Significance
Muscle wasting as occurs with disuse, spinal injuries, aging, and many diseases (including cancer, sepsis, and renal failure) results primarily from the accelerated destruction of the myofibrillar apparatus, although the molecular mechanisms for this effect are largely unclear. To gain insight into the sequence of events leading to myofibril destruction, we studied the atrophy induced by denervation. We found that phosphorylation and ubiquitination of the desmin cytoskeleton precede its depolymerization, which eventually causes myofibril destruction. We further uncovered a delayed phase in the atrophy process, which involves the induction of genes that facilitate myofibril breakdown, including the AAA-ATPase p97/VCP, by the transcription factor paired box 4 (PAX4). Consequently, desmin phosphorylation, p97/VCP, and PAX4 may represent new therapeutic targets to reduce myofibril breakdown during atrophy.
Keywords: PAX4, muscle atrophy, desmin, ubiquitin, Trim32
Abstract
A hallmark of muscle atrophy is the excessive degradation of myofibrillar proteins primarily by the ubiquitin proteasome system. In mice, during the rapid muscle atrophy induced by fasting, the desmin cytoskeleton and the attached Z-band–bound thin filaments are degraded after ubiquitination by the ubiquitin ligase tripartite motif-containing protein 32 (Trim32). To study the order of events leading to myofibril destruction, we investigated the slower atrophy induced by denervation (disuse). We show that myofibril breakdown is a two-phase process involving the initial disassembly of desmin filaments by Trim32, which leads to the later myofibril breakdown by enzymes, whose expression is increased by the paired box 4 (PAX4) transcription factor. After denervation of mouse tibialis anterior muscles, phosphorylation and Trim32-dependent ubiquitination of desmin filaments increased rapidly and stimulated their gradual depolymerization (unlike their rapid degradation during fasting). Trim32 down-regulation attenuated the loss of desmin and myofibrillar proteins and reduced atrophy. Although myofibrils and desmin filaments were intact at 7 d after denervation, inducing the dissociation of desmin filaments caused an accumulation of ubiquitinated proteins and rapid destruction of myofibrils. The myofibril breakdown normally observed at 14 d after denervation required not only dissociation of desmin filaments, but also gene induction by PAX4. Down-regulation of PAX4 or its target gene encoding the p97/VCP ATPase reduced myofibril disassembly and degradation on denervation or fasting. Thus, during atrophy, the initial loss of desmin is critical for the subsequent myofibril destruction, and over time, myofibrillar proteins become more susceptible to PAX4-induced enzymes that promote proteolysis.
Atrophy of skeletal muscle is associated with motor-neuron diseases (e.g., ALS), many myopathies, disuse, denervation (e.g., spinal cord injuries), fasting, excessive glucocorticoids, and many systemic diseases (e.g., diabetes, cancer cachexia) (1–3). This loss of mass leads to reduced contractile force owing to the excessive destruction of myofibrillar proteins, primarily by the ubiquitin proteasome system (UPS) (4), and reduced endurance resulting from the degradation of mitochondria by autophagy (5, 6). Although it has long been known that myofibril breakdown during atrophy is mediated primarily by the UPS (4), the roles of specific ubiquitin ligases in this process and their regulation are only beginning to be understood.
Previous microarray studies on mouse muscles identified a set of genes (termed “atrogenes”) that are induced in diverse wasting conditions (7–9) by Forkhead box O (FoxO) transcription factors (10, 11), the activation of which is sufficient to cause wasting (10). Among these atrogenes are multiple components of the UPS (10) and many autophagy genes (6, 12, 13). These findings suggested that similar mechanisms promote myofibril destruction and loss of muscle mass in various forms of atrophy. Two ubiquitin ligases—muscle RING-finger 1 (MuRF1) and atrogin1/MAFbx—were found to be dramatically and rapidly induced in all types of atrophy (14, 15). and their deletion was shown to attenuate muscle wasting (14). We and others have shown that during the atrophy induced by denervation, MuRF1 first ubiquitinates the thick filament-stabilizing proteins myosin binding protein-C and myosin light chains 1 and 2, and subsequently myosin heavy chain, leading to proteasomal degradation (16, 17). In subsequent studies, we demonstrated that during atrophy induced by fasting, Z-band and thin filament components are degraded by a distinct mechanism requiring the ubiquitin ligase, tripartite motif-containing protein 32 (Trim32), and that their loss is preceded by the Trim32-dependent destruction of the desmin cytoskeleton (18).
Unlike MuRF1, Trim32 is expressed throughout the body, and mutations in this enzyme cause limb-girdle muscular dystrophy 2H (LGMD-2H) (19). Trim32 is critical for normal neuronal and muscle development, as demonstrated by the neurologic defects and myopathies in Trim32-null mice (20, 21). However, in normal postnatal muscles, Trim32 seems to limit fiber growth, and suppression of its activity alone can induce muscle hypertrophy and reduce fiber atrophy (22, 23). We show here that Trim32-dependent depolymerization of the desmin cytoskeleton precedes and promotes the subsequent loss of myofibrils.
Desmin intermediate filaments (IFs) are critical for muscle architecture and function, because they connect the Z-lines of adjacent myofibrils laterally to the sarcolemma, mitochondria, and nuclear membrane (24, 25). Desmin-deficient mouse muscles exhibit misaligned sarcomeres and disorganized myofibrils (26). Furthermore, desmin mutations cause a cardiomyopathy characterized by compromised desmin IF assembly, desmin aggregation, and severe disturbances in the ordered alignment of sarcomeres (27). The expression of mutant desmin (E245D) in cardiomyocytes leads to displacement of endogenous desmin from the Z-lines and perturbation of actin filament architecture (28). Thus, desmin IFs are important for the integrity of myofibrils, especially of thin filaments.
Desmin consists of a central α-helical rod domain flanked by non–α-helical amino-terminal head and carboxyterminal tail domains (29). The head domain is important for filament stability and polymerization, and phosphorylation of serine residues within this domain promotes IF depolymerization (18, 30, 31). We previously showed that during fasting, phosphorylation in this domain promotes ubiquitination by Trim32 and destruction of desmin filaments (18). The present study demonstrates that myofibril breakdown during atrophy requires not only desmin IF phosphorylation, ubiquitination, and depolymerization, but also gene expression by the transcription factor paired box 4 (PAX4).
PAX4 is a member of the paired box (PAX) family of transcription factors, which play important roles in fetal development (32). Mammalian cells contain nine members of this family, PAX1–PAX9, all of which have a paired domain that recognizes specific DNA sequences (33). PAX3 and PAX7 are important for skeletal muscle development and regeneration and for the maintenance of satellite cells (34, 35); however, PAX4 plays a critical role in differentiation of insulin-producing β cells in the pancreas (36). PAX4-null mice lack pancreatic β cells (36), and mutations in the PAX4 paired DNA-binding domain cause type 2 diabetes in human (37, 38). We show here that PAX4 is expressed in skeletal muscle and is of prime importance in the enhancement of protein degradation during atrophy induced by denervation or fasting. This transcription factor induces certain genes that promote myofibril destruction, including the AAA-ATPase p97/VCP, in a second phase of gene expression during atrophy, long after induction of the major atrogenes.
Results
Depolymerization of Desmin Filaments Promotes Myofibril Breakdown.
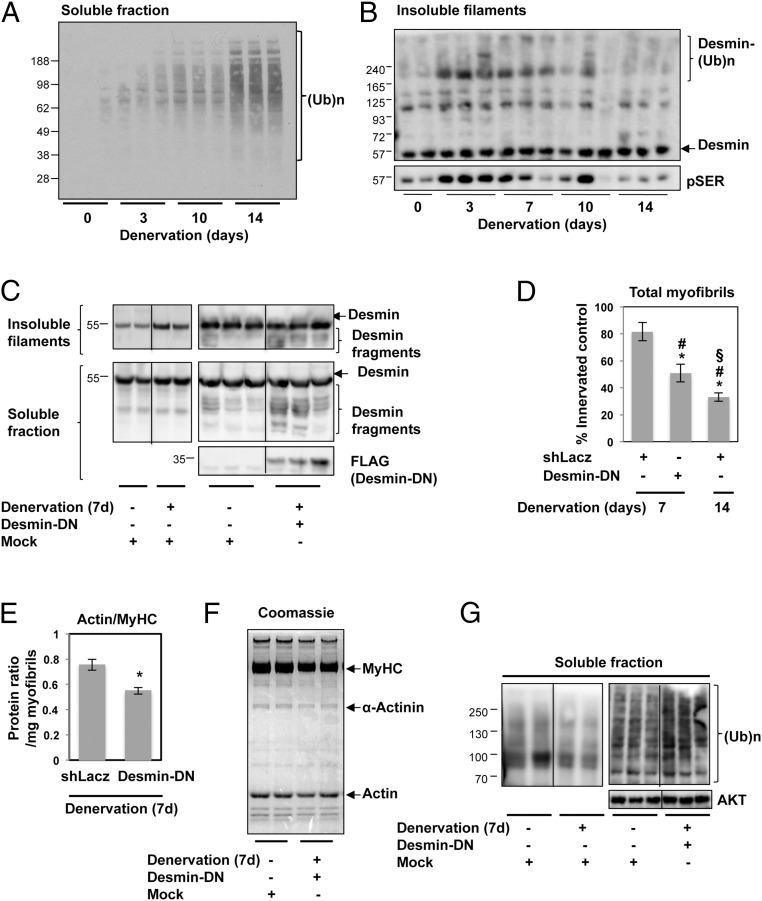
To dissect the order of events leading to myofibril destruction during atrophy, we investigated the wasting induced by denervation. The sciatic nerve on one limb of wild-type mice was sectioned, and gastrocnemius muscles were dissected 3, 10, and 14 d later. At 14 d after denervation, there was a 40% decrease in the average weight of the denervated muscles below the levels measured in contralateral innervated control mice (32.7 ± 2.8 mg vs. 55 ± 4.3 mg). At this time, myofibrillar proteins are undergoing rapid degradation (16), and the muscle content of ubiquitin conjugates is dramatically increased (Fig. 1A).
Fig. 1.
Depolymerization of desmin by desmin-DN promotes myofibril breakdown at 7 d after denervation. (A) At 14 d after denervation, there is a marked accumulation of ubiquitinated proteins. Soluble fractions of muscle extracts at 3, 10, or 14 d after denervation were analyzed by immunoblotting with an antibody against ubiquitin conjugates. (B) On denervation, desmin phosphorylation precedes its degradation. The time course of the phosphorylation and loss of desmin was analyzed by immunoblotting of equal amounts of myofibrils from TA muscles denervated for 0, 3, 7, 10, and 14 d. (C–G) To test whether disassembly of desmin filaments influences myofibril stability, TA muscles were electroporated with an empty plasmid (mock) or one encoding a dominant negative mutant of desmin (desmin-DN) to induce filament disassembly. Muscles were denervated at the time of electroporation and were dissected 7 d later. Data are compared with that of the contralateral innervated limb. (C) Desmin-DN enhances disassembly of desmin filaments in muscles at 7 d after denervation. Pellets (6,000 g) and soluble fractions from transfected muscles were analyzed by SDS/PAGE and immunoblotting. (D) Disassembly of desmin filaments by desmin-DN triggers myofibril destruction. The mean content of myofibrils per electroporated muscle is presented as the percentage of innervated control. n = 4. *P < 0.0005 vs. shLacz; #P < 0.05 vs. innervated control; §P < 0.05 vs. Desmin-DN. (E) Desmin depolymerization by desmin-DN accelerates the loss of actin relative to myosin. Equal amounts of myofibrillar fraction (2.5 μg) from TA muscles denervated for 7 d expressing desmin-DN or shLacz were analyzed by SDS/PAGE and Coomassie blue staining. The intensity of myosin and actin protein bands was measured by densitometry and is expressed as the mean ratios of actin to myosin. n = 4. *P < 0.05 vs. shLacz. (F) Desmin depolymerization facilitates the degradation of both myosin and actin. Equal amounts (2.5 μg) of myofibrillar fraction from innervated TA muscles and TA muscles denervated for 7 d expressing desmin-DN or shLacz were analyzed by SDS/PAGE and Coomassie blue staining. (G) Depolymerization of desmin by desmin-DN causes an increase in total ubiquitin conjugates at 7 d after denervation. Soluble fraction of innervated and 7-d denervated muscles expressing control plasmid or desmin-DN, respectively, were analyzed by immunoblotting with an antibody against ubiquitin conjugates.
Our previous findings suggested that the destruction of thin filaments and Z-bands in mouse muscle during fasting is preceded by depolymerization of the desmin cytoskeleton (18). To determine whether the depolymerization of desmin IFs leads to myofibril breakdown, we first determined the time course of phosphorylation and degradation of desmin IFs after denervation. Immunoblotting of equal amounts of insoluble filaments from control tibialis anterior (TA) muscles and those at 3, 7, 10, and 14 d after denervation showed that by 3 d, desmin IFs were phosphorylated; however, the amount of phosphorylated desmin filaments was markedly reduced between 10 d and 14 d after denervation (Fig. 1B), when myofibrillar proteins were ubiquitinated and degraded (Fig. 1A) (16). Surprisingly, the total amount of desmin filaments did not change over this period; thus, after denervation, only a small fraction of cytoskeletal desmin was phosphorylated and gradually depolymerized (Fig. 1B). Interestingly, this rapid increase in desmin phosphorylation was accompanied by increased ubiquitination (Fig. 1B), presumably by Trim32 (see below) (18). Thus, the phosphorylation of desmin IF precedes its disassembly, and the depolymerization of desmin filaments precedes the degradation of myofibrils (18).
Consequently, we tested more directly whether this depolymerization of desmin filaments triggers myofibril destruction. To promote the disassembly of desmin filaments, we used a dominant negative inhibitor of desmin assembly (desmin-DN) (18, 39). TA muscles were electroporated with an empty plasmid (mock) or desmin-DN at the time of denervation, and the soluble and insoluble fractions were analyzed 7 d later by immunoblotting. At 7 d after denervation of muscles expressing empty plasmid, the desmin filaments were intact, even though their phosphorylation increased (Fig. 1B); however, the overexpression of desmin-DN enhanced the depolymerization of desmin filaments, as demonstrated by the appearance of desmin fragments in the insoluble fraction and an increase in the amount of soluble desmin fragments above levels in the control muscles (Fig. 1C). These fragments were likely formed by the desmin-DN–mediated inhibition of desmin assembly and the incomplete degradation of desmin monomers, perhaps by proteases (e.g., caspases) and the proteasome (39).
Interestingly, at 7 d after denervation, when desmin IFs were not depolymerized (Fig. 1B) and there was no significant loss of Z-band and myofibrillar proteins (16), the content of myofibrils was similar to that in innervated muscles (Fig. 1D). In contrast, the enhanced disassembly of desmin IFs by desmin-DN accelerated myofibril destruction, which was now evident at 7 d after denervation, and the content of myofibrils in muscles was reduced to 50% below that in innervated muscles (expressing shLacz control) (Fig. 1D). This marked loss of myofibrillar components was less than that observed in muscles denervated for 14 d, where myofibril content was reduced by >70% below levels in control muscles (Fig. 1D), likely because other factors are important for promoting myofibril breakdown in addition to desmin IF dissociation (see below).
Along with reducing the total amount of myofibrillar proteins, disassembly of desmin IFs by desmin-DN also reduced the actin:myosin ratio by 38% (from 0.8 to 0.5) below the levels in muscles denervated for 7 d (Fig. 1E), as well as the amount of α-actinin (Fig. 1F). These findings indicate an important role of desmin IFs in thin filament and Z-band stability. Interestingly, desmin IF integrity seems to be critical for myosin stability as well, because the forced dissociation of desmin filaments led to a reduction in the amount of myofibrillar myosin heavy chain (MyHC) (Fig. 1F), perhaps by promoting structural changes in the A-band, where actin and myosin interdigitate to generate force. This accelerated destruction of myofibrillar proteins caused an accumulation of ubiquitinated proteins in the soluble fraction of the denervated muscles expressing desmin-DN (Fig. 1G). Thus, disassembly of the desmin cytoskeleton appears to be a critical step in promoting myofibril breakdown during atrophy.
The Transcription Factor PAX4 Induces Genes That Promote Myofibril Destruction at a Late Phase After Denervation.
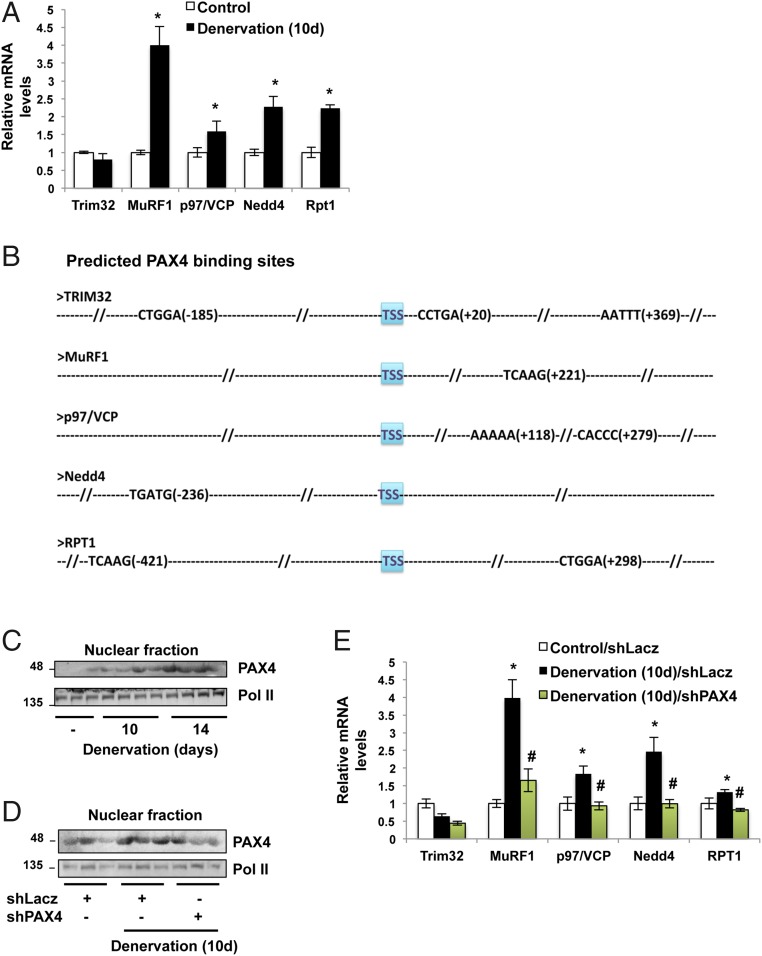
These findings imply that myofibril breakdown is accelerated by enzymes present in the muscles at 7 d after denervation, provided that there is a mechanism promoting the disassembly of desmin filaments. However, to account for the dramatic loss of myofibrillar proteins at 14 d after denervation, it seemed likely that in addition to desmin IF dissociation, other enzymes may become activated to promote myofibril destruction. At this time, expression of the p97/VCP ATPase complex and the proteasomal subunit Rpt1 increases (40). Furthermore, during atrophy induced by inactivity, the levels of the ubiquitin ligases Trim32 and Nedd4 also rise (41, 42). These enzymes may contribute to the myofibril breakdown at 14 d after nerve section, which is 8–12 d after the rapid rise in expression of most atrogenes (9, 14). Therefore, we explored whether they are induced before the rapid degradation of myofibrillar proteins, e.g., at 10 d after denervation. Interestingly, the levels of mRNA for p97/VCP, the proteasome subunit Rpt1, Nedd4, and, surprisingly, the atrogene MuRF1 increased at this time (Fig. 2A). Thus, their induction seems to represent a second phase of gene expression that likely promotes proteolysis in the inactive muscles.
Fig. 2.
On denervation, PAX4 induces distinct genes that promote proteolysis. (A) The genes for MuRF1, Nedd4, p97/VCP, and Rpt1 are induced at 10 d after denervation. RT-qPCR data of mRNA preparations from denervated (10 d) and control muscles using primers for the indicated genes are plotted as the mean fold change relative to innervated control. n = 7. *P < 0.05 vs. control. (B) Predicted PAX4-binding sites in the promoter regions of the genes for Trim32, MuRF1, Nedd4, p97/VCP, and Rpt1 and their distances from TSS. Slashes represent areas of the unbound promoter regions. (C) PAX4 enters the nucleus at 10 and 14 d after denervation. Nuclear fractions from innervated muscles and muscles at 10 and 14 d after denervation were analyzed by SDS/PAGE and immunoblotting with the indicated antibodies. (D) shRNA-mediated knockdown of PAX4 in denervated muscles. Nuclear fractions from muscles expressing shPAX4 or control shRNA at 10 d after denervation were analyzed by Western blot analysis. (E) PAX4 induces the genes MuRF1, Nedd4, p97/VCP, and Rpt1 at 10 d after denervation. RT-qPCR was performed on mRNA preparations from denervated (10 d) and control muscles expressing shLacz or shPAX4 using primers for the indicated genes. Data are plotted as the mean fold change relative to the innervated control. n = 6. *P < 0.05 vs. control; #P < 0.05 vs. denervated shLacz.
To identify the transcription factor responsible for the induction of MuRF1, p97/VCP, Nedd4, and Rpt1, we searched for potential transcription factor-binding sites in the promoter regions of all four of these genes using the TRANSFAC Match algorithm (Table S1). Potential transcription factor-binding motifs were chosen based on two criteria: a matrix match score >0.9 and a sequence match score >0.8. Four different weighted matrices predicted binding motifs for the transcription factor PAX4 in all four genes with the highest scores for both criteria (Fig. 2B). Although the TRIM32 promoter also harbors PAX4-binding motifs, surprisingly, the expression of Trim32 did not increase at this time, as we also found during fasting (18), even though it plays a critical role in the accompanying destruction of thin filament proteins (Fig. 4) (18).
Table S1.
Predictions of PAX4-binding sites using the TRANSFAC Match algorithm
| Gene | Motif matrix | Position | Matrix match | Sequence match | Binding site |
| TRIM32 | V$PAX4_01 | −185 | 0.979 | 0.801 | gccgaatcgctCCTGAgcccg |
| V$PAX4_01 | 20 | 0.979 | 0.794 | taatacctgctCCTGAatgcg | |
| V$PAX4_02 | 369 | 0.987 | 0.935 | gtaAATTTatc | |
| MuRF1 | V$PAX4_01 | 223 | 0.986 | 0.808 | cccagTCAAGagtcaaatacc |
| Rpt1 | V$PAX4_01 | 298 | 1 | 0.801 | gaactTCATGcctcaggcagg |
| V$PAX4_02 | −421 | 0.936 | 0.878 | tatCAATTggg | |
| Nedd4 | V$PAX4_01 | −236 | 1 | 0.751 | tgattTGATGcttaattacat |
| p97/VCP | V$PAX4_03 | 279 | 1 | 0.981 | ctgcacCACCCc |
| V$PAX4_04 | 118 | 1 | 0.835 | AAAAAtcaacctactttctatataaacacc |
Summary of motif matrices used for the prediction of PAX4-binding sites, the position of the motif from TSS, matrix match and sequence match, and the actual binding site. The core of the binding site is shown in uppercase letters.
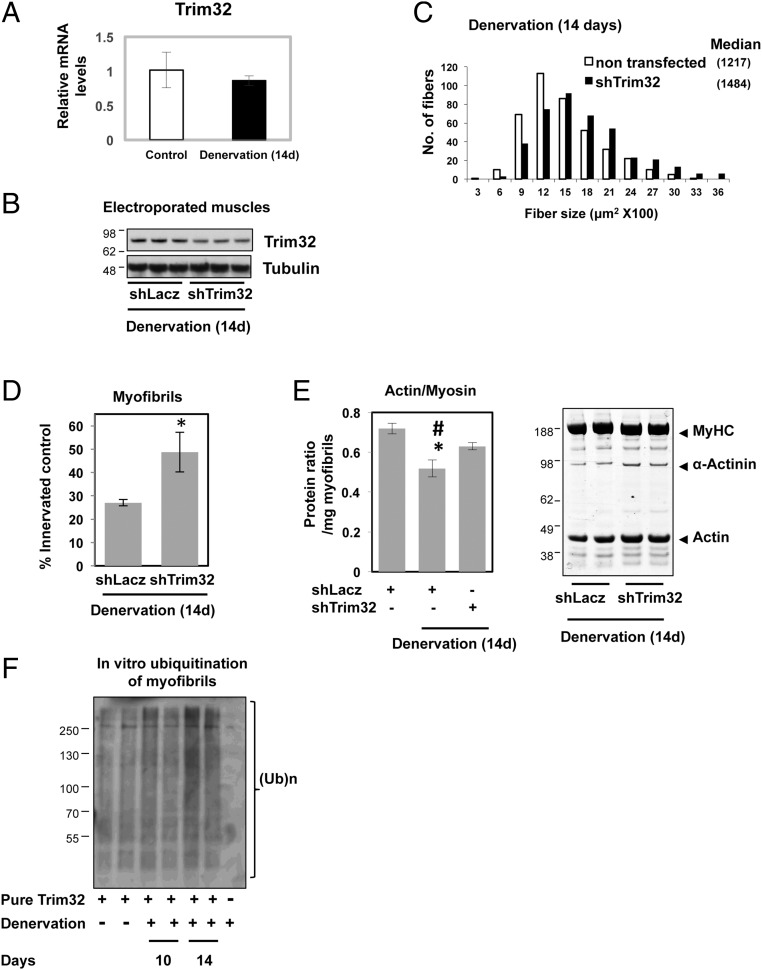
Fig. 4.
On denervation, down-regulation of Trim32 attenuates atrophy and the loss of thin filaments. TA muscles were electroporated with shRNA plasmids against Trim32 (shTrim32) or Lacz (shLacz). Denervation was performed at the time of electroporation, and muscles were dissected at 14 d after denervation. Electroporation of shLacz into mouse muscles did not affect fiber size (10). Error bars represent SEM. (A) Trim32 is not induced in TA muscles at 14 d after denervation. RT-qPCR was performed on mRNA preparations from denervated (14 d) and control muscles using primers for Trim32. Data are plotted as the mean fold change relative to innervated control. (B) shRNA-mediated knockdown of Trim32 in denervated muscles. Soluble extracts from denervated muscles expressing shTrim32 or shLacz were analyzed by immunoblotting with specific antibodies. (C) Down-regulation of Trim32 in denervated muscles reduces fiber atrophy. Shown is the measurement of cross-sectional areas of 500 fibers transfected with shTrim32 (and expressing GFP; black bars) vs. 500 nontransfected fibers (open bars) in the same muscle. Data were acquired from four mice. (D) Down-regulation of Trim32 attenuates myofibril destruction in denervated muscle. The mean content of myofibrils per electroporated muscle is presented as the percentage of innervated controls. n = 5. *P < 0.05 vs. shLacz. (E) Down-regulation of Trim32 decreases the loss of actin after denervation. (Right) Equal amounts of myofibrillar fractions from innervated TA muscles and TA muscles denervated for 14 d expressing shTrim32 or shLac were analyzed by SDS/PAGE and Coomassie blue staining. (Left) The intensity of myosin and actin protein bands was measured by densitometry and expressed as the mean ratios of actin to myosin. n = 5. *P < 0.05 vs. innervated control; #P < 0.05 vs. shTrim32. (F) At 14 d after denervation, myofibril constituents are efficiently ubiquitinated by Trim32. Myofibrillar proteins were ubiquitinated by recombinant Trim32 in isolated myofibrils from control muscles and muscles denervated for 10 or 14 d using UbcH5 and His-tagged ubiquitin. His-ubiquitinated proteins were purified with a nickel column and detected by immunoblotting with anti-ubiquitin.
To test whether PAX4 is in fact essential for the induction of these genes on denervation, we first determined whether its cellular distribution changes during atrophy. Interestingly, PAX4 translocated into the nucleus at 10 and 14 d after nerve section (Fig. 2C), which coincided with the time of induction of MuRF1, p97/VCP, Nedd4, and Rpt1. To clarify the role of PAX4, we suppressed its expression by electroporation into mouse TA of shRNA plasmid (shPAX4; shPAX4-1 in Table S2), which efficiently reduced PAX4 protein levels in the nucleus below the levels in denervated muscles (Fig. 2D). Similar results have been obtained by down-regulating PAX4 with shPAX4-2 (Table S2). The down-regulation of PAX4 with shPAX4 resulted in a marked decrease in the expression of MuRF1, p97/VCP, Nedd4, and Rpt1 (Fig. 2E). Thus, on denervation, PAX4 is required for the induction of genes that promote ubiquitination and protein breakdown long after the expression of most atrogenes (9).
Table S2.
qPCR primers and shRNA oligos used in the present study
| DNA | Gene | Sequence (5′to 3′) |
| shRNA 1 | Trim32 | GGCTGATTGGTGTCACTGATA |
| shRNA 2 | Trim32 | AGCTGCTGGTCTTGGACTGTT |
| shRNA 1 | PAX4 | GCAGATTGTGCAGCTAGCAAT |
| shRNA 2 | PAX4 | CCCAGTGTGTCCTCTATCAAT |
| PCR primer, forward | Trim32 | GGCAAATCAGCCACTTCTTC |
| PCR primer, reverse | Trim32 | ATGCCCACTGGACAGGTAAG |
| PCR primer, forward | MuRF1 | GTCCATGTCTGGAGGTCGTT |
| PCR primer, reverse | MuRF1 | AGGAGCAAGTAGGCACCTCA |
| PCR primer, forward | Nedd4 | CGGAGGACGAGGTATGGGAG |
| PCR primer, reverse | Nedd4 | GGCCTATGCCAGCTATAACCT |
| PCR primer, forward | Rpt1 | TGAAGTGCAGAGAACCATGC |
| PCR primer, reverse | Rpt1 | ATTGAGCGAGCGTGAATCTT |
| PCR primer, forward | p97/VCP | GAGGTGACACGGTGTTGCTA |
| PCR primer, reverse | p97/VCP | ATTGCCAGTGATGCCTTCCA |
| PCR primer, forward | RPLO | GCGACCTGGAAGTCCAACTA |
| PCR primer, reverse | RPLO | ATCTGCTTGGAGCCCACAT |
| PCR primer, forward | PAX4 | TTGCCTGTGCTGGAACCATT |
| PCR primer, reverse | PAX4 | CAATCAGATGATGCACAGGAT |
Down-Regulation of PAX4 or Its Target Gene p97/VCP Attenuates Myofibril Disassembly on Denervation or Fasting.
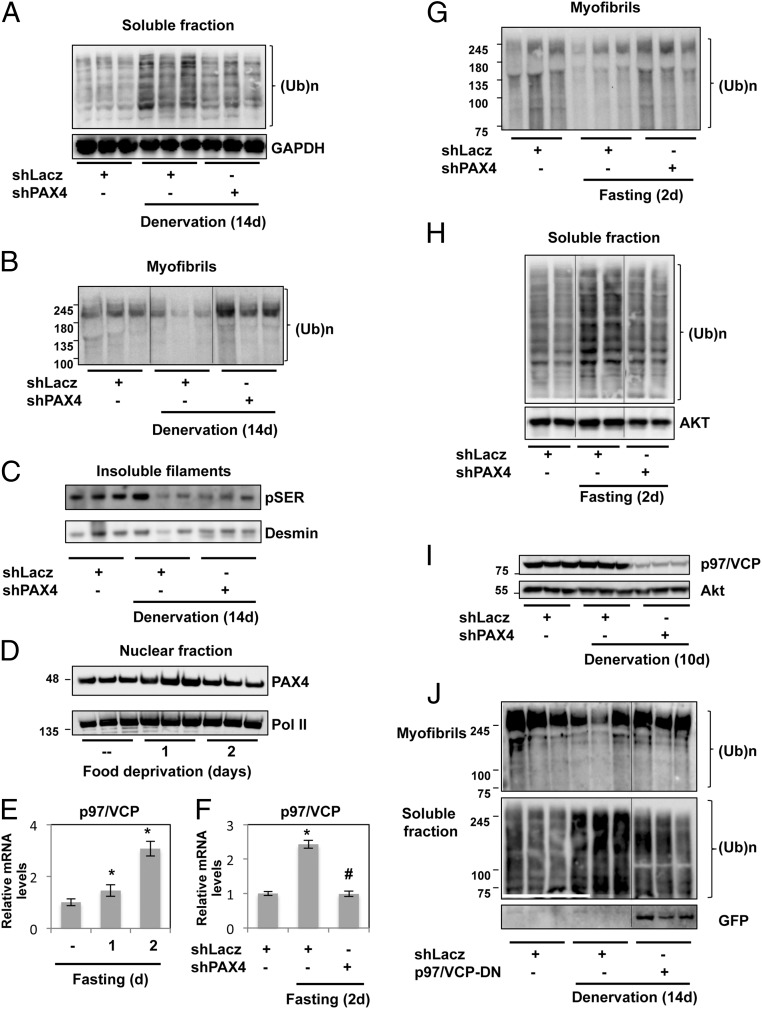
These observations make it likely that in atrophy, PAX4 is important for the induction of enzymes that catalyze myofibril degradation. Therefore, we determined the effects of PAX4 down-regulation on the total amount of ubiquitinated proteins in the soluble and insoluble fractions of denervated muscles. As noted above (Fig. 1A), the levels of ubiquitinated proteins increased in the soluble fraction at 14 d after denervation (Fig. 3A), although the levels of ubiquitinated proteins in the myofibrillar fraction decreased (Fig. 3B), suggesting solubilization of ubiquitinated myofibrillar components. However, down-regulation of PAX4 and the subsequent reduced expression of its various target genes prevented the increase in soluble ubiquitin conjugates (Fig. 3A). Instead, ubiquitinated proteins accumulated as insoluble components in the myofibrillar pellet (Fig. 3B). This pellet also contained phosphorylated desmin filaments, which were degraded in muscles denervated for 14 d and did not accumulate when PAX4 was down-regulated (Fig. 3C). These findings strongly suggest that at 14 d after denervation, ubiquitinated myofibrillar proteins are released into the soluble fraction for degradation, and that this step requires one or more proteins with expression catalyzed by PAX4.
Fig. 3.
During atrophy, PAX4 and its target gene, p97/VCP, promote solubilization of myofibrillar proteins. (A) Down-regulation of PAX4 in denervated muscles reduces the levels of cytosolic ubiquitinated proteins. The soluble fractions from innervated and denervated (14 d) muscles expressing shLacz or shPAX4 were analyzed by SDS/PAGE and immunoblotting with an antibody against ubiquitin conjugates. (B) On denervation, down-regulation of PAX4 prevents the release of ubiquitinated proteins from the myofibrils. Equal fractions of myofibrils (0.1% of the total amount of myofibrils) from denervated muscles expressing shPAX4 or shLacz were analyzed by SDS/PAGE and anti-ubiquitin antibody. (C) Phosphorylated desmin filaments are degraded in muscles expressing shPAX4 at 14 d after denervation. Equal amounts (2.5 μg) of myofibrillar fraction from innervated TA muscles and ones denervated for 14 d expressing shPAX4 or shLacz were analyzed by SDS/PAGE and immunoblotting with an anti-phosphoserine antibody. (D) PAX4 enters the nucleus at 1 d after food deprivation. Nuclear fractions from control and atrophying muscles from fed mice and mice deprived of food for 1 and 2 d were analyzed by SDS/PAGE and immunoblotting with the indicated antibodies. (E) The gene p97/VCP is induced in TA muscles during fasting. RT-qPCR was performed on mRNA preparations from muscles from fed mice or mice deprived of food for 1 or 2 d using specific primers. Data are plotted as the mean fold change relative to the fed control. n = 3. *P < 0.05 vs. control. (F) PAX4 induces the gene p97/VCP during fasting. RT-qPCR was performed on mRNA preparations from muscles expressing shLacz or shPAX4 from fed mice or mice deprived of food for 2 d using specific primers. Data are plotted as the mean fold change relative to the fed control. n = 3. *P < 0.001 vs. control, #P < 0.01 vs. fasting shLacz. (G) PAX4 down-regulation in atrophying muscles from fasted mice blocks disassembly of ubiquitinated myofibrils. Equal fractions of myofibrils (0.1% of total myofibrils) from normal and atrophying muscles expressing shPAX4 or shLacz from fasted mice were analyzed by SDS/PAGE and anti-ubiquitin antibody. (H) During fasting, PAX4 down-regulation prevents the accumulation of ubiquitinated proteins in the cytosol. Soluble fractions of control and atrophying muscles from fasted mice were analyzed by SDS/PAGE and immunoblotting with an antibody against ubiquitin conjugates. (I) PAX4 is essential for induction of p97/VCP in denervated muscles. Soluble fractions of denervated muscles electroporated with shLacz of shPAX4 were analyzed by SDS/PAGE and immunoblotting with anti-p97 antibody. An Akt blot served as a loading control. (J) The p97/VCP ATPase complex promotes myofibril disassembly after denervation. Soluble and insoluble fractions from innervated and 14-d denervated muscles expressing shLacz or p97/VCP dominant negative (p97/VCP-DN) were analyzed by SDS/PAGE and immunoblotting. Ubiquitinated proteins were detected with ubiquitin antibody.
To learn whether PAX4 serves a similar role in other types of atrophy, we down-regulated this transcription factor in mouse muscles that were atrophying due to food-deprivation and analyzed the effects on protein ubiquitination and myofibril solubilization. As we had observed after denervation, PAX4 translocated into the nucleus already after 1 d of food deprivation (Fig. 3D), when p97/VCP was induced (Fig. 3E). Furthermore, PAX4 was required for p97/VCP induction, because its down-regulation by electroporation of shPAX4 into muscles from fasted mice resulted in a marked decrease in p97/VCP expression (Fig. 3F). In these muscles, myofibril disassembly was attenuated (Fig. 3 G and H). Although ubiquitinated proteins in the myofibrillar fraction decreased after fasting, when PAX4 was down-regulated (Fig. 3G), they increased in the myofibrils and decreased in the soluble fraction (Fig. 3H). Thus, PAX4-dependent gene expression seems to enable myofibrillar disassembly in multiple forms of atrophy, specifically to allow the extraction and degradation of ubiquitinated myofibrillar components.
This PAX4-dependent accumulation of ubiquitinated proteins in the cytosol at 14 d after denervation (Fig. 3A), when myofibrillar components are undergoing rapid degradation (16), coincided with the induction of proteins that can promote myofibril disassembly and degradation, especially the ATPase complex p97/VCP. This complex acts as a protein “segregase,” which extracts ubiquitinated proteins from larger structures, such as the endoplasmic reticulum (ER) membrane in the ER-associated protein degradation (ERAD) pathway, before proteasomal degradation (43). Piccirillo and Goldberg (40) presented evidence that the p97/VCP complex is important for the overall increase in protein breakdown during atrophy and is necessary for extraction of ubiquitinated components from the myofibrils. As shown above (Fig. 2E), its expression at 10 d after denervation is completely dependent on the function of PAX4, the absence of which prevented specifically the step at which p97/VCP was proposed to act (Fig. 3I).
To determine whether PAX4-mediated p97/VCP induction is required for the disassembly and degradation of myofibrillar proteins, we inhibited p97/VCP function in the denervated muscle by electroporation of a dominant negative (40, 44). By 14 d after nerve section, there was a reduction in the levels of ubiquitinated myofibrillar proteins, whereas ubiquitin conjugates increased in the cytosol (Fig. 3J). However, as we observed with shPAX4 (Fig. 3 A, B, G, and H), inhibition of p97/VCP by the expression of a dominant negative subunit that lacks ATPase activity prevented myofibril solubilization, and ubiquitinated myofibrillar proteins accumulated in the insoluble fraction (Fig. 3J). Thus, during atrophy, a second late phase of PAX4-dependent induction of p97/VCP and other components of the UPS is required to accelerate myofibril breakdown.
Trim32 Down-Regulation Attenuates Denervation Atrophy and Loss of Thin Filament.
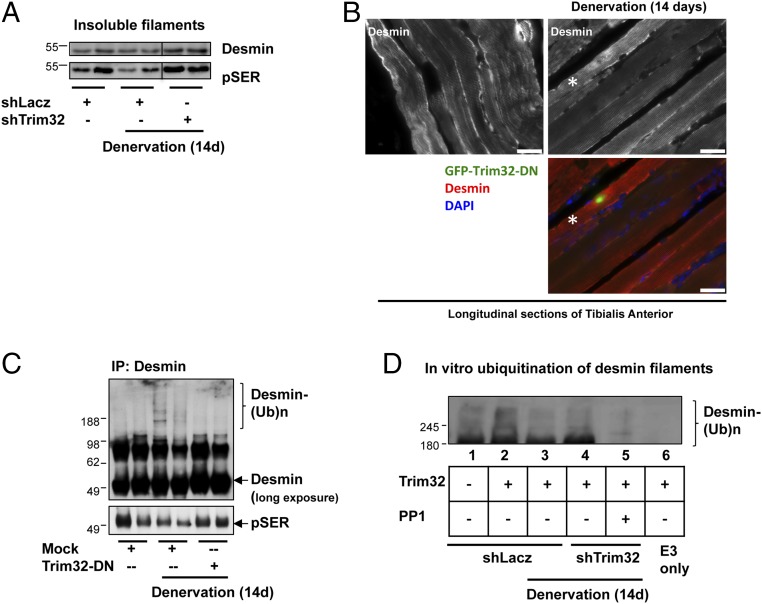
Although PAX4-mediated gene induction is required for myofibril destruction, enyzmes that show no rise in expression after denervation may play a critical role as well. In fact, Trim32 is not induced during atrophy induced by food deprivation, although this enzyme is clearly essential for the loss of desmin, Z-band, and thin filaments (18). After denervation, Trim32 expression also did not increase in the muscles (Figs. 2A and 4A) even though the Trim32 promoter harbors PAX4-binding motifs (Fig. 2B). To determine whether Trim32 is essential for the loss of mass on denervation, we first analyzed the effects of its down-regulation on fiber atrophy and thin filament content. After electroporation with shTrim32, its expression was lower in denervated muscles (at 14 d) than in the denervated muscles electroporated with shLacz (Fig. 4B). This decrease in Trim32 levels was sufficient to attenuate fiber atrophy, because the cross-sectional area of nontransfected, denervated fibers was smaller than that of those fibers expressing shTrim32 (Fig. 4C). This reduction in fiber atrophy occurred largely through the attenuation of myofibrillar destruction, as demonstrated by the higher content of myofibrillar proteins in the denervated muscles expressing shTrim32 compared with the denervated muscles expressing shLacz (Fig. 4D).
To learn whether Trim32 preferentially catalyzes the loss of thin filaments and Z-bands after denervation, we analyzed the effects of Trim32 down-regulation on the content of actin relative to myosin in the atrophying muscles. In denervated muscles, the actin:myosin ratio decreased by 30% (from 0.7 to 0.5) below levels in the contralateral innervated limbs (Fig. 4E); thus, actin is lost to a greater extent than myosin. Trim32 down-regulation with shRNA clearly attenuated this differential loss of actin, because the actin content relative to myosin was 0.63 (Fig. 4E), even though many fibers were not electroporated. Furthermore, in the denervated muscles expressing shTrim32, the loss of the Z-band protein α-actinin, the degradation of which seemed to be linked to the loss of thin filaments and desmin IF (18), was also attenuated (Fig. 4E, Right). Thus, although Trim32 is not induced on denervation, this enzyme is clearly essential for myofibril breakdown by primarily catalyzing the loss of thin filament components and Z-bands.
On Denervation, Trim32 Promotes Depolymerization of Phosphorylated Desmin Filaments.
The foregoing findings suggest that at 14 d after denervation, when the muscle content of ubiquitinated proteins is markedly increased (Fig. 1A) and desmin filaments are depolymerized (Fig. 1B), myofibrils may be loosened such that their constituents are more susceptible to ubiquitination by Trim32 or other ubiquitin ligases. To test this idea, we compared ubiquitination in vitro of myofibrils isolated from these muscles by recombinant Trim32 and UbcH5 (Fig. 4F). At 10 d after denervation, when the degradation of myofibrillar actin and myosin is slow (15), Trim32 ubiquitinated at similar rates as myofibrils isolated from the contralateral denervated and innervated muscles (Fig. 4F). In contrast, myofibrils from muscles denervated for 14 d, when PAX4 target genes are highly expressed (Fig. 2) and myosin and actin are rapidly degraded (16), were more sensitive to ubiquitination by Trim32 in vitro (Fig. 4F). Thus, when desmin is solubilized and protein ubiquitination is accelerated, myofibrils are more sensitive to Trim32, and perhaps to other ubiquitin ligases as well.
To learn whether Trim32 is required for this loss of phosphorylated desmin filaments on denervation, which precedes PAX4-mediated gene induction and the resulting myofibril destruction (Fig. 1), we analyzed the effects of Trim32 down-regulation on desmin filaments. At 14 d after denervation, when myofibrillar proteins are rapidly degraded (Fig. 1D) (16), the total amount of desmin filaments in the denervated muscles was similar to that in controls (Fig. 5A), in sharp contrast to the marked loss of desmin in muscles on fasting (18). However, immunoblotting with an anti-phosphoserine antibody revealed the presence of phosphorylated desmin filaments in the innervated muscles, the level of which decreased in the contralateral denervated muscles (Fig. 5A). The fraction of desmin IFs phosphorylated and degraded after denervation must be small, because the total content of desmin filaments was not reduced significantly (Figs. 1B and 5A).
Fig. 5.
On denervation, Trim32 catalyzes the loss of phosphorylated desmin filaments. (A) Trim32 promotes the disassembly and degradation of phosphorylated desmin filaments after denervation. Desmin filaments were isolated from innervated and denervated muscles expressing shLacz or shTrim32 and then analyzed by SDS/PAGE and immunoblotting. Phosphorylated desmin was detected with anti-phosphoserine antibody. (B) Desmin is degraded in atrophy induced by denervation. Paraffin-embedded longitudinal sections of innervated and denervated TA muscles expressing Trim32-DN-GFP were stained with an antibody against desmin. Asterisk indicates transfected fiber expressing GFP-Trim32-DN. (Scale bar: 35 µm.) (C) On denervation, desmin is ubiquitinated, but when Trim32 is inhibited, phosphorylated species accumulate in the cytosol. Desmin was immunoprecipitated from soluble fractions of control or denervated muscles expressing shLacz or Trim32-DN. Precipitates were analyzed by immunoblotting using antibodies against desmin and phosphoserine. (D) On denervation, phosphorylation of desmin filaments facilitates their ubiquitination by Trim32. Isolated desmin filaments from normal muscles (lanes 1 and 2) and atrophying muscles (lanes 3–5) expressing shLacz (lanes 1–3) or shTrim32 (lanes 4 and 5) were treated with protein phosphatase 1 (PP1; lane 5) or left untreated (lanes 1–4), and then subjected to ubiquitination by recombinant Trim32 and UbcH5 using 6His-tagged ubiquitin. Ubiquitinated desmin was purified with a nickel column and analyzed by SDS/PAGE and immunoblotting using anti-desmin.
This loss of phosphorylated desmin filaments at 14 d after denervation was dependent on Trim32, which was completely blocked on knockdown or inhibition of this enzyme (Fig. 5 A and B). Instead, desmin accumulated as an insoluble phosphorylated species (Fig. 5A). Furthermore, desmin immunoprecipitation from the soluble fraction of denervated and innervated muscles indicated that on denervation, desmin is released from the cytoskeleton into a soluble form as a phosphorylated species and is most likely ubiquitinated and degraded, but not when Trim32 was inhibited by transfection of a dominant negative (Trim32-DN) (Fig. 5C). Moreover, the in vitro ubiquitination of isolated desmin filaments by Trim32 and UbcH5 revealed that Trim32 can efficiently ubiquitinate desmin filaments isolated from normal muscle (Fig. 5D, lane 2), but not desmin filaments isolated from denervated atrophying muscles, in which Trim32 was active and catalyzing the degradation of phosphorylated desmin (Fig. 5D, lane 3). In other words, the muscles in which Trim32 functioned in vivo contained less phosphorylated desmin IF and were modified less by purified Trim32 in vitro.
In contrast, the desmin filaments from the atrophying muscles lacking functional Trim32, in which phosphorylation levels were similar to those in fed controls (Fig. 5A), were extensively ubiquitinated by Trim32. However, when the desmin filaments were pretreated with the phosphatase PP1 (Fig. 5D, compare lanes 4 and 5), little or no ubiquitination occurred, indicating that phosphorylation of desmin filaments enhances their recognition and ubiquitination by Trim32. Thus, on denervation, phosphorylation of desmin filaments facilitates their ubiquitination by this E3, which is essential for the gradual solubilization of this cytoskeletal network and the resulting myofibril destruction by Trim32, together with p97/VCP and other enzymes induced by PAX4.
Discussion
Recent biochemical studies of atrophying muscles have generally emphasized a single common mechanism that stimulates net protein degradation by the UPS throughout the process of atrophy (3, 8, 45, 46). However, our studies of mRNA changes during the disuse atrophy induced by denervation (Fig. 2) or spinal isolation (9) and on degradation of myofibrillar proteins (16, 18, 22) clearly indicate that patterns of mRNA, protein synthesis, and proteolysis change markedly with time and follow a specific sequence during the atrophy process. As shown here, after sciatic nerve section, certain enzymes are induced long after the major atrogenes are expressed maximally (14, 15). This second phase of transcription coincides with a marked accumulation in the soluble fraction of ubiquitinated proteins that appear to be intermediates in the disassembly and rapid degradation of myofibrillar proteins.
In this second phase of gene expression, occurring at 10–14 d after section of the sciatic nerve, when protein loss and muscle atrophy are most pronounced, desmin IFs are depolymerized, actin and myosin are rapidly lost from the myofibril (16), and there is a marked accumulation of ubiquitinated proteins in the soluble cytosol (Fig. 1A). In addition, we show that the expression and content of the p97/VCP ATPase complex, which seem important for myofibril disassembly and degradation, increase after denervation or fasting, just when the degradation of myofibrillar components is accelerated. The increased expression of p97/VCP, as well as proteasome components (Fig. 2) (40), the ubiquitin ligases Nedd4 and MuRF1 (Fig. 2), and probably others represent a second phase of the induction of key factors promoting proteolysis by the UPS. Moreover, at this time, myofibrils isolated from the atrophying muscles were more susceptible to ubiquitination in vitro by Trim32 (Fig. 4F). These myofibrils likely are more loosely organized, allowing for more effective modification by ubiquitin ligases. The reduced structural integrity of desmin filaments on denervation (Fig. 5) is likely the key step in the destabilization of Z-band proteins and thin filaments during atrophy, leading to the enhanced susceptibility to ubiquitination.
Disassembly of Desmin Filaments Triggers Myofibril Destruction During Atrophy.
Whereas denervation eliminates all contractile work in specific muscles and causes them to atrophy, muscle wasting occurs systemically during fasting and in many systemic diseases (2), resulting from low levels of insulin and insulin-like growth factor 1 and increased levels of glucocorticoids (3). However, the mechanisms for disassembly and degradation of myofibrillar proteins in these two types of atrophy seem similar and involve the phosphorylation and depolymerization of the desmin cytoskeleton by Trim32. Reducing Trim32 function by either shRNA or a dominant-negative inhibitor markedly attenuated the decrease in fiber diameter on denervation (Fig. 4C) or fasting (18). Despite this essential role in atrophy, Trim32 function alone is not sufficient to induce muscle wasting or to accelerate protein degradation (Fig. S1). Thus, the accelerated proteolysis during atrophy seems to require the induction of additional cofactors (e.g., by PAX4) to function with Trim32 and to enhance the susceptibility of the myofibrils to this enzyme. For example, the substrates of Trim32 may be modified by phosphorylation before their ubiquitination by this enzyme, as we found for desmin (Fig. 5D) (18).
Fig. S1.
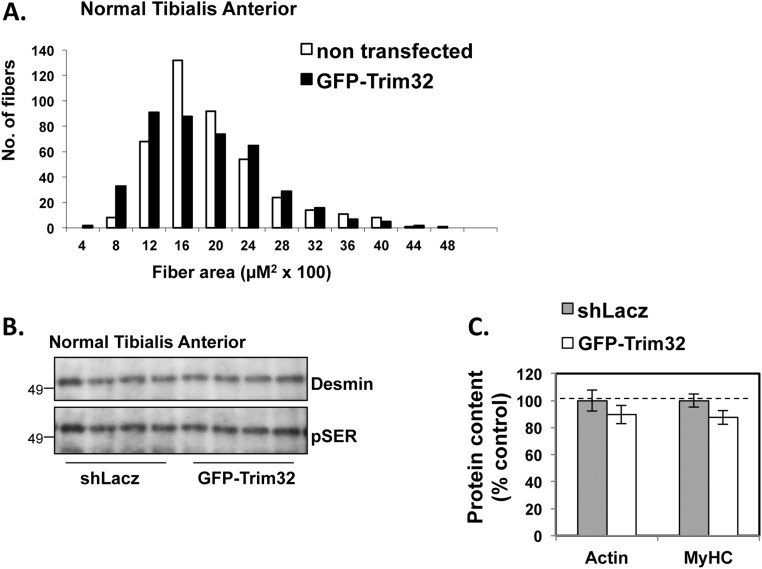
Overexpression of Trim32 in normal muscle does not trigger atrophy. (A) Overexpression of Trim32 in TA muscles does not affect fiber size. Shown are measurements of a cross-sectional area of 500 fibers transfected with GFP-Trim32 (black bars) vs. 500 nontransfected fibers (open bars) in the same muscle. Data were acquired from five mice. (B) Overexpression of Trim32 in TA muscle does not affect levels of desmin or its phosphorylation. Insoluble fractions from normal TA muscles expressing shLacz or GFP-Trim32 were analyzed by SDS/PAGE and immunoblotting. Phosphorylated desmin was detected with anti-phosphoserine antibody. (C) Overexpression of Trim32 in TA muscles does not affect the levels of myofibrillar actin and myosin. Equal amounts of myofibrilar fraction from shLacz-expressing (shaded) or GFP-Trim32–expressing muscles (open) were analyzed by SDS/PAGE and Coomassie blue staining. The intensities of myosin and actin protein bands were measured by densitometry, and the mean intensity is expressed as percentage of shLacz-expressing muscles. n = 5.
In related studies, we found that Trim32 activity also promotes atrophy via inhibition of anabolic signaling by the PI3K-AKT pathway (21). These various observations on the effects of selective down-regulation of Trim32 in muscles of adult mice differ from the findings reported by Spencer et al. (20) in knockout mice lacking Trim32 in all cells. Those mice exhibited multiple neurologic defects, mild myopathies, and reduced muscle growth and body size (21, 47). Surprisingly, muscles from those mice atrophy to a similar extent on hind-limb suspension or fasting as muscles from wild-type mice (47). In contrast, knockdown of Trim32 causes a reduction in atrophy on fasting (18) and denervation (Fig. 4) and causes hypertrophy in normal muscles (22, 23). Thus, the loss of Trim32 during development likely elicits compensatory responses that replace Trim32 in its many roles.
Our findings that (i) down-regulation of Trim32 in denervated muscle inhibits depolymerization of desmin IF (Fig. 5) and the loss of myofibrillar proteins (Fig. 4) and (ii) that depolymerization of desmin IF during fasting (18) or on denervation (Fig. 1) accelerates myofibril breakdown support previous suggestions that the integrity of desmin filaments is critical for myofibril stability (18, 28, 48). Desmin knockout mice exhibit a cardiomyopathy and muscular dystrophy characterized by disorganized myofibrils (26, 49); however, human desmin-related myopathies are characterized not by a total lack of desmin, but rather by an accumulation of desmin aggregates (50). These various human mutations in the desmin gene cause cardiac and skeletal muscle lesions, muscle weakness, arrhythmias, and congestive heart failure (51–53). The present study provides further evidence that desmin is of crucial importance for the integrity of cardiac and skeletal muscle cells, especially for maintenance of the contractible apparatus.
Accordingly, in denervated muscles deficient in Trim32, the loss of desmin and myofibrillar proteins was reduced. During atrophy induced by denervation or fasting, the increased phosphorylation of desmin filaments enhanced their ubiquitination by Trim32 and subsequent solubilization and degradation (Fig. 5D) (18). Several kinases, including protein kinase A, protein kinase C (PKC), Ca2+/calmodulin kinase II, cdc2 kinase, glycogen synthase kinase 3 (GSK3), and rho-kinase, can phosphorylate desmin within the head domain in vitro and affect filament structure (54, 55). There are no data on whether any of these kinases is activated and phosphorylates desmin during atrophy in vivo. PKC and GSK3 are strong candidates, given that phosphorylation of desmin by these kinases has been proposed to promote myofibril disarray in cardiomyocytes (48, 56). Other good candidates are AMP-activated protein kinase and p38 mitogen-activated protein kinase, which are activated during muscle atrophy and may promote proteolysis, perhaps by phosphorylating desmin IFs (57, 58).
In mice deprived of food for 2 d, desmin was virtually absent (18). In contrast, at 14 d after denervation, there was no reduction in the total amount of desmin IFs, even though the muscle content of phosphorylated desmin filaments was markedly decreased. Presumably, during this slower atrophy of the inactive muscles, only a small fraction of desmin IFs are phosphorylated and degraded, but this modification or loss is sufficient to promote myofibril breakdown. Thus, unlike in fasting, where up to 30% of muscle mass can be lost in 2 d, in denervated muscles desmin IFs are depolymerized only slowly, which is sufficient to promote myofibril destruction.
The precise mechanism for disassembly of the desmin cytoskeleton during disuse atrophy remains unclear and is an important question for future research. If desmin filaments are continuously being formed and depolymerized, then phosphorylation within the desmin head domain, like the expression of a truncated desmin (desmin-DN), should inhibit desmin polymerization and favor its disassembly. In fact, it was previously suggested that desmin depolymerization can be stimulated by the phosphorylation of various serine residues in its head domain, which can lead to fragmentation and dissociation of the cytoskeletal network (59). Consistently, the enhanced solubilization of desmin IFs by desmin-DN at 7 d after denervation led to the accumulation of soluble desmin fragments (Fig. 1C), which cannot represent short desmin filaments (which are oligomers) (60), because these fragments were of lower molecular weight than monomeric desmin (53 kDa). Once phosphorylated, the soluble desmin cannot assemble into filaments (61) and is rapidly degraded by the proteasome in a Trim32-dependent fashion (Fig. 5) (18). Thus, these desmin fragments were likely formed by the incomplete degradation of phosphorylated, ubiquitinated desmin monomers by the proteasome. By 3 d after nerve section, there was an increase in desmin IF phosphorylation and ubiquitination (Fig. 1B), likely owing to Trim32 (Fig. 5). The basal levels of this enzyme must suffice to catalyze the rapid dissociation of desmin filaments, given that the atrophy resulting from food deprivation (18) or denervation (Figs. 4 and 5), the loss of desmin occurred with no increase in Trim32 content. Because the depolymerization of desmin occurred long after its phosphorylation, the dissociation of this cytoskeletal network likely requires the activation of additional factors.
This loss of phosphorylated desmin (Fig. 1) and depolymerization preceded significant loss of myosin and actin (16). In fact, the strongest evidence that desmin IF disassembly promotes and accelerates myofibril destruction was our finding that at 7 d after denervation, when desmin filaments are intact and before any clear loss of myofibrillar proteins is seen, expression of a truncated desmin (desmin-DN) promoted disassembly of desmin filaments, dramatically accelerated myofibril breakdown, and caused a marked accumulation of ubiquitinated proteins in the soluble fraction (Fig. 1). This marked loss of myofibrils (∼50%) was less than that observed at 14 d after denervation (∼70%; Fig. 1D), however, presumably because additional factors (e.g., PAX4 gene induction) besides desmin IF disassembly are required to promote myofibril destruction. Clearly, the dissociation of the desmin cytoskeleton enhances the destruction of Z-bands and thin filaments, because in muscles expressing desmin-DN to promote desmin IF depolymerization, actin and α-actinin were degraded to a greater degree than myosin (Fig. 1E). Furthermore, in denervated muscles deficient in Trim32, in which desmin IFs are stabilized, actin degradation was slower than myosin degradation (Fig. 4E). Although degradation of both thin and thick filaments can be accelerated by dissociation of the cytoskeleton (Fig. 1), Trim32, by promoting desmin IF depolymerization, cannot by itself be important for the loss of thick filaments, because thick filaments are protected completely from degradation in muscles lacking functional MuRF1 (16). Thus, taken together, the foregoing findings and our previous results on fasting identify desmin depolymerization as an early critical step for overall protein degradation during atrophy.
PAX4 Is Critical in Promoting Myofibril Breakdown During Atrophy.
The present study identifies a role for PAX4 in the induction of genes that promote myofibril disassembly after denervation or fasting. To our knowledge, the functions of PAX4 in skeletal muscle have not been investigated previously, although its role in the development of insulin-producing pancreatic β cells is well established (36). The PAX4-dependent induction of MuRF1 at 10 d after denervation is surprising, given that the expression of this atrogene is stimulated by FoxO transcription factors at an early phase in various types of atrophy (10). This second phase of MuRF1 induction by PAX4 at 10 d after denervation occurs just as degradation of myofibrils is accelerated and is likely required to maintain high MuRF1 protein levels in the cytosol to facilitate the destruction of thick filament components (16, 17). Whether PAX4 is also required for the early induction of MuRF1, and whether there are indeed two phases of MuRF1 induction or if its level remains high throughout the atrophy process, require further study.
Previous work has emphasized how the various types of muscle atrophy share a common transcriptional program (3, 45, 46), leading to enhanced proteolysis by the UPS (8, 9) as well as autophagy (5, 6). However, after food deprivation, the rapid rate of muscle loss in rodents makes it impossible to determine whether there are distinct initial and later phases of transcription in the muscles. It will be of interest to explore whether there is also a delayed rapid loss of myofibrils along with a second phase of gene induction in more prolonged systemic wasting conditions (e.g., cancer cachexia, caloric restriction, renal or cardiac failure).
Whereas PAX4 is critical for the induction of Nedd4, p97/VCP, MuRF1, and Rpt1 and thus can affect muscle mass, these findings raise some intriguing questions for future research about PAX4 regulation, such as what facilitates its translocation into the nucleus during fasting and late after denervation. Although the Trim32 promoter harbors PAX4-binding motifs, this enzyme is not induced after denervation or fasting, even though it clearly plays a critical role in atrophy. After denervation, this enzyme catalyzes the initial loss of desmin filaments, and then at a later phase, it seems to act on the myofibrillar apparatus together MuRF1 and other enzymes induced by PAX4 (e.g., ubiquitin ligases, the p97/VCP complex). Down-regulation of PAX4 or its target gene p97/VCP prevented the release of ubiquitinated proteins from the myofibrils to the cytosol, where they presumably are more susceptible to proteasomal degradation. These findings thus strongly suggest that PAX4-mediated gene induction at the later phase is required for the excessive proteolysis after the Trim32-dependent loss of the desmin cytoskeleton.
Materials and Methods
In Vivo Transfection.
All animal experiments were performed in accordance with Israeli Council on Animal Experiments guidelines, the institutional regulations on animal care and use, and ethical guidelines. Animal care was provided by specialized personnel in the institution’s animal facility. Muscle denervation was performed in adult CD-1 male mice (∼30 g) by sectioning the sciatic nerve on one limb, with the other leg serving as a control (16). In vivo electroporation experiments were performed at the time of denervation as described previously (22). In brief, 20 μg of plasmid DNA was injected into adult mouse TA muscles, and a mild electric pulse was applied by two electrodes (12V, five pulses, 200-ms intervals).
Excised muscles were snap-frozen in isopentane, and cross-sections were fixed in 4% PFA. Cross-sectional areas of transfected (expressing GFP) and adjacent nontransfected fibers in the same 10-µm muscle section were measured using Metamorph (Molecular Devices). Data collected from at least 500 fibers from six mice were plotted. Individual fiber size was determined in the entire muscle cross-section. Images were collected using a Nikon Ni-U upright fluorescence microscope with a Plan Fluor 20× 0.5 NA objective lens with a Hamamatsu C8484-03 cooled CCD camera and MetaMorph software.
Antibodies and Materials.
Anti-phosphoserine, laminin, GAPDH, and tubulin were purchased from Sigma-Aldrich; anti-desmin was obtained from Abcam, anti-Akt was obtained from Cell Signaling Technology; and anti-ubiquitin (clone FK2) was purchased from Biomol. The Trim32 antibody was kindly provided by J. Schwamborn, University of Luxembourg. The UbcH5 clone was provided by K. Iwai, Osaka City University, and the mammalian expression vector encoding the N-terminal region of desmin was provided by V. Cryns, Northwestern University (18, 39). The dominant negative p97/VCP plasmid, p97(K524A)-GFP, was a generous gift from A. Kakizuka, Kyoto University (44). The shRNA oligos against PAX4, Trim32, and Lacz were generated using the Invitrogen BLOCK-iT RNAi Expression Vector Kit as described previously (22).
Fractionation of Muscle Tissue.
Muscles were homogenized in cold buffer (20 mM Tris⋅HCl pH 7.2, 5 mM EGTA, 100 mM KCl, 1% Triton X-100, and protease and phosphatase inhibitor mixtures), and myofibrils were isolated by centrifugation at 3,000 × g for 30 min at 4 °C (16). The myofibrillar pellet was washed twice in wash buffer (20 mM Tris⋅HCl pH 7.2, 100 mM KCl, and 1 mM DTT), and after the final centrifugation (3,000 × g for 10 min at 4 °C) was resuspended in storage buffer (20 mM Tris⋅HCl, pH 7.2, 100 mM KCl, 1 mM DTT, and 20% vol/vol glycerol) and kept at −80 °C.
To isolate the nuclear fraction, the myofibrillar pellet (6,000 × g) was washed twice in buffer C (20 mM Tris⋅HCl, pH 7.6, 100 mM KCl, 5 mM EDTA, 1 mM DTT, 1 mM sodium orthovanadate, 1 mM PMSF, 50 mM NaF, and protease inhibitor mixture), the obtained pellet was then resuspended in buffer N (20 mM Hepes pH 7.9, 1.5 mM MgCl2, 500 mM NaCl, 5 mM EDTA, 20% glycerol, 1% Triton X-100, 1 mM sodium orthovanadate, 10 μg/mL leupeptin, 3 mM benzamidine, 1 mM PMSF, and 50 mM NaF), and following incubation on ice for 30 min and a final centrifugation (9,000 × g for 30 min at 4 °C), the supernatant was collected as an isolated nuclear fraction.
To extract myofibrils and purify desmin filaments, myofibrillar pellets (equivalent to 0.1% of total myofibrils/muscle) were resuspended in the extraction buffer (0.6 M KCl, 1% Triton X-100, 2 mM EDTA, 1 mM DTT, 2 mM PMSF, 10 μg/mL leupeptin, 3 mM benzamidine, 1 μg/mL trypsin inhibitor, and 1× PBS) for 10 min at 4 °C, spun at 3,000 × g for 10 min at 4 °C, and then washed briefly with 20 mM Tris buffer.
Protein Analysis.
All assays were performed as described previously (16, 18, 22). To determine the total number of myofibrils per muscle, the concentration of myofibrillar proteins was multiplied by the total volume of myofibrils. To determine the ratio of actin to MyHC, equal amounts (2.5 μg) of isolated myofibrils from transfected muscles were analyzed by SDS/PAGE and Coomassie blue staining, the intensity of the specific bands was measured by densitometry, and the ratio of densities was graphed.
For immunoblotting, soluble or myofibrillar fractions from TA muscles as well as the in vitro ubiquitination reactions were resolved by SDS/PAGE, transferred onto PVDF membranes, and immunoblotted with specific antibodies.
In Vitro Ubiquitination Assay.
The capability of Trim32 to ubiquitinate washed myofibrils (5 μg, as described above) was assayed for 90 min at 37 °C in 20-μL mixtures containing 22.5 nM E1, 0.75 μM UbcH5, 0.4 μM Trim32, and 59 μM His-ubiquitin in reaction buffer (2 mM ATP, 20 mM Tris⋅HCl pH 7.6, 20 mM KCl, 5 mM MgCl2, and 1 mM DTT). The ubiquitinated filaments were analyzed by SDS/PAGE and immunoblotting with anti-ubiquitin (FK2 clone).
This assay was also used to determine the ability of Trim32 to ubiquitinate isolated desmin filaments. Specifically, before the ubiquitination reaction, desmin filaments were purified as described above, treated with 25 U of protein phosphatase 1 (P0754S; BioLabs) for 1 h at 30 °C or left untreated, and washed three times with reaction buffer.
Immunofluorescence Labeling of Paraffin-Embedded Muscle Sections.
Paraffin-embedded longitudinal sections of innervated and denervated TA mouse muscles mice were cut at 10 µm. To remove paraffin, slides were immersed in xylene for 5 min and then gradually rehydrated in 100%, 95%, 50%, 25%, and 0% ethanol/PBS. Immunofluorescence analysis of the rehydrated sections was performed using a 1:50 dilution of desmin antibody and a 1:1,000 dilution of Alexa Fluor 555- conjugated secondary antibody, both diluted in blocking solution (50 mg/mL BSA/PBST). Images were collected at room temperature using a Nikon Ni-U upright fluorescence microscope with a Plan Fluor 40× 1.3 NA objective lens, a 545/30-nm excitation filter and 620/60-nm emission filter, a Hamamatsu C8484-03 cooled CCD camera, and MetaMorph software.
Real-Time qPCR.
Total RNA was isolated from muscle using TRI reagent (T9424; Sigma-Aldrich) and served as a template for the synthesis of cDNA by reverse transcription. Real-time qPCR was performed on mouse target genes using specific primers (Table S2) and the Perfecta SYBR Green qPCR Kit (95073-012; Quanta Biosciences) according to the manufacturer’s protocol, and PCR using the Red Load Taq Master Kit (PCR-108L; LAROVA).
Computational Analysis.
Promoter sequences 500 bp upstream and 500 bp downstream of transcription start sites (TSS) of the genes p97/VCP, MuRF1, Nedd4, Rpt1, and Trim32 were downloaded from the UCSC Genome Browser (62), genome version mm9. The sequence was then used as a template for predication of transcription factor-binding sites using the Match algorithm from TRANSFAC (63), filtered by mouse preferences. PAX4-binding sites were identified using four different weighted matrices: V$PAX4_01 (ngn[a/c/g]gTCANGcgtgnn[g/c]nn[c/t]n), V$PAX4_02 (naa[A/T]AATTan[g/c]), V$PAX4_03 (nnnnn[c/t]CACCC[c/g/t]), and V$PAX4_04 ([A/G]AAAAwtannnnnnnnnnnnnnnycacncc). Binding sites were chosen based on Match scores based on the criteria of a matrix Match score >0.9 and a sequence match >0.8.
Statistical Analysis and Image Acquisition.
Data are presented as mean ± SEM. The statistical significance was accessed using the paired Student’s t test. Muscle sections were imaged at room temperature with an upright fluorescent microscope (Nikon Ni-U) and a monochrome camera (Hamamatsu C8484-03). Image acquisition and processing were performed using MetaMorph software. Black and white images were processed with Adobe Photoshop CS3, version 10.0.1.
Acknowledgments
This project was supported by a Israeli Ministry of Science, Technology, and Space Grant; a Dr. Bernard and Bobbie Lublin Cancer Research Award; and a Malat Family Award (to S.C.). The data presented in Fig. 4 were supported by grants from the National Institute for General Medical Sciences (R01 GM051923), the Muscular Dystrophy Association, and Project ALS (to A.L.G.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1753.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612988114/-/DCSupplemental.
References
- 1.Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287(4):C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- 2.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 3.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015;14(1):58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 4.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271(43):26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: Response to food deprivation. FASEB J. 2002;16(13):1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 8.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 9.Sacheck JM, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21(1):140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 10.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Brault JJ, Schild A, Goldberg AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy. 2008;4(3):378–380. doi: 10.4161/auto.5633. [DOI] [PubMed] [Google Scholar]
- 13.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285(25):19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 15.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke BA, et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198(4):575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frosk P, et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin ligase gene. Am J Hum Genet. 2002;70(3):663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudryashova E, Wu J, Havton LA, Spencer MJ. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet. 2009;18(7):1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudryashova E, Struyk A, Mokhonova E, Cannon SC, Spencer MJ. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum Mol Genet. 2011;20(20):3925–3932. doi: 10.1093/hmg/ddr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen S, Lee D, Zhai B, Gygi SP, Goldberg AL. Trim32 reduces PI3K-Akt-FoxO signaling in muscle atrophy by promoting plakoglobin-PI3K dissociation. J Cell Biol. 2014;204(5):747–758. doi: 10.1083/jcb.201304167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, et al. Tripartite motif 32 prevents pathological cardiac hypertrophy. Clin Sci (Lond) 2016;130(10):813–828. doi: 10.1042/CS20150619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazarides E, Hubbard BD. Immunological characterization of the subunit of the 100 A filaments from muscle cells. Proc Natl Acad Sci USA. 1976;73(12):4344–4348. doi: 10.1073/pnas.73.12.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarides E. The distribution of desmin (100 A) filaments in primary cultures of embryonic chick cardiac cells. Exp Cell Res. 1978;112(2):265–273. doi: 10.1016/0014-4827(78)90209-4. [DOI] [PubMed] [Google Scholar]
- 26.Milner DJ, Weitzer G, Tran D, Bradley A, Capetanaki Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J Cell Biol. 1996;134(5):1255–1270. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muñoz-Mármol AM, et al. A dysfunctional desmin mutation in a patient with severe generalized myopathy. Proc Natl Acad Sci USA. 1998;95(19):11312–11317. doi: 10.1073/pnas.95.19.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conover GM, Henderson SN, Gregorio CC. A myopathy-linked desmin mutation perturbs striated muscle actin filament architecture. Mol Biol Cell. 2009;20(3):834–845. doi: 10.1091/mbc.E08-07-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisler N, Weber K. The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J. 1982;1(12):1649–1656. doi: 10.1002/j.1460-2075.1982.tb01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sihag RK, Inagaki M, Yamaguchi T, Shea TB, Pant HC. Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp Cell Res. 2007;313(10):2098–2109. doi: 10.1016/j.yexcr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inagaki M, Nishi Y, Nishizawa K, Matsuyama M, Sato C. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature. 1987;328(6131):649–652. doi: 10.1038/328649a0. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi S, Rocancourt D, Buckingham M, Relaix F. Lack of in vivo functional compensation between Pax family groups II and III in rodents. Mol Biol Evol. 2011;28(10):2787–2798. doi: 10.1093/molbev/msr114. [DOI] [PubMed] [Google Scholar]
- 33.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122(9):2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 34.Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: Roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73(1):1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 35.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 36.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386(6623):399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 37.Shimajiri Y, et al. A missense mutation of Pax4 gene (R121W) is associated with type 2 diabetes in Japanese. Diabetes. 2001;50(12):2864–2869. doi: 10.2337/diabetes.50.12.2864. [DOI] [PubMed] [Google Scholar]
- 38.Mauvais-Jarvis F, et al. PAX4 gene variations predispose to ketosis-prone diabetes. Hum Mol Genet. 2004;13(24):3151–3159. doi: 10.1093/hmg/ddh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen F, Chang R, Trivedi M, Capetanaki Y, Cryns VL. Caspase proteolysis of desmin produces a dominant-negative inhibitor of intermediate filaments and promotes apoptosis. J Biol Chem. 2003;278(9):6848–6853. doi: 10.1074/jbc.M212021200. [DOI] [PubMed] [Google Scholar]
- 40.Piccirillo R, Goldberg AL. The p97/VCP ATPase is critical in muscle atrophy and the accelerated degradation of muscle proteins. EMBO J. 2012;31(15):3334–3350. doi: 10.1038/emboj.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 42.Koncarevic A, Jackman RW, Kandarian SC. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J. 2007;21(2):427–437. doi: 10.1096/fj.06-6665com. [DOI] [PubMed] [Google Scholar]
- 43.Rabinovich E, Kerem A, Fröhlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22(2):626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T, Tanaka K, Inoue K, Kakizuka A. Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J Biol Chem. 2002;277(49):47358–47365. doi: 10.1074/jbc.M207783200. [DOI] [PubMed] [Google Scholar]
- 45.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49(1):59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280(17):4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 47.Kudryashova E, Kramerova I, Spencer MJ. Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J Clin Invest. 2012;122(5):1764–1776. doi: 10.1172/JCI59581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X, et al. Protein kinase C-mediated desmin phosphorylation is related to myofibril disarray in cardiomyopathic hamster heart. Exp Biol Med (Maywood) 2002;227(11):1039–1046. doi: 10.1177/153537020222701113. [DOI] [PubMed] [Google Scholar]
- 49.Li Z, et al. Desmin is essential for the tensile strength and integrity of myofibrils but not for myogenic commitment, differentiation, and fusion of skeletal muscle. J Cell Biol. 1997;139(1):129–144. doi: 10.1083/jcb.139.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goebel HH. Desmin-related neuromuscular disorders. Muscle Nerve. 1995;18(11):1306–1320. doi: 10.1002/mus.880181114. [DOI] [PubMed] [Google Scholar]
- 51.Goldfarb LG, et al. Missense mutations in desmin associated with familial cardiac and skeletal myopathy. Nat Genet. 1998;19(4):402–403. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]
- 52.Li D, et al. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation. 1999;100(5):461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 53.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Desmin myopathy. Brain. 2004;127(Pt 4):723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 54.Inagaki N, et al. Visualization of protein kinase activities in single cells by antibodies against phosphorylated vimentin and GFAP. Neurochem Res. 1996;21(7):795–800. doi: 10.1007/BF02532302. [DOI] [PubMed] [Google Scholar]
- 55.Kawajiri A, et al. Functional significance of the specific sites phosphorylated in desmin at cleavage furrow: Aurora-B may phosphorylate and regulate type III intermediate filaments during cytokinesis coordinatedly with Rho-kinase. Mol Biol Cell. 2003;14(4):1489–1500. doi: 10.1091/mbc.E02-09-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agnetti G, et al. Desmin modifications associate with amyloid-like oligomers deposition in heart failure. Cardiovasc Res. 2014;102(1):24–34. doi: 10.1093/cvr/cvu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li YP, et al. TNF-alpha acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19(3):362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. AMP-activated protein kinase agonists increase mRNA content of the muscle-specific ubiquitin ligases MAFbx and MuRF1 in C2C12 cells. Am J Physiol Endocrinol Metab. 2007;292(6):E1555–E1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 59.Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119(7):1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Köster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. Intermediate filament mechanics in vitro and in the cell: from coiled coils to filaments, fibers and networks. Curr Opin Cell Biol. 2015;32:82–91. doi: 10.1016/j.ceb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geisler N, Weber K. Phosphorylation of desmin in vitro inhibits formation of intermediate filaments: Identification of three kinase A sites in the aminoterminal head domain. EMBO J. 1988;7(1):15–20. doi: 10.1002/j.1460-2075.1988.tb02778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]