Significance
Activated PI3K Delta Syndrome (APDS) is a primary immunodeficiency disease caused by activating mutations in phosphoinositide 3-kinases (PI3Kδ). Activating mutations in either the p110δ catalytic or the p85α regulatory subunit of PI3Kδ result in APDS. Mutations in p85α leading to APDS are surprising, as other p85α-activating mutations are oncogenic when bound to the PI3Kα isoform. Using hydrogen–deuterium exchange mass spectrometry, we determined the molecular mechanisms by which APDS mutations in p110δ or p85α activate PI3Kδ and reveal why the p85α APDS2 mutant primarily activates PI3Kδ. All APDS mutants are potently inhibited by the PI3Kδ-specific inhibitor idelalisib. Together, the biophysical and biochemical data reveal insights into PI3Kδ regulation and provide a possible therapeutic strategy for treating patients with APDS.
Keywords: PIK3CD, PIK3R1, HDX-MS, phosphoinositides, PI3K/AKT
Abstract
Activated PI3K Delta Syndrome (APDS) is a primary immunodeficiency disease caused by activating mutations in either the leukocyte-restricted p110δ catalytic (PIK3CD) subunit or the ubiquitously expressed p85α regulatory (PIK3R1) subunit of class IA phosphoinositide 3-kinases (PI3Ks). There are two classes of APDS: APDS1 that arises from p110δ mutations that are analogous to oncogenic mutations found in the broadly expressed p110α subunit and APDS2 that occurs from a splice mutation resulting in p85α with a central deletion (Δ434–475). As p85 regulatory subunits associate with and inhibit all class IA catalytic subunits, APDS2 mutations are expected to similarly activate p110α, β, and δ, yet APDS2 largely phenocopies APDS1 without dramatic effects outside the immune system. We have examined the molecular mechanism of activation of both classes of APDS mutations using a combination of biochemical assays and hydrogen–deuterium exchange mass spectrometry. Intriguingly, we find that an APDS2 mutation in p85α leads to substantial basal activation of p110δ (>300-fold) and disrupts inhibitory interactions from the nSH2, iSH2, and cSH2 domains of p85, whereas p110α is only minimally basally activated (∼2-fold) when associated with mutated p85α. APDS1 mutations in p110δ (N334K, E525K, E1021K) mimic the activation mechanisms previously discovered for oncogenic mutations in p110α. All APDS mutations were potently inhibited by the Food and Drug Administration-approved p110δ inhibitor idelalisib. Our results define the molecular basis of how PIK3CD and PIK3R1 mutations result in APDS and reveal a potential path to treatment for all APDS patients.
The class I phosphoinositide 3-kinases (PI3Ks) are responsible for the generation of the key lipid-signaling molecule phosphatidylinositol (3–5) Tris-phosphate (PIP3), which is vital for the recruitment of effector proteins containing PIP3-binding domains, leading to transduction of extracellular signals at the plasma membrane. PI3Ks are activated downstream of a number of signaling inputs, including receptor tyrosine kinases (RTKs), G-protein–coupled receptors, and the Ras superfamily of GTPases (1). The spatiotemporal production of PIP3 is tightly controlled, regulating downstream pathways involved in cell growth, death, and proliferation (2). Misregulation of class I PI3K activity through either activating or inactivating mutations underlies a number of human diseases including cancer (3), developmental disorders (4), and primary immunodeficiencies (5–7).
The three class IA PI3Ks are obligate heterodimers composed of a catalytic subunit (p110α, p110β, or p110δ) and an associated regulatory subunit (p85α, p85β, p50α, p55α, or p55γ). The different p110 catalytic subunits have distinct tissue expression profiles, with p110α and p110β being ubiquitously expressed and p110δ being primarily expressed in immune cells (8). Binding of the ubiquitously expressed regulatory subunit p85 to the p110 catalytic subunit plays three key roles: (i) it stabilizes the catalytic subunit, (ii) it inhibits catalytic activity, and (iii) it mediates activation downstream of phosphorylated RTKs and their adaptors (9). Both p110 catalytic and p85 regulatory subunits are large, multidomain proteins. All class IA p110 subunits are composed of an adaptor-binding domain (ABD) that mediates binding to p85, a Ras-binding domain (RBD), a C2 domain, a helical domain, and a bilobal kinase domain. All p85 regulatory subunits contain two SH2 domains (nSH2 and cSH2) linked by a coiled-coil region referred to as the inter-SH2 domain (iSH2). The class IA p110 catalytic subunits are differentially inhibited by p85, with p110α containing inhibitory contacts between the nSH2 domain of p85 and the C2, helical, and kinase domains of p110α, as well as between the iSH2 domain of p85 and the C2 domain of p110α (10). Both p110β and p110δ contain an additional regulatory contact between the cSH2 of p85 and the kinase domain (11–13). Recruitment of the SH2 domains to phosphorylated receptors and their adaptors disrupts both nSH2 and cSH2 inhibitory contacts. The catalytic domain also contains intramolecular inhibitory interactions between the ABD and kinase domains. Activating oncogenic mutations in p110α mediate activation by disruption of these inhibitory interfaces, a process that normally occurs during the catalytic cycle of the enzyme on membranes (14).
The complex of p110δ with p85α (referred to hereafter as PI3Kδ) plays a critical role in regulating the immune system (5, 15). The importance of PI3Kδ is highlighted by mutations that either activate or inactivate PI3Kδ in patients with immunodeficiency. Activated PI3K Delta Syndrome (APDS), class 1 or class 2, is a primary immunodeficiency characterized by heterozygous, gain-of-function mutations in PI3Kδ. Class 1 APDS (APDS1) mutations occur in the PIK3CD gene encoding the p110δ catalytic subunit, resulting in single-amino-acid substitutions throughout the primary sequence (6, 7). They occur in regions analogous to oncogenic mutations in p110α. Class 2 APDS mutations occur in the PIK3R1 gene (APDS2), encoding the p85α regulatory subunit. They lead to a splice site mutation that excludes exon 11, resulting in a deletion within the N terminus of the iSH2 coiled-coil domain (Δ434–475) (16, 17). Patients with either form of APDS have increased PIP3 levels, defects in B- and T-cell functions, recurrent respiratory infections, and increased susceptibility to herpes viruses (5). The clinical phenocopy of APDS1 patients with mutations in p110δ by APDS2 patients with mutations in p85α is surprising, as p85α can associate with any of the class IA catalytic isoforms, and it would be expected that activation of PI3Kα would lead to oncogenic transformation or overgrowth syndromes. Because p85α mutations leading to increased p110 lipid kinase activity were shown to be oncogenic when associated with p110α (18, 19), it could be expected that if p110α were activated by p85 APDS2 mutations, this could lead to oncogenesis.
To understand the molecular mechanism for how APDS mutations activate PI3Kδ, we examined both the conformational dynamics and the lipid kinase activity for both APDS1 and APDS2 mutations using hydrogen–deuterium exchange mass spectrometry (HDX-MS) and biochemical assays. The APDS2 p85α splice variant was examined with both p110α and p110δ, and unexpectedly we found that this variant of p85α leads to a selective activation of PI3Kδ with only a minimal effect on PI3Kα activity. HDX-MS revealed that the p85α splice variant disrupted all inhibitory interactions between p85α and p110δ, whereas only a partial disruption of p85-mediated inhibitory interactions was observed in p110α. HDX-MS experiments carried out on wild-type (WT) PI3K and APDS1 mutants revealed that activation of PI3Kδ occurs by a mechanism similar to oncogenic mutations in PI3Kα (14). We also found that all APDS1 and APDS2 mutants are similarly inhibited by the potent p110δ inhibitor idelalisib (20). Our results provide molecular insights into the conformational mechanisms by which PI3Kδ is activated in primary immunodeficiencies and reveal how mutations in PIK3R1 (p85α) can specifically phenocopy gain-of-function mutations in PIK3CD (p110δ).
Results
Lipid Kinase Activity of APDS Mutations.
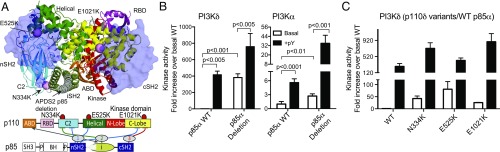
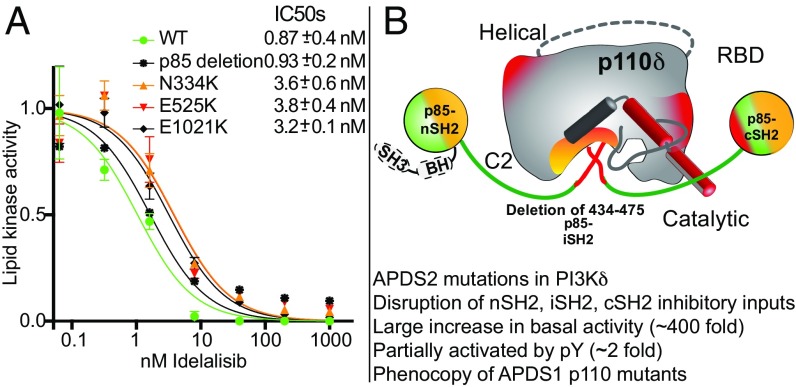
To understand how clinical mutations in the catalytic (p110δ) or regulatory subunits (p85α) alter the function of PI3Kδ, we characterized the lipid kinase activity of both APDS1 and APDS2 mutants. Because the p85α subunit pairs with all class IA p110 subunits, we characterized the differential effects of the APDS2 splice variant [p85α (Δ434–475)] on p110α versus p110δ (Fig. 1). The APDS2 deletion removes the first 42 residues of helix α1 in the iSH2 coiled-coil. In WT PI3K complexes, this region interacts with helices α2 and α3 in the iSH2 coiled-coil and makes contacts with the C2 and kinase domains of the catalytic subunit (SI Appendix, Fig. S1D). The APDS1 mutations studied include three previously described mutations in the C2 domain, the helical domain, and the kinase domain (N334K, E525K, and E1021K, respectively). N334K and E525K mutations are located at the C2–iSH2 and helical–nSH2 interfaces, respectively (SI Appendix, Fig. S1C). E1021K is located in the catalytically important C terminus of the kinase domain near the lipid-binding region and the kinase–cSH2 inhibitory interface with p85 (SI Appendix, Fig. S1C). Lipid kinase experiments on PI3K complexes were carried out on phosphatidylserine vesicles containing 5% PIP2 in both the presence and the absence of a phosphopeptide (referred afterward as pY) to mimic activation by RTKs.
Fig. 1.
APDS1 and APDS2 mutants lead to increased basal and pY-activated lipid kinase activity compared with WT. (A) The location of APDS mutations in p110δ/p85α is mapped on a model of the PI3Kδ complex based on the structure of the p110δ/p85α iSH2 complex [Protein Data Bank (PDB): 5DXU (37)] with the nSH2 modeled from the PI3Kα structure [PDB: 3HHM (38)] and the cSH2 modeled from the PI3Kβ structure [PDB: 2Y3A (13)]. The nSH2 and cSH2 are shown as transparent, with APDS1 mutations shown as pink spheres. The APDS2 truncation in the iSH2 is white. Full molecular details of the APDS mutants are shown in SI Appendix, Fig. S1. A domain schematic of p110δ/p85α is included with inhibitory interfaces highlighted. (B) Fold activation of APDS2 mutation in the context of PI3Kα and PI3Kδ. Lipid kinase assays of WT PI3Kδ or PI3Kα with WT p85α or the APDS2 p85α splice variant [p85α (Δ434–475)] in the presence (+pY) or absence (basal) of a stimulating RTK-derived phosphopeptide (1 μM). Specific activity was normalized to WT (p110δ/α-p85α). (C) Fold activation of APDS1 mutations in p110δ in the presence (+pY) or absence (basal) of a stimulating RTK-derived phosphopeptide. Assays measured the production of ADP in the presence of 0.1–100 nM of enzyme, 100 μM ATP, and 5% PIP2/95% PS vesicles. Kinase assays were performed in triplicate (error shown as SD; n = 3).
Lipid kinase activity of APDS2 splice variants.
Intriguingly, the APDS2 splice variant showed a very large isoform-specific difference in basal lipid kinase activity. The APDS2 p85α splice variant with p110δ showed an ∼400-fold increase in activity over the WT p110δ/p85α complex (Fig. 1C). However, the APDS2 p85α splice variant with p110α was minimally activated (approximately twofold) compared with WT p110α/p85α (Fig. 1C). The PI3Kδ complex with the APDS2 p85α splice variant appeared to be close to fully active in the absence of stimulation (−pY) and showed only limited activation upon pY stimulation (approximately twofold). In contrast, the PI3Kα complex with the APDS2 p85α splice variant was further activated in the presence of pY, with an ∼10-fold increase in lipid kinase activity upon pY stimulation. This indicates that the SH2 inhibitory interfaces from p85α are almost fully disrupted in PI3Kδ, whereas the nSH2–helical inhibitory interface is likely partially conserved in the PI3Kα APDS2 complex. The PI3Kα APDS2 p85α complex was more active than WT PI3Kα in the presence of pY.
Lipid kinase activity of APDS1 mutants.
The basal lipid kinase activity (−pY) of all APDS1 mutants results in a significant activation compared with WT, with E525K showing the highest level of activation (∼60-fold), followed by N334K (∼40-fold), and E1021K (∼20-fold) (Fig. 1D). The activation of basal lipid kinase activity by p110δ mutants is very similar to the activation of p110α by oncogenic mutations in similar positions (14), with the exception of E1021K, which is not as potently activated as the H1047R mutation in p110α. The E525K mutant disrupts the inhibitory interface between the nSH2 and the p110δ helical domains, mimicking activation downstream of pY stimulation, so it would be expected that this mutation would cause the largest increase in activity. The equivalent position in p110α, E545K, leads to full activation, and no further stimulation is achieved by the addition of pY. The lack of full activation in p110δ is consistent with the additional inhibitory cSH2–kinase domain interface present in p110δ (11, 12).
Both the APDS1 mutants and WT PI3Kδ were activated by pY, with all APDS1 mutants being hyperactivated compared with WT. E1021K showed the highest level of hyperactivation (∼3-fold), followed by N334K (∼2-fold), with E525K being the least hyperactivated (∼1.5-fold). The overall trend in activation of RTK-stimulated activity of PI3K by APDS1 mutants closely matches the results for oncogenic mutations in p110α; however, a much lower activation fold was displayed by E1021K in p110δ compared with the similar H1047R mutation in p110α (14).
HDX-MS of PI3Kδ.
To understand the conformational dynamics of how both APDS1 and APDS2 mutations mediate activation of PI3Kδ, we carried out HDX-MS experiments. HDX-MS is a powerful bio-analytical technique that measures the exchange of amide hydrogens with deuterated solvent. As the main determinant of amide exchange is involvement in secondary structure (21), HDX-MS acts as a powerful readout of protein conformational dynamics. It has been applied to study protein–protein (22), protein–membrane (23), and allosteric conformational changes in large macromolecular complexes (24). We have previously applied it to study the molecular basis of oncogenic mutations in PI3Kα and have revealed a set of conformational changes involved in the activation of PI3Kα activity (14), as well as revealing a cSH2-mediated inhibition of PI3Kδ (12). We carried out HDX-MS under three states for WT and APDS1 mutations of PI3Kδ: basal, pY peptide-activated, and pY-activated bound to vesicles mimicking the plasma membrane. Experiments on APDS2 mutations in PI3Kδ and PI3Kα were carried out under basal conditions. A representative set of raw and processed HDX-MS data are shown for a single peptide in SI Appendix, Fig. S2.
Our HDX-MS data for the WT p110δ/p85α and its activation by both pY and membrane binding were consistent with previous results (SI Appendix, Fig. S3) (11, 12). These data are consistent with the previously proposed mechanism of activation requiring four distinct conformational steps: disengagement of the p85 SH2 domains upon pY binding from the p110 helical and kinase domains; disruption of the p110 C2–p85 iSH2 interface; reorientation of the p110 ABD relative to the p110 kinase domain; and, finally, interaction of the kinase domain with the membrane and resident PIP2 substrate.
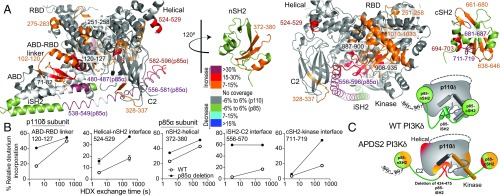
HDX-MS Reveals That APDS2 Mutations in PIK3R1 Lead to Disruption of Inhibitory Interfaces in PI3Kδ.
To investigate the molecular mechanisms by which APDS2 mutations in PIK3R1 led to considerable activation of PI3Kδ, we used HDX-MS to compare dynamics of WT p110δ/p85α and WT p110δ/Δ434–475 p85α. The full set of all peptides analyzed for both p110δ and p85α is shown in SI Appendix, Fig. S4. A number of regions in both p110δ and p85α of the APDS2 mutant PI3Kδ showed significant changes (defined as greater than 7% and 0.7 Da change in deuterium incorporation at any time point) compared with WT (Fig. 2). Within p85, peptides spanning the nSH2–helical interface and cSH2–kinase interface had large increases in exchange, suggesting that the APDS2 p85 deletion disrupts both the nSH2 and the cSH2 inhibitory inputs. Apart from the iSH2 portion that contacts the ABD and remained unaffected, all of the iSH2 showed considerable increases in exchange (>50%) and were fully deuterated at 3 s of exposure with D2O, indicating a lack of secondary structure. This includes the inhibitory contacts between the iSH2 and C2 domains.
Fig. 2.
HDX-MS reveals that APDS2 mutation in p85α leads to disruption of inhibitory interactions in PI3Kδ. (A) Peptides in p110δ and p85α that showed differences in HDX both greater than 0.7 Da and 7% in the APDS2 p85α mutation compared with the WT are highlighted on the structural model from Fig. 1A according to the legend. nSH2 and cSH2 are shown disconnected from the catalytic subunit as the HDX data suggest that these interfaces are disrupted. (B) Time course of deuterium incorporation for a selection of peptides in both p110δ and p85α with differences in HDX in the APDS2 p85α mutant (error shown as SD; n = 3). (C) Schematic model of WT PI3Kδ and conformational changes that occur in the complex with the APDS2 splice variant of p85.
Several regions in the p110δ catalytic subunit showed increases in HDX in the APDS2 mutant compared with the WT (Fig. 2), including many regions with similar increases in HDX upon either pY binding or membrane binding in the WT. This included the ABD-RBD linker and the ABD domain at the interface with helix α1 of the iSH2, as well as the interface with the kinase domain (SI Appendix, Fig. S4). Every inhibitory interface with the p85 subunit showed increases in HDX, including the C2–iSH2, helical–nSH2, and kinase–cSH2 interfaces. A network of connected secondary structure elements in every domain of the catalytic subunit had increased HDX, suggesting a possible destabilization of the catalytic subunit. Together, the HDX results for p110δ bound to the APDS2 splice variant of p85 implied that the removal of the first helix α1 of the iSH2 leads to a disruption of all inhibitory interfaces on the kinase domain of p110δ.
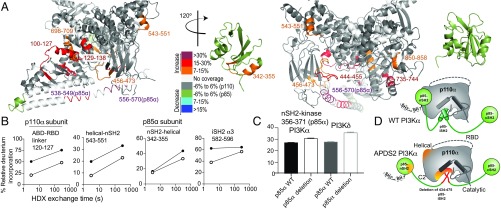
HDX-MS Reveals That APDS2 Mutations in PIK3R1 Lead to Partial Disruption of Inhibitory Interfaces in PI3Kα.
To understand how the APDS2 splice variant of p85α led to only a partial activation of PI3Kα compared with PI3Kδ, we carried out HDX-MS experiments on the complex of WT p110α/p85α and a complex containing p110α with the p85α APDS2 splice variant (Fig. 3). The full set of all peptides analyzed for both p110α and p85α is shown in SI Appendix, Fig. S5. Compared with WT p85α, association with the p85α APDS2 splice variant caused large increases in HDX in the p110α catalytic subunit in the ABD-RBD linker, and the p85 inhibitory C2–iSH2, C2–nSH2, and helical–nSH2 interfaces, similar to regions that showed increases in exchange either upon pY or membrane binding in the WT (14). Within peptides spanning p85α there were numerous regions that showed increases in HDX. The iSH2 coiled-coil outside of the ABD interface showed very large increases in HDX, similar to the APDS2 PI3Kδ complex, suggesting that in both complexes the p85α splice variant leads to large portions of the iSH2 becoming disordered. Intriguingly, regions of the nSH2 in contact with the catalytic subunit showed increases in exchange, suggesting that part of the interface is disrupted. However, these changes were less pronounced than those seen in the APDS2 PI3Kδ complex. This is best highlighted by a region in the nSH2 that directly contacts the kinase domain (residues 356–371) that showed a larger increase in exchange in the APDS2 PI3Kδ complex compared with the APDS2 PI3Kα complex (Fig. 3C).
Fig. 3.
HDX-MS reveals that APDS2 mutation in p85α leads to partial disruption of inhibitory interactions in PI3Kα. (A) Peptides in p110α and p85α that showed differences in HDX both greater than 0.7 Da and 7% in the APDS2 p85α mutation compared with the WT are highlighted on the structure of p110α bound to the nSH2 and iSH2 of p85α [PDB: 3HHM (38)]. The nSH2 domain is shown disconnected from the catalytic subunit, as the HDX data suggest that this interface is partially disrupted. (B) Time course of deuterium incorporation for a selection of peptides in both p110α and p85α with HDX differences in the APDS2 PIK3R1 mutant. (C) HDX exchange levels for the APDS2 complex of PI3Kα and PI3Kδ compared with WT for a peptide in the nSH2 of p85α located at the nSH2–kinase interface at the 300-s time point. Error bars in all graphs represent SD (n = 3). (D) Schematic model of WT PI3Kα and conformational changes that occur in the complex with the APDS2 splice variant of p85α.
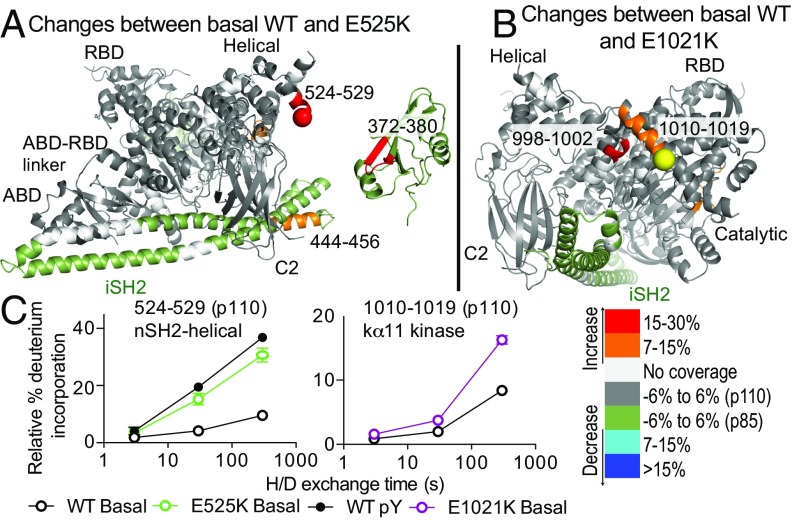
HDX-MS Reveals That APDS1 Mutations in p110δ Mimic the Activation Mechanism of Oncogenic Mutations in p110α.
To understand the molecular basis for how APDS1 mutations in the catalytic subunit of p110δ modify the dynamic regulation of PI3Kδ, we carried out HDX-MS experiments in three states: basal, pY-activated, and pY-activated bound to membranes for WT PI3Kδ and PI3Kδ APDS1 mutants (E525K and E1021K). The full set of peptides for both p110δ and p85α under all conditions and time points is shown in SI Appendix, Fig. S6, with differences between states for highlighted peptides shown in SI Appendix, Figs. S7 and S8. The differences in exchange between WT PI3Kδ and APDS1 mutants showed that APDS1 mutants mimic and enhance conformational dynamics seen in the activation of the WT enzyme, a mechanism that is similar to the activation of p110α by oncogenic mutations (Fig. 4). For E525K, there were increases in exchange at both sides of the helical–nSH2 interface, similar to the change seen in pY-binding experiments. The E1021K mutant showed increases in exchange in the C terminus of the kinase domain. An unanswered question for E1021K was whether this mutation also led to disruption of the cSH2–kinase interface, as this mutation is located within 6 Å of the cSH2; however, there were no differences in exchange within the cSH2 between WT and PI3Kδ-E1021K, suggesting that this interface is preserved. All APDS1 mutants showed larger decreases in exchange upon membrane binding compared with the WT, suggesting that the increased lipid kinase activity upon pY stimulation for these mutants is at least partially due to enhanced membrane binding (SI Appendix, Figs. S6–S8).
Fig. 4.
HDX-MS reveals that APDS1 mutations in p110δ mimic the activated form of WT PI3Kδ. (A and B) Peptides in p110δ and p85α with HDX differences both greater than 0.7 Da and 7% in the APDS1 mutants E525K and E1021K in PIK3CD compared with the WT are highlighted on the structural model as in Fig. 2. (C) Time course of deuterium incorporation for a selection of peptides in both p110δ and p85α under a variety of conditions (basal, pY-phosphopeptide bound) with HDX differences in the APDS1 mutants. Error bars represent SD (n = 3).
Both APDS1 and APDS2 Mutations in PI3Kδ Are Potently Inhibited by the PI3Kδ-Specific Inhibitor Idelalisib.
To determine if conformational differences identified in both APDS1 and APDS2 mutants of PI3Kδ would lead to differential inhibition by ATP competitive kinase inhibitors, we carried out lipid kinase assays in the presence or the absence of the clinically approved PI3Kδ inhibitor idelalisib (25). For all APDS1 and APDS2 mutants in p110δ and p85α, respectively, there were very similar IC50 values compared with the WT (Fig. 5A), indicating that all might be targetable by idelalisib.
Fig. 5.
Inhibition of APDS1 and APDS2 mutants by the potent PI3Kδ inhibitor idelalisib and model for activation of PI3Kδ and PI3Kα by APDS2 mutations in PIK3R1. (A) Inhibition of WT, APDS1, and APDS2 complexes of PI3Kδ by the potent PI3Kδ inhibitor idelalisib. Lipid kinase activity was normalized to kinase activity in the absence of inhibitors. IC50 values were generated from triplicate independent inhibitor dilutions, and error is shown as SD. (B) Summary of the proposed mechanism for activation of the APDS2 PI3Kδ complex mapped on a schematic model with conformational changes and kinase activity data summarized.
Discussion
Understanding the molecular mechanisms by which APDS1 and APDS2 mutations in PIK3CD and PIK3R1 activate PI3Kδ is essential in the design of novel therapeutic strategies aimed at treating patients suffering from APDS. APDS1 mutations were initially identified in patients with primary immunodeficiencies in PIK3CD, with mutations identified at E1021K (7), as well as at C416R (26), N334K, and E525K (6) in the catalytic p110δ subunit. This discovery was rapidly followed by the discovery of a APDS2 mutation in PIK3R1 associated with primary immunodeficiency (16, 17). That mutation resulted in a deletion within the N terminus of the iSH2 domain. Since then, over 100 primary immunodeficiency patients have been identified with either APDS1 or APDS2 mutations (27, 28). Patients with APDS exhibit a heterogeneous set of clinical features, including symptoms of both immune deficiency and immune dysfunction (5).
PI3Kδ is activated in immune cells downstream of phosphorylated receptors, including cytokine receptors, toll-like receptors, FceR, and indirectly downstream of the T-cell or B-cell receptor (15). Mutations in these subunits leading to either gain or loss of function in PI3Kδ result in defects in the immune system. Mice lacking the regulatory p85α subunit had defects in B-cell development and proliferation (29), and mice containing a kinase-dead mutation in the p110δ catalytic subunit showed impaired immune function and deficient antigen receptor signaling in both B and T cells (30). Patients with immune cell defects had loss-of-function mutations in PIK3R1 (31) and PIK3CD (32). The fact that patients with immune diseases have been identified with mutations leading to either gain or loss of function of PI3Kδ shows that PIP3 levels must be precisely controlled for proper immune cell function. Consistent with this, primary immunodeficiency patients have been identified with loss-of-function mutations in the phosphatase PTEN (33) that antagonizes the PI3K pathway.
One of the most complicated questions regarding APDS mutations identified so far is why germline APDS2 mutations in PIK3R1 (p85α) primarily seem to phenocopy the activating APDS1 mutations in PIK3CD, but do not directly affect oncogenicity as PIK3CA mutations. The p85α subunit is ubiquitously expressed, and several somatic point mutations in the iSH2 domain have been identified as oncogenic (19). A variety of somatic mutations resulting in in-frame deletions and insertions in the region from 434 to 475 in the iSH2 have been identified in endometrial carcinomas (12 in total, with Δ450–451 and Δ457–461 in p85α as examples) with two patients identified with the Δ434–475 mutant in p85α (18). However, in a cohort study of APDS2 patients with germline mutations resulting in a Δ434–475 p85α, the main increased oncogenic risk appeared to be lymphoma (34), with so far no evidence that patients with the APDS2 splice variant have any increased cancer risk outside the immune system. The only nonimmune defect so far identified is growth retardation (27), which has also been identified in SHORT syndrome, a disorder that is caused by heterozygous mutations in PIK3R1 that prevent RTK binding, leading to altered PI3K signaling (4).
Our biochemical and structural studies show that the APDS2 mutant of p85α leads to a substantial basal activation of PI3Kδ at a level close to the fully activated state when bound to phosphopeptide mimicking RTK stimulation. The APDS2 complex of PI3Kα was only weakly basally activated and was hyperactivated by RTK pY. HDX-MS results showed a disruption of the C2–iSH2 and reorientation of the ABD in the PI3Kα APDS2 complex, and this provides a possible mechanism for RTK pY hyperactivation compared with WT. The APDS2 PI3Kδ complex was only weakly hyperactivated by RTK pY, possibly suggesting that the ABD and C2–iSH2 interfaces are already broken in the absence of RTK pY. The HDX-MS results revealed that in the APDS2 PI3Kδ complex all inhibitory interfaces between the nSH2, iSH2, and cSH2 of p85α were disrupted (Figs. 2 and 5B); however, in PI3Kα the inhibitory interface with the nSH2 is only partially disrupted. Previous HDX-MS comparisons between PI3Kα and PI3Kδ showed that the nSH2 is more tightly bound to p110α compared with p110δ (11). Both the HDX-MS and kinase assays indicate that an intact coiled-coil is required to maintain the nSH2 and cSH2 interactions in PI3Kδ, whereas the tighter interaction between nSH2 and p110α is able to maintain a partial level of inhibition in the absence of the full iSH2 coiled-coil. This difference in activation level in the APDS2 complex of PI3Kδ compared with PI3Kα provides a possible molecular mechanism for how APDS2 mutations in PIK3R1 phenocopy the APDS1 mutations in PIK3CD. The basal partial activation in p110α/Δ434–475 p85α may still be relevant as an oncogene due to its discovery as being somatically mutated in endometrial carcinomas (18). The hyperactivation of the APDS2 complex of PI3Kα by pY compared with WT further underscores this potential relevance; however, further study will be required to examine the expression and stability of the APDS2 PI3Kα complex to understand the difference between oncogenicity in germline and somatic mutations.
The p110α isoform is frequently mutated in cancer (3), and mutations are distributed throughout the primary sequence. The APDS1 mutations identified in p110δ are located in similar locations to oncogenic mutations that have been identified in p110α (35). One of the major questions is whether mutations in p110δ lead to activation of lipid kinase activity by a mechanism similar to oncogenic mutations in p110α. HDX-MS experiments on WT PI3Kδ showed that there are four conformational differences upon pY activation and membrane binding: (i) disruption of the nSH2 and cSH2 inhibitory contacts, (ii) disruption of the C2–iSH2 interface, (iii) reorientation of the ABD relative to the rest of the catalytic subunit, and (iv) interaction of the kinase domain with membranes. All of the tested p110δ mutations mimicked or enhanced one of these conformational differences. The E525K mutant led to disruption of the nSH2–helical interface, similar to the level seen upon WT binding to phosphopeptide. The E1021K mutant led to conformational changes within the kinase domain in regions that interact with the membrane surface, with similar effects to those seen in the H1047R p110α mutant. The location of the cSH2–kinase interface near the E1021K mutation had alluded to a mechanism of activation through disruption of the cSH2 interface that increases membrane affinity (7); however, we find no conformational differences in the cSH2, implying that this inhibitory interface is maintained. The likely mechanism for activation of E1021K is increased membrane recruitment, similar to H1047R in p110α. As APDS1 mutations in p110δ and oncogenic mutations in p110α both stimulate PI3K activity by similar mechanisms, we could expect mutations in analogous regions to oncogenic mutants in p110α in p110δ will cause APDS.
Many of the APDS1 and APDS2 mutants in PI3Kδ led to conformational changes in the active site of the enzyme. Treatment of APDS patients with inhibitors that target either PI3Kδ or downstream signaling molecules has been proposed, with clinical trials for both oral and inhaled PI3K inhibitors having been announced (NCT02435173, NCT02593539). Inhibitor assays carried out with the clinically approved PI3Kδ inhibitor idelalisib showed that WT, APDS1, and APDS2 variants had very similar IC50 values. This is an important validation that APDS mutations can be targeted by p110δ-specific inhibitors and supports a potential therapeutic option for all APDS patients.
Materials and Methods
Lipid Kinase Assays.
Lipid kinase assays monitoring hydrolysis of ATP were carried out using the Transcreener ADP2 FI assay (Bellbrook Laboratories), according to previously published protocols (11, 13), as described in full in SI Appendix, SI Materials and Methods. In short, assays were carried out using lipid vesicles composed of 5% C8 PIP2, 95% brain phosphatidylserine (PS) (wt/vol) at a final concentration of 0.45 mg/mL, and 100 μM ATP. Experiments with phosphopeptide [mouse platelet-derived growth factor receptor (PDGFR) residues 735–767 with pY740 and pY751] were carried out at a final concentration of 1 µM.
Deuterium Exchange Measurements.
HDX experiments were set up as described previously (36). In brief, HDX exchange reactions were initiated by the addition of PI3K complexes (1 µM final concentration) to either blank or pY peptide solution (5 µM final concentration) or a solution containing pY and lipid vesicles mimicking the composition of the plasma membrane (5% PIP2, 30% PS, 50% phosphatidylethanolamine (PE), 15% phosphatidylcholine (PC), final concentration 100 µg/mL) for 1–2 min, followed by addition of 98% D2O solution (10 µM Hepes, pH 7.5, 100 mM NaCl), to a final concentration of 70–80% D2O. Lipid vesicles were prepared as described in SI Appendix, SI Materials and Methods. Three time points of on exchange (3, 30, and 300 s) at 23 °C were carried out followed by addition of a quench buffer (final concentration 0.8% formic acid, 0.5 M guanidine–HCl). Samples were then immediately frozen in liquid nitrogen, and stored for no more than 1 wk at −80 °C until mass analysis.
Complete materials and methods are described in full in SI Appendix.
Supplementary Material
Acknowledgments
J.E.B. is supported by a new investigator grant from the Canadian Institutes of Health Research; a discovery research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC-2014-05218), as well as a small project health research grant from the British Columbia Proteomics Network. C.L.L. is supported by Yale University and a National Heart, Lung, and Blood Institute R00HL125668 Award.
Footnotes
Conflict of interest statement: C.L.L. collaborates with Novartis on related studies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617244114/-/DCSupplemental.
References
- 1.Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem Sci. 2015;40(2):88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11(5):329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 3.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 4.Dyment DA, et al. FORGE Canada Consortium Mutations in PIK3R1 cause SHORT syndrome. Am J Hum Genet. 2013;93(1):158–166. doi: 10.1016/j.ajhg.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kδ and primary immunodeficiencies. Nat Rev Immunol. 2016;16(11):702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angulo I, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kok K, Geering B, Vanhaesebroeck B. Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem Sci. 2009;34(3):115–127. doi: 10.1016/j.tibs.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011;4(195):re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 10.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317(5835):239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 11.Burke JE, Williams RL. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS) Adv Biol Regul. 2013;53(1):97–110. doi: 10.1016/j.jbior.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke JE, et al. Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α. Structure. 2011;19(8):1127–1137. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41(5):567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc Natl Acad Sci USA. 2012;109(38):15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deau M-C, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J Clin Invest. 2015;125(4):1764–1765. doi: 10.1172/JCI81746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas CL, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J Exp Med. 2014;211(13):2537–2547. doi: 10.1084/jem.20141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urick ME, et al. PIK3R1 (p85α) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 2011;71(12):4061–4067. doi: 10.1158/0008-5472.CAN-11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaiswal BS, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16(6):463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopal AK, et al. PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2014;370(11):1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner JJ, Lim WK, Bédard S, Black BE, Englander SW. Protein dynamics viewed by hydrogen exchange. Protein Sci. 2012;21(7):996–1005. doi: 10.1002/pro.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPhail JA, Ottosen EH, Jenkins ML, Burke JE. The molecular basis of aichi virus 3A protein activation of phosphatidylinositol 4 kinase IIIβ, PI4KB, through ACBD3. Structure. 2017;25(1):121–131. doi: 10.1016/j.str.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Vadas O, Burke JE. Probing the dynamic regulation of peripheral membrane proteins using hydrogen deuterium exchange-MS (HDX-MS) Biochem Soc Trans. 2015;43(5):773–786. doi: 10.1042/BST20150065. [DOI] [PubMed] [Google Scholar]
- 24.Harrison RA, Engen JR. Conformational insight into multi-protein signaling assemblies by hydrogen-deuterium exchange mass spectrometry. Curr Opin Struct Biol. 2016;41:187–193. doi: 10.1016/j.sbi.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somoza JR, et al. Structural, biochemical, and biophysical characterization of idelalisib binding to phosphoinositide 3-kinase δ. J Biol Chem. 2015;290(13):8439–8446. doi: 10.1074/jbc.M114.634683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crank MC, et al. Mutations in PIK3CD can cause hyper IgM syndrome (HIGM) associated with increased cancer susceptibility. J Clin Immunol. 2014;34(3):272–276. doi: 10.1007/s10875-014-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olbrich P, et al. Activated PI3Kδ syndrome type 2: Two patients, a novel mutation, and review of the literature. Pediatr Allergy Immunol. 2016;27(6):640–644. doi: 10.1111/pai.12585. [DOI] [PubMed] [Google Scholar]
- 28.Coulter TI, et al. Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: A large patient cohort study. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fruman DA, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 1999;283(5400):393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 30.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297(5583):1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 31.Conley ME, et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85α subunit of PI3K. J Exp Med. 2012;209(3):463–470. doi: 10.1084/jem.20112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang KJ, Husami A, Marsh R, Jordan MB. Identification of a phosphoinositide 3‐kinase (PI‐3K) p110δ (PIK3CD) deficient individual. J Clin Immunol. 2013;33:673–674. [Google Scholar]
- 33.Tsujita Y, et al. Phosphatase and tensin homolog (PTEN) mutation can cause activated phosphatidylinositol 3-kinase δ syndrome-like immunodeficiency. J Allergy Clin Immunol. 2016;138(6):1672–1680.e10. doi: 10.1016/j.jaci.2016.03.055. [DOI] [PubMed] [Google Scholar]
- 34.Elkaim E, et al. Clinical and immunologic phenotype associated with activated phosphoinositide 3-kinase δ syndrome 2: A cohort study. J Allergy Clin Immunol. 2016;138(1):210–218.e9. doi: 10.1016/j.jaci.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Rudd ML, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17(6):1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vadas O, Jenkins ML, Dornan GL, Burke JE. Using Hydrogen–Deuterium Exchange Mass Spectrometry to Examine Protein–Membrane Interactions. Methods in Enzymology. Elsevier; Amsterdam: 2016. [DOI] [PubMed] [Google Scholar]
- 37.Heffron TP, et al. The rational design of selective benzoxazepin inhibitors of the α-isoform of phosphoinositide 3-kinase culminating in the identification of (S)-2-((2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326) J Med Chem. 2016;59(3):985–1002. doi: 10.1021/acs.jmedchem.5b01483. [DOI] [PubMed] [Google Scholar]
- 38.Mandelker D, et al. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci USA. 2009;106(40):16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.