Significance
This study shows that the hormonal environment in which a tumor originates may affect a hormone receptor’s enhancer usage. We further show that enhancer function is less tissue specific than previously thought. By implementing ChIP sequencing in a unique patient cohort, we compared estrogen receptor α (ERα) profiles in endometrial tumors that developed in different hormonal environments and integrated these comparisons with transcriptomic data. Our data show that tumors associated with therapeutic intervention have a distinct ERα DNA-binding signature with regulatory potentials that resemble ERα-binding patterns in breast cancer. These results highlight the value of cistromic analyses in clinical specimens, which enabled us to distinguish subtypes of tumors on the level of transcriptional regulation.
Keywords: endometrial cancer, tamoxifen, breast cancer, estrogen receptor, ChIP-seq
Abstract
The DNA-binding sites of estrogen receptor α (ERα) show great plasticity under the control of hormones and endocrine therapy. Tamoxifen is a widely applied therapy in breast cancer that affects ERα interactions with coregulators and shifts the DNA-binding signature of ERα upon prolonged exposure in breast cancer. Although tamoxifen inhibits the progression of breast cancer, it increases the risk of endometrial cancer in postmenopausal women. We therefore asked whether the DNA-binding signature of ERα differs between endometrial tumors that arise in the presence or absence of tamoxifen, indicating divergent enhancer activity for tumors that develop in different endocrine milieus. Using ChIP sequencing (ChIP-seq), we compared the ERα profiles of 10 endometrial tumors from tamoxifen users with those of six endometrial tumors from nonusers and integrated these results with the transcriptomic data of 47 endometrial tumors from tamoxifen users and 64 endometrial tumors from nonusers. The ERα-binding sites in tamoxifen-associated endometrial tumors differed from those in the tumors from nonusers and had distinct underlying DNA sequences and divergent enhancer activity as marked by histone 3 containing the acetylated lysine 27 (H3K27ac). Because tamoxifen acts as an agonist in the postmenopausal endometrium, similar to estrogen in the breast, we compared ERα sites in tamoxifen-associated endometrial cancers with publicly available ERα ChIP-seq data in breast tumors and found a striking resemblance in the ERα patterns of the two tissue types. Our study highlights the divergence between endometrial tumors that arise in different hormonal conditions and shows that ERα enhancer use in human cancer differs in the presence of nonphysiological endocrine stimuli.
Estrogen receptor α (ERα) is a steroid hormone receptor that behaves as a transcription factor by interacting with the DNA. The DNA-binding profile (cistrome) of ERα is dependent on context and tissue type (1). The hormonal environment of the cell greatly influences this cistrome because estrogen activates ERα by binding its ligand-binding domain. Upon activation, ERα’s structural conformation changes to interact with cofactors at the DNA (2) and to regulate a transcriptional program that drives cell proliferation (3). Hence, the hormonal environment modulates the ERα cistrome and thereby rewires downstream effects.
Endocrine therapies, as exemplified by tamoxifen, manipulate the DNA-binding capacities of the steroid hormone receptor ERα. Tamoxifen, a small-molecule inhibitor that competes with estrogens to bind ERα, is a major endocrine agent used in treating ERα-positive breast cancer patients. Studies in the breast cancer cell line MCF-7 show that prolonged tamoxifen exposure shifts the ERα cistrome, which consequently changes gene expression (4–6).
Tamoxifen is a well-known selective estrogen receptor modulator (SERM) with tissue-selective physiological action. Early reports on tamoxifen’s effects on transplanted MCF-7 cells in athymic mice revealed decreased tumor cell growth but also increased uterine weight in response to drug treatment (7). A species-selective action of tamoxifen could not be excluded at this stage. Later, however, growth-stimulatory effects of tamoxifen on human endometrial carcinomas were shown in a nude mouse model (8) and were reported in breast cancer patients, in whom tamoxifen treatment increased endometrial thickness as well as the risk of endometrial cancer in postmenopausal women by two- to sevenfold, depending upon treatment duration (9–15).
Another study directly compared the contrasting actions of tamoxifen in athymic mice, transplanting endometrial EnCa101 tumors and MCF-7–derived tumors within the same mouse (16). Tamoxifen blocked tumor growth in the MCF-7 tumor while stimulating growth in the EnCa101 tumor. The two tumor types however, had qualitatively very similar patterns of tamoxifen metabolites, precluding differential tamoxifen metabolism as potential explanation for the observed tissue-selective effects.
Cell line data illustrated that tamoxifen affected gene regulation differently in the endometrium than in the breast (17). These data showed the agonistic effects of tamoxifen on ERα using the endometrial cancer cell line Ishikawa, but only for a handful of binding sites and related genes (17). Our previously published data revealed thousands of ERα-binding sites in multiple endometrial tumors from tamoxifen-treated patients and showed remarkable overlap with the ERα cistrome in breast cancer (18), but we lacked data on the tumors of patients who had never received endocrine treatment.
We hypothesize that the ERα cistrome differs in ERα-positive tumors that arise in the presence or absence of tamoxifen and expect this difference will have consequences for the tumor’s transcriptome. The TAMARISK (Tamoxifen-Associated Malignancies: Aspects of Risk) study, which consists of endometrial tumors from patients who had a history of breast cancer, (half of whom had received tamoxifen), provides an opportunity to investigate this hypothesis. We combined ChIP coupled with massive parallel sequencing (ChIP-seq) and gene-expression data in endometrial tumors from this cohort and used bioinformatic analysis to investigate differences in endometrial tumors that originated in different hormonal environments (i.e., tamoxifen vs. no tamoxifen).
We found that tamoxifen-associated endometrial tumors have a distinct ERα DNA-binding signature that differs from that of endometrial tumors that develop in a hormonal environment without tamoxifen. The differentially enriched ERα sites were associated with differences in gene expression, and the enriched ERα sites in tamoxifen-associated endometrial tumors resembled the ERα-binding patterns in breast cancer. Our data thus suggest that the hormonal environment in which a tumor arises is associated with differential enhancer use of ERα.
Materials and Methods
Patient Material.
The design of the TAMARISK study has been described previously as a nation-wide population-based prospective cohort of patients who developed uterine corpus cancer after breast cancer (13, 19). Here we present the prospective part of this study, in which samples were obtained from patients who developed uterine corpus cancer between 2003 and 2006 after previously being diagnosed for breast cancer. Residual endometrial samples of anonymized patients, who signed an informed consent, from the TAMARISK study were used. This study was performed in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in The Netherlands (https://www.federa.org) and has been approved by the local medical ethics committee of The Netherlands Cancer Institute. Endometrial samples were derived from patients who had a history of breast cancer. Fresh-frozen (frozen within 30 min after surgery and stored at −80 °C) endometrial tumor specimens were collected and used for ChIP-seq and microarray. Clinicopathological parameters of these endometrial samples can be found in Table 1. Detection of microsatellite instability was performed in the retrospective part of the cohort (Table S1), as described previously (13, 18).
Table 1.
Clinicopathological parameters of endometrioid adenocarcinomas
| Parameter | ChIP-seq | Gene expression | ||
| Tamoxifen user | Nonuser | Tamoxifen user | Nonuser | |
| N | 9 | 6 | 47 | 64 |
| Tamoxifen use in years, median (range) | 3.2 (1.4–5.6) | N/A | 4.35 (2.0–15.0) | N/A |
| Interval between breast and endometrial cancer in years, median (range) | 3.4 (2.0–4.7) | 5.6 (0–17.1) | 5.0 (2.1–26.5) | 6.6 (−0.1 to 29.9) |
| Median age in years at breast cancer diagnosis (range) | 60.5 (51.0–83.2) | 55.6 (44.2–67.3) | 60.7 (35.9–83.2) | 57.4 (35.2–77.3) |
| Median age in years at endometrial cancer diagnosis (range) | 64.2 (53.6–87.6) | 59.4 (54.3–75.0) | 69.5 (53.6–89.0) | 68.0 (49.9–84.6) |
| Menopausal status, n (%)* | ||||
| Postmenopausal | 9 (100) | 6 (100) | 43 (91.5) | 60 (93.7) |
| Perimenopausal | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.6) |
| Unknown | 0 (0.0) | 0 (0.0) | 4 (8.5) | 3 (4.7) |
| Tamoxifen use, n (%) | ||||
| Former user | 0 (0.0) | 0 (0.0) | 9 (19.1) | 0 (0.0) |
| Recent user (last 12 mo) | 0 (0.0) | 0 (0.0) | 11 (23.4) | 0 (0.0) |
| Current user | 9 (100) | 0 (0.0) | 23 (48.9) | 0 (0.0) |
| Nonuser | 0 (0.0) | 6 (100) | 0 (0.0) | 64 (100) |
| Unknown | 0 (0.0) | 0 (0.0) | 4 (8.5) | 0 (0.0) |
| Histological type, n (%) | ||||
| Endometrioid adenocarcinoma and variants | 9 (100) | 6 (100) | 47 (100.0) | 64 (100.0) |
| Use of other hormonal therapy, n (%) | 0 (0.0) | 0 (0.0) | 4 (8.5) | 6 (9.4) |
| FIGO stage, n (%) | ||||
| I | 7 (77.7) | 5 (83.3) | 34 (72.3) | 47 (73.4) |
| II | 1 (11.1) | 0 (0.0) | 5 (10.6) | 8 (12.5) |
| III/IV | 1 (11.1) | 1 (16.7) | 4 (8.5) | 6 (9.4) |
| Unknown | 0 (0.0) | 0 (0.0) | 4 (8.5) | 3 (4.7) |
| Chemotherapy, n (%) | ||||
| Yes | 4 (44.4) | 2 (33.3) | 10 (21.3) | 9 (14.1) |
| No | 5 (55.6) | 4 (66.7) | 34 (72.3) | 52 (81.3) |
| Unknown | 0 (0.0) | 0 (0.0) | 3 (6.4) | 3 (4.7) |
There are no significant differences between tamoxifen users and nonusers with regard to interval between the diagnosis of breast and diagnosis of endometrial cancer, the age at the diagnosis of breast cancer, or the age at the diagnosis of endometrial cancer (t test) or in FIGO stage or use of chemotherapy (χ2 test). FIGO, International Federation of Gynecologists and Obstetricians.
At diagnosis of endometrial cancer.
Table S1.
Clinicopathological parameters of the endometrioid adenocarcinomas from the retrospective part of the TAMARISK study
| Parameter | Tamoxifen user | Nonuser |
| N | 67 | 129 |
| Tamoxifen use in years, median (range) | 3.8 (2.0–17.9) | N/A |
| Interval between breast and endometrial cancer in years, median (range) | 7.2 (2.1–18.7) | 5.8 (1.0–21.5) |
| Age at breast cancer diagnosis, median (range) | 64.0 (34.9–79.5) | 59.9 (37.4–87.9) |
| Age at endometrial cancer diagnosis, median (range) | 70.8 (52.1–86.6) | 68.0 (44.1–93.5) |
| Menopausal status, n (%)* | ||
| Postmenopausal | 66 (98.5) | 120 (93.0) |
| Perimenopausal | 1 (1.5) | 5 (3.9) |
| Premenopausal | 0 (0.0) | 3 (2.3) |
| Unknown | 0 (0.0) | 1 (0.8) |
| Tamoxifen use, n (%) | ||
| Former user | 20 (29.9) | 0 (0.0) |
| Recent user (last 12 mo) | 47 (70.1) | 0 (0.0) |
| Current user | 0 (0.0) | 0 (0.0% |
| Nonuser | 0 (0.0) | 129 (100.0) |
| Unknown | 0 (0.0) | 0 (0.0) |
| Histological type, n (%) | ||
| Endometrioid adenocarcinoma and variants | 67 (100.0) | 129 (100.0) |
| Use of other hormonal therapy, n (%)* | 6 (9.0) | 9 (7.0) |
| FIGO stage, n (%) | ||
| I | 43 (64.2) | 99 (76.7) |
| II | 3 (4.5) | 7 (5.4) |
| III/IV | 1 (1.5) | 0 (0.0) |
| Unknown | 19 (28.4) | 21 (0.0) |
| MSI status, n (%) | ||
| Negative or low | 42 (62.7) | 74 (57.4) |
| High | 9 (13.4) | 25 (19.4) |
| Unknown | 16 (23.9) | 30 (23.3) |
| Chemotherapy, n (%) | ||
| Yes | 7 (10.4) | 12 (9.3) |
| No | 60 (89.6) | 115 (89/1) |
| Unknown | 0 (0.0) | 2 (1.6) |
There are no significant differences between tamoxifen users and nonusers with regard to the interval time between breast and endometrial cancer, the age at diagnosis of breast cancer endometrial cancer, according to t test; there are no differences in International Federation of Gynecologists and Obstetricians (FIGO) stage, use of chemotherapy, or microsatellite instability (MSI) status, according to χ2 test.
At diagnosis of endometrial cancer.
Sections were stained with H&E and ERα as previously described (13). H&E staining of all tumors used (for both ChIP-seq and microarray) was reviewed by multiple gynecologic pathologists in terms of classification and grade (WHO classification 1994). ERα staining was done using mouse monoclonal antibodies (MCA1799) in a dilution of 1:20 (Serotec).
ChIP-Seq.
ChIP-seq was performed as previously described (20, 21) on 16 endometrioid adenocarcinomas from the prospective TAMARISK cohort (Table 1). Thirty 30-µm cryosections of fresh-frozen endometrioid adenocarcinomas that contained at least 50% tumor tissue were fixed with 1% formaldehyde for 20 min and were processed for sonication. For each ChIP, 10 μg of antibody and 100 μL of Protein A magnetic beads (Invitrogen) were used. Antibodies raised to detect ERα (SC-543; Santa Cruz) and H3K27ac (ab4729; Abcam) were used.
High-Throughput Sequencing and Processing.
Single-end 51-bp (ERα ChIP-seq) and 65-bp [histone 3 containing the acetylated lysine 17 (H3K27ac)] ChIP-seq reads were generated using the Illumina HiSeq. 2000 Genome Analyzer and were aligned to the hg19 human genome using Burrow–Wheeler Aligner (BWA) version 0.5.9 with default parameters. Reads that were poorly aligned or that mapped to multiple locations were filtered out based on the mapping quality. Only reads with mapping quality scores (MAPQ) >20 were retained for further peak calling and analysis. The number of mapped and filtered reads is listed in Table S2.
Table S2.
Number of sequencing reads, mapped reads, and called peaks
| Sample ID | Factor | Group | Total reads | Mapped reads | % mapped reads | Filtered reads | % filtered reads | No. peaks |
| wz154 | ERα | Tamoxifen user | 15,331,544 | 14,207,826 | 92.67 | 12,342,539 | 86.87 | 1,589 |
| wz483 | ERα | Tamoxifen user | 18,754,169 | 17,283,692 | 92.16 | 1,5005,839 | 86.82 | 11,560 |
| wz484 | ERα | Tamoxifen user | 19,287,976 | 18,138,678 | 94.04 | 15,509,489 | 85.51 | 2,355 |
| wz489 | ERα | Tamoxifen user | 18,569,672 | 16,528,291 | 89.01 | 14,129,722 | 85.49 | 714 |
| wz487 | ERα | Tamoxifen user | 23,924,152 | 15,740,024 | 65.79 | 13,543,920 | 86.05 | 100 |
| WZ485 | ERα | Tamoxifen user | 32,296,216 | 26,465,862 | 81.95 | 22,901,235 | 86.53 | 504 |
| wz439 | ERα | Tamoxifen user | 19,603,948 | 17,976,811 | 91.7 | 15,752,509 | 87.63 | 21,800 |
| wz152 | ERα | Tamoxifen user | 14,930,170 | 12,101,826 | 81.06 | 10,116,835 | 83.6 | 27,915 |
| wz170 | ERα | Tamoxifen user | 20,680,117 | 19,906,669 | 96.26 | 17,433,136 | 87.57 | 1,631 |
| wz438 | ERα | Tamoxifen user | 15,611,205 | 14,725,046 | 94.32 | 12,681,132 | 86.12 | 123 |
| wz490 | ERα | Nonuser | 20,899,419 | 18,917,776 | 90.52 | 16,402,313 | 86.7 | 455 |
| wz488 | ERα | Nonuser | 19,444,847 | 18,274,374 | 93.98 | 160,19,957 | 87.66 | 26,714 |
| wz486 | ERα | Nonuser | 30,403,020 | 18,373,438 | 60.43 | 15,788,112 | 85.93 | 19,055 |
| wz470 | ERα | Nonuser | 32,936,222 | 29,363,575 | 89.15 | 24,987,722 | 85.1 | 1,312 |
| wz503 | ERα | Nonuser | 14,880,365 | 13,780,675 | 92.61 | 11,885,851 | 86.25 | 391 |
| wz504 | ERα | Nonuser | 22,411,738 | 21,007,108 | 93.73 | 18,174,472 | 86.52 | 3,223 |
| wz984 | H3K27ac | Tamoxifen user | 23,175,814 | 22,016,196 | 95 | 20,519,524 | 93.2 | |
| wz985 | H3K27ac | Tamoxifen user | 22,900,854 | 22,227,909 | 97.06 | 20,621,089 | 92.77 | |
| wz565 | H3K27ac | Tamoxifen user | 41,850,397 | 40,395,208 | 96.52 | 36,913,144 | 91.38 | |
| wz990 | H3K27ac | Tamoxifen user | 29,613,393 | 28,327,999 | 95.66 | 26,416,294 | 93.25 | |
| wz991 | H3K27ac | Tamoxifen user | 24,977,421 | 23,739,843 | 95.05 | 21,921,727 | 92.34 | |
| wz992 | H3K27ac | Tamoxifen user | 31,443,859 | 29,791,112 | 94.74 | 27,645,455 | 92.8 | |
| wz1368 | H3K27ac | Tamoxifen user | 20,886,714 | 19,953,687 | 95.53 | 18,656,222 | 93.5 | |
| wz1045 | H3K27ac | Tamoxifen user | 25,954,861 | 24,892,026 | 95.91 | 23,468,485 | 94.28 | |
| wz981 | H3K27ac | Tamoxifen user | 26,601,717 | 25,544,357 | 96.03 | 23,749,000 | 92.97 | |
| Tam_5 | H3K27ac | Nonuser | 27,210,981 | 26,584,367 | 97.7 | 25,114,577 | 94.47 | |
| Tam_33 | H3K27ac | Nonuser | 28,796,446 | 27,937,355 | 97.02 | 26,006,528 | 93.09 | |
| Tam_38 | H3K27ac | Nonuser | 23,348,596 | 22,763,584 | 97.49 | 21,272,824 | 93.45 | |
| Tam_55 | H3K27ac | Nonuser | 29,667,180 | 28,638,563 | 96.53 | 26,233,736 | 91.6 | |
| Tam_102 | H3K27ac | Nonuser | 22,491,570 | 21,890,598 | 97.33 | 20,508,226 | 93.69 | |
| Tam_119 | H3K27ac | Nonuser | 22,791,103 | 22,188,485 | 97.36 | 20,885,046 | 94.13 |
Peak Calling.
Two algorithms were used for peak calling of ERα ChIP-seq data: MACS 1.4 (22) and DFilter v1 (23). We used MACS with default parameter settings, except for the P value cutoff, which we set at 10−7. We used DFilter with parameter settings as recommended for transcription factor ChIP-seq peak calling (bs = 50, ks = 30, refine, nonzero). Only peaks called with both peak-calling algorithms were considered for further analyses. The number of called peaks is listed in Table S2. The raw and processed data have been deposited in the Gene Expression Omnibus database (accession no. GSE94031).
DNA Copy Number Calling.
We used the CopywriteR R package (24) to extract DNA copy number information from off-target (background) reads of ChIP-seq data. We used the package with the default parameters.
ChIP-Seq Data Analysis.
The DiffBind R package (6) was used to identify genomic regions differentially bound by ERα in two groups of endometrial cancer. Peaks present in at least half of the patients in one of the groups were considered for the analysis. Differential read count analysis was performed without control read subtraction; the significance threshold was set at a false-discovery rate (FDR) <0.1. Heatmaps visualizing raw the ChIP-seq signal in peaks were built using seqMINER 1.3.3 software (25). Snapshots of the ChIP-seq signal and average signal profiles in peaks were generated using the TransView R package (26).
Annotation of ChIP-seq peaks relative to the nearest gene was performed using the cis-regulatory element annotation (CEAS) tool (27) with default settings. Motifs enriched at ERα-binding sites were identified using SeqPos tools (with default settings) available through Galaxy Cistrome (28). Genes that had an ERα peak within the gene body or 20 kb upstream of the transcription start site (TSS) were identified as potential targets of the corresponding ERα-binding sites. For functional enrichment of the genes, we used QIAGEN’s Ingenuity Pathway Analysis (IPA; www.ingenuity.com). Gene ontology (GO) analysis of the potential target genes was performed using the PANTHER gene-classification database (29).
Public ChIP-Seq Data Processing and Analysis.
We previously published the data on endometrial tumors of tamoxifen users that we included in this study with National Center for Biotechnology Information (NCBI) Gene Expression omnibus (GEO) accession numbers GSM2144746, GSM2144758, and GSM2144760 (18). We also used publicly available ERα ChIP-seq data from primary breast cancer tissue from two cohorts of patients. The data were obtained from NCBI GEO [accession numbers GSE32222 (6) and GSE40867 (21)]. Raw FASTQ files were aligned to the hg19 genome with BWA. The TransView R package was used to generate average signal profiles and to calculate reads per kilobase of transcript per million reads mapped (RPKM) values in peaks. The publicly available cell line ChIP-seq data that were used are presented in Table S3. For data provided by the Encode project (30), bed files were downloaded from https://www.encodeproject.org. Intersection of peak lists from two replicates was created; only peaks shared by the two replicates were used where applicable. For ERα and forkhead box A1 (FOXA1) ChIP-seq in the breast cancer cell line MCF-7, the data from Hurtado et al. (4) were used. Raw FASTQ files were downloaded from https://www.ebi.ac.uk/arrayexpress/; alignment and peak calling were performed as described above. The intersection of peak lists from multiple replicates was used.
Table S3.
Publicly available cell line ChIP-seq data
| Cell line | Factor | Source | Accession no. |
| MCF7 | CTCF | Encode | ENCSR000DMS |
| MCF7 | CTCF | Encode | ENCSR000DMR |
| Ishikawa | CTCF | Encode | ENCSR000BQE |
| T47D | CTCF | Encode | ENCSR000BNO |
| MCF7 | TCF12 | Encode | ENCSR000BUN |
| Ishikawa | TCF12 | Encode | ENCSR000BUV |
| MCF7 | FOXM1 | Encode | ENCSR000BUJ |
| Ishikawa | FOXM1 | Encode | ENCSR000BUS |
| MCF7 | REST | Encode | ENCSR000BSP |
| Ishikawa | REST | Encode | ENCSR000BUU |
| MCF7 | EGR1 | Encode | ENCSR000BUX |
| Ishikawa | EGR1 | Encode | ENCSR000BSQ |
| MCF7 | MAX | Encode | ENCSR000BUL |
| Ishikawa | MAX | Encode | ENCSR000BTY |
| MCF7 | EP300 | Encode | ENCSR000BTR |
| Ishikawa | EP300 | Encode | ENCSR000BUE |
| T47D | EP300 | Encode | ENCSR000BLM |
| MCF7 | RAD21 | Encode | ENCSR000BTQ |
| Ishikawa | RAD21 | Encode | ENCSR000BTU |
| MCF7 | CEBPB | Encode | ENCSR000BSR |
| Ishikawa | CEBPB | Encode | ENCSR000BTT |
| MCF7 | TEAD4 | Encode | ENCSR000BUO |
| Ishikawa | TEAD4 | Encode | ENCSR000BSW |
| MCF7 | SRF | Encode | ENCSR000BVA |
| Ishikawa | SRF | Encode | ENCSR000BTD |
| MCF7 | TAF1 | Encode | ENCSR000AHF |
| Ishikawa | TAF1 | Encode | ENCSR000BTO |
| MCF7 | POLR2 | Encode | ENCSR000DMN |
| MCF7 | POLR2 | Encode | ENCSR000DMT |
| MCF7 | POLR2 | Encode | ENCSR000DMK |
| Ishikawa | POLR2 | Encode | ENCSR000BKI |
| Ishikawa | ESR1 | Encode | ENCSR000BIY |
| T47D | ESR1 | Encode | ENCSR000BKN |
| Ishikawa | FOXA1 | Encode | ENCSR000BKW |
| T47D | FOXA1 | Encode | ENCSR000BKY |
| MCF7 | FOXA1 | ArrayExpress | ERR022028/ERR022029 |
| MCF7 | ESR1 | ArrayExpress | ERR022052/ERR022053/ERR022048/ERR022049/ERR022026 |
RNA Isolation, cDNA Synthesis, and RNA Amplification and Labeling for Microarray.
Microarray data were generated early after tissue collection as part of the prospective TAMARISK study. We included endometrial tumors of the endometrioid adenocarcinoma subtype from 47 patients who had been on tamoxifen for at least 2 years and from 64 patients who never used tamoxifen (Table 1). Thirty 30-µm cryosections of fresh-frozen endometrial tumors that were at least 50% tumorigenic were used for RNA isolation using TRIzol (15596-026; Invitrogen) according to the manufacturer’s instructions. RNA was purified using the RNeasy Mini Kit (74104; Qiagen) and was treated with DNase using the RNase-Free DNase Set (79254; Qiagen). The concentration and purity of the RNA were measured on a nanodrop spectrophotometer (Isogen Life-Science), and the integrity of the RNA was determined by agarose gel. Next, cDNA was synthesized. First- and second-strand cDNA synthesis was performed using T7-(dT)24 primer and RT SuperScript III (18064-022; Invitrogen). The cDNA was purified using a QIAquick PCR Purification Column (Qiagen) and was checked on a 1% agarose gel. Amplified RNA from the cDNA was obtained using T7-mRNA amplification (Invitrogen SuperScript RNA Amplification System, L1016-001). Amplified RNA was labeled with Cy5 or Cy3 (EA-006; Kreatech Biotechnology). The labeled amplified RNA was checked on a nanodrop spectrophotometer (Isogen Life-Science) and pooled with the same amount of reverse-color Cy-labeled RNA from the reference. As a reference, a pool of RNAs was made that consisted mostly of RNA from endometrial tumors of patients who had never used tamoxifen and RNA from a few patients who had received tamoxifen; this pool reflected the ratio of endometrial subtypes as occurs within the population. The labeled amplified RNA was then fragmented using RNA fragmentation reagents (8740; Ambion) and mixed with blocking solution containing Poly d(A) (27-7836-01; Pharmacia), Cot-1 DNA (15279-011; Invitrogen), and yeast tRNA (109 495; Roche). Each tumor sample contained a replicate because the tumor samples were profiled once with Cy5 and once with Cy3. Labeled amplified RNAs were kept at 42 °C until use and then mixed at 42 °C with preheated 2× F-hybridization buffer that contained formamide (F 7503-1000; Sigma Aldrich) and 20× SCC (19812323; BioSolve BV) at a 1:1 ratio and 0.1% SDS (51232; BioWhittaker).
Gene-Expression Profiling with Microarrays.
Spotted oligo microarrays with the Operon V3.0 library, human 35K oligo array (Operon Biotechnologies) were manufactured by the Netherlands Cancer Institute. A hybridization chamber (10040; Ambion) was used. The microarray was prehybridized at 42 °C for 1 h using a buffer (5× SSC, 0.1% SDS, and 1% BSA) and then washed with distilled water for 10 min, and washed again for 5 min. Hybridization occurred at 42 °C overnight. Washes were performed at 42 °C with the following solutions: 5× SSC and 0.1% SDS for 30 s, 2× SSC and 0.1% SDS for 30 s, and 1× SSC for 5 s. Two other washes were performed at room temperature with the solutions 0.2× SSC for 2 min and 0.05× SSC for 20 s. The hybridized array was scanned on a DNA Microarray Scanner (Model G2505B, serial no. US22502518; Agilent Technologies). The fluorescence intensities were measured using ImaGene software (Biodiscovery).
Gene-Expression Analysis.
After background correction, the intensities from the Cy5 and the Cy3 channel were used to calculate log2-transformed ratios. These ratios were normalized using the locally weighted scatterplot smoothing (LOWESS) subarray method (31). The normalized data were analyzed further in R. To create one dataset, experiments done in dye swap were combined to generate gene-expression log-ratios for patients who had not received tamoxifen and for patients who had received the drug for more than 2 years (Table 1). Only the probes with an assigned gene symbol and statistically significant log-ratios (P < 0.05) in at least 40 patients were retained (n = 3,734). Differential gene expression in endometrial tumors from tamoxifen users and nonusers was assessed using the limma R package (32). Fold changes from limma analysis were used to rank genes for gene set enrichment analysis (GSEA) (software.broadinstitute.org/gsea/index.jsp) (33). For pathway analyses we used curated hallmarks and oncogenic signature gene set collections from Molecular Signatures Database (mSigDB) version 5.0 (33).
The pathway enrichment network was generated using the Enrichment Map (34) application from Cytoscape (35). To generate gene sets of up- and down-regulated genes in the breast cancer cell line MCF-7, we used publicly available gene-expression data from Zwart et al. (20). Gene-expression data were processed with the BeadArray R package (36). After quantile normalization, differential gene expression in the vehicle and estradiol conditions was determined using limma workflow: After fitting of the genewise linear model, empirical Bayes statistics were estimated. P values were adjusted for multiple testing using the Benjamini–Hochberg procedure. Genes with an adjusted P value below 0.05 and an absolute log-fold change above 1 were considered to be differentially expressed upon estradiol stimulation. For the endometrial cancer cell line Ishikawa, gene-expression data for vehicle- and estradiol-stimulated cells were downloaded from Gertz et al. (37). RPKM values were processed using the limma package to identify genes differentially expressed in estradiol- and vehicle-stimulated cells as described above. Genes with an adjusted P value below 0.05 and an absolute log-fold change above 1 were used to construct up- and down-regulated gene sets.
Data from the Cancer Genome Atlas.
The Cancer Genome Atlas (TCGA) pan-cancer processed and normalized gene expression (38) was downloaded from https://tcga-data.nci.nih.gov/docs/publications/tcga?. We used the limma R package to generate fold changes in gene expression between endometrial (endometrioid adenocarcinoma subtype) and breast (ERα-positive subtype) cancers. These fold changes were used to rank genes for GSEA.
Results
ERα Binds DNA Differentially in Endometrial Tumors of Tamoxifen Users and Nonusers.
Tamoxifen is a ligand that binds ERα, and increases the risk for endometrial cancer in postmenopausal women. Tamoxifen-associated endometrioid adenocarcinomas are morphologically indistinguishable from endometrioid adenocarcinomas that arise in a tamoxifen-free environment (Fig. 1A), and they cannot be distinguished by DNA copy number profile (19). Because tamoxifen targets ERα, we tested if ERα binding to the DNA differed in endometrial tumors from tamoxifen users and nonusers.
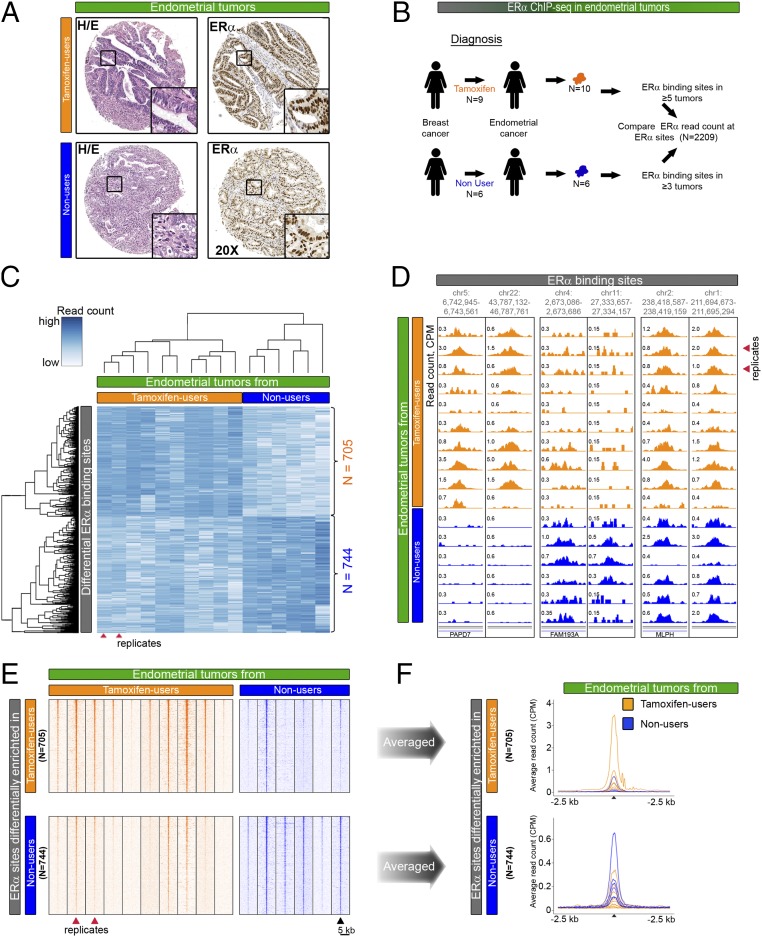
Fig. 1.
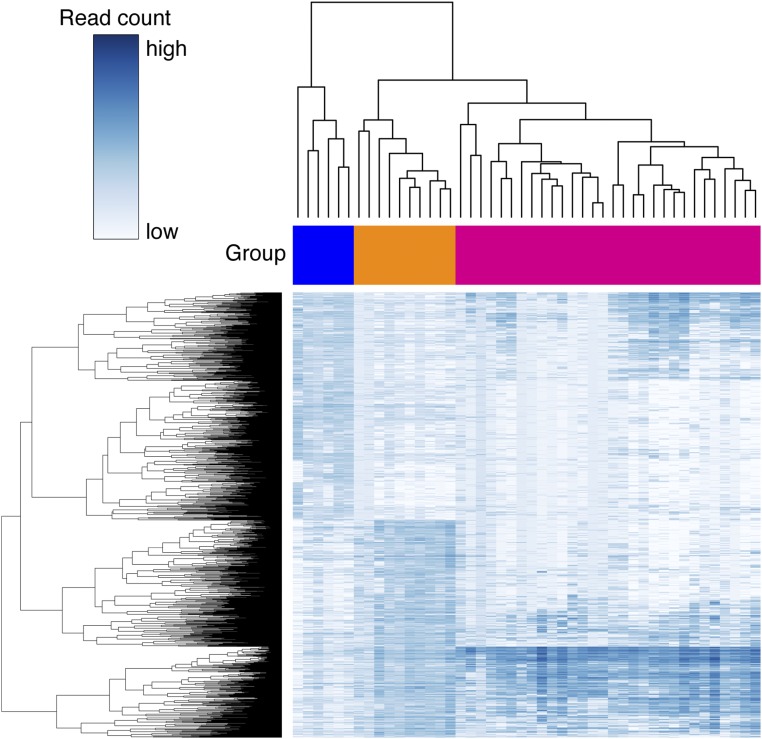
Comparative analysis of ERα binding in endometrial tumors from tamoxifen users and nonusers. (A) H&E staining and ERα immunohistochemistry staining in endometrial tumors from tamoxifen users and nonusers. (Magnification: 20×.) (B) Experimental set-up of ChIP-seq analyses in endometrial cancers. The analysis compares ERα binding in 10 endometrial tumors from nine tamoxifen users (orange) and ERα binding in six patients who never received endocrine therapy for breast cancer treatment (blue). Patient characteristics are described in Table 1. (C) Hierarchical clustering based on the results of differential binding analysis. (Upper) ERα-binding sites (705) that have a higher read count in tamoxifen-associated endometrial tumors (orange). (Lower) ERα-binding sites (744) that have a higher read count in endometrial tumors from patients who never used tamoxifen (blue). Red arrowheads indicate two tumors that originated from one patient. (D) Snapshots depicting ERα-binding sites in 16 endometrial tumors at the indicated genomic locations. Read counts were normalized [in counts per million sequenced reads (cpm)]. (E) Heatmap visualizing raw read count intensity of ERα at differential binding sites in tamoxifen-associated endometrial tumors (orange) and endometrial tumors from patients who never used tamoxifen (blue). Upper and lower panels show differential ERα-binding sites as described in C. The ChIP-seq signal aligns on the center of the peaks with a window of 5 kb. (F) Averaged read counts for ERα ChIP-seq data in tumors from tamoxifen users (orange) and nonusers (blue) at differential ERα-binding sites. Data align on the center of ERα peaks with a 2.5-kb window.
To investigate the ERα cistrome in endometrial tumors from tamoxifen users and nonusers, we used 16 fresh-frozen clinical specimens from the prospective TAMARISK cohort. Patients from the TAMARISK series had suffered from breast cancer (half of whom had received tamoxifen) and subsequently developed endometrial cancer. To compare the ERα cistrome in endometrial tumors from tamoxifen users and nonusers, we performed ERα ChIP-seq on 16 endometrial tumors. We previously published the results for endometrial tumors of tamoxifen users (and compared biological replicates of ERα ChIP-seq data) (18) that we also include in this study. All tested tumors were of the most common subtype, endometrioid adenocarcinoma, as determined by our pathologists (Table 1). No differences in clinicopathological parameters, including prior chemotherapy use (Table 1) or microsatellite instability (Table S1), were evident in endometrial tumors from tamoxifen users vs. nonusers.
Of the 16 endometrial tumors that we used for ChIP-seq, six endometrial cancer samples arose in patients who had never received endocrine treatment for their breast cancer (nonusers). The remaining 10 tumors came from nine tamoxifen users, who had used tamoxifen until the day of surgery (Fig. 1B). We included two specimens from one patient to provide a replicate experiment.
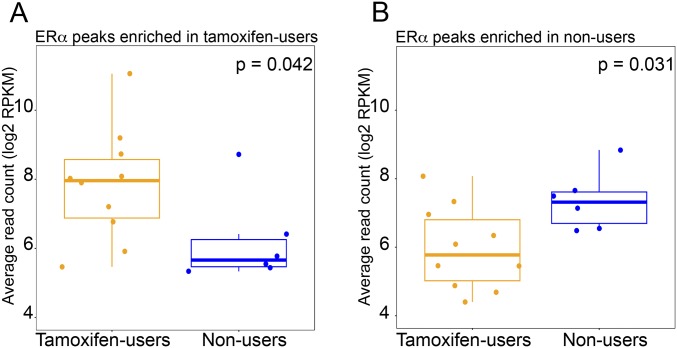
To identify ERα chromatin-binding sites that differed in the two tumor groups, we performed differential binding analysis (6). We included sites that were present in at least half of the tumors in each group and performed the analysis on the union of those sites (n = 2,209) (Fig. 1B). In total, we identified 1,449 binding sites as significantly different (FDR < 0.1) in the two groups (P < 0.00013 based on 8,008 available group label permutations). The ERα read count is higher at 705 sites and lower at 744 sites in endometrial tumors from tamoxifen-treated patients than in endometrial tumors from patients who never used tamoxifen (Fig. 1C and Dataset S1). Importantly, the two specimens from the same patient clustered together. Snapshots of the ERα signal exemplify both differential and nondifferential ERα sites (Fig. 1D). Analysis of the ERα ChIP-seq data shows the raw (Fig. 1E) and average (Fig. 1F and Fig. S1) read counts in the ERα peaks that are enriched differentially in the two groups.
Fig. S1.
Boxplots visualizing the normalized average read count of ERα sites differentially enriched in endometrial tumors of tamoxifen users (A) and nonusers (B). P values are according to the Mann–Whitney test.
Differential ERα-Binding Sites in Tumors from Tamoxifen Users and Nonusers Have Distinct Underlying DNA Sequences and Potential Activity.
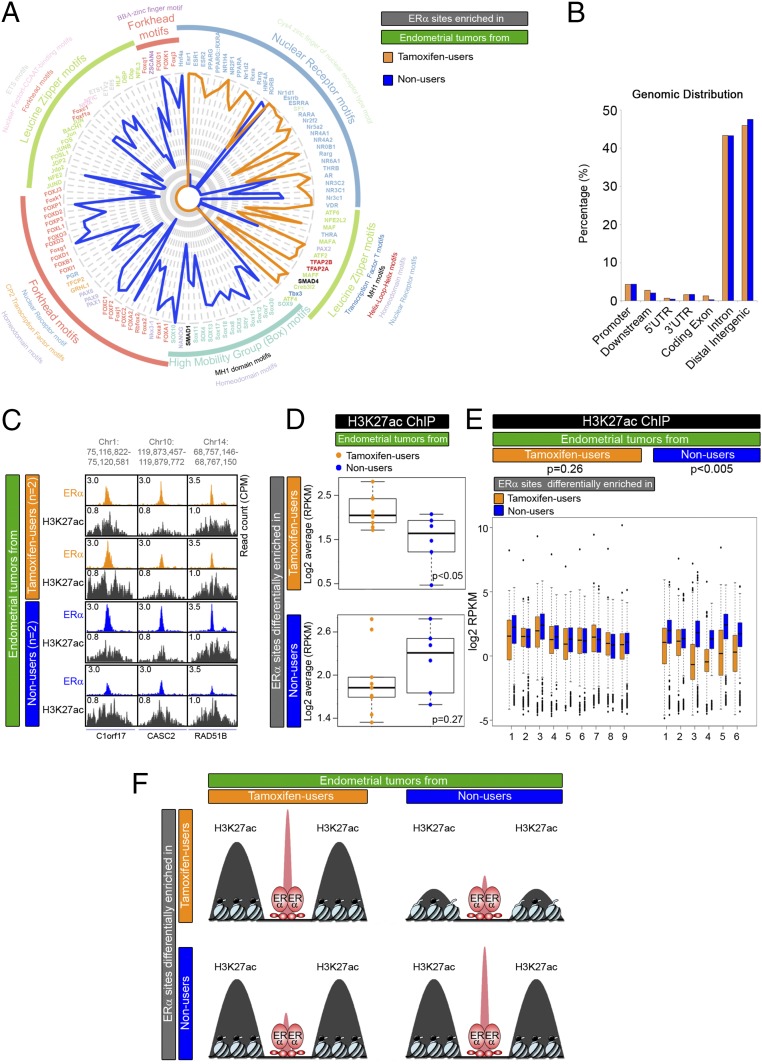
ERα binds the DNA differently in tamoxifen-associated endometrial tumors compared to endometrial tumors from patients who never received tamoxifen treatment (Fig. 1). To investigate the genomic features of these differential binding sites, we characterized their DNA sequences, their genomic distribution, and their regulatory activity (Fig. 2).
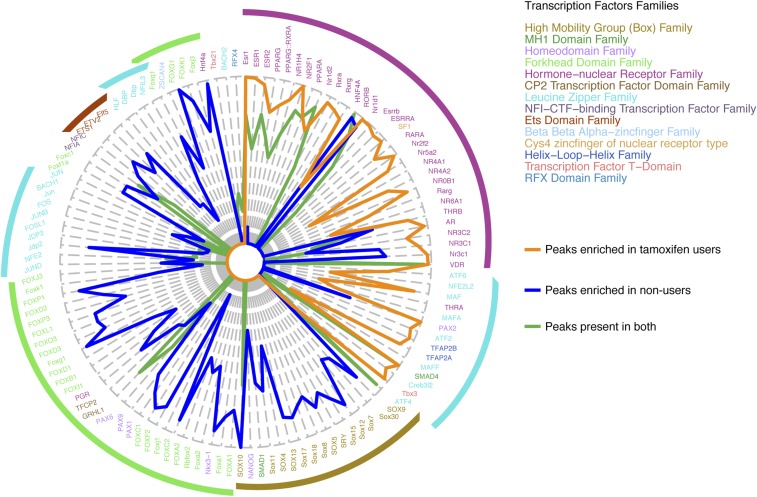
Fig. 2.
Characterization of ERα sites differentially bound in endometrial tumors from tamoxifen users and nonusers. (A) Radar plot visualizing DNA motif enrichment at genomic ERα sites differentially enriched in endometrial tumors from tamoxifen users (orange) and nonusers (blue). Lengths of radii correspond to the fraction of peaks that contain the identified motif. Motif colors correspond to transcription factor families. (B) Genomic distribution of ERα sites that are differentially enriched in endometrial tumors from tamoxifen users (orange) and nonusers (blue), relative to the nearest gene. (C) Snapshots depicting the H3K27ac ChIP-seq signal at ERα-binding sites in endometrial tumors from tamoxifen users (orange) and nonusers (blue) at the indicated genomic locations. Read counts were normalized [counts per million sequenced reads (cpm)]. (D) Boxplots showing the average normalized H3K27ac read count in endometrial tumors from tamoxifen users (orange dots) and nonusers (blue dots) at differential ERα-binding sites. (E) Boxplots showing normalized H3K27ac read counts at ERα differential binding sites in endometrial tumors from tamoxifen users (orange) and nonusers (blue). P values of the paired t test for each tumor group are shown. (F) Model for the intensity of H3K27ac mark (black) at differential ERα-binding sites (red) in the two tumor groups.
Sequence motif analysis revealed that the DNA motifs in the enriched ERα-binding sites of tamoxifen-associated endometrial tumors are different from those in the enriched ERα-binding sites in nonusers (Fig. 2A). Tamoxifen-associated endometrial tumors exhibited enriched ERα-binding sites that mostly contained motifs of estrogen receptor 1 (ESR1) and other well-known hormone receptors such as androgen receptor, glucocorticoid receptor, and thyroid hormone receptor. In contrast, ERα-binding sites enriched in endometrial tumors from nonusers mostly included motifs of the forkhead domain family and the high-mobility Box family group. This last group contains well-known stem cell markers, such as sex-determining region Y-box 4 (SOX4) and Nanog homeobox (NANOG), which associate with endometrial cancer (39, 40). Both groups contained motifs for leucine zipper proteins at the differential ERα-binding sites.
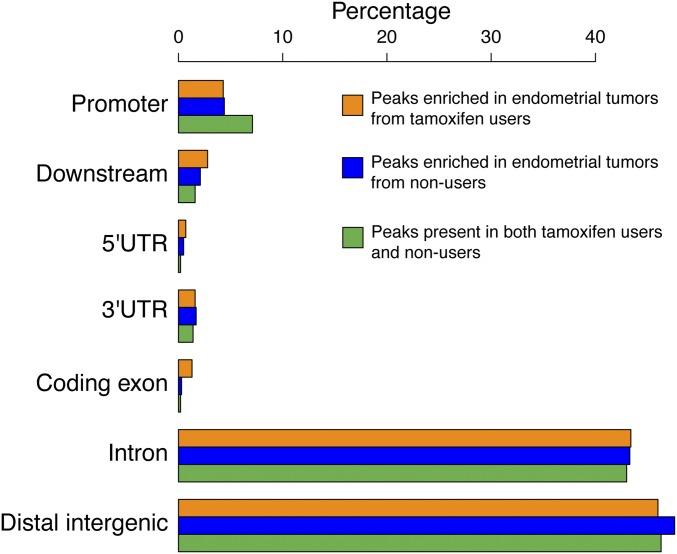
Differential ERα sites, enriched in endometrial tumors from either tamoxifen users or nonusers, locate mainly at distal intergenic regions and gene introns (Fig. 2B). This localization corresponds with previously described data on the distribution of ERα peaks and is characteristic for enhancer-binding transcription factors (18, 41). We also defined a set of ERα-binding sites that do not differ in the two tumor groups (absolute log-fold change <0.5, n = 423) (Dataset S1). These sites show enhancer-like genomic distribution (Fig. S2) and harbor estrogen receptor (ER) motifs (Fig. S3).
Fig. S2.
Genomic distribution of ERα sites that are shared (green) or differentially enriched in endometrial tumors from tamoxifen users (orange) and nonusers (blue), relative to the nearest gene.
Fig. S3.
Radar plot visualizing DNA motif enrichment at genomic ERα sites that are shared (green) or are differentially enriched in endometrial tumors from tamoxifen users (orange) or nonusers (blue). Lengths of radii correspond to the fraction of peaks that contain the identified motif. Motif colors correspond to transcription factor families.
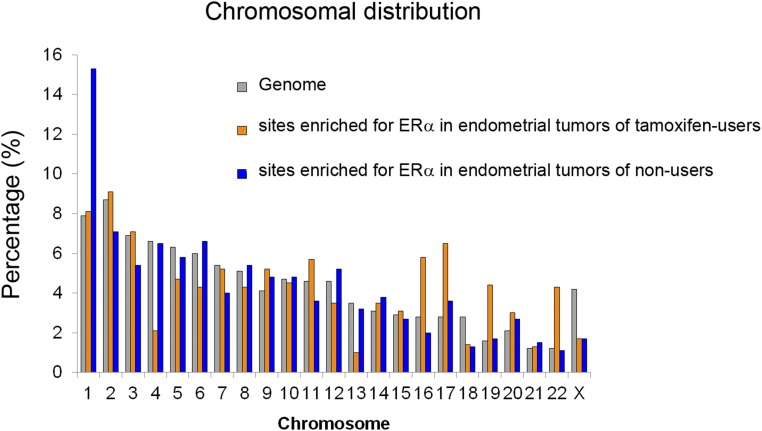
The chromosomal distribution of the differential ERα-binding sites varied in the endometrial tumors of tamoxifen users and nonusers (Fig. S4). We tested if this difference could be caused by distinct chromosomal aberrations present in the two tumor groups. In agreement with previous reports (19), we could not distinguish tamoxifen-associated endometrial tumors from endometrial tumors that arose in a tamoxifen-free environment based on their copy number profiles (Fig. S5). From this observation we conclude that the apparent bias of ERα binding to specific chromosomes is not caused by differences in chromosomal copy number.
Fig. S4.
Chromosomal distribution of ERα-binding sites differentially enriched in endometrial tumors from tamoxifen users (orange) or nonusers (blue).
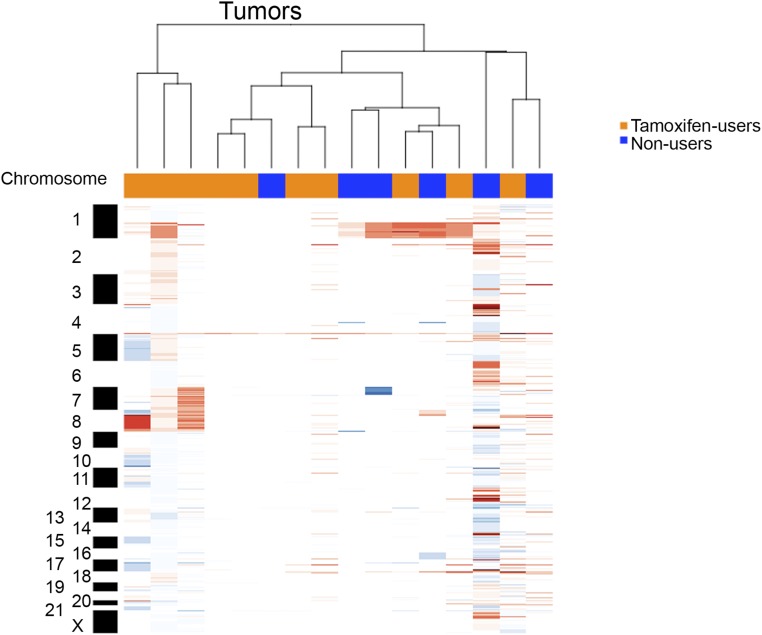
Fig. S5.
Copy number variations analysis of endometrial cancers used for ChIP-seq (red indicates genomic gains; blue indicates loss). Copy number profiles are clustered based on correlation; endometrial tumors from tamoxifen users and nonusers do not cluster according to tumor group.
We next investigated if differential ERα-binding sites harbored the H3K27ac histone mark, which would indicate active enhancers. Visual inspection of H3K27ac ChIP-seq data revealed a strong signal at ERα-binding sites (Fig. 2C and Fig. S6). We observed that the higher binding of ERα in tamoxifen users was accompanied by a more prominent H3K27ac signal at those regions than in nonusers (Fig. 2D, Upper). At the sites where ERα-binding was enriched in nonusers, H3K27ac was equally present in both tamoxifen users and nonusers (Fig. 2D, Lower). Similarly, in tamoxifen-associated endometrial tumors, the H3K27ac signal was comparable at ERα sites that were enriched in both tamoxifen users and nonusers (Fig. 2E, Left). In contrast, in tumors of nonusers there is a difference in enhancer activity at the differential sites, with enriched ERα sites in nonusers exposing higher H2K27ac (Fig. 2E, Right).
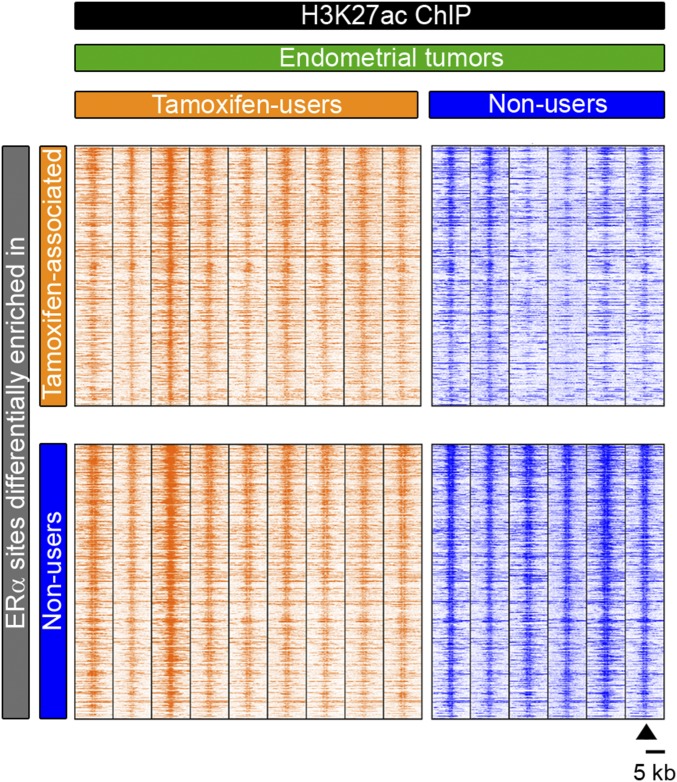
Fig. S6.
Heatmap showing raw read count intensity of H3K27ac in endometrial tumors from tamoxifen users (orange) and nonusers (blue) at DNA sites that are differentially bound by ERα per group.
Taken together, these data reveal that the differential ERα-binding sites in endometrial tumors from tamoxifen users and nonusers are enriched for different DNA motifs and distributed mostly to active enhancers. As evaluated by H3K27Ac levels, the activity of the ERα sites that are enriched in nonusers does not differ between the two groups of endometrial tumors. In contrast, H3K27Ac levels at tamoxifen-associated ERα sites are higher in the group of tamoxifen users (Fig. 2F).
Differential ERα-Binding Sites in Tamoxifen Users and Nonusers Affect Gene Regulation Differently.
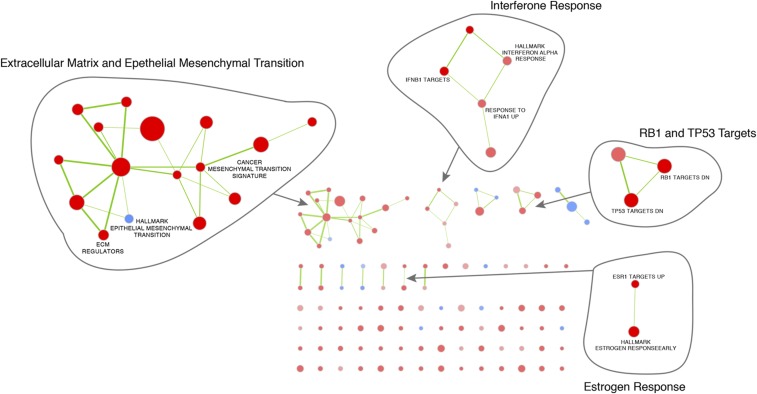
Differences in ERα profiles and H3K27ac signals in endometrial tumors from tamoxifen users and nonusers suggest deviations in corresponding gene expression (Fig. 2). To link tamoxifen treatment with gene activity, we generated microarray data from endometrial tumors (endometrioid adenocarcinoma subtype, largely ER-positive) (42) of 47 tamoxifen users and 64 nonusers in the TAMARISK series (Table 1). Pathway analysis revealed a number of biological processes associated with the genes differentially expressed in the two tumor groups (Dataset S1). Network representation of these pathways shows that, in addition to ERα targets, genes related to extracellular matrix and mesenchymal transition (EMT), genes down-regulated by retinoblastoma susceptibility 1 (RB1) and tumor protein p53 (TP53), and IFN targets are up-regulated in the group of tamoxifen users (Fig. S7).
Fig. S7.
The networks illustrate the results of the pathway enrichment analysis (with GSEA) of differential gene expression in tamoxifen users and nonusers. Nodes represent pathways that link overlapping genes (overlap coefficient cutoff 0.5). Red nodes illustrate pathways up-regulated in tamoxifen users, and blue nodes represent pathways up-regulated in nonusers.
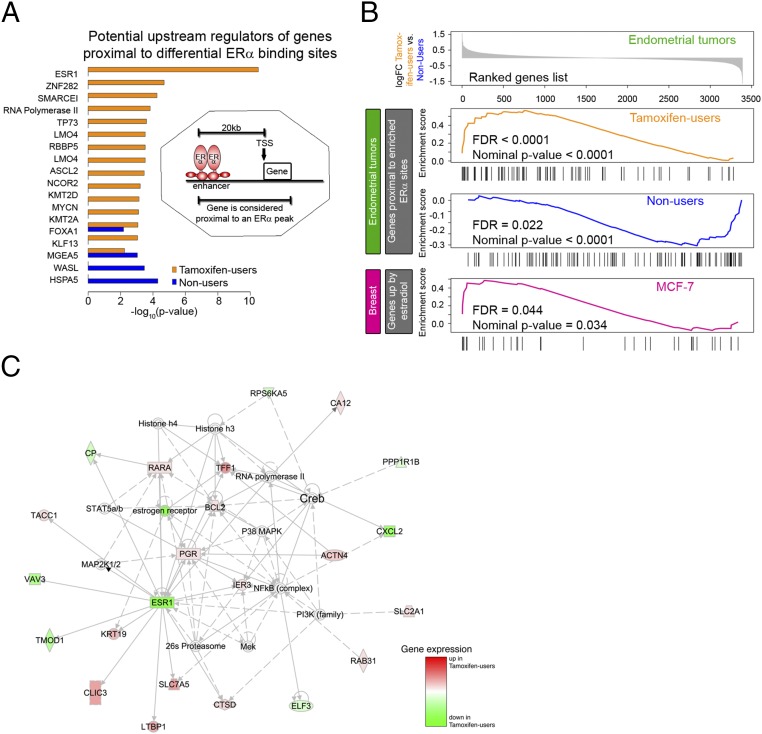
To understand possible functions of the identified differential ERα-binding sites in gene expression, we first characterized their potential target genes (388 genes for tamoxifen-associated endometrial tumors and 402 genes for endometrial tumors of nonusers; Dataset S1). As in a previous study (5), we considered a gene to be a potential target if an ERα peak was positioned within the gene body or within 20 kb upstream of the TSS. Using IPA, we found that ESR1 was a potential upstream regulator of genes proximal to ERα-binding sites that were enriched in endometrial tumors of tamoxifen users but not in tumors of nonusers. Instead, the top potential upstream regulator for genes proximal to ERα-binding sites in endometrial tumors of nonusers was HSPA5, a heat shock protein (Fig. 3A). These data suggest that in the differential ERα sites of both groups, only the binding sites that are enriched in tamoxifen-associated endometrial tumors regulate known target genes of ERα. GO analysis revealed a number of biological processes that were specific for the genes potentially targeted by ERα peaks enriched in nonusers, including negative regulation of collagen biosynthesis and metabolism and negative regulation of the multicellular organismal metabolic process and the phosphorus metabolic process (Tables S4 and S5).
Fig. 3.
ERα-mediated gene regulation in endometrial tumors from tamoxifen users and nonusers. (A) The bar plot shows potential upstream regulators of genes proximal to differential ERα-binding sites according to IPA. The Inset shows how potential target genes are defined. (B) GSEA based on differential gene expression in endometrial tumors of tamoxifen users and nonusers from the TAMARISK cohort. (Upper) Ranked log-fold change in gene expression between the two cancer groups. (Lower) Enrichment scores versus gene rank in three significantly enriched gene sets: genes proximal to the binding sites enriched in tamoxifen-associated tumors, genes proximal to the binding sites enriched in tumors of nonusers, and genes up-regulated by estradiol in the MCF-7 breast cancer cell line. Patient characteristics are described in Table 1. (C) Top network from IPA based on genes identified as potential targets of ERα by combined analysis of gene expression and ChIP-seq data.
Table S4.
GO biological processes enriched among the genes potentially targeted by ERα peaks enriched in tamoxifen users
| GO biological process | No. of genes in the reference set | No. of genes in the uploaded list | Expected no. of genes | Enrichment (over/under) | Fold enrichment | Enrichment P value |
| System development (GO:0048731) | 4,042 | 111 | 69.38 | + | 1.6 | 9.17e-04 |
| Negative regulation of biological process (GO:0048519) | 4,622 | 122 | 79.34 | + | 1.54 | 1.41e-03 |
| Single multicellular organism process (GO:0044707) | 5,417 | 137 | 92.99 | + | 1.47 | 1.98e-03 |
| Negative regulation of cellular process (GO:0048523) | 4,281 | 114 | 73.49 | + | 1.55 | 2.90e-03 |
| Single-organism process (GO:0044699) | 12,544 | 260 | 215.33 | + | 1.21 | 5.02e-03 |
| Animal organ development (GO:0048513) | 2,888 | 84 | 49.57 | + | 1.69 | 6.01e-03 |
| Developmental process (GO:0032502) | 5,333 | 133 | 91.54 | + | 1.45 | 7.69e-03 |
| Multicellular organism development (GO:0007275) | 4,640 | 119 | 79.65 | + | 1.49 | 1.02e-02 |
| Single-organism cellular process (GO:0044763) | 11,217 | 237 | 192.55 | + | 1.23 | 1.12e-02 |
| Biological process (GO:0008150) | 17,072 | 325 | 293.05 | + | 1.11 | 1.95e-02 |
| Single-organism developmental process (GO:0044767) | 5,239 | 129 | 89.93 | + | 1.43 | 2.55e-02 |
| Anatomical structure development (GO:0048856) | 4,986 | 124 | 85.59 | + | 1.45 | 2.71e-02 |
Table S5.
GO biological processes enriched among the genes potentially targeted by ERα peaks enriched in nonusers
| Complete GO biological process | No. of genes in the reference set | No. of genes in the uploaded list | Expected no. of genes | Enrichment (over/under) | Fold enrichment | Enrichment P value |
| Single-multicellular organism process (GO:0044707) | 5,417 | 159 | 99.19 | + | 1.6 | 1.88e-07 |
| Multicellular organism development (GO:0007275) | 4,640 | 142 | 84.96 | + | 1.67 | 2.47e-07 |
| Anatomical structure development (GO:0048856) | 4,986 | 146 | 91.29 | + | 1.6 | 2.87e-06 |
| Single-organism process (GO:0044699) | 12,544 | 286 | 229.68 | + | 1.25 | 9.51e-06 |
| Single-organism developmental process (GO:0044767) | 5,239 | 148 | 95.93 | + | 1.54 | 2.66e-05 |
| Anatomical structure morphogenesis (GO:0009653) | 1,956 | 73 | 35.81 | + | 2.04 | 3.59e-05 |
| Developmental process (GO:0032502) | 5,333 | 149 | 97.65 | + | 1.53 | 4.93e-05 |
| System development (GO:0048731) | 4,042 | 120 | 74.01 | + | 1.62 | 1.27e-04 |
| Multicellular organismal process (GO:0032501) | 6,482 | 168 | 118.69 | + | 1.42 | 6.79e-04 |
| Single-organism cellular process (GO:0044763) | 11,217 | 255 | 205.38 | + | 1.24 | 1.55e-03 |
| Animal organ development (GO:0048513) | 2,888 | 90 | 52.88 | + | 1.7 | 2.03e-03 |
| Negative regulation of collagen biosynthetic process (GO:0032966) | 10 | 5 | 0.18 | + | 27.31 | 1.20e-02 |
| Biological_process (GO:0008150) | 17,072 | 346 | 312.59 | + | 1.11 | 1.51e-02 |
| Negative regulation of collagen metabolic process (GO:0010713) | 11 | 5 | 0.2 | + | 24.82 | 1.90e-02 |
| Phosphorus metabolic process (GO:0006793) | 2,192 | 70 | 40.14 | + | 1.74 | 2.69e-02 |
| Negative regulation of multicellular organismal metabolic process (GO:0044252) | 12 | 5 | 0.22 | + | 22.76 | 2.89e-02 |
We further investigated the regulatory link between differential ERα-binding sites and gene expression by ranking genes from the microarray data according to their differential expression in endometrial tumors from tamoxifen users and nonusers. We used six gene sets for GSEA: (i) genes proximal to ERα-binding sites enriched in tamoxifen-associated endometrial cancer; (ii) genes proximal to ERα-binding sites enriched in endometrial tumors of nonusers; (iii) genes up-regulated by estradiol in the MCF-7 breast cancer cell line; (iv) genes down-regulated by estradiol in the MCF-7 breast cancer cell line; (v) genes up-regulated by estradiol in the endometrial cancer cell line Ishikawa; and (vi) genes down-regulated by estradiol in the endometrial cancer cell line Ishikawa. RNA expression levels of genes proximal to enriched ERα sites are higher in the corresponding tumor group than in the tumor group in which the ChIP-seq signal at these ERα sites is less pronounced. Genes up-regulated by estradiol in the breast cancer cell line MCF-7 are among the genes that are more highly expressed in endometrial tumors from tamoxifen users (Fig. 3B).
To focus further on the transcriptional effects of the differential ERα-binding sites, we narrowed the list of potential target genes by combining gene-expression and ChIP-seq data by means of GSEA: Only the genes that contributed to the leading edge (core enrichment) in the GSEA analysis were taken (Tables S6 and S7). IPA revealed a strong enrichment of ESR1-regulated genes (15 of 70 target genes are described as ESR1-regulated, P = 4.7e-13) and constructed a functional network that is centered around ERα and includes well-known targets such as progesterone receptor (PGR), retinoic acid receptor alpha (RARA), trefoil factor 1 (TFF1) and vav guanine nucleotide exchange factor 3 (VAV3), and others (Fig. 3C).
Table S6.
Potential target genes of ERα peaks enriched in tamoxifen users as identified by combined analysis of gene expression and ChIP-seq
| Gene symbol | Core enrichment |
| ACTN4 | Yes |
| ANO1 | Yes |
| B4GALT1 | Yes |
| BCL2 | Yes |
| C16orf45 | Yes |
| CA12 | Yes |
| CGNL1 | Yes |
| CLIC3 | Yes |
| CTSD | Yes |
| DCXR | Yes |
| DOCK1 | Yes |
| HS6ST1 | Yes |
| IER3 | Yes |
| IGFBP4 | Yes |
| KLC1 | Yes |
| KRT19 | Yes |
| LAMC1 | Yes |
| LTBP1 | Yes |
| MPST | Yes |
| NBN | Yes |
| NOSIP | Yes |
| PGR | Yes |
| RAB31 | Yes |
| RAB5B | Yes |
| RARA | Yes |
| RASGRF1 | Yes |
| SETD7 | Yes |
| SLC2A1 | Yes |
| SLC47A1 | Yes |
| SLC7A5 | Yes |
| SLCO2A1 | Yes |
| SMOC2 | Yes |
| SPINT2 | Yes |
| TACC1 | Yes |
| TFF1 | Yes |
| TST | Yes |
| ZFHX3 | Yes |
| ZNF185 | Yes |
Genes in the list contain an ERα peak 20 kb upstream of the TSS or in the gene body and contribute strongly to the enrichment among up-regulated genes in GSEA.
Table S7.
Potential target genes of ERα peaks enriched in nonusers as identified by combined analysis of gene expression and ChIP-seq
| Gene symbol | Core enrichment |
| AKAP7 | Yes |
| AQP9 | Yes |
| AUNIP | Yes |
| C10orf11 | Yes |
| CADPS2 | Yes |
| CP | Yes |
| CRYL1 | Yes |
| CXCL2 | Yes |
| DLGAP5 | Yes |
| ELF3 | Yes |
| ESR1 | Yes |
| EYA2 | Yes |
| FAM107B | Yes |
| FAM129A | Yes |
| FGF18 | Yes |
| GCNT2 | Yes |
| GRAMD3 | Yes |
| HGD | Yes |
| HIGD1A | Yes |
| HOMER2 | Yes |
| MGP | Yes |
| PDE4DIP | Yes |
| PIGR | Yes |
| PLLP | Yes |
| PPP1R1B | Yes |
| RBM47 | Yes |
| RPS6KA5 | Yes |
| TMEM101 | Yes |
| TMOD1 | Yes |
| UGDH | Yes |
| VAV3 | Yes |
| WWTR1 | Yes |
Genes in the list contain an ERα peak 20 kb upstream of the TSS or in the gene body and contribute strongly to the enrichment among up-regulated genes in GSEA.
Taken together, these data reveal that the differential ERα-binding sites in endometrial tumors from tamoxifen users and nonusers regulate gene expression differently. Gene expression of ERα targets in tamoxifen-associated endometrial tumors resembles that of estradiol-responsive genes in the MCF-7 cell line, suggesting an ERα cistrome potentially similar to breast tumors.
ERα Binding to DNA in Tamoxifen-Associated Endometrial Tumors Resembles ERα Chromatin Binding in Breast Cancer.
In contrast to endometrial tumors of nonusers, ERα-binding sites enriched in tamoxifen-associated endometrial tumors are proximal to known ER targets in breast cancer (Fig. 3). Because tamoxifen has been reported to stimulate cell growth in the endometrium in postmenopausal patients, similar to estrogen in breast, we compared our findings with publicly available ChIP-seq data on ERα and other transcription factors in 30 primary breast tumors (Fig. 4A).
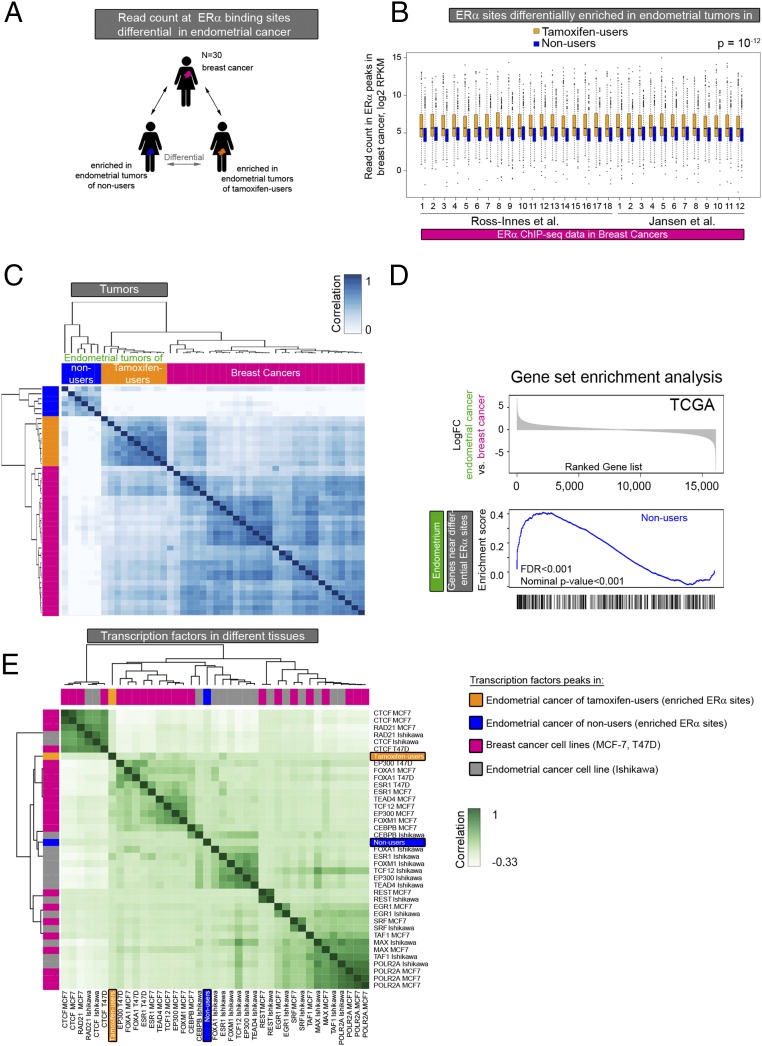
Fig. 4.
Comparative analysis of ERα-binding sites in endometrial and breast tumor tissue. (A) Analysis set-up. ERα binding in breast cancer at the differential sites in the two endometrial cancer groups was evaluated. (B) Boxplot showing the normalized ERα ChIP-seq read count in breast cancers at the ERα-binding sites differentially enriched in endometrial tumors from tamoxifen users (orange boxplots) and nonusers (blue boxplots). The P value of the paired t test is P = 10−12. (C) Heatmap visualization of the correlation matrix based on ERα ChIP-seq read count at differential ERα-binding sites in endometrial tumors of tamoxifen users (orange) and nonusers (blue) and in breast tumors (pink). (D) GSEA based on differential gene expression in endometrial and breast cancers from the TCGA pan-cancer project. (Upper) Ranked log-fold change in gene expression in endometrial cancer vs. breast cancer. (Lower) Enrichment scores vs. gene rank in the significantly enriched gene set: genes proximal to the binding sites enriched in tumors of nonusers. (E) Hierarchical clustering of the correlation between transcription factor genomic occupancy (peaks) from publicly available ChIP-seq data in the breast cancer cell lines MCF-7 and T47D (pink), the endometrial cancer cell line Ishikawa (gray), and the ERα sites enriched in endometrial tumors from tamoxifen users (orange) and nonusers (blue).
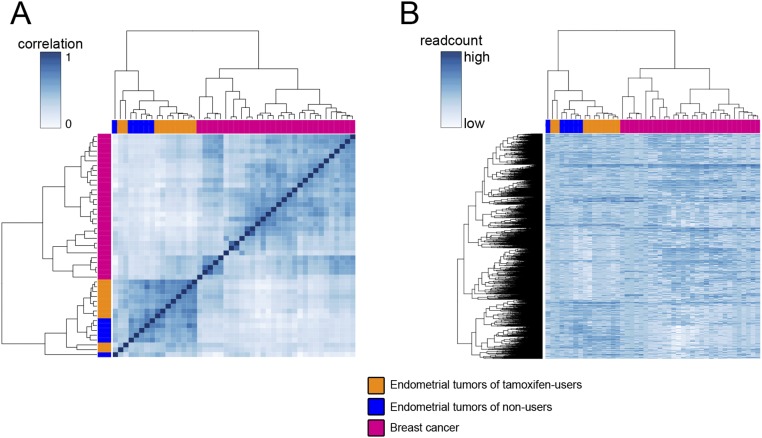
To group the tumors according to similarity in ERα ChIP-seq signal, we performed hierarchical clustering of the two tumor types (breast and endometrial). We first analyzed global ERα ChIP-seq signal in the two endometrial groups (tamoxifen users and nonusers) and breast cancer at ERα-binding sites present in at least five of 46 tumors analyzed (n = 16,516). Based on the ERα ChIP-seq read count, the tumors clustered by tumor type (Fig. S8).
Fig. S8.
(A) Heatmap visualization of the correlation matrix based on ERα ChIP-seq read count at ERα-binding sites in endometrial tumors of tamoxifen users (orange), nonusers (blue), and in breast tumors (pink). Only peaks found in at least five tumors were included. (B) Hierarchical clustering of ERα ChIP-seq signal in breast tumors (pink) and endometrial tumors (tamoxifen users are orange; nonusers are blue) in peaks present in at least five tumors.
Next, we focused on the sites that are differentially bound by ERα in endometrial tumors of tamoxifen users and nonusers. We analyzed the ERα ChIP-seq data of the 30 breast tumors at genomic sites that are differentially bound by ERα in tamoxifen-associated endometrial cancer. The ERα ChIP-seq signal in breast tumors is significantly higher at the sites that are enriched in tamoxifen-associated endometrial tumors than at the binding sites enriched in endometrial tumors from nonusers (Fig. 4B). In addition, unsupervised hierarchical clustering showed that the ERα read count at differential ERα sites (between endometrial tumors of tamoxifen users and nonusers) correlated most between breast tumors and tamoxifen-associated endometrial tumors (Fig. 4C for the correlation heatmap and Fig. S9 for the readcount at differential sites).
Fig. S9.
Hierarchical clustering of the ERα ChIP-seq signal in endometrial tumors from tamoxifen users (orange) and nonusers (blue) and in breast tumors (pink) in peaks that are differential in endometrial tumors of tamoxifen users and nonusers.
To investigate whether differential ERα-binding sites might regulate gene expression in a variable manner in endometrial cancer and breast cancer, we used gene-expression data from the TCGA pan-cancer project (38) for GSEA. We ranked genes from the TCGA data according to fold change in expression between endometrioid adenocarcinoma and ER-positive breast cancer and used two gene sets for the analysis: (i) genes proximal to ERα sites enriched in endometrial tumors from tamoxifen users and (ii) genes proximal to ERα sites enriched in endometrial tumors from nonusers. We found that one gene set (i.e., genes proximal to the ERα sites that are enriched in nonusers) was more highly expressed in endometrial tumors than in breast tumors (Fig. 4D). This analysis suggests that the ERα sites enriched in tumors from nonusers indeed are involved in the execution of transcriptional programs specific for endometrial cancer.
To investigate the transcription factor network in which the differential ERα-binding sites function, we used publicly available cell line data. We correlated the differentially enriched ERα-binding sites of endometrial tumors from tamoxifen users and nonusers with binding sites of several transcription factors from the endometrial cancer cell line Ishikawa and the breast cancer cell lines MCF-7 and T47D (Fig. 4E). Transcription factors involved in DNA looping [such as CTCCF-binding factor (CTCF) and the double-strand break repair protein rad21 homolog (RAD21)] and also transcription factors that bind promoters [RNA polymerase (POL2RA), serum response factor (SRF), and transcription initiation factor TFIID subunit 1 (TAF1)] cluster together irrespective of cell line or tissue type. In contrast, enhancer-binding transcription factors (ESR1, FOXA1, and EP300) cluster according to tissue type. In accordance with Fig. 2B, the differential ERα-binding sites of endometrial tumors cluster with enhancer-binding transcription factors rather than with promoter-binding factors. The ERα-binding sites enriched in tumors from nonusers clustered with the transcription factor-binding sites in Ishikawa, whereas the ERα-binding sites enriched in tamoxifen-associated endometrial tumors clustered with the transcription factor-binding sites in breast cancer cell lines. Taken together, these data illustrate a resemblance between breast cancer and tamoxifen-associated endometrial cancer at sites that are enriched for ERα in endometrial tumors of tamoxifen users compared with nonusers. In contrast, genomic regions enriched for ERα in endometrial tumors from nonusers correspond to ERα enhancers in the endometrial cancer cell line Ishikawa and potentially regulate the expression of genes more specific for endometrial tumors.
Discussion
We found that even though endometrioid adenocarcinomas of tamoxifen users and nonusers are indistinguishable on a morphological level, a large part of the ERα cistrome and its downstream transcriptional programs differ. The differential ERα-binding sites have distinct underlying DNA sequences and potential regulatory function. Interestingly, ERα binding to the DNA in tamoxifen-associated endometrial tumors resembles ERα chromatin binding in breast cancer, highlighting a conserved ERα pathway between the two tumor types from different organs despite different ligands.
Studies in the breast cancer cell line MCF-7 show that prolonged tamoxifen exposure shifts the ERα cistrome (4–6), which consequently changes gene expression (4), possibly by changing its interactome (2, 43–45). These data are hard to translate between tissues because there are far fewer models to study ERα in endometrial cancer. Thus far, there is no model of tamoxifen-associated endometrial cancer; the only model for the effects of tamoxifen in endometrial tissue is the endometrial cancer cell line Ishikawa, which is derived from a tumor of a nonuser (46). The effects of tamoxifen on the ERα cistrome in this model lack genome-wide data (37, 47). Our previous study was the first to show genome-wide ERα-binding sites in tamoxifen-associated endometrial cancer, but it lacked data to identify ERα sites differentially expressed in endometrial tumors of tamoxifen users compared to nonusers (18).
Using patient samples from the unique TAMARISK study (Table 1), we now show that the ERα cistrome differs in endometrial tumors that originate from different hormonal backgrounds (tamoxifen-rich vs. tamoxifen-free), and that these tumors therefore are epigenetically distinguishable. Prior chemotherapy for the treatment of breast cancer did not differ in the two patient groups (Table 1), precluding differences in systemic therapy beyond tamoxifen use as a potential confounder. Furthermore, we excluded a genetic predisposition in the form of Lynch syndrome in either patient group by showing that microsatellite instability was comparable in tamoxifen users and nonusers (Table S1) (48). However, unknown genetic predispositions cannot be excluded at this point.
Although several studies report the effects of ligands, including tamoxifen, on the conformation of ERα, other determinants of the ERα cistrome in endometrial tissue remain obscure. The motifs we found hint at proteins involved at ERα/chromatin interactions at differential ERα-binding sites in endometrial tumors of tamoxifen users and nonusers. These motifs indicate a role for stem cell markers, such as SOX4 (39) and NANOG (40) in nonusers, and for members of the nuclear receptor family, including the androgen receptor, glucocorticoid, and thyroid hormone receptor in tamoxifen users.
Compared with endometrial tumors of nonusers, tamoxifen-associated endometrial tumors showed up-regulation of genes involved in pathways that contribute to cancer progression such as EMT, RB1, TP53, and IFN targets. These data suggest that endometrial tumors that originate in presence of tamoxifen may have an intrinsically different tumor biology, resulting in different tumor drivers in this setting. Our previous immunohistochemical studies, which show that longer tamoxifen exposure equates with worse survival, higher TP53 expression, and lower ESR1 expression (13), are in line with our current results.
Previous studies have shown that enhancer activity differs in various tissues (49). Our data show that ERα profiles in tamoxifen-associated endometrial tumors resemble those found in breast tumors, suggesting that endocrine stimuli reprogram this pathway in endometrial tissue.
To conclude, our study sheds light on the ERα cistrome and on the regulation of gene expression in endometrial tumors and indicates that the two kinds of endometrioid adenocarcinomas that we investigated in this report, albeit morphologically identical, are clearly divergent on a cistromic and transcriptional level. Our results pave the way for further discoveries in endometrial cancer and highlight the added value of cistromic analyses in clinical specimens, especially in settings where model systems are not available. By functionally distinguishing tumors on the level of transcriptional regulation, novel subtypes may be revealed with further clinical and prognostic implications.
Supplementary Material
Acknowledgments
We thank Ron Kerkhoven, Shan Baban, and Marja Nieuwland from The Netherlands Cancer Institute (NKI) genomics facility for sample processing; Arno Velds for bioinformatics support; Lisette Hoogendoorn for clinical data collection; Koen van de Vijver for help with pathological analyses; and the NKI Core Facility Molecular Pathology and Biobanking (CFMPB) for supplying NKI Biobank material and laboratory support. This work was supported by Grant NKI 2002-586 from the Dutch Cancer Society and by Pink Ribbon.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE94031).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615233114/-/DCSupplemental.
References
- 1.Droog M, Mensink M, Zwart W. The estrogen receptor α-cistrome beyond breast cancer. Mol Endocrinol. 2016;30(10):1046–1058. doi: 10.1210/me.2016-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 3.Flach KD, Zwart W. The first decade of estrogen receptor cistromics in breast cancer. J Endocrinol. 2016;229(2):R43–R56. doi: 10.1530/JOE-16-0003. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Severson TM, et al. Neoadjuvant tamoxifen synchronizes ERα binding and gene expression profiles related to outcome and proliferation. Oncotarget. 2016;7(23):33901–33918. doi: 10.18632/oncotarget.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osborne CK, Hobbs K, Clark GM. Effect of estrogens and antiestrogens on growth of human breast cancer cells in athymic nude mice. Cancer Res. 1985;45(2):584–590. [PubMed] [Google Scholar]
- 8.Satyaswaroop PG, Zaino RJ, Mortel R. Estrogen-like effects of tamoxifen on human endometrial carcinoma transplanted into nude mice. Cancer Res. 1984;44(9):4006–4010. [PubMed] [Google Scholar]
- 9.van Leeuwen FE, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343(8895):448–452. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- 10.Fisher B, et al. Endometrial cancer in tamoxifen-treated breast cancer patients: Findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86(7):527–537. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- 11.Lahti E, et al. Endometrial changes in postmenopausal breast cancer patients receiving tamoxifen. Obstet Gynecol. 1993;81(5 ( Pt 1)):660–664. [PubMed] [Google Scholar]
- 12.Bergman L, et al. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Comprehensive Cancer Centres’ ALERT Group. Assessment of Liver and Endometrial cancer Risk following Tamoxifen. Lancet. 2000;356(9233):881–887. doi: 10.1016/s0140-6736(00)02677-5. [DOI] [PubMed] [Google Scholar]
- 13.Hoogendoorn WE, et al. Comprehensive Cancer Centers TAMARISK-group Prognosis of uterine corpus cancer after tamoxifen treatment for breast cancer. Breast Cancer Res Treat. 2008;112(1):99–108. doi: 10.1007/s10549-007-9823-1. [DOI] [PubMed] [Google Scholar]
- 14.Jones ME, et al. Endometrial cancer survival after breast cancer in relation to tamoxifen treatment: Pooled results from three countries. Breast Cancer Res. 2012;14(3):R91. doi: 10.1186/bcr3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swerdlow AJ, Jones ME. British Tamoxifen Second Cancer Study Group Tamoxifen treatment for breast cancer and risk of endometrial cancer: A case-control study. J Natl Cancer Inst. 2005;97(5):375–384. doi: 10.1093/jnci/dji057. [DOI] [PubMed] [Google Scholar]
- 16.Gottardis MM, Robinson SP, Satyaswaroop PG, Jordan VC. Contrasting actions of tamoxifen on endometrial and breast tumor growth in the athymic mouse. Cancer Res. 1988;48(4):812–815. [PubMed] [Google Scholar]
- 17.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295(5564):2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 18.Droog M, et al. Comparative cistromics reveals genomic cross-talk between FOXA1 and ERα in tamoxifen-associated endometrial carcinomas. Cancer Res. 2016;76(13):3773–3784. doi: 10.1158/0008-5472.CAN-14-1813. [DOI] [PubMed] [Google Scholar]
- 19.Fles R, et al. Genomic profile of endometrial tumors depends on morphological subtype, not on tamoxifen exposure. Genes Chromosomes Cancer. 2010;49(8):699–710. doi: 10.1002/gcc.20781. [DOI] [PubMed] [Google Scholar]
- 20.Zwart W, et al. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011;30(23):4764–4776. doi: 10.1038/emboj.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen MP, et al. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. 2013;73(22):6632–6641. doi: 10.1158/0008-5472.CAN-13-0704. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar V, et al. Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol. 2013;31(7):615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 24.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye T, et al. seqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39(6):e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller J. 2016 TransView: Read Density Map Construction and Accession. Visualization of ChIPSeq and RNASeq Data Sets. R package version 1.16.0. Available at bioconductor.org/packages/release/bioc/html/TransView.html. Accessed January 30, 2017.
- 27.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: Cis-regulatory element annotation system. Nucleic Acids Res. 2006;34(Web Server issue):W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YH, et al. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isserlin R, Merico D, Voisin V, Bader GD. Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000 Res. 2014;3:141. doi: 10.12688/f1000research.4536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunning MJ, Smith ML, Ritchie ME, Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23(16):2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 37.Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Res. 2012;22(11):2153–2162. doi: 10.1101/gr.135681.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinstein JN, et al. Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang YW, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69(23):9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubbard SA, et al. Evidence for cancer stem cells in human endometrial carcinoma. Cancer Res. 2009;69(21):8241–8248. doi: 10.1158/0008-5472.CAN-08-4808. [DOI] [PubMed] [Google Scholar]
- 41.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Nyholm HC, Nielsen AL, Lyndrup J, Norup P, Thorpe SM. Biochemical and immunohistochemical estrogen and progesterone receptors in adenomatous hyperplasia and endometrial carcinoma: Correlations with stage and other clinicopathologic features. Am J Obstet Gynecol. 1992;167(5):1334–1342. doi: 10.1016/s0002-9378(11)91712-8. [DOI] [PubMed] [Google Scholar]
- 43.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11(2):316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 44.Brzozowski AM, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389(6652):753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 45.Mohammed H, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Reports. 2013;3(2):342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishida M, Kasahara K, Kaneko M, Iwasaki H, Hayashi K. [Establishment of a new human endometrial adenocarcinoma cell line, Ishikawa cells, containing estrogen and progesterone receptors] Nippon Sanka Fujinka Gakkai Zasshi. 1985;37(7):1103–1111. [PubMed] [Google Scholar]
- 47.Gertz J, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Risinger JI, et al. Genetic instability of microsatellites in endometrial carcinoma. Cancer Res. 1993;53(21):5100–5103. [PubMed] [Google Scholar]
- 49.Andersson R, et al. FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.