Significance
The centromere is a highly specialized genomic locus playing a critical role in chromosome segregation during cell division and is made of characteristic repetitive DNA sequences. Due to their repetitive nature, it has been difficult to study the organization of human centromeres. Here, we present a way to monitor stability of the human centromeres using chromosome-orientation fluorescent in situ hybridization and superresolution microscopy. We show that the centromere-specific histone variant CENP-A and the centromere-associated proteins CENP-C and CENP-T contribute to maintenance of centromere integrity and present evidence of centromere disregulation in cancer and during cellular senescence, which is related to aging. Our study reveals the existence of a centromere-specific mechanism to organize the repetitive structure and prevent human centromeres from suffering illegitimate rearrangements.
Keywords: centromere, genome integrity, α-satellite repeats, cancer, CO-FISH
Abstract
Centromeres are highly specialized chromatin domains that enable chromosome segregation and orchestrate faithful cell division. Human centromeres are composed of tandem arrays of α-satellite DNA, which spans up to several megabases. Little is known about the mechanisms that maintain integrity of the long arrays of α-satellite DNA repeats. Here, we monitored centromeric repeat stability in human cells using chromosome-orientation fluorescent in situ hybridization (CO-FISH). This assay detected aberrant centromeric CO-FISH patterns consistent with sister chromatid exchange at the frequency of 5% in primary tissue culture cells, whereas higher levels were seen in several cancer cell lines and during replicative senescence. To understand the mechanism(s) that maintains centromere integrity, we examined the contribution of the centromere-specific histone variant CENP-A and members of the constitutive centromere-associated network (CCAN), CENP-C, CENP-T, and CENP-W. Depletion of CENP-A and CCAN proteins led to an increase in centromere aberrations, whereas enhancing chromosome missegregation by alternative methods did not, suggesting that CENP-A and CCAN proteins help maintain centromere integrity independently of their role in chromosome segregation. Furthermore, superresolution imaging of centromeric CO-FISH using structured illumination microscopy implied that CENP-A protects α-satellite repeats from extensive rearrangements. Our study points toward the presence of a centromere-specific mechanism that actively maintains α-satellite repeat integrity during human cell proliferation.
The centromere is a specialized genomic locus, which forms the primary constriction in the mitotic chromosomes, and supports kinetochore assembly for faithful transmission of chromosomes to dividing cells (1–3). Whereas DNA sequences of the centromere are highly diverged across phyla, the repetitive nature of centromeres is common, from fungi to plants and animals (1, 2). Human centromeres are characterized by the presence of head-to-tail tandem arrays of a ∼171 bp AT-rich α-satellite DNA (3–5). These tandem units are further organized into multimeric higher-order repeat (HOR) arrays that are repeated multiple times to form a core centromeric region up to 5 Mb in size (2, 6). The HORs of all but the human Y chromosome contain a 17-bp motif called the CENP-B box for the specific recruitment of the centromere protein B, CENP-B (Fig. 1A) (7, 8). Despite the accumulating knowledge of HOR organization, the repetitive nature of the α-satellite has hindered assembly of a contiguous centromere sequence for each chromosome (9) and has made it difficult to understand how the centromeric DNAs are organized at the primary constriction.
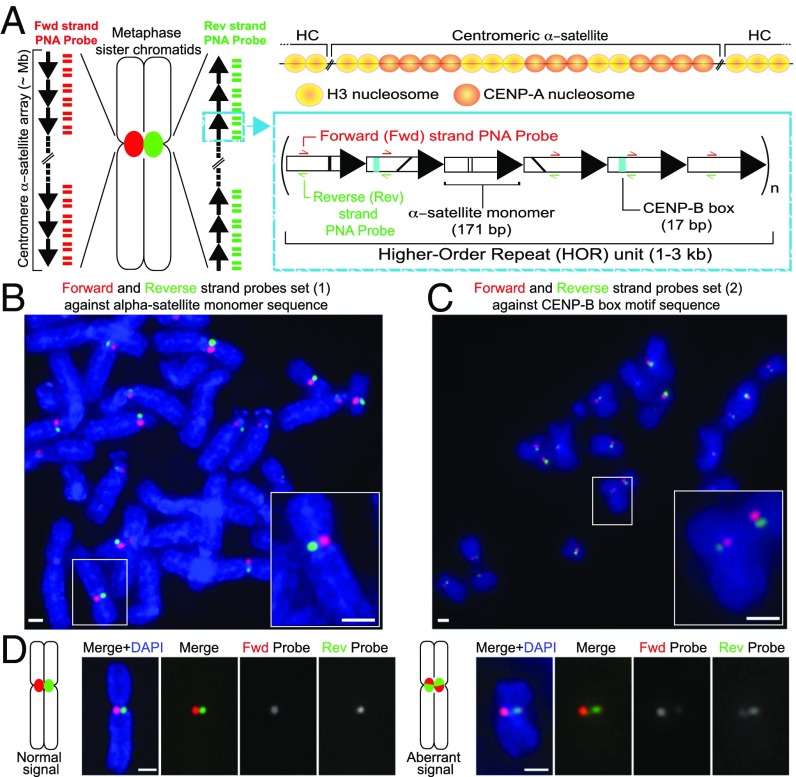
Fig. 1.
CO-FISH application to monitor human centromeric α-satellite repeats. (A) Diagram of the human centromere. (Left) The centromere locus of metaphase sister chromatids is depicted in red or green due to hybridization by unidirectional PNA probes, differentially labeled to detect the forward or reverse strands of one sister chromatid. Each black arrow symbolizes a HOR in the α-satellite array. The blue box gives a zoom in into a schematic example of the HOR tandem repeat monomers arranged head to tail. Represented on each monomer are black lines indicating divergence of sequence, teal box where the CENP-B box sequence is found, and the regions where the probes hybridize. (Above) The localization of CENP-A nucleosomes shown at the core centromere repeats, interspersed with canonical H3 nucleosomes. Outside of the core domain into the pericentromeric heterochromatin (HC), only H3 nucleosomes are found. (B) Unidirectional PNA probe set 1 against α-satellite or (C) PNA probe set 2 against the CENP-B box. White box indicates zoomed-in Insets. (D) Diagrammatic figures and example images of c-CO-FISH using probe set 1 in hTERT-RPE1 cells showing separated signal, where only one probe hybridizes on each sister chromatid (normal) and aberrant c-CO-FISH pattern, where the signal has exchanged between the two sister chromatids and probes overlap. (All scale bars, 1 μm.)
Inheritance and functions of centromeric chromatin is epigenetically defined by the centromere-specific histone H3 variant, CENP-A (10). Whereas CENP-A normally associates with the HOR of human centromeres (1, 2, 11) where it marks kinetochore assembly, the fact that neocentromeres, which contain CENP-A but lack α-satellite sequence, are stably maintained during cell divisions (9, 12–15), poses the question of why repetitive sequences are widely conserved between organisms. This question represents one of the paradoxes of centromere biology (16).
Another conandrum is the extent to which α-satellite repeats undergo recombination. Centromeric sequence presents evidence of mutational processes during evolution, including recombination, amplification, replication slippage, and crossovers, that have contributed to the formation of identical tandemly arranged repeats (17–20). Suppression of meiotic recombination at centromeres is established in eukaryotes, including human (19, 21), whereas active mitotic recombination at the centromere has been reported in budding yeast, fission yeast, and mice (22–25). In mice, DNA methylation by the DNA methyltransferases DNMT3a/b and chromatin remodeler α-thalassemia mental retardation X-linked protein (ATRX) contruibutes to suppression of centromere-associated sister chromatid exchanges (C-SCEs) (24, 25), but it remains unclear whether human centromeres actively undergo recombination, and whether a direct mechanism exists to suppress illegitimate recombination within the core centromeric HOR.
In this study, we applied chromosome-orientation fluorescent in situ hybridization (CO-FISH) to centromeres and found that human centromeres can undergo rearrangements in proliferating tissue culture cells. Levels of centromere aberrations increase upon depletion of CENP-A, CENP-C, and CENP-T/W, during replicative senescence, and in cancer cells. Our study reveals a mechanism dependent on CENP-A and its associated proteins that maintains centromere integrity in human cells.
Results
Detection of Human Centromeric α-Satellite Repeats by CO-FISH.
To monitor active recombination at human centromeres, we used CO-FISH (hereafter referred to as c-CO-FISH), using strand-specific probes (26) that differentially label the parental forward and reverse strand (3′–5′) of α-satellite DNAs (Fig. 1A). First, tissue culture cells were labeled with the nucleotide analogs bromodeoxyuridine (BrdU) and bromodeoxycytidine (BrdC) through a single round of DNA replication before being arrested in the following metaphase. Newly synthesized DNA that is labeled with BrdU and BrdC and subsequently UV nicked is enzymatically digested to reveal the forward and reverse parental strands, which are then hybridized by two strand-specific peptide nucleic acid (PNA) probes differentially labeled with Cy3 (red) and Alexa Fluor 488 (green), respectively. This technique takes advantage of the repetitive nature of the centromere and yielded the expected staining pattern, with each signal, green or red, specifically localized within one parental strand of one chromatid (Fig. 1 A–C and SI Appendix, Fig. S1A).
We used two sets of short, unidirectional PNA probes: probe set 1, against a 18-bp sequence found in α-satellite DNA monomers of 18 HORs (Fig. 1B and SI Appendix, Fig. S1 B–D) or probe set 2, complementary to the CENP-B box sequence present within a subset of α-satellite DNAs (Fig. 1C and SI Appendix, Fig. S2 A and B). When hybridized in telomerase-immortalized female retinal pigmented epithelial 1 (hTERT-RPE1) cells, the fluorescence intensity of PNA probe set 1 (SI Appendix, Fig. S1B) hybridized to the centromeres of 14 human chromosomes (SI Appendix, Fig. S1D), largely consistent with the representation of probe set 1 sequence against the Human Reference (HuRef) Genome (27) (SI Appendix, Fig. S1C), except for chromosomes 8 (D8Z2), 10 (D10Z1), and 11 (D11Z1) HOR satellite arrays (SI Appendix, Fig. S1 C and D). This discrepancy may reflect nucleotide divergence across different cell lines. Probe set 2, against the CENP-B box (SI Appendix, Fig. S2A), hybridized to most chromosomes in the hTERT-RPE1 cells (SI Appendix, Fig. S2B). Differences in CENP-B box probe intensity are likely due to the heterogeneous size of the CENP-B box-containing HOR region between chromosomes and the variance in CENP-B box sequences. c-CO-FISH was also able to detect cases of aberrant staining patterns where the sister chromatids are stained with both probes, likely reflecting C-SCE events (Fig. 1D; quantified in Fig. 2C).
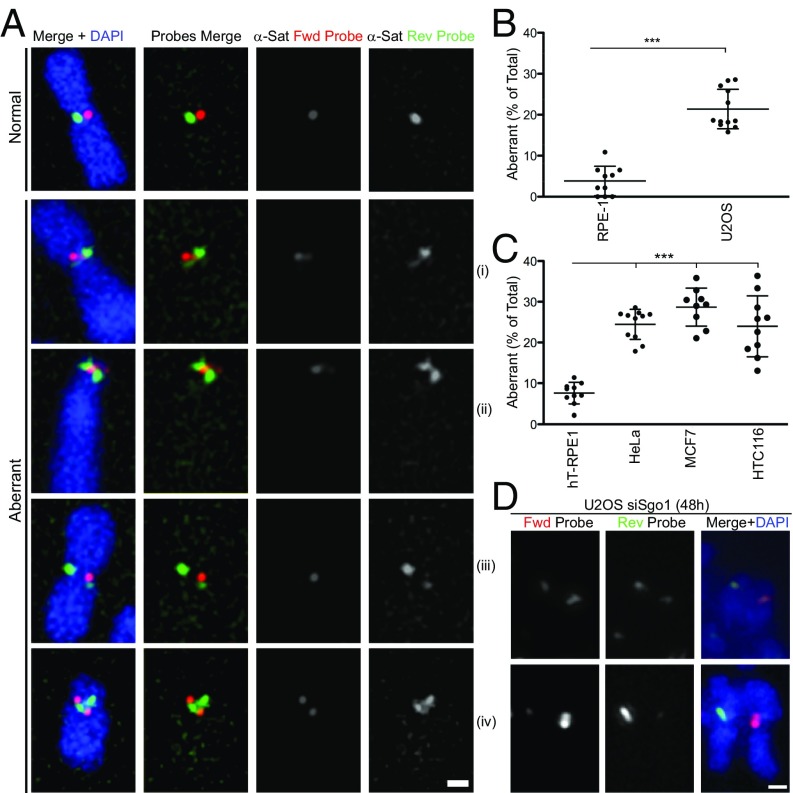
Fig. 2.
Centromere instability monitored by c-CO-FISH in human cancer cells. (A) c-CO-FISH of human cancer cells using probe set 1. Example of normal signal (Top) and different types of centromeric α-satellite repeat rearrangements identified using the technique in U2OS cells. (Scale bar, 1 μm.) (B and C) Frequency of aberrant c-CO-FISH patterns detected by probe set 2 in indicated cell lines. n ≥ 10. Error bars ± SD, ***P ≤ 0.0005. (D) Two examples of aberrant c-CO-FISH signal of prematurely separated sister chromatids after siSgo1 for 48 h in U2OS cells. (Scale bar, 1 μm.)
Monitoring Integrity of Centromeric α-Satellite Repeats by c-CO-FISH in Cancer Cells.
We first examined whether the frequency of the aberrant c-CO-FISH patterns vary among different cell lines. In primary RPE1 cells, more than 90% of chromosomes exhibit stereotypical c-CO-FISH patterns, yet the human bone cancer line U2OS showed a three- to fourfold increase in aberrant c-CO-FISH patterns, where both forward and reverse probes hybridize to the same chromatid (Fig. 2 A and B and SI Appendix, Fig. S3 B and F). We found different types of centromere aberrations in U2OS (Fig. 2A, i–iv), in line with evidence suggesting that satellite changes are driven by different mutational processes, from recombination to unequal exchange and translocation (18, 28). The majority of rearrangements observed were as in Fig. 2A examples i and ii, where each c-CO-FISH probe continuously stains centromeres of both sister chromatids. We interpret this pattern as a C-SCE generated during a single prior cell cycle, some of which result in unequal crossovers. More rarely, we observed cases where one of the sister chromatids contains two distinct regions stained by two different probes and the sum of these regions is larger than the other sister chromatid’s centromere (Fig. 2A, iii), which may have resulted from an unequal C-SCE or duplicated inversion of the α-satellite cluster. About 1–2% of the total aberrations in the cancer line U2OS appeared to be combinations of both C-SCE and inversion or chromosome fusion (Fig. 2A, iv). We excluded from our quantification, chromosomes showing two sets of inverted α-satellite clusters separated by an intervening noncentromeric DNA region (SI Appendix, Fig. S3E), likely caused by chromosome fusions at genomic loci outside the centromeric domain during past generations. To exclude the possibility that the aberrant c-CO-FISH pattern was due to a geometrical anomaly of centromeric DNAs rather than DNA rearrangements, Sgo1 was down-regulated by siRNA in U2OS cells to induce premature dissociation of sister chromatids (29). The CO-FISH signals of both forward and reverse probes can be seen in a single chromatid (Fig. 2D), suggesting that this aberration is caused by DNA rearrangements. Aberrant c-CO-FISH patterns were found in other human cancer cell lines, including high chromosome instability (CIN) lines (human epithelial cervical and breast cancer cells HeLa and MCF7, respectively), like U2OS, and low CIN lines (human colon cancer cells HCT116) (Fig. 2C and SI Appendix, Fig. S3 C, D, and F). Collectively, our data indicate that centromere stability is compromised in several cancer cell lines. Interestingly, we found that chromatin-bound CENP-A was decreased in these cancer cell lines (HeLa, MCF7, and HTC116; SI Appendix, Fig. S3G), except for U2OS defective in the ATRX pathway (30) (Discussion).
CENP-A Maintains α-Satellite Repeat Stability in Human Cells.
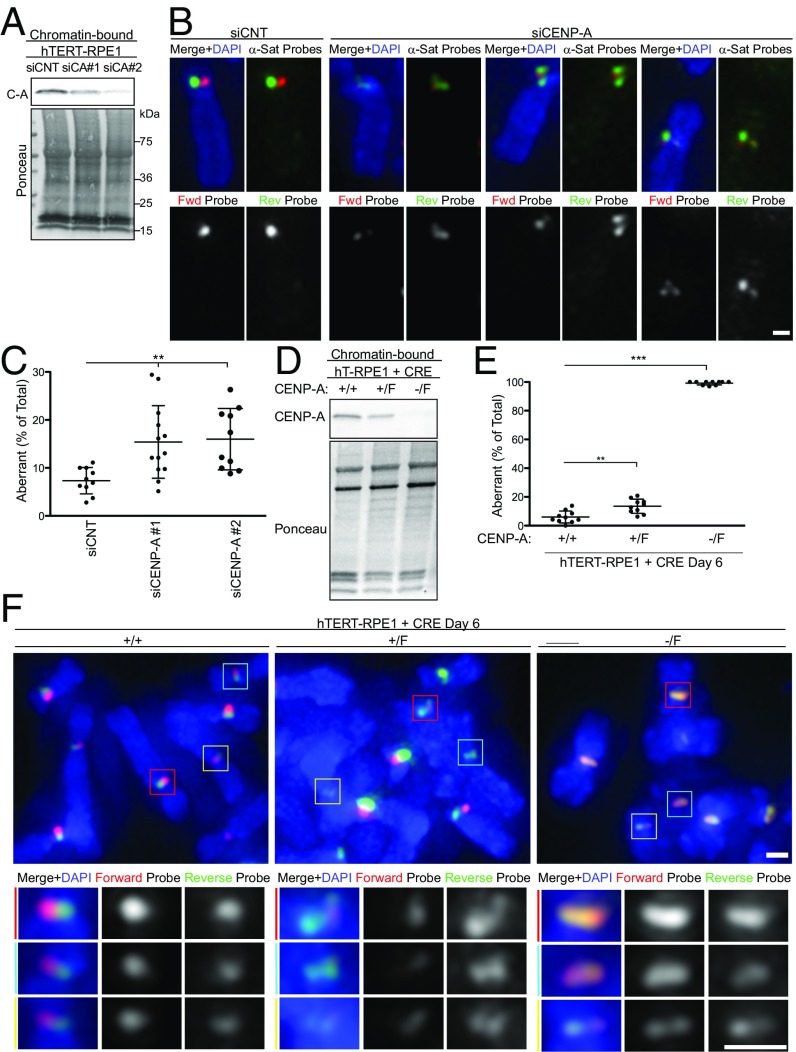
To test the possibility that CENP-A contributes to the mechanism that suppresses centromere rearrangements, we depleted CENP-A using two different siRNAs (Fig. 3A and SI Appendix, Fig. S4A). Reduced recruitment of CENP-A to chromatin by either siRNA (Fig. 3A) led to an increase in centromere aberrations (Fig. 3 B and C and SI Appendix, Fig. S4B). To achieve long-term removal of CENP-A from human cells, we used the diploid hTERT-RPE1 line where the CENP-A gene can be conditionally knocked out (31). The +/F line contains a wild-type allele and a CENP-A allele flanked by flox recombination sequences, which is removed by viral expression of Cre recombinase, generating the CENP-A+/− genotype. In the −/F line, where the wild-type allele is deleted, Cre-mediated recombination resulted in CENP-A−/− (Fig. 3D). At the time of harvesting for CO-FISH 6 d post-Cre induction, the CENP-A−/− cells retained their ability to cycle and incorporate BrdU (SI Appendix, Fig. S4H). As previously shown (31), removal of CENP-A led to a slower cell cycle progression; therefore, we adjusted the length of CO-FISH treatments accordingly. In the CENP-A null cells (−/−), more than 90% of centromeres in each metaphase exhibited aberrant c-CO-FISH patterns (Fig. 3 E and F and SI Appendix, Fig. S4 C and D). The CENP-A+/− line also gave a statistically significant increase in centromere aberrations compared with wild-type hTERT-RPE1 cells (Fig. 3 E and F and SI Appendix, Fig. S4 C and D). c-CO-FISH of CENP-A null cells showed broadly colocalized forward and reverse probes (SI Appendix, Fig. S5). In addition, excision of centromeres, where signal was localized outside of DAPI-stained chromosomes, was also observed (SI Appendix, Fig. S4C). However, telomere-associated SCEs did not increased in these cells (SI Appendix, Fig. S4E), suggesting that these rearrangements are restricted to centromeres.
Fig. 3.
CENP-A maintains the integrity of the α-satellite repeats. (A) hTERT-RPE1 cells were treated with control siRNA (siCNT) or two different siRNA, #1 and #2 targeting to CENP-A for 72 h. Chromatin-bound proteins were analyzed by Western blotting using anti–CENP-A antibody (Top) and ponceau (Bottom). (B) Examples of aberrant c-CO-FISH patterns in siCENP-A cells using probe set 1. (C) Quantification of aberrant c-CO-FISH. n ≥ 10. Error bars ± SD, **P ≤ 0.005. (D) hTERT-RPE1 lines upon deletion of the CENP-A floxed allele by infection with Adeno-CRE for 6 d. +/+ hTERT-RPE1; +/F one floxed allele; −/F one deleted and one floxed allele. Chromatin-bound proteins were analyzed by Western blotting using anti–CENP-A antibody (Top) and ponceau (Bottom). (E) c-CO-FISH (probe set 2) quantification of centromere instability in CENP-A flox hTERT-RPE1 cell lines as in D; n ≥ 10. Error bars ± SD, ***P ≤ 0.0005. (F) c-CO-FISH of a metaphase spread from cell lines as in D. (Scale bars, 1 μm.) Three centromeres from each figure marked by the colored boxes are zoomed in and shown in the panel Below, as indicated.
To exclude the possibility that the aberrant c-CO-FISH pattern seen in CENP-A–depleted cells was due to a geometrical reorganization of centromeric DNAs rather than bona fide rearrangements, we performed Sgo1 RNAi for 48 h in cells knocked out for CENP-A to accumulate prematurely dissociated sister chromatid (29). The CO-FISH signals of both forward and reverse probes can be seen in a single chromatid (SI Appendix, Fig. S4F), suggesting that this aberration is unlikely to be caused by geometrical reorganization of the α-satellite and are instead indicative of C-SCE. This aberrant hybridization pattern depends on the BrdU:C incorporation and exonuclease steps of the CO-FISH procedure (SI Appendix, Fig. S4G), ruling out the possibility that a change in the frequency of DNA damage or ssDNAs affects the hybridization. These results point toward a pivotal role for CENP-A in stabilizing highly repetitive α-satellite DNA.
Superresolution Microscopy of Centromere Organization Reveals Disruption of α-Satellites Structure upon CENP-A Depletion.
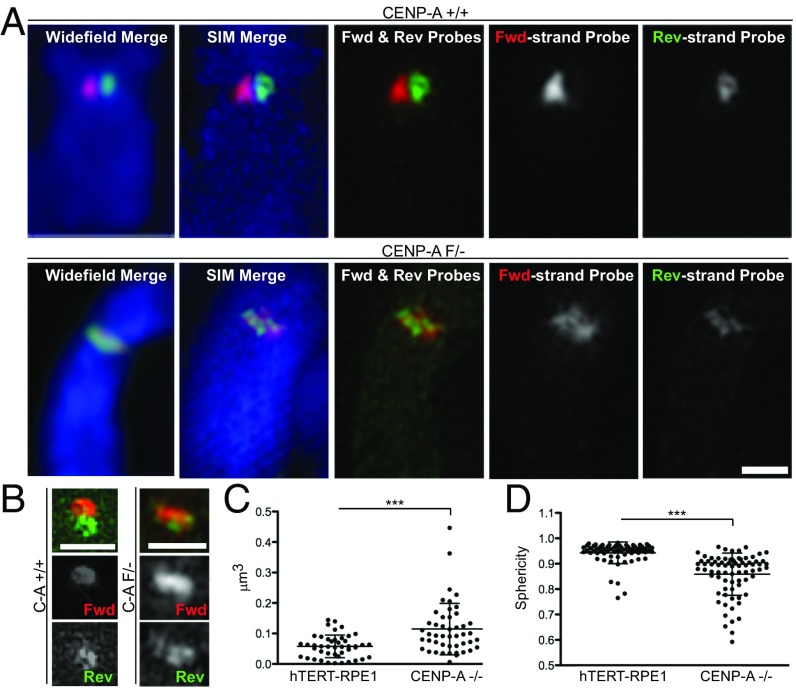
To gain better insight into the severe centromeric aberration in cells depleted of CENP-A, we applied 3D structured illumination microscopy (SIM). In CENP-A knockout cells, whereas both forward and reverse probes were broadly distributed over the primary constriction, the two probes occupy distinct regions, which often do not overlap (Fig. 4 A and B and SI Appendix, Fig. S6A). These arrangements may reflect multiple crossover events at the centromere. Furthermore, we found an overall change in distribution, size, and shape of the α-satellites, with both CO-FISH probes occupying a broader centromere region in CENP-A knockout cells than in control (Fig. 4 C and D and SI Appendix, Fig. S6A). These data indicate that CENP-A not only suppresses extensive rearrangement of α-satellites but also plays an important role in structural organization of α-satellites at the primary constriction.
Fig. 4.
Superresolution imaging of centromeric CO-FISH in CENP-A knockout cells. (A) Conventional deconvolution microscopy (Left) and 3D-SIM images of the same chromosome stained by c-CO-FISH (probe set 1) in human hTERT-RPE1 (Top) or CENP-A knockout cells (Bottom). (B) A 3D-SIM example of centromeres in hTERT-RPE1 (C-A; CENP-A+/+) or CENP-A knockout (C-A−/−). (Scale bars, 1 μm.) (C) Volume and (D) sphericity in cells as in A. Error bars ± SD, ***P ≤ 0.0005. Plotted are single centromeres, calculated using the green reverse-strand probe fluorescence.
Centromere Instability During the Course of Replicative Senescence.
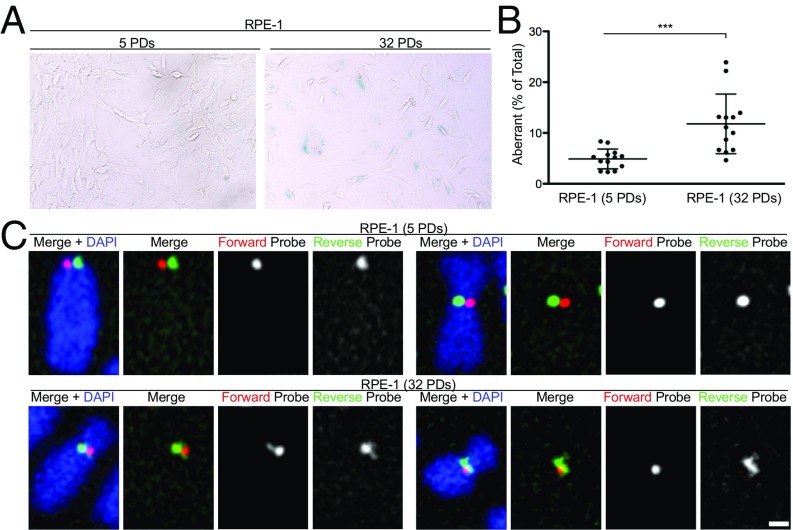
Because a physiological decline in the level of CENP-A protein has been reported in senescent cells (32) and in islet cells during organismal aging (33), we investigated whether centromere stability deteriorated with cellular senescence. To senesce cells, we passaged primary RPE1 cells that do not express telomerase over multiple population doublings (PDs), until a subset of cells entered replicative senescence as determined by staining with the senescence-associated marker β-galactosidase (Fig. 5A). c-CO-FISH was then performed on the early passage (5 PDs) and late passage (32 PDs) cells. The number of aberrant CO-FISH patterns significantly increased in late passage cells (Fig. 5 B and C), indicating that replicative senescence destabilizes not only telomeres but also centromeres.
Fig. 5.
Senescence-associated centromere instability in RPE1 cells. (A) Senescence-associated β-galactosidase staining in RPE1 cells at different population doublings (PDs): young (5 PDs) versus old (32 PDs). (B) Quantification of aberrant c-CO-FISH patterns. Error bars ± SD, ***P ≤ 0.0005. (C) c-CO-FISH examples of normal c-CO-FISH pattern (Top) using probe 1 found in young cells and aberrant patterns (Bottom) seen in presenescence cells in old RPE1.
Depletion of Constitutive Centromere-Associated Network Components, CENP-C and CENP-T/W, Causes α-Satellite Repeat Instability.
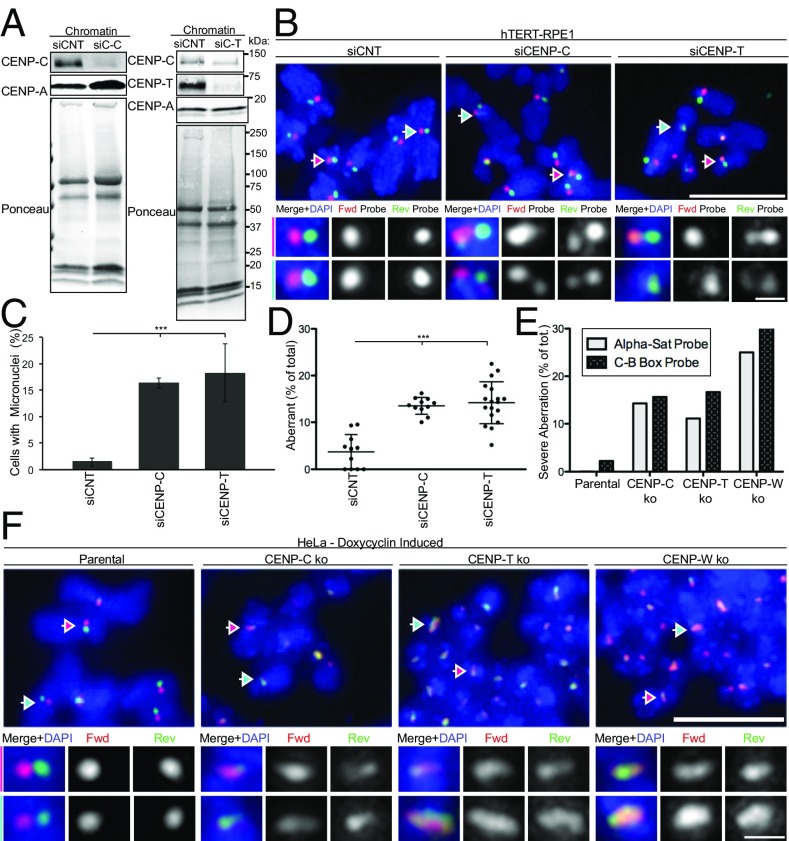
To better understand the mechanism by which CENP-A suppresses centromeric rearrangements, we investigated the importance of CENP-C and the CENP-T/W complex, which are basal members of the constitutive centromere-associated network (CCAN) and are directly recruited to CENP-A nucleosomes throughout the cell cycle (34). Acute down-regulation of CENP-C or CENP-T by siRNA in hTERT-RPE1 caused micronuclei formation due to chromosome missegregation (Fig. 6 A and C and SI Appendix, Fig. S7B), whereas it did not affect the level of chromatin- or centromere-associated CENP-A (Fig. 6A and SI Appendix, Fig. S7B). c-CO-FISH analyses using centromeric probe set 2 (Fig. 6D) and probe set 1 (SI Appendix, Fig. S7C) revealed that siRNA treatment against either CENP-C or CENP-T led to a significant increase in aberrant c-CO-FISH patterns to a level comparable to cells treated with CENP-A siRNA (Fig. 6 B and D and SI Appendix, Fig. S7C).
Fig. 6.
CCAN components CENP-C and CENP-T/W contribute to centromere stability. (A) hTERT-RPE1 cells were treated with control siRNA (siCNT) or siRNA targeting to CENP-C and CENP-T for 72 h, Left and Right, respectively. Western blotting using antibodies as indicated and ponceau staining of chromatin. Molecular weights (in kilodaltons) are indicated. (B) Representative c-CO-FISH images upon CENP-C and CENP-T RNAi. (Scale bar, 5 μm.) Colored arrows indicate the centromeres that were zoomed in and the channels separated in the tiles below. (Scale bar, 0.5 μm.) (C) Quantification of micronuclei in cells as in A. Error bars ± SD, ***P ≤ 0.0005. (D) Quantification of aberrant centromeres. n ≥ 10. Error bars ± SD, **P ≤ 0.005. (E) Inducible knockout HeLa cells with Cas9 under control of a stably integrated Tet-On promoter. Doxycycline was added for 5 d for CENP-C and CENP-T knockouts and 3 d for CENP-W knockout. Cells were then harvested for c-CO-FISH and quantified for the percentage of metaphases where more than 80% of chromosomes show aberrant c-CO-FISH pattern. n = 40. (F) Examples of parental HeLa and CENP-C, CENP-T, and CENP-W knockouts showing the severe c-CO-FISH aberrations quantified in E. (Scale bar, 5 μm.) Colored arrows indicate the centromeres that were zoomed in and the channels separated in the tiles Below. (Scale bar, 0.5 μm.)
To verify these results, we used HeLa cell lines where CENP-C, CENP-T, and CENP-W can be conditionally knocked out by doxycycline-mediated induction of Cas9 (35). At 3 d (CENP-W) or 5 d (CENP-C or CENP-T) after doxycycline induction, chromatin-association and c-CO-FISH pattern were monitored. As previously reported (35), all knockout treatments led to decrease of chromatin- or centromere-associated CENP-T without affecting CENP-A, whereas CENP-C knockout, but not CENP-T or CENP-W knockout, decreased chromatin-associated CENP-C (SI Appendix, Fig. S7 D and E). Chronic knockout of CENP-W, CENP-C, and CENP-T caused a significant increase in aberrant c-CO-FISH patterns (SI Appendix, Fig. S7F). We found that a subset of metaphase cells exhibit severe centromeric aberration, where over 80% of CO-FISH positive centromeres are aberrant (Fig. 6F), resembling the phenotype seen upon CENP-A knockout (Fig. 3F). Quantifications of these severe centromere aberrations amounted to about 15–25% of the total knockout metaphase cells, whereas fewer than 2% of parental metaphase cells exhibited this phenotype (Fig. 6E). Our data show that CCAN components CENP-C and CENP-T/W that act downstream of CENP-A are also involved in maintaining stability of the α-satellite repeats.
CENP-A–Dependent Mechanism to Maintain Centromere Integrity Acts Independently of Chromosome Segregation.
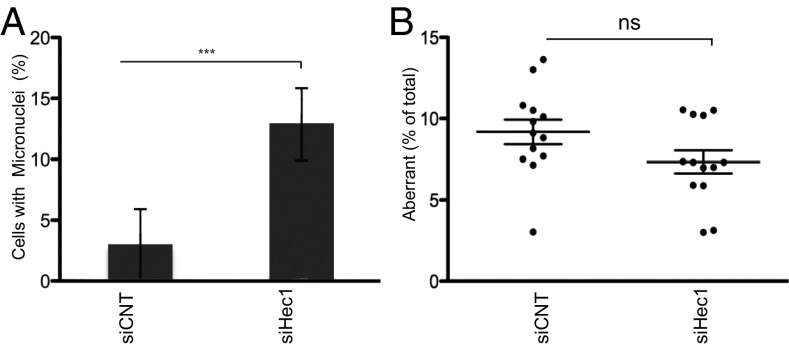
Perturbation of CENP-A (SI Appendix, Fig. S8A and ref. 31), CENP-C, and CENP-T (Fig. 6C) causes chromosome missegregation due to failure in mitotic kinetochore assembly. To examine the possibility that centromere rearrangements are a byproduct of chromosome missegregation, the Hec1 subunit of Ndc80, the mitosis-specific kinetochore complex critical for end-on microtubule attachment and correct chromosome segregation, was depleted by siRNA (36, 37) in hTERT-RPE1 cells, and c-CO-FISH was performed (SI Appendix, Fig. S8 B–D). Although Ndc80 depletion induced chromosome missegregation and an increase in interphase cells containing one or more micronuclei (Fig. 7A), frequency of aberrant c-CO-FISH patterns was not affected (Fig. 7B), suggesting that induction of centromeric rearrangements is not due to chromosome missegregation. Additionally, we used an alternative method to cause chromosome missegragation by performing a monastrol-washout experiment in hTERT-RPE1 (SI Appendix, SI Methods). Monastrol is an inhibitor of Eg5 that causes mitotic arrest with monopolar spindles (38). When monastrol is washed out, many kinetochores form merotelic and syntelic attachments, causing chromosome missegregation (39–41). Monastrol caused micronuclei formation (SI Appendix, Fig. S8E) but no increase in aberrant c-CO-FISH patterns in these cells (SI Appendix, Fig. S8F). These results indicate that chromosome missegregation during aberrant mitotic division is not causally linked to centromere repetitive DNA instability. Collectively, our data point toward the existence of a CENP-A–dependent mechanism that suppresses centromeric DNA recombination and maintains repeats stability independent of its role on chromosome segregation.
Fig. 7.
Depleting the Ndc80 complex does not cause centromere DNA instability. (A) Quantification of micronuclei in hTERT-RPE1 cells, treated with control siRNA (siCNT) or siRNA targeting to Hec1 for 72 h. Error bars ± SD, ***P ≤ 0.0005. (B) Quantification of aberrant c-CO-FISH pattern. n = 13. Error bars ± SD. Ns, nonsignificant.
Discussion
DNA rearrangements, including unequal exchange, aberrant recombination, amplification, and excision, have been predicted to drive centromere evolution (18). This prediction raises the question of whether there is an active mechanism to preserve repetitive DNAs at the centromere. Here, we present evidence toward the existence of a CENP-A/CCAN-dependent mechanism that maintains centromere integrity.
Consistent with the role of CENP-A in preserving the integrity of α-satellite DNA repeats, reduction of the repeat has been observed at the spontaneously inactivated centromere in the chromosome that gained a neocentromere (42), although it is not clear whether repeat reduction is a consequence or cause of centromere inactivation. It is possible that removal of CENP-A may induce centromere fragility by dechromatinizing the α-satellites. However, histone H3.3 has been reported to act as a placeholder for CENP-A loading (43), suggesting that depletion of CENP-A from the centromere may not be sufficient to disrupt nucleosomes. In addition, we show that knockout of CENP-C and CENP-T/W, which depend on CENP-A for their recruitment, also causes severe centromeric aberrations without affecting chromatin-bound CENP-A levels. This suggests that the phenotype we report is unlikely due to reduced nucleosome occupancy at centromeres depleted of CENP-A but rather through CENP-A–dependent recruitment of specific components to centromeres.
As CENP-A and the CCAN associate with the centromere throughout the cell cycle, the mechanism that protects centromere integrity may operate outside of mitosis, particularly in S phase. Just as telomere-binding proteins are required for replication fork progression at the telomere repeats (44), it is possible that they may contribute to efficient replication at α-satellite DNA, preventing stalled replication forks, subsequent induction of homologous recombination, and centromere instability. Consistent with this idea, a recent study in Xenopus egg extracts suggests that the human α-satellite repeats exhibit a relatively slow rate of replication fork progression (45). Alternatively, in line with CENP-A being implicated in the DNA damage response (46), presence of CENP-A on chromatin may directly facilitate the repair of lesions at centromeric repeats. It is also possible that chromatin compaction status may affect centromere integrity. Our 3D-SIM analysis suggests that CENP-A depletion causes an apparent increase in the chromatin volume occupied by α-satellites (Fig. 6F), in agreement with a previous report showing CENP-A knockout mouse embryos have a more diffuse centromeric domain as stained by CREST (human calcinosis, Raynaud's phenomenon, esophageal dysfunction, sclerodactyly, and telangiectasia) antiserum (47).
Physiologically, CENP-A level decreases during cellular senescence and organismal aging (32, 33). Consistent with this fact, we found enhanced centromere instability during the course of cellular senescence. An age-related decrease in CENP-A levels may represent a safety mechanism to prevent additional cell division in the presence of centromere instability. Future studies will be required to establish whether the compromised CENP-A and α-satellite DNA repeats can be recovered upon transformation or during generation of induced pluripotent cells.
We found that several cancer cell lines exhibit enhanced rearrangements of the α-satellite DNAs. The centromere instability was not limited to CIN cancer cell lines (U2OS, HeLa, and MCF7) but also found in karyotypically stable HCT116 cells (41), which is yet defective in intra–S-checkpoint activation during replication and in DNA damage repair (48). Except for U2OS, cancer cell lines used in this study showed a decrease in chromatin-bound CENP-A. Though its functional significance remains to be tested, the reduction in chromatin-bound CENP-A raises a possibility that centromere instability in cancer cells may be in part due to compromised functionality of CENP-A and CCAN proteins. In U2OS cells, ATRX is deleted and telomere is maintained via the alternative lengthening of telomeres (ALT) pathway (30). It has been recently shown that C-SCE is enhanced in mouse embryos lacking ATRX, and suggested that this hyperrecombination may result in chromosome breakage at the centromere and fusion (25). Thus, it is likely that multiple mechanisms drive centromere instability in cancer cells.
Our study using the c-CO-FISH unveiled the function of CENP-A and its associated protein in suppressing rearrangements of α-satellite DNA repeats at the human centromere. Further dissection of the mechanism that contributes to the centromere integrity will help in understanding the consequence of enhanced rearrangements at the centromere during aging and cancer development.
Methods
CO-FISH at centromere was adapted from ref. 44 (SI Appendix, SI Methods). Centromere probes from PNABio were as follows: pair 1 against the α-satellite, F3003 CENT-Cy3 (AAACTAGACAGAAGCATT) and reverse complement CENT-RC-488 labeled with Alexa Fluor 488 (AATGCTTCTGTCTAGTTT) or pair 2 against the CENP-B box, F3002 CENP-B Cy3 (ATTCGTTGGAAACGGGA) and reverse complement CENP-B-RC-488 labeled with Alexa Fluor 488 (TCCCGTTTCCAACGAAT). Quantification to score for c-CO-FISH aberrations was done by counting the number of abnormal signals, and the percentage of aberrant signals over the total number of centromere pairs for each metaphase was plotted as a single dot in Prism. P values were calculated by Mann–Whitney. Error bars are ±SD.
Additional methods, extended data display, and discussion are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank N. Bosco for technical support and providing helpful insights; F. Lottersberger for advice with the CO-FISH technique; K. Miga for α-satellite genomic sequence analysis; T. Davoli, and T. de Lange for sharing reagents and advice; I. Cheeseman, D. Cleveland, W. Earnshaw, D. Fachinetti, and G. Goshima for providing reagents; the Bio-Imaging and Flow Cytometry Resource Centers, and S. J. K. Thomas for helping with data acquisition; and N. Bosco, K. Miga, M. Wheelock, and D. Wynne for critical reading of the manuscript. S.G. was supported by the American-Italian Cancer Foundation and Women in Science Rockefeller fellowships. This work was supported in part by Grant 2013-049 from the Tri-Institutional Stem Cell Initiative (to H.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615133114/-/DCSupplemental.
References
- 1.Steiner FA, Henikoff S. Diversity in the organization of centromeric chromatin. Curr Opin Genet Dev. 2015;31:28–35. doi: 10.1016/j.gde.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Fukagawa T, Earnshaw WC. The centromere: Chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30(5):496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo KHA. The Centromere. Oxford Univ Press; Oxford, UK: 1997. [Google Scholar]
- 4.Waye JS, et al. Chromosome-specific alpha satellite DNA from human chromosome 1: Hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics. 1987;1(1):43–51. doi: 10.1016/0888-7543(87)90103-0. [DOI] [PubMed] [Google Scholar]
- 5.Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19(6):1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294(5540):109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 7.Earnshaw WC, et al. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104(4):817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109(5):1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miga KH. Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosome Res. 2015;23(3):421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- 10.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144(4):471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden KE, et al. Sequences associated with centromere competency in the human genome. Mol Cell Biol. 2013;33(4):763–772. doi: 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am J Hum Genet. 1993;52(6):1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 13.Choo KH. Centromere DNA dynamics: Latent centromeres and neocentromere formation. Am J Hum Genet. 1997;61(6):1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334(6056):686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 15.Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82(2):261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 17.McFarlane RJ, Humphrey TC. A role for recombination in centromere function. Trends Genet. 2010;26(5):209–213. doi: 10.1016/j.tig.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371(6494):215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 19.Mahtani MM, Willard HF. Physical and genetic mapping of the human X chromosome centromere: Repression of recombination. Genome Res. 1998;8(2):100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 20.Biscotti MA, Olmo E, Heslop-Harrison JSP. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015;23(3):415–420. doi: 10.1007/s10577-015-9499-z. [DOI] [PubMed] [Google Scholar]
- 21.Jackson MS, See CG, Mulligan LM, Lauffart BF. A 9.75-Mb map across the centromere of human chromosome 10. Genomics. 1996;33(2):258–270. doi: 10.1006/geno.1996.0190. [DOI] [PubMed] [Google Scholar]
- 22.Liebman SW, Symington LS, Petes TD. Mitotic recombination within the centromere of a yeast chromosome. Science. 1988;241(4869):1074–1077. doi: 10.1126/science.3137657. [DOI] [PubMed] [Google Scholar]
- 23.Minet M, Grossenbacher-Grunder AM, Thuriaux P. The origin of a centromere effect on mitotic recombination : A study in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1980;2(1):53–60. doi: 10.1007/BF00445694. [DOI] [PubMed] [Google Scholar]
- 24.Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J Cell Biol. 2008;181(6):885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De La Fuente R, Baumann C, Viveiros MM. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development. 2015;142(10):1806–17. doi: 10.1242/dev.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey SM, Williams ES, Cornforth MN, Goodwin EH. Chromosome orientation fluorescence in situ hybridization or strand-specific FISH. Methods Mol Biol. 2010;659:173–183. doi: 10.1007/978-1-60761-789-1_12. [DOI] [PubMed] [Google Scholar]
- 27.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalitsis P, Griffiths B, Choo KHA. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci USA. 2006;103(23):8786–8791. doi: 10.1073/pnas.0600250103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118(5):567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Lovejoy CA, et al. ALT Starr Cancer Consortium Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8(7):e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fachinetti D, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15(9):1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010;30(9):2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S-H, Itkin-Ansari P, Levine F. CENP-A, a protein required for chromosome segregation in mitosis, declines with age in islet but not exocrine cells. Aging (Albany NY) 2010;2(11):785–790. doi: 10.18632/aging.100220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 2016;17(1):16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinley KL, et al. The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol Cell. 2015;60(6):886–898. doi: 10.1016/j.molcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297(5590):2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 37.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7(1):45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286(5441):971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 39.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180(4):665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153(3):517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci USA. 2011;108(44):17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasson D, et al. Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma. 2011;120(6):621–632. doi: 10.1007/s00412-011-0337-6. [DOI] [PubMed] [Google Scholar]
- 43.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2(2):146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol. 2016;18(6):684–691. doi: 10.1038/ncb3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeitlin SG, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci USA. 2009;106(37):15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97(3):1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takemura H, et al. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J Biol Chem. 2006;281(41):30814–30823. doi: 10.1074/jbc.M603747200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.