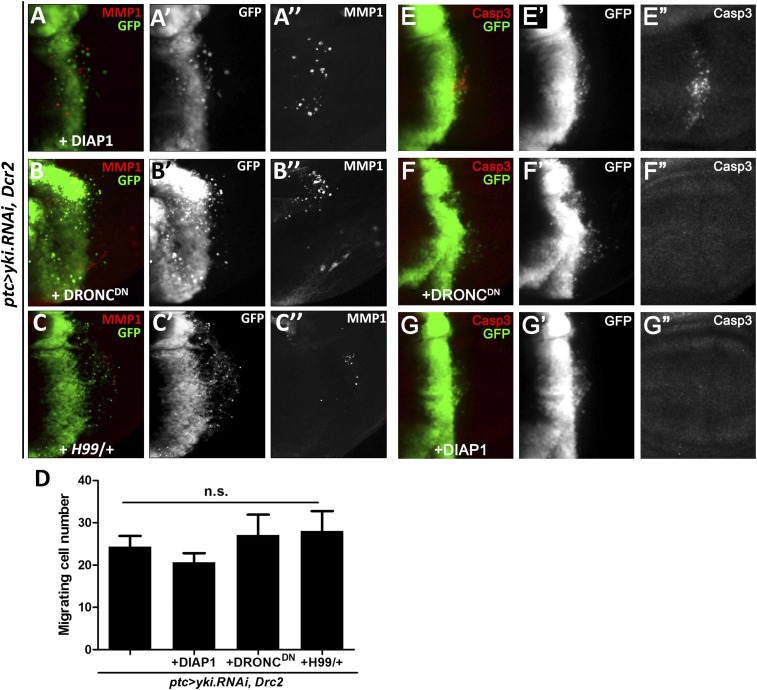
Hippo pathway activation regulates apoptosis through transcriptional regulation of DIAP1 (4, 5, 21). In accordance with this finding, we found that loss of yki induced strong apoptosis (Fig. S2B). Given that cell invasion is frequently accompanied with apoptosis (22), to test if Hippo pathway-induced cell invasion is a result of apoptosis, we blocked caspase activity by coexpressing DIAP1 or a dominant-negative form of the caspase-9 homolog Drosophila Nedd-2-like caspase (DRONC), or by the deficiency Df(3L)H99 that deletes three proapoptotic genes, reaper (rpr), head involution defective (hid), and grim. Interestingly, none of these alternations can significantly suppress loss of yki-induced cell invasion and MMP1 expression (Fig. S3 A–D), despite a complete inhibition of apoptosis (Fig. S3 E–G), confirming that the invasion behavior is not simply a secondary effect of apoptosis.
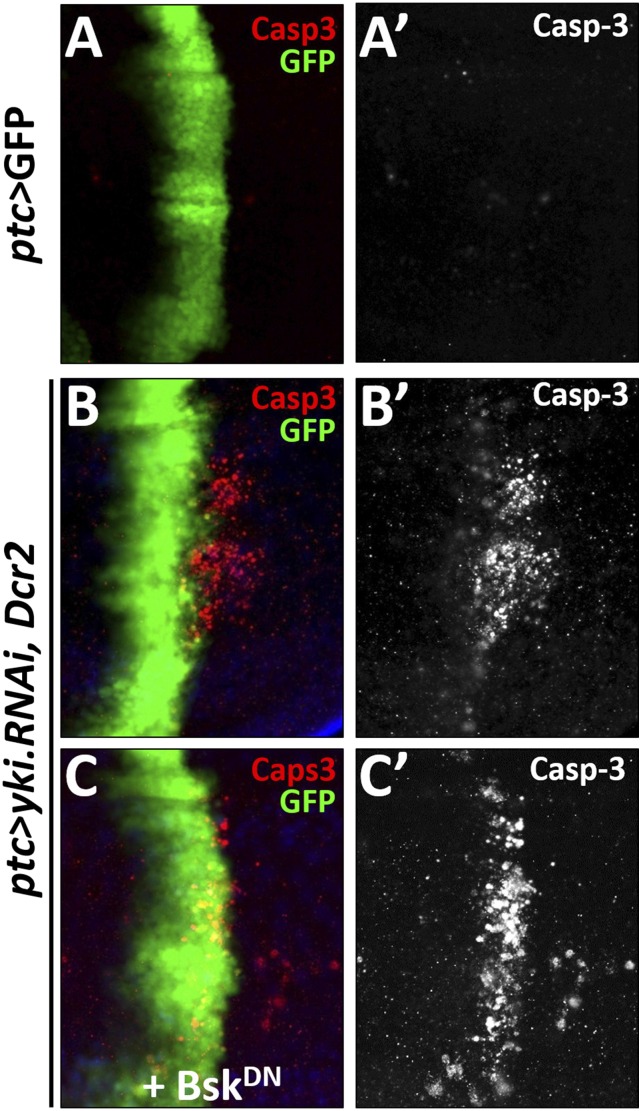
Fig. S2.
Inhibition of JNK signaling cannot suppress loss of yki-induced apoptosis. Compared with the control (A), loss of yki induced apoptosis, as indicated by caspase 3 staining (B), and remained unaffected by blocking JNK signaling (C). Genotypes: (A) ptc-Gal4 UAS-GFP/+. (B) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+. (C) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-BskDN. (Magnification: 20×.)
Fig. S3.
Hpo activation-induced cell invasion is not a secondary effect of cell death. Fluorescence micrographs of wing discs are shown, anterior is to the Left, and cells are labeled with GFP expression. (A–C) Depletion of yki induced cell invasion and MMP1 activation cannot be suppressed by coexpression of DIAP1 (A), DRONCDN (B), or losing one copy of all three apoptosis regulating genes, including rpr, hid, and grim (C). (D) Quantification of cell invasion phenotype induced by loss of yki. Results are shown as mean + SEM. P values were calculated using a one-way ANOVA by GraphPad Prism 5 (GraphPad Software Inc.) n.s. no significant difference. (E–G) Blocking Caspase activity via expressing DRONCDN or DIAP1-suppressed loss of yki-induced apoptosis. [Magnification: (A–C and E–G) 20×.] Genotypes: (A, G) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-DIAP1. (B, F) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-DRONCDN. (C) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/H99. (E) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+.
The induced MMP1 expression does not fully colocalized with GFP-labeled Hippo pathway-activating cells (Fig. 1 B–D), suggesting that the Hippo pathway triggers MMP1 activation both autonomously and nonautonomously. To confirm this theory, we blocked apoptosis by expressing p35 in yki mutant clones using the mosaic analysis with a repressible cell marker (MARCM) technique (23). We observed protrusions, like structure and distinct MMP1 activation, both cell autonomously (Fig. 1J, arrow) and nonautonomously (Fig. 1J, arrowhead). Taking these data together, we conclude that Hippo activation induces both autonomous and nonautonomous MMP1 activation and invasive behavior.
JNK Is Required for Hippo Activation-Induced Cell Invasion.
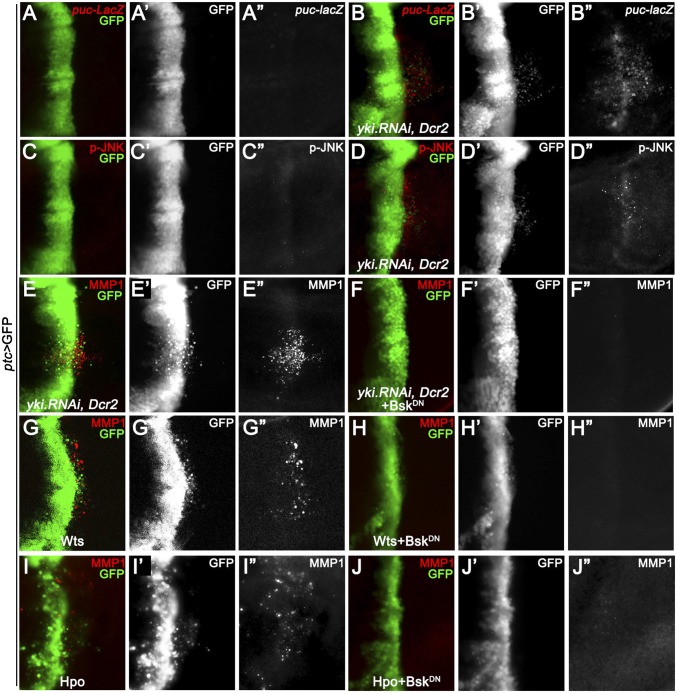
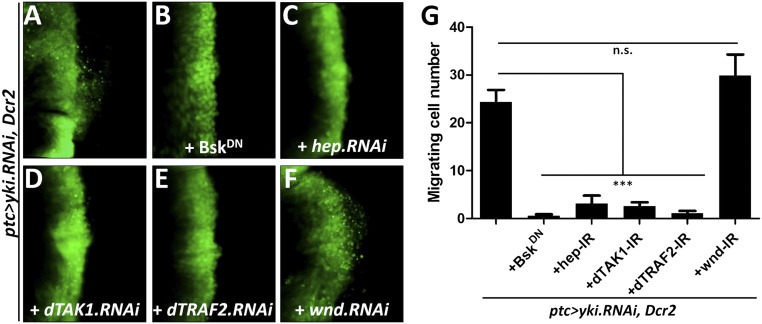
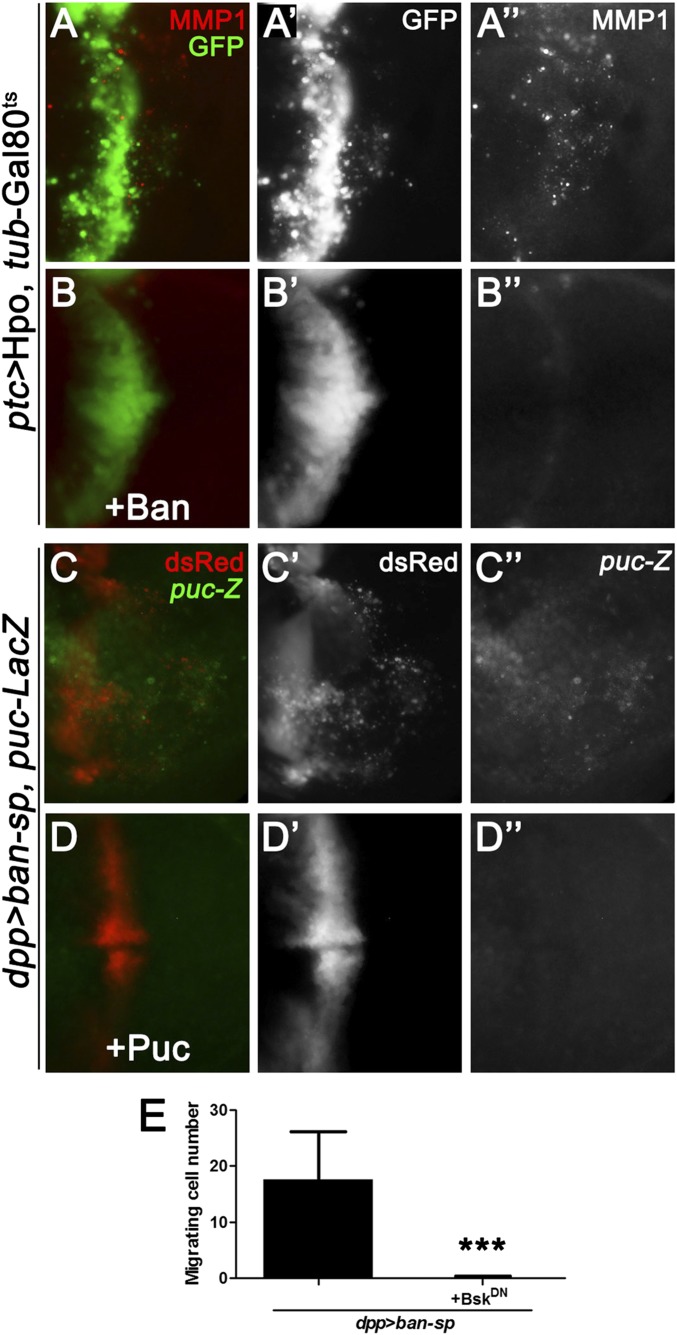
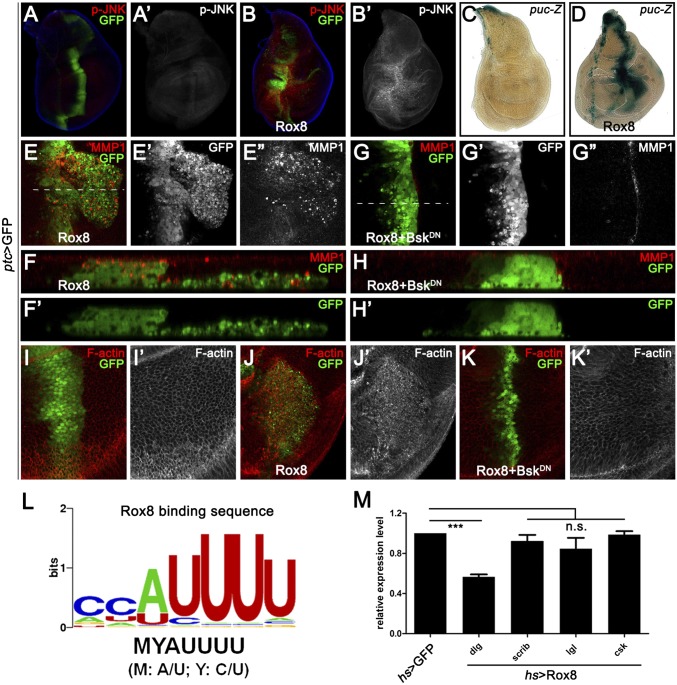
MMP1 is a direct transcriptional target of JNK signaling (24), and JNK activation has been shown to play a critical role in modulating cell invasion (17, 18, 24–26). Thus, we hypothesized that the Hippo pathway might regulate cell invasion through activating JNK signaling. To test this idea, we first checked the expression of puckered (puc), a transcriptional target and readout of JNK pathway activation (27). Compared with the control (Fig. 2A), knocking down Yki by ptc-Gal4 resulted in a significant up-regulation of puc-LacZ (Fig. 2B). Next, we examined JNK activation directly using an antibody specific to the phosphorylated JNK (p-JNK). Elevated p-JNK staining was detected upon Yki depletion (Fig. 2 C and D). Furthermore, we found blocking JNK signaling by expressing a dominant-negative form of basket (BskDN, bsk encodes the Drosophila JNK) completely impeded yki depletion, Wts or Hpo overexpression-induced cell invasion behavior, and MMP1 expression (Fig. 2 E–J). In addition, our genetic epistasis analysis showed that compromised JNK signaling by knocking down the JNK kinase hemipterous (hep) (27), JNKK kinase dTAK1 (28), or TNF receptor-associated factors 2 (dTRAF2) (29) all significantly suppressed Yki loss-induced invasion phenotype (Fig. S4 A–E and G), indicating Yki functions upstream of dTRAF2. As a negative control, inactivation of wallenda (wnd), a recently identified JNKK kinase that positively regulates cell invasion (18), failed to suppress the invasive behavior (Fig. S4 F and G). Intriguingly, blocking JNK signaling is not sufficient to suppress loss of yki-induced apoptosis (Fig. S2C). Together, these results suggest that JNK signaling is essential for activated Hippo signaling-induced cell invasion, but not apoptosis.
Fig. 2.
Bsk is essential for Hpo activation-induced cell invasion. Fluorescence micrographs of wing discs are shown, anterior is to the Left, and cells are labeled with GFP expression. (A–D) Depletion of yki by ptc-Gal4 up-regulates puc transcription (B) and JNK phosphorylation (D). (E–J) Expression of BskDN completely suppressed loss of yki (E), expression of Wts (G) or Hpo (I) induced cell invasion and MMP1 expression (F, H, and J). (Magnification: 20×.) Genotypes: (A) ptc-Gal4 UAS-GFP/+; pucE69/+; (B) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/pucE69; (C) ptc-Gal4 UAS-GFP/+; (D and E) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+; (F) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-BskDN; (G) ptc-Gal4 UAS-GFP/+; UAS-Wts/tub-Gal80ts; (H) ptc-Gal4 UAS-GFP/+; UAS-Wts/UAS-BskDN, tub-Gal80ts; (I) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/+; and (J) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/UAS-BskDN.
Fig. S4.
JNK signaling is required for Hippo activation-induced cell invasion. Fluorescence micrographs of wing discs are shown, anterior in all panels to the Left, and cells are labeled with GFP expression. (A–F) Inhibiting bsk (B), hep (C), dTAK1 (D), dTRAF2 (E), but not that of wnd (F), suppress loss of yki-induced cell invasion. (G) Quantification of cell invasion phenotype induced by loss of yki. Results are shown as mean + SEM. P values were calculated using a one-way ANOVA. ***P < 0.001; n.s., no significant difference. Genotypes: (A) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+. (B) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-BskDN. (C) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-hep.RNAi. (D) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-dTAK1.RNAi. (E) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-dTRAF2.RNAi. (F) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-wnd.RNAi. (Magnification: 20×.)
JNK Is Required for Hippo Activation-Induced Border Cell Migration.
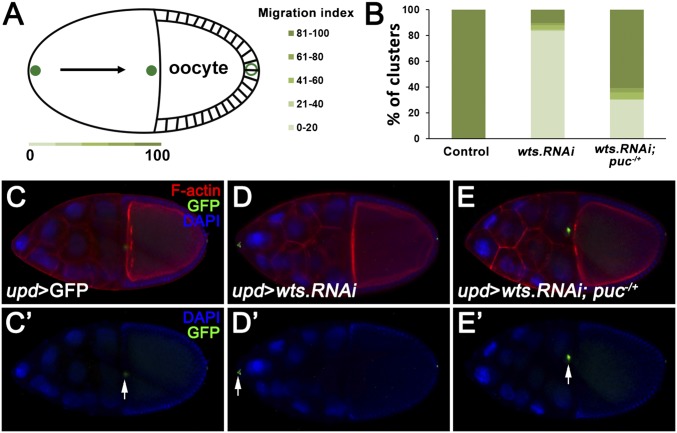
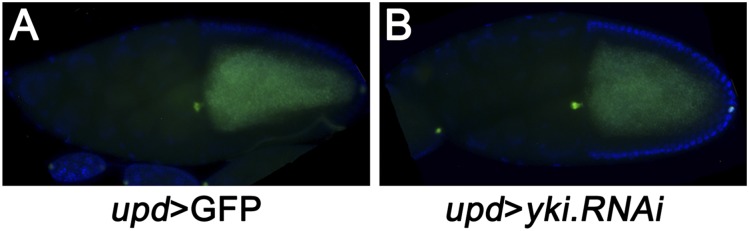
The epistasis data we present above compellingly suggest that Hippo modulates cell migration via JNK activation. Next, to investigate the physiological role of Hippo-JNK cross-talk in regulating cell migration, we turn to oogenesis, a developmental process where both JNK and Hippo are required for correct border cell migration (30–32). During normal development, the border cell cluster arrives at the nurse cell–oocyte boundary by stage 10 (Fig. 3A) (33), so we selected stage-10 egg chambers to test their genetic interactions. Consistent with previous data (31), we found knocking down wts expression in polar cells by upd-Gal4 severely disrupted border cell migration (Fig. 3 B–D). Nevertheless, enhancing JNK signaling by simultaneously deleting one copy of puc (34) significantly rescued the border cell migration defect (Fig. 3 B and E), suggesting that JNK signaling also acts downstream of the Hippo pathway in regulating border cell migration. Interestingly, despite that Yki overexpression phenocopies wts knockdown-induced migratory defect (31), we found inhibition of Yki activity under upd promoter is not sufficient to accelerate border cell migration (Fig. S5 A and B), which is consistent with a previous study (31).
Fig. 3.
Hippo pathway promotes JNK-dependent border cell migration in oogenesis. upd-Gal4 was used to overexpress or knockdown genes specifically in polar cells. (A) Stage-10 migration index for quantification of border cell migration. (B) Quantification of stage-10 migration index for the following genotypes: control (n = 43), upd > wts.RNAi (n = 109), and upd > wts.RNAi + puc−/+ (n = 109). (C–E) Compared with controls, knockdown of wts induced border cell migration defect (D) was rescued by deleting one copy of puc (E). (Magnification: 20×.) Genotypes: (C) upd-Gal4, UAS-GFP/+; (D) upd-Gal4, UAS-GFP/+; UAS-wts.RNAi; and (E) upd-Gal4, UAS-GFP/+; UAS-wts.RNAi/pucE69.
Fig. S5.
(A and B) yki inhibition does not accelerate border cell migration. Genotypes: (A) upd-Gal4, UAS-GFP/+. (B) upd-Gal4, UAS-GFP/+ ;UAS-yki.RNAi UAS-Dcr2/+. (Magnification: 20×.)
ban Is Essential for Loss of yki-Induced Cell Invasion.
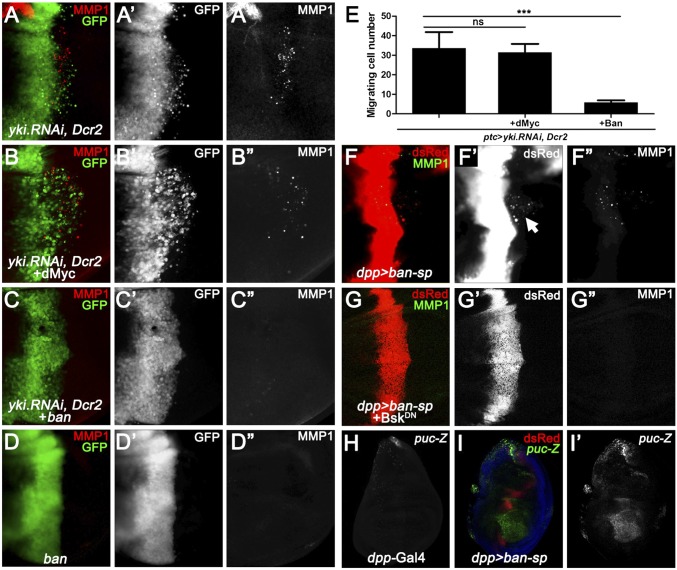
To investigate the molecular mechanism by which JNK mediates Hippo activation-induced cell invasion, we dissected the role of Yki target genes individually, including Diap1, dmyc, and ban (21, 35–37). Overexpression of DIAP1 or Myc fails to suppress loss of yki-induced invasion (Fig. 4 A, B, and E and Fig. S3 A and D), whereas ectopic ban expression strongly impedes ptc > yki.RNAi and ptc > Hpo-induced invasive phenotype and MMP1 expression (Fig. 4 C and E and Fig. S6 A and B), and expression of ban alone has no obvious invasive phenotype (Fig. 4D). On the other hand, when ban activity was reduced along the anterior/posterior boundary, significant number of cells migrated toward the posterior part (Fig. 4F′ and Fig. S6E), coupled with increased MMP1 expression (Fig. 4F′′) and JNK activation (Fig. 4 H and I), phenocopied loss of yki induced invasive behavior. More importantly, the cell invasion, MMP1 activation, and JNK activation phenotypes were all completely suppressed when JNK signaling was blocked (Fig. 4G and Fig. S6 C–E).
Fig. 4.
ban is essential for loss of yki-induced cell invasion. Fluorescence micrographs of wing discs are shown, anterior in all panels is to the Left, and cells are labeled with GFP expression. (A–D) Overexpression of ban (C), but not dMyc (B), impedes loss of yki-induced cell invasion and MMP1 expression, whereas expression of ban alone gives no obvious invasion phenotype (D). (E) Quantification data of cell invasion phenotype in A–C. Data are presented as mean + SEM. P values were calculated using a one-way ANOVA. ***P < 0.001; n.s., no significant difference. (F–H) Expression of ban sponge driven by dpp promoter induces mild cell invasion, MMP1 expression (F), and intensive puc transcription (I). (G) Blocking JNK activity dramatically suppresses loss of ban induced cell invasion and MMP1 activation. [Magnification: (A–D and F–I) 20×.] Genotypes: (A) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/+; (B) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-dMyc; (C) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/banEP3622; (D) ptc-Gal4 UAS-GFP/+; banEP3622/+; (F) dpp-Gal4 UAS-ban-sponge/+; (G) dpp-Gal4 UAS-ban-sponge/UAS-BskDN; (H) dpp-Gal4/pucE69; and (I) dpp-Gal4 UAS-ban-sponge/pucE69.
Fig. S6.
Ban is essential for Hpo-induced cell invasion. Fluorescence micrographs of wing discs are shown, anterior is to the Left, and cells are labeled with GFP (A and B) or dsRed (C and D). Ectopic Hippo expression induced cell invasion (A′) and MMP1 activation (A′′) were both dramatically suppressed by coexpression of Ban (B–B′′). Loss of ban induced puc activation (C) were completely suppressed by inhibiting JNK signaling (D). (E) Quantification of Fig. 4 F′, G, and G′. Results are shown as mean + SEM. P values were calculated using Student’s t test. ***P < 0.001. Genotypes: (A) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/+. (B) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/UAS-Ban. (C) dpp-Gal4, UAS-ban-sp/pucE69. (D) dpp-Gal4, UAS-ban-sp/UAS-Puc, pucE69. (Magnification: 20×.)
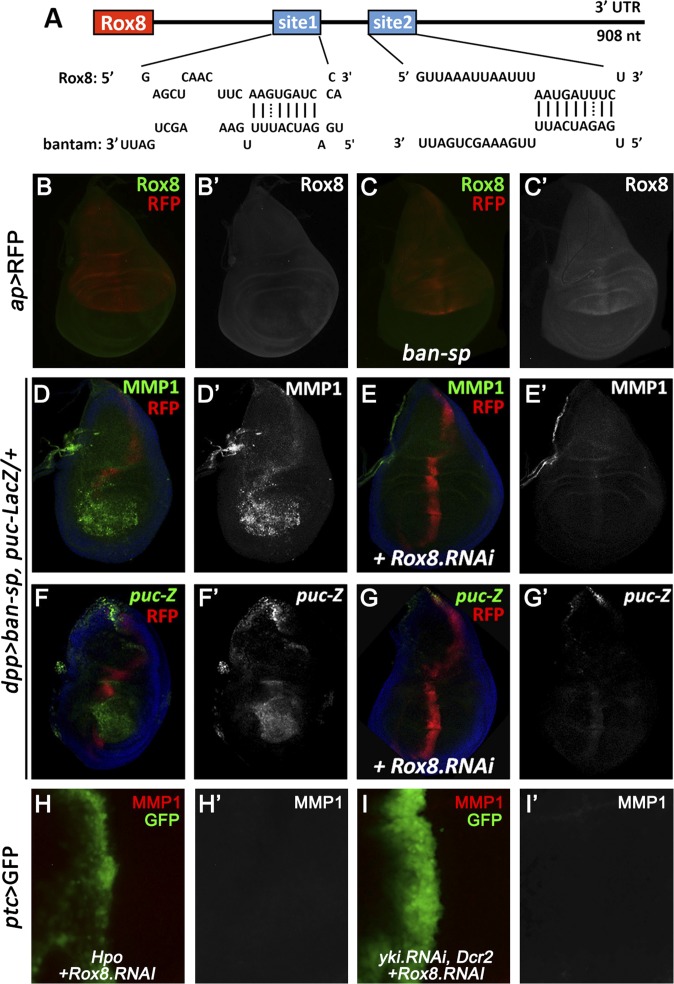
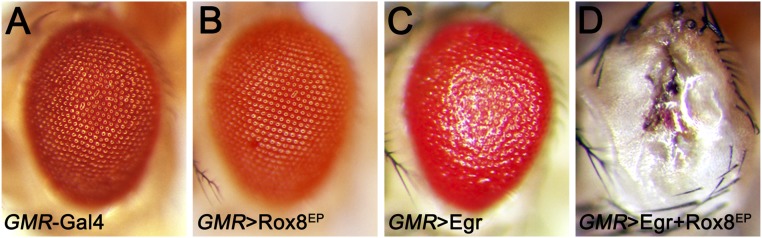
Next, to explore the detailed mechanism by which ban miRNA regulates JNK-mediated cell invasion, we checked the predicted ban binding targets by using an available algorithm called microRNA.org (38). Among all of the candidates, we called specific attention to one gene, Rox8, which harbors two potential ban binding targets in its 3′UTR region (Fig. 5A). Rox8 encodes a RNA-binding protein that controls important aspects of development, including alternative splicing and stress granule formation (39, 40). In addition, we have previously performed an unbiased genetic screen for factors modulating JNK signaling (41), and identified Rox8 as a positive regulator of JNK signaling for Rox8 expression synergistically enhances Egr-induced JNK-dependent cell death (Fig. S7). Importantly, consistent with the computational prediction, we found knocking down ban significantly up-regulates Rox8 protein level (Fig. 5 B and C). Furthermore, depletion of Rox8 dramatically suppressed loss of ban-induced cell invasion, MMP1 expression, and JNK activation (Fig. 5 D–G), as well as Hippo pathway activation-induced cell invasion and MMP1 expression (Fig. 5 H and I). These data indicate that ban-Rox8 signaling constitutes an essential module downstream of Yki in regulating JNK-mediated cell invasion.
Fig. 5.
ban down-regulates Rox8 to regulate cell invasion. (A) Schematic drawing of the 3′UTR regions of Rox8 gene highlighting the ban seed sites. (B and C) Compared with controls (B), knocking down ban by ap-Gal4 significantly up-regulated Rox8 protein level (C). (D–I) Fluorescence micrographs of wing discs are shown, anterior in all panels is to the Left. Loss of ban induced MMP1 expression (D′) and JNK activation (F′) were both completely suppressed by knocking down Rox8 activity (E′ and G′). Reducing Rox8 activity impeded Hpo overexpression or loss of yki-induced cell invasion (H and I). (Magnification: 20×.) Genotypes: (B) ap-Gal4 UAS-RFP/+; (C) ap-Gal4 UAS-RFP/+; UAS-ban-sponge; (D and F) dpp-Gal4 UAS-ban-sponge/pucE69; (E and G) dpp-Gal4 UAS-ban-sponge/UAS-Rox8.RNAi, pucE69; (H) ptc-Gal4 UAS-GFP/UAS-Hpo; tub-Gal80ts/UAS-Rox8.RNAi; and (I) ptc-Gal4 UAS-GFP/+; UAS-yki.RNAi, UAS-Dcr2/UAS-Rox8.RNAi.
Fig. S7.
Rox8 is a positive regulator of JNK signaling in Drosophila. Light micrographs of adult Drosophila eye are shown. Compared with the GMR-Gal4 control (A), mild JNK activation in the eye by ectopic Egr expression results in cell death and a rough eye phenotype (C). Although expression of Rox8 alone has no obvious phenotype (B), it dramatically enhanced the GMR > Egr eye phenotype (D), Genotypes: (A) GMR-Gal4/+. (B) GMR-Gal4/+; Rox8EP/+. (C) GMR-Gal4/+; UAS-Egr/+. (D) GMR-Gal4/+; UAS-Egr/Rox8EP. (Magnification: 10×.)
YAP Negatively Regulates TIA1 and Suppress Cell Invasion.
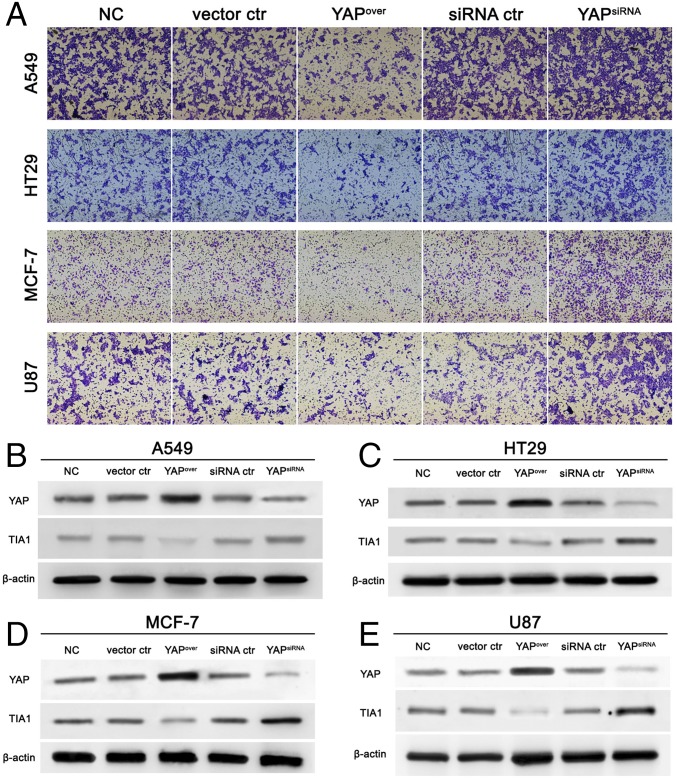
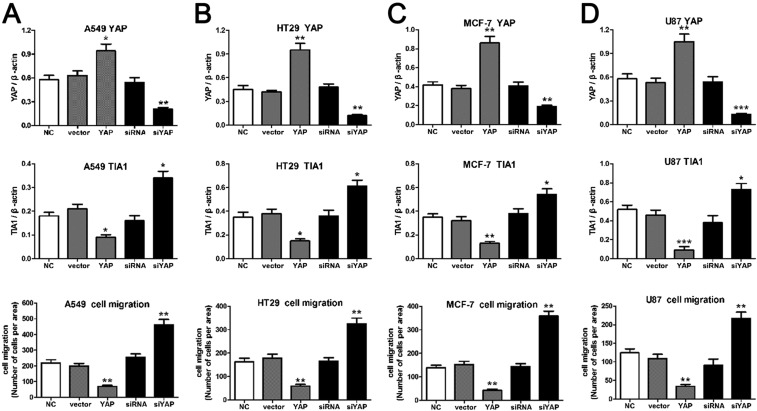
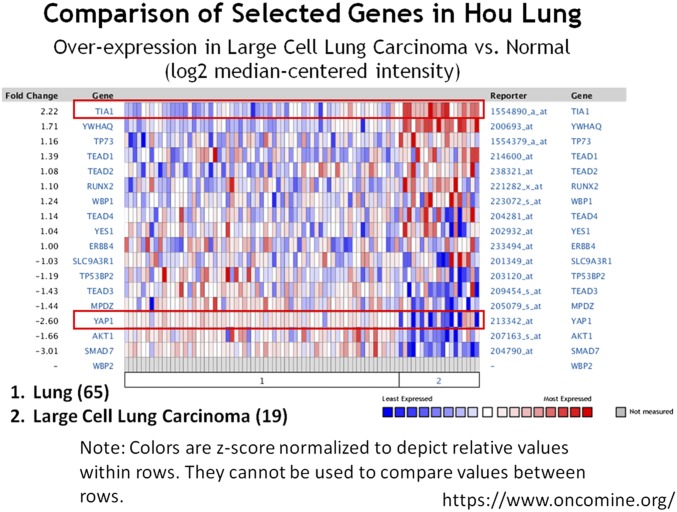
Having demonstrated that Hippo activation promotes cell invasion through inhibiting Rox8 in Drosophila, we next asked whether the Hippo pathway retains a conserved role in mammals. We examined various cancer cell lines of different origins, including lung (A549), colon (HT29), breast (MCF-7), and brain (U87), and generated stable cell lines with increased or decreased YAP expression using lentivirus (Fig. 6 and Fig. S8). We found YAP overexpression significantly decreases cell invasion in all cancer cell lines, as shown by a transwell assay (Fig. 6 and Fig. S8). Conversely, inhibition of YAP activity significantly increases invasion (Fig. 6 and Fig. S8). More importantly, we further showed that ectopic YAP significantly decreases, whereas YAP knockdown increases TIA1 (Rox8 ortholog) protein level (Fig. 6 and Fig. S8). Intriguingly, after analyzing tumor microarray data from the ONCOMINE database (https://www.oncomine.org/index.jsp), we also found a negative correlation between YAP1 and TIA1 levels in both normal lung cells and large cell lung carcinoma (Fig. S9). Together, these data suggest that apart from its tumor-promoting role, YAP can also function as an invasion suppressor.
Fig. 6.
YAP negatively regulates cell invasion and TIA1 expression. Stable YAP overexpression increased, whereas YAP depletion decreased cell migration (A) and TIA1 protein level (B–E) in cancer cell lines A549, HT29, MCF-7, and U87. NC, negative control; siRNA ctr: control siRNA lentivirus vector; vector ctr, empty lentivirus vector; YAPover, YAP expression lentivirus vector; YAPsiRNA: YAP knockdown lentivirus vector. [Magnification: (A) 100×.]
Fig. S8.
YAP negatively regulates cell invasion and TIA1 expression. (A–D) Quantification data of Western blot and cell invasion shown in Fig. 6. Results are shown as mean + SEM. P values were calculated using a one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. S9.
Negative correlation between YAP and TIA1. Heat map of coexpression profile of TIA1 and Hippo pathway genes in large cell lung carcinoma with normal lung study (Oncomine database). TIA1 and YAP1 are highlighted in red boxes.
Rox8 Induces JNK-Dependent Cell Invasion.
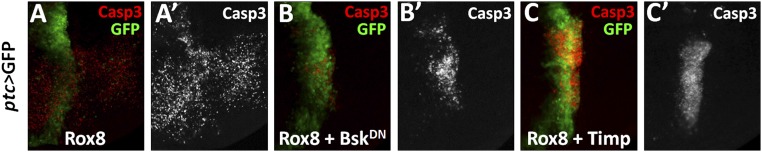
In accordance with the physiological role of Rox8 in yki loss-induced cell invasion, we found overexpression of Rox8 is sufficient to induce JNK activation (Fig. 7 A–D), MMP1 expression (Fig. 7E), and dramatic basal side invasion of the wing epithelium (Fig. 7F), which can be strongly suppressed by blocking JNK signaling (Fig. 7 G and H). Consistent with the notion that invasive behavior is associated with disruption of epithelial integrity, Rox8-expressing cells exhibited increased actin accumulation (Fig. 7J), which was also suppressed by BskDN expression (Fig. 7K). Conversely, we found Rox8-triggered apoptosis remained unaffected by BskDN or expression of Timp (MMP inhibitor), even though the cell invasion behavior was completely impeded (Fig. S10), indicating that Rox8-induced cell invasion is also uncoupled from cell death. Taken together, these data suggest that JNK signaling is indispensable for Rox8 induced cell invasion, but not cell death.
Fig. 7.
Rox8 induces JNK-dependent cell invasion. (A–D) Expression of Rox8 activates JNK phosphorylation (B) and puc transcription (D). (E–H) Ectopic Rox8-induced MMP1 expression (E′) and basal cell invasion (F) and F-actin are dramatically suppressed by coexpression of BskDN (G, and H). Compared with control (I), Rox8 expression-induced F-actin accumulation (J) is completely suppressed by JNK inhibition (K). (L) Rox8 binding sequence is shown. (M) Heat shock-induced Rox8 expression decreases dlg mRNA level, whereas scrib, lgl, and Csk mRNA levels remained unchanged, expression data were normalized to one using hs > GFP as the control. Results are shown as mean + SEM. P values were calculated using a one-way ANOVA. ***P < 0.001; n.s., no significant difference. [Magnification: (A–D) 10×; (E–K) 20×.] Genotypes: (A and I) ptc-Gal4 UAS-GFP/+; (B, E, F, J, and M–O) ptc-Gal4 UAS-GFP/+; UAS-Rox8/+; (C) ptc-Gal4 UAS-GFP/+; pucE69/+; (D) ptc-Gal4 UAS-GFP/+; UAS-Rox8/pucE69; and (G, H, and K) ptc-Gal4 UAS-GFP/+; UAS-Rox8/UAS-BskDN.
Fig. S10.
Rox8-induced apoptosis is independent of JNK signaling and cell invasion. Fluorescence micrographs of wing discs are shown, anterior is to the Left, and cells are labeled with GFP expression. Ectopic Rox8 expression induced apoptosis (A) remained unaffected by inhibiting JNK signaling (B) or coexpression of Timp (C), although cell invasion behavior was dramatically suppressed. Genotypes: (A) ptc-Gal4 UAS-GFP/+; UAS-Rox8/+. (B) ptc-Gal4 UAS-GFP/+; UAS-Rox8/UAS-BskDN. (C) ptc-Gal4 UAS-GFP/+; UAS-Rox8/UAS-Timp. (Magnification: 20×.)
Given RNA-binding proteins can directly bind to a specific sequence in mRNA to regulate its stabilization and translation to affect cancer progression (42), and taking into account that Rox8 is a positive regulator of JNK signaling, we cautiously examined the 3′UTR region of several well-known negative regulators of JNK signaling, including cell polarity complex components, disk large (dlg), lethal giant larvae (lgl), scrib, as well as C-terminal Src kinase (Csk). Interestingly, we identified five Rox8 putative binding sites (Fig. 7L) (CISBP-RNA Database) (43) in the 3′UTR region of dlg, two in scirb, and one in lgl and Csk, indicating that Rox8 may also regulate mRNA level of those genes. To test this theory, we expressed Rox8 under the heat shock (hs) promoter in Drosophila and examined the mRNA level of these candidate genes. Remarkably, we found only the dlg mRNA level was significantly decreased after Rox8 expression, whereas scrib, lgl, and Csk mRNA levels remained unaffected (Fig. 7M). Because it has been shown previously that loss of dlg under the ptc promoter can also induce JNK-dependent cell invasion (44), we conclude that Rox8 expression decreases Dlg, which in turn activates JNK-mediated cell invasion.
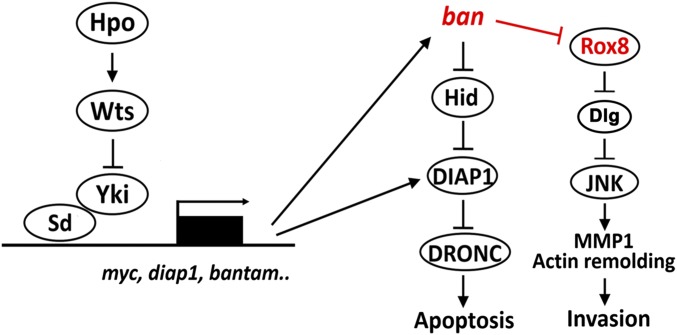
Drosophila has been widely considered as an excellent organism to address cancer-related problems for the past decade (45, 46), several in vivo cell invasion and metastasis models were established to dissect genetic details of cancer progression (22, 47–49). Here, using Drosophila wing epithelium as a major model, we bring forward an interesting model that elevated Hippo signaling positively regulates cell migration/invasion through ban-Rox8 module-mediated JNK activation (Fig. S11). Moreover, we also demonstrated a conserved role of YAP in regulating cell invasion and TIA1 expression (Fig. 6). Consistent with our data, both TIA1 and YAP have nucleo-cytoplasmic shuttling ability (7, 50, 51). Interestingly, recent studies found that hpo and wts are both required for border cell migration during oogenesis (31, 32), highlighting the importance of cell migration promoting roles of the Hippo pathway. On the basis of that finding, our data further demonstrate that JNK acts downstream of Hippo pathway in inducing normal border cell migration (Fig. 3). Finally, given the fact that YAP is being considered as an important drug target (8, 52–54), our evidence presented herein offers a wake-up call for the therapeutic interventions of Hippo pathway-related cancers because inhibiting Yki (YAP) activity may paradoxically accelerate cell invasion.
Fig. S11.
A model illustrating how Hippo pathway regulates JNK-dependent cell invasion.
Materials and Methods
Drosophila Strains and Husbandry.
All crosses were reared on standard Drosophila media at 25 °C first; 1 d after egg laying, the F1 generations were shifted to a 29 °C incubator unless indicated otherwise. For experiments involving Hpo and Wts overexpression, tub-Gal80ts was used, flies were first raised at 18 °C to restrict Gal4 activity for 5 d, then shifted to 29 °C for 2 d to inactivate Gal80ts. The following strains were used for this study: ptc-Gal4, UAS-GFP, UAS-Dcr2, tub-Gal80ts, nd Rox8EP were obtained from the Bloomington Stock Center; UAS-yki RNAi (#40497) was obtained from the Vienna Drosophila RNAi Center; UAS-Rox8 (GS17980) was a GS line obtained from the Kyoto Drosophila Genetic Resource Center; UAS-DIAP1, UAS-DRONCDN, Df(3L)H99, UAS-hep RNAi, UAS-BskDN (41), UAS-Egr, UAS-dTAK1 RNAi, UAS-dTRAF2 RNAi (17), UAS-wnd RNAi (18), UAS-Ban (55) were previously described; and UAS-dMyc (gift from Peter Gallant, University of Würzburg, Wuerzburg, Germany), UAS-ban-sp (bantam sponge, gift from Marco Milán, The Barcelona Institute of Science and Technology, Barcelona), UAS-Hpo and UAS-Wts (gifts from Shian Wu, Nankai University, Tianjin, China), upd > GFP (gift from Erika A. Bach, New York University School of Medicine).
Clonal Analysis.
yki mutant clones were generated by crossing hs-FLP; FRT 42D, tub-Gal80; tub-Gal4, UAS-GFP with FRT 42D, ykiB5; UAS-p35/SM6-TM6B. Flp-out ectopic expression clones were generated by crossing UAS-transgenes with y w hs-FLP;act > y+ >Gal4 UAS-GFP. Clones were induced at the second instar: heat shock for 6 min at 37 °C 48–72 h after egg laying, and dissection was performed 36 h or 72 h after clone induction.
Immunostaining and X-Gal Staining.
Third-instar larvae wing discs were dissected in cold PBS and fixed in freshly made 4% (wt/vol) paraformaldehyde and stained as described previously (18). A detailed description of antibodies used in this study is provided in the SI Materials and Methods.
Analysis of Border Cell Migration.
Stage-10 egg chambers were selected and analyzed as previously described (31). As an index for migration, stage-10 egg chambers were categorized based on the location of the border cell cluster as depicted in Fig. 3A. Fig. 3B was generated with Excel (Microsoft).
Cell Invasion Assay.
A total of 50,000 cells in suspension with trypsin treatment were added to the upper well of transwell chambers and incubated at 37 °C in 5% CO2 for 48 h. The bottom chamber contained medium with 10% (vol/vol) FBS to serve as a chemoattractant. Cells that had invaded to the lower surface were fixed in 10% (vol/vol) formalin at room temperature for 30 min, stained with 0.05% Crystal violet, and counted by light microscopy. Mean invasion cells and SD were calculated. Invasion assays were performed in triplicate in three independent experiments. Mammalian cell culture, Western blot analysis, and quantitative real-time PCR are described in SI Materials and Methods.
Statistical Analysis.
Quantification of the data was presented in bar graphs created with Graphpad Prism 5 (GraphPad Software). Data represent mean values + SD. P values were calculated using a one-way ANOVA with corrected Bonferroni multiple comparison tests to calculate statistical significance.
SI Materials and Methods
Immunostaining and X-Gal Staining.
Primary antibodies used: mouse anti-MMP1 [1:100; Developmental Studies Hybridoma Bank (DSHB)], mouse anti–β-Gal (1:100; DSHB), rat anti–DE-cadherin (1:100; DSHB), rabbit anti-Laminin B1 (1:40; Abcam), rabbit anti–phospho-JNK (1:200; Calbiochem), rabbit antiactive Caspase 3 (1: 400; Cell Signaling Technology), rabbit anti-Rox8 (kind gift from Henri-Marc Bourbon, Université Paul Sabatier Toulouse III, Auvergne, France). Secondary antibodies: anti-rabbit Alexa (1:1,000; Cell Signaling Technology), anti-mouse Cy3 and anti-Rat Cy3 (1:1,000; Jackson Immuno Research).
For X-Gal staining, wing discs were dissected from third-instar larvae in PBST (1× PBS pH 7.0, 0.1% Triton X-100) and stained for β-galactosidase activity (Tinagen Biotech).
Mammalian Cell Culture.
The lung cancer cell line A549, colorectal adenocarcinoma cell line HT29, breast cancer cell line MCF-7, and the glioblastoma cell line U87 were cultured in RPMI medium 1640 (Invitrogen) containing 10% FBS (HyClone) at 37 °C in a humidified incubator of 5% CO2. For lentiviral packaging, HEK-293T cells were cotransfected with the lentiviral vector, and packaging vectors psPAX2 and PMD2G using LipoFiter Liposomal Transfection Reagent (Hanbio). YAP overexpression and YAP shRNA stable cell lines were made by transduction with pHBL-CMV-ZsGreen-Puro vector (Hanbio). The targeting sequence of control shRNA was CTAGGTATACTCCAGCTGGCA, and YAP shRNAwas CAGGTGATACTATCAACC AAA.
Western Blot Analysis.
Cells were lysed in RIPA Extraction reagents (Pierce) and total protein in the cell lysate was quantified using a BCA protein quantification kit (Pierce). Equal amounts of protein (20 μg) from each condition were separated by 10% SDS/PAGE and electrotransferred onto polyvinylidene difluoride membrane (Amersham Bioscience). The membrane was blocked with 5% nonfat dry milk in TBST buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature, then washed four times in TBST and incubated with the primary antibodies overnight at 4 °C. The membrane was washed three times in PBST and then incubated with secondary antibody. β-Actin was used as a loading control. Protein bands were visualized using ECL chemiluminescent reagent (#34080 and #34095; Thermo Fisher) and detected by the ChemiDOC MP Imaging System (Bio-Rad) and quantified using ImageJ (NIH). The following primary antibodies were used: TIA1 (1:1,500; Abcam, # ab140595), YAP (1:2,000; Abcam, # ab52771), β-actin (1:2,000; Cell Signaling Technology, # ab119716). The following secondary antibody is used: anti-rabbit IgG (1:2,000; Abcam, # ab6720).
Quantitative Real-Time PCR.
Larvae of indicated genotypes were heat shock at 37 °C for 30 min and recovered for 2 h at room temperature, total RNA was then extracted with wing discs of indicated genotype using PureLink RNA Mini Kit (Ambion). Total RNA was reverse-transcribed into cDNA with the PrimeScript RT Master Mix (Takara); and quantitative PCR was performed with SYBR Premix ExTaq II (Takara) and quantified by the Stratagene MX3000P system (Stratagene). RP49 was used as an internal control. The following primer sequences were used for real-time PCR.
lgl: TGACGACCACCACTTTGTGT, CTTTGTTGGTTTGCGTGCTA
Dlg: ACCTGGAGAACGTAACGCAC, ATGCACCTGACTTTGGCTCT
Csk: GCAGAAGTTTCTGGCTGAGG, GATGCTTGCTGGTGAAGACA
Scrib: GTTTCACGCAATGATATACCCGA, GGTCAGGTTCTTTAGCTGGGA
Rp49: CCACCAGTCGGATCGATATGC, CTCTTGAGAACGCAGGCGACC
Acknowledgments
We thank Duojia Pan, Shian Wu, Peter Gallant, Marco Milán, Henri-Marc Bourbon, the Bloomington Stock Center, Vienna Drosophila RNAi Center, Kyoto Drosophila Genetic Resource Center and the Core Facility of Drosophila Resource and Technology, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, for providing fly stocks and reagents; Dr. Hilal Kazan for helping finding the Rox8 binding motif; and Duojia Pan and Tian Xu. This work was supported by the National Natural Science Foundation of China (31371490, 31571516, 31601024, 81572501, and 81572626) and the Shanghai Committee of Science and Technology (09DZ2260100). Part of the work was conducted in Duojia Pan laboratory at the Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, and in the Tian Xu laboratory at Department of Genetics, Howard Hughes Medical Institute, Yale University School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621359114/-/DCSupplemental.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9(5):534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 3.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121(4):1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 4.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114(4):457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 5.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114(4):445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 6.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5(10):914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122(3):421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103(33):12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamar JM, et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci USA. 2012;109(37):E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629. doi: 10.1038/ncomms5629. [DOI] [PubMed] [Google Scholar]
- 12.Nallet-Staub F, et al. Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol. 2014;134(1):123–132. doi: 10.1038/jid.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau AN, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 2014;33(5):468–481. doi: 10.1002/embj.201386082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan M, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15(11):1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- 16.Cottini F, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20(6):599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X, et al. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death Differ. 2014;21(3):407–415. doi: 10.1038/cdd.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X, et al. Rho1-Wnd signaling regulates loss-of-cell polarity-induced cell invasion in Drosophila. Oncogene. 2015;35(7):846–55. doi: 10.1038/onc.2015.137. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, et al. Src42A modulates tumor invasion and cell death via Ben/dUev1a-mediated JNK activation in Drosophila. Cell Death Dis. 2013;4:e864. doi: 10.1038/cddis.2013.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14(3):388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006;10(1):33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 24.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006;25(22):5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16(11):1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 26.Ma X, et al. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Dev Biol. 2013;380(2):211–221. doi: 10.1016/j.ydbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Agnès F, Suzanne M, Noselli S. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development. 1999;126(23):5453–5462. doi: 10.1242/dev.126.23.5453. [DOI] [PubMed] [Google Scholar]
- 28.Takatsu Y, et al. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol Cell Biol. 2000;20(9):3015–3026. doi: 10.1128/mcb.20.9.3015-3026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue L, et al. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Llense F, Martín-Blanco E. JNK signaling controls border cell cluster integrity and collective cell migration. Curr Biol. 2008;18(7):538–544. doi: 10.1016/j.cub.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Lin TH, Yeh TH, Wang TW, Yu JY. The Hippo pathway controls border cell migration through distinct mechanisms in outer border cells and polar cells of the Drosophila ovary. Genetics. 2014;198(3):1087–1099. doi: 10.1534/genetics.114.167346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas EP, et al. The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J Cell Biol. 2013;201(6):875–885. doi: 10.1083/jcb.201210073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 34.Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5(5):811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 35.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126(4):767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Ziosi M, et al. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6(9):e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19(4):507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katzenberger RJ, Marengo MS, Wassarman DA. Control of alternative splicing by signal-dependent degradation of splicing-regulatory proteins. J Biol Chem. 2009;284(16):10737–10746. doi: 10.1074/jbc.M809506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khong A, Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J Virol. 2011;85(4):1439–1451. doi: 10.1128/JVI.02220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, et al. NOPO modulates Egr-induced JNK-independent cell death in Drosophila. Cell Res. 2012;22(2):425–431. doi: 10.1038/cr.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordero JB, et al. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev Cell. 2010;18(6):999–1011. doi: 10.1016/j.devcel.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastor-Pareja JC, Xu T. Dissecting social cell biology and tumors using Drosophila genetics. Annu Rev Genet. 2013;47:51–74. doi: 10.1146/annurev-genet-110711-155414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willoughby LF, et al. An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis Model Mech. 2013;6(2):521–529. doi: 10.1242/dmm.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dar AC, Das TK, Shokat KM, Cagan RL. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature. 2012;486(7401):80–84. doi: 10.1038/nature11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302(5648):1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 49.Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;22(21):5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang T, Delestienne N, Huez G, Kruys V, Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J Cell Sci. 2005;118(Pt 23):5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- 51.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan D. YAPing Hippo forecasts a new target for lung cancer prevention and treatment. J Clin Oncol. 2015;33(20):2311–2313. doi: 10.1200/JCO.2015.61.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen HY, et al. R331W missense mutation of oncogene YAP1 is a germline risk allele for lung adenocarcinoma with medical actionability. J Clin Oncol. 2015;33(20):2303–2310. doi: 10.1200/JCO.2014.59.3590. [DOI] [PubMed] [Google Scholar]
- 54.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161(4):1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]