Abstract
mRNA stability appears to play a key role in the ontogenic regulation of the apical sodium dependent bile acid transporter (ASBT). The RNA binding proteins, Hu antigen R (HuR) and Tristetraprolin (TTP), stabilize and destabilize ASBT mRNA, respectively. Potential HuR binding sites were assessed by sequence analysis in the context of prior in vitro functional analyses of the rat ASBT 3′UTR. Wild type and mutant binding sites were investigated by gel shift analysis using IEC-6 cell extracts. The functional consequences of binding site mutations were assessed using two different hybrid reporter constructs linking the 3′UTR element to a either a luciferase or a β-globin coding mRNA sequence. A specific metastasis associated gene 1 cis-element (MTA1) was identified in the ASBT 3′UTR that became associated with proteins in IEC-6 cell extracts and could be supershifted by HuR or TTP antibodies. Mutation of this cis-element abrogated the gel shift of IEC-6 proteins. Furthermore hybrid constructs containing a mutant MTA1 element had reduced responses to modulation of HuR or TTP. For the first time we have identified a single specific sequence element in the 3′UTR of the rat ASBT mRNA that mediates counter-regulatory changes in mRNA abundance in response to both HuR and TTP.
Keywords: bile acid, intestine, mRNA decay, RNA abundance, RNA binding protein
INTRODUCTION
Bile acids are synthesized from cholesterol in the liver and secreted into the small intestine where they facilitate absorption of long chain fatty acids and fat-soluble vitamins. The apical sodium-dependent bile acid transporter (ASBT) is the major carrier protein involved in the ileal reabsorption of bile acids [1]. Considering its central role in the enterohepatic circulation, ASBT may play a role in the pathogenesis or clinical presentation of a variety of gastrointestinal problems and diseases, including primary bile acid malabsorption [2], hypertriglyceridemia [3], cholelithiasis [4], Crohn disease [5], necrotizing enterocolitis [6]; amongst others. Bile acids may also serve as signaling molecules and as such their reabsorption in the terminal ileum is implicated in glucose and lipid metabolism [7].
The expression of ASBT is highly regulated in part due to its central role in varied homeostatic processes. An extensive literature exists describing transcriptional regulation of ASBT expression [5, 8–14]. Investigations have begun to examine post-transcriptional regulatory processes including mRNA stability in the regulation of ASBT. Descriptive analyses of the ontogeny of ASBT in rat ileum and kidney suggested that ASBT expression may be controlled in part by regulated changes in mRNA stability [15, 16]. Recent studies have revealed a novel role for HuR and TTP in the regulation of ASBT at the level of mRNA turnover. HuR stabilizes while TTP destabilizes ASBT mRNA [17]. Counter-regulatory control of mRNA stability by HuR and TTP has been described for cyclooxygenase 2 in colon cancer [18]. We suspect that HuR/TTP based counter-regulation may be a more generalized paradigm for gene regulation and thus we have focused efforts to understand the molecular mechanisms of that counter-regulation. As an initial step we sought to identify a potential cis-element in the 3′untranslated region of ASBT that mediates HuR or TTP response using published potential HuR target RNA motifs [17, 19]. The focus of these investigations is on one of these HuR target motifs, MTA1, which was identified in a 0.3 kb region of the rat ASBT 3′UTR that mediated changes in mRNA stability in response to HuR.
MATERIALS AND METHODS
Cell lines, siRNA, expression constructs
Rat IEC-6 intestinal epithelial cells (American Type Culture Collection, Manassas, VA) were maintained in Minimum Essential Medium (MEM) containing 10% fetal bovine serum (FBS). The HuR and TTP small interfering RNAs (siRNA) were purchased from Santa Cruz Biotechnologies, Inc. (Santa Cruz, CA). Exogenous HuR was expressed in cells following transfection of the plasmid construct pcDNA3.1/mHuRcoding-3′ untranslated region (UTR)/Flag (generous gift from Dr. Beth S. Lee, Ohio State University, Columbus, OH) (15). TTP was overexpressed in cells by transfection of cells with the plasmid construct pCMV/hTTP that was prepared from an original clone (Clone ID: LIFESEQ3352575) (Thermo Scientific Life Science Research, Waltham, MA).
Generation of recombinant plasmid constructs
The hybrid construct pGL3-rASBT3′/0.3 was generated from an SV-40 driven luciferase reporter plasmid pGL3-Promoter (Promega, Madison, WI), as described previously (13). The construct contains 0.3kb rat (2114–2434, GenBank NM_017222) ASBT 3′UTR sequence downstream of a firefly luciferase coding sequence. The hybrid constructs Globin/rASBT3′ was prepared from a c-fos promoter driven β-globin reporter plasmid, where the same 0.3 kb rat ASBT 3′UTR is downstream of the globin coding sequence (13).
Site-directed mutagenesis of ASBT 3′UTR sequence
The constructs pGL3-rASBT3′/0.3 and Globin/rASBT3′ were used as templates for site directed mutagenesis to make point mutations in the ASBT mRNA stability-related region using the QuickChange™ Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) [8]. The specific nucleotides for mutagenesis in the MTA1 cis-element were chosen on the basis of prior studies and comparison of the 3′UTR of ASBT in multiple species (Figure 1A) and on predicted structure derived at http:\\rna.tbi.univie.ac.at. (Figures 1B and 1C, see results and discussion). Two highly conserved nucleotides in the predicted loop of the MTA1 cis-element were targeting in the MTA1 cis-element, positioned from 2366 to 2383 in the rat ASBT 3′UTR. A DNA oligonucleotide primer was synthesized, with the sequences of 5′-CAGCTCATAGGAGGTGCCATACCAGCCACCACG-3′, which contained mutations at the site as shown by bold nucleotides (T to C, G to A). The PCR products, pGL3-rASBT3′/0.3-MTA1mu and Globin/rASBT3′-MTA1mu were examined by DNA sequencing for the point mutations contained within the ASBT 3′UTRs.
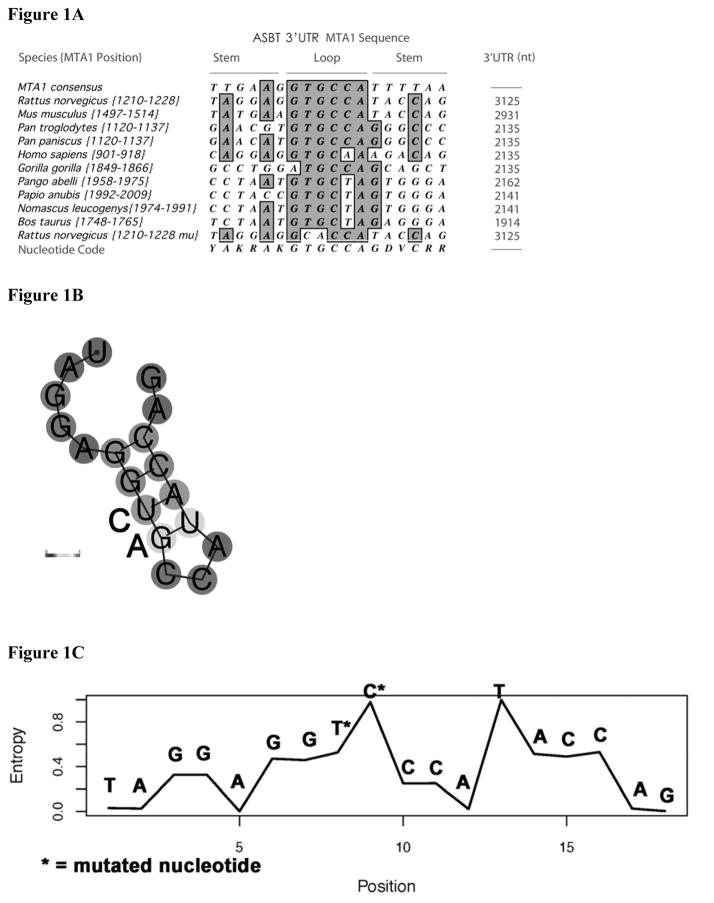
Figure 1. Alignment of the potential HuR MTA1 binding site in the 3′untranslated region of the apical sodium dependent bile acid transporter from multiple species.
(A) The 3′ UTR of the apical sodium dependent bile acid transporter was interrogated for the presence of a possible MTA1 cis-element (potential HuR binding site) using the published consensus sequence [19]. The sequence of various species is shown. The number in brackets represents the position of the sequence within the 3′UTR and the total length of the 3′UTR is shown in the last column. The consensus sequence for the MTA1 site based upon these identified elements is shown in the last line. The sequence of the mutated rat MTA1 site {1210–1228 mu} is shown in the second from bottom sequence (note the conserved TG has been changed to CA). (B) Predicted secondary structure of rat MTA1 site based upon structure performed at http:\\rna.tbi.univie.ac.at. Nucleotide substitutions are shown. (C) Entropy plot of specific nucleotides in the rat MTA1 site based upon calculations at http:\\rna.tbi.univie.ac.at. Position of specific nucleotide substitutions are indicated by an asterix.
Transient transfection and dual luciferase assay
IEC-6 cells (5x105/well) were transfected with 3μg of the rat ASBT3′UTR-luciferase construct of interest and 0.1μg of a quantification control plasmid pRL-TK containing a thymidine kinase-promoter-driven Renilla luciferase gene (Promega, Madison, WI). Co-transfection was performed with the overexpression vector or silencing RNA of interest. Dual luciferase reporter assay system was performed according to manufacturer’s instructions. Results are expressed as relative lights units (RLU), the ratio of firefly to Renilla luciferase activities. Reporter activity was normalized to the wild type construct, set as 100%.
Gel shift analysis
Gel shift analysis was performed with 20 μg of cytoplasmic proteins prepared as previously described [17]. Proteins were incubated at 37°C for 30 minutes with 25 pM of 32P-labelled RNA probe in 15 mmol/L KCl, 5 mmol/L MgCl2, 0.25 mmol/L EDTA, 0.25 mmol/L dithiothreitol, 12 mmol/L Hepes (pH 7.9), 10% glycerol, and 200 ng/μL Escherichia coli transfer RNA. The 32P-labeled RNA probes of the 0.3 kb rASBT 3′UTR was prepared by in vitro transcription of 0.5 μg of the construct pSPT18-rASBT3′/0.3, using an SP6/T7 in vitro transcription kit (Roche, Nutley, NJ). Transcripts were purified using native polyacrylamide gel electrophoresis. All RNA oligonucleotides used in the studies were in vitro synthesized (Invitrogen, Carlsbad, CA). RNA oligonucleotides were 3′-end labeled with 32P-CTP and SP6 using Ambion’s protocol (Ambion, Austin, TX). Unincorporated label was removed using a NucAway spin column (Ambion, Austin, TX). Gel supershift assays were performed with 2 μg of HuR or TTP antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Unbound RNA probes were digested for 30 minutes with RNase T1 (Invitrogen, Carlsbad, CA). Nonspecific protein binding was reduced by adding 5 mg/mL heparin to the mixture for a 10-minute incubation at room temperature. Samples were then subjected to electrophoresis in a 7% native polyacrylamide gel with 0.5X Tris-borate-EDTA running buffer. The gels were dried and exposed to Kodak BIOMAX MS films (Kodak, Rochester, NY) at −80°C.
Northern blot analysis and mRNA half-life assay
IEC-6 cells were plated on a 150cm2 culture vessel and transfected with 60μg of Globin/rASBT3′ or Globin/rASBT3′-muMTA1 3′UTR-β-globin hybrid construct plus or minus siHuR or TTP wild type expression vector. Eighteen hours after transfection, cells were incubated in 10% minimum essential media without serum and with 1% penicillin-streptomycin at 8% CO2 for 24 hours to force the cells to enter a quiescent state. After serum starvation for 24 hours culture medium was removed and fresh media with 10% serum was added for another 24 hours [20]. Serum was then withdrawn to arrest transcription of new mRNA and cells were harvested at desired time points using TRIzol reagent. Twenty micrograms of total cellular RNA were analyzed by Northern blotting with 32P-labeled β-globin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. Signal was quantified using a Phosphorimager (Bio-Rad, Hercules, CA). mRNA half-life was determined by linear regression analysis. The decay rate constant is obtained from the slope of a semi-logarithmic plot of mRNA as a function of time. The half-life was then calculated with equation: t½= ln2/k decay.
Statistical analysis
Statistical analysis was performed using In-Stat software (GraphPad Software, Inc., San Diego CA). Unless otherwise stated, means were compared using the Tukey-Kramer multiple comparisons test, all values were mean ± SD, and a value of p<0.05 was considered statistically significant. Graph analysis was performed using PRISM (GraphPad Software, Inc., San Diego CA).
RESULTS AND DISCUSSION
There are a variety of potential mechanisms by which RNA binding proteins can counter-regulate gene expression including competitive binding at the same RNA cis-element binding site. We sought to explore the biology of the regulation of ASBT to provide insight into this question. Initial searches for possible HuR binding sites were informed by the predictions generated by the studies of Lopez de Silanes [19]. Interrogation of the 3′UTR of ASBT identified several candidate binding sites [17]. The MTA1 site had a high likelihood for biologic relevance for the regulation of ASBT. This site was identified in the metastasis associated gene 1 mRNA, which has been shown to have potential clinical importance for breast, gastric, colorectal, pancreatic and hepatocellular cancers [21–23]. A potential MTA1 site was identified in a 300 base pair fragment of the rat ASBT 3′UTR [17]. The MTA1 site was the only potential HuR binding site found in this 300 base pair sequence. This small fragment had been shown to impart RNA destabilizing effects in heterologous constructs and the effects could be altered by changes in HuR expression [17]. In addition, the binding site was found in the 3′UTR of ASBT in a variety of higher eukaryotic species (Figure 1A). A highly conserved TG dinucleotide was identified, which is predicted to be in the loop of a stem-loop structure (Figures 1B and 1C) [19]. This conserved dinucleotide was mutated in the functional assays in this study.
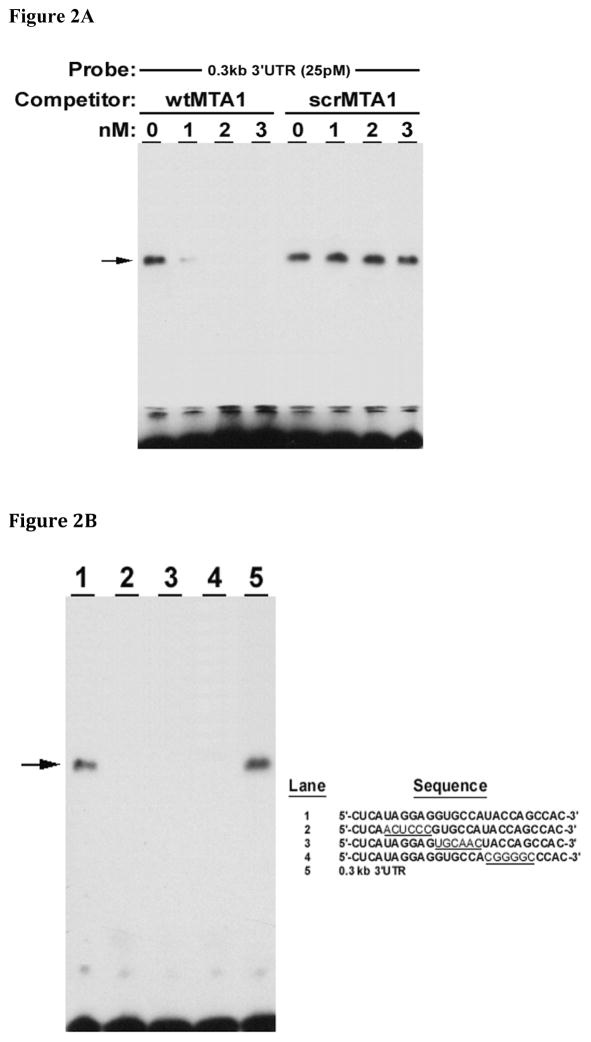
Initial investigations relied upon competition gel-shift analyses that were performed using extracts from rat intestinal derived, IEC-6 cells, incubated with oligonucleotide probes based upon the potential binding site, MTA1. wtMTA1 but not scrMTA1 competed for binding of IEC-6 extracts with the rat ASBT 0.3kb 3′UTR construct (Figure 2A). Gel shift analysis of IEC-6 cell extracts and MTA1 oligonucleotides was performed. Gel shift was observed with IEC-6 cell extracts incubated with either the ASBT 0.3kb 3′ UTR RNA or the wild type MTA1 oligonucleotide. A shifted complex was not observed when IEC-6 cell extracts were incubated with the three different mutated 3′UTR constructs. These studies suggest that an intact MTA1 site is required for HuR binding (Figure 2B). Supershifting was observed with either HuR or TTP antibodies and either the 0.3 kb rat ASBT 3′UTR or the MTA1 oligonucleotide (Figure 2C) indicating that the MTA1 site is bound by both HuR and TTP.
Figure 2. HuR and TTP binding is dependent upon an intact MTA1 site.
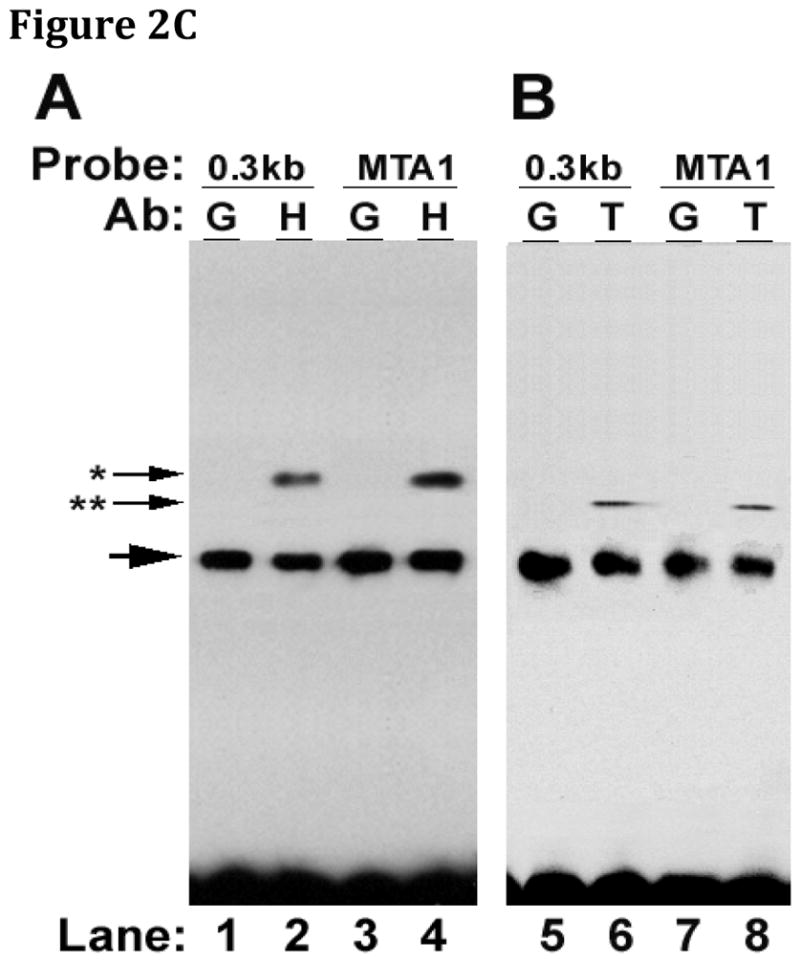
Gel shift assays were performed using cytoplasmic extracts from IEC-6 cells and various oligonucleotides with supershifting induced by HuR or TTP antibodies. (A) Competition gel shift analysis of IEC-6 cell extracts, the rat 0.3 kb 3′UTR fragment and MTA1 oligonucleotide. A wild type probe (wtMTA1, 5′-CUCAUGGAGGUGCCAUACCAGCCAC-3′) and scrambled control (scrMTA1) were incubated with IEC-6 cell extracts. The extracts were incubated with the radiolabeled 0.3 kb rat ASBT 3′UTR and analyzed by gel electrophoresis. wtMTA1 but not scrMTA1 competes for binding of IEC-6 extracts with the rat 0.3 kb ASBT 3′UTR construct. (B) Gel shift analysis of IEC-6 cell extracts and MTA1 oligonucleotides. Wild type MTA 1 oligonucleotide (lane 1), three different mutant MTA1 oligonucleotides (lanes 2 –4) and the 0.3 kb rat ASBT 3′UTR (lane 5) radiolabeled probes were incubated with IEC-6 cell extracts. Only wild type MTA1 and the 0.3 kb rat ASBT 3′UTR bind the IEC-6 cell extracts. (C) Supershift assay of the rat ASBT 0.3 kb 3′UTR or MTA1 oligonucleotide. IEC-6 cell extracts were incubated with either the 0.3 kb rat ASBT 3′UTR fragment (0.3 kb) or the MTA1 oligonucleotide (MTA1). Supershifting was assessed with growth hormone antibody (G – as a negative control), HuR antibody (H – panel A), or TTP antibody (T – panel B). Supershifting was observed with either HuR (*) or TTP (**) antibodies and with either the 0.3 kb rat ASBT 3′UTR or the MTA1 oligonucleotide.
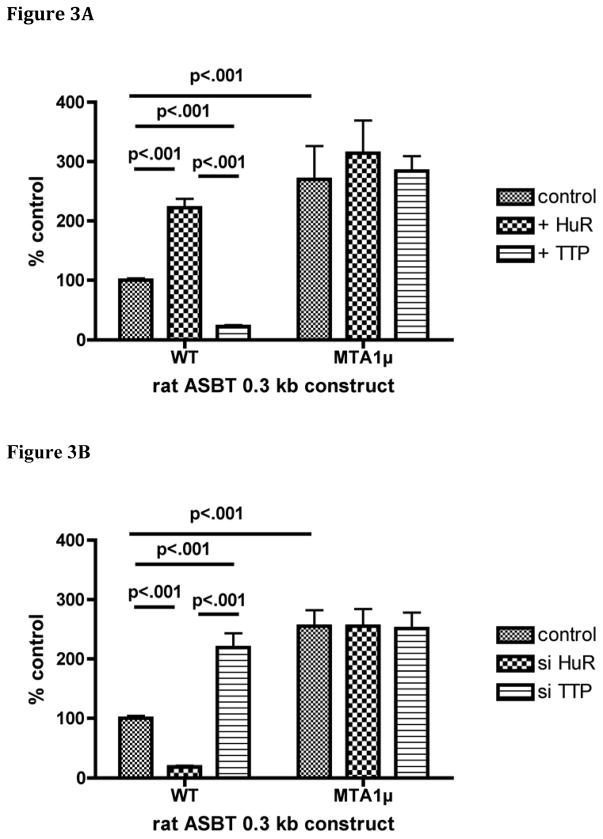
The functional role of the MTA1 site in mediating changes in RNA stability was first studied using the luciferase reporter construct containing the 0.3 kb rat ASBT 3′ UTR with a native and mutated MTA1 site. Lucifererase activity in this assay is dependent upon both mRNA abundance and translation of that mRNA. Basal activity of the MTA1 mutant was increased compared to the wild type construct suggesting that in IEC-6 cells the destabilizing effect of TTP predominates over HuR and thus the presence of an MTA1 site imparts reduced expression (Figures 3A and 3B). A complete abrogation of the response to HuR or TTP was observed when the MTA1 site was mutated [(wt: 100 ± 3; +HuR 222 ± 15; +TTP 22 ± 3; p<0.001), (muMTA1: 270 ± 56; +HuR 314 ± 55, +TTP 284 ± 25; p>0.05)]; [(wt: 100 ± 4; +siHuR 18 ± 2; +siTTP 219 ± 24; p<0.001), (muMTA1: 255 ± 27; +siHuR 255 ± 29, + siTTP 251 ± 27; p>0.05)] (Figures 3A and 3B). Thus, site directed mutagenesis of the two conserved and apparently critical nucleotides in the MTA-1 element abrogated changes in luciferase reporter activity in response to alterations in the expression of either HuR or TTP.
Figure 3. An intact MTA-1 cis-element is required for responses of a luciferase reporter of the 3′ ASBT UTR modulation of HuR and TTP expression.
0.3 kb of the rat ASBT 3′UTR was inserted downstream of the luciferase coding sequence and used as a reporter for effects of changes in that sequence on expression. The wild type (WT) construct contained the natural rat ASBT sequence, while the MTA1 mutant (MTA1μ) was altered as shown in Figure 1. (A) Modulation by overexpression of HuR or TTP. IEC-6 cells were transfected with the indicated luciferase construct and either a HuR or TTP expression construct. Data are represented relative to control, which is the wild type 3′UTR that is untreated with an overexpression vector and is set to 100%. The basal activity of the MTA1μ construct is higher than the WT (p<0.001). WT is activated by HuR and repressed by TTP (p< 0.001), whereas the MTAμ is unaffected by overexpression of either HuR or TTP (p >0.05). (B) Modulation by silencing of HuR or TTP. As in Figure 1A, the basal activity of the MTA1μ construct is higher than the WT (p<0.001). WT is repressed in response to silencing HuR and activated by silencing TTP (p<0.001), whereas the MTAμ is unaffected by silencing of either HuR or TTP (p>0.05).
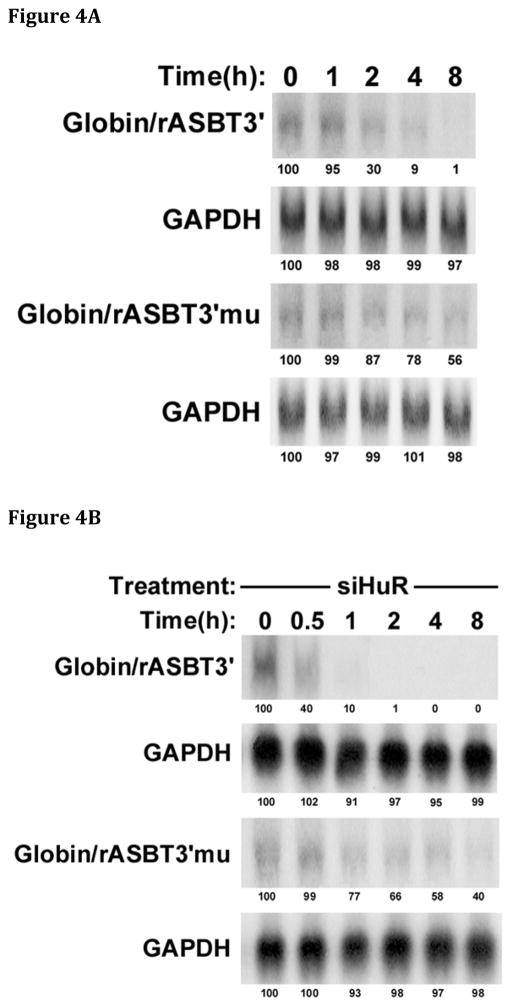
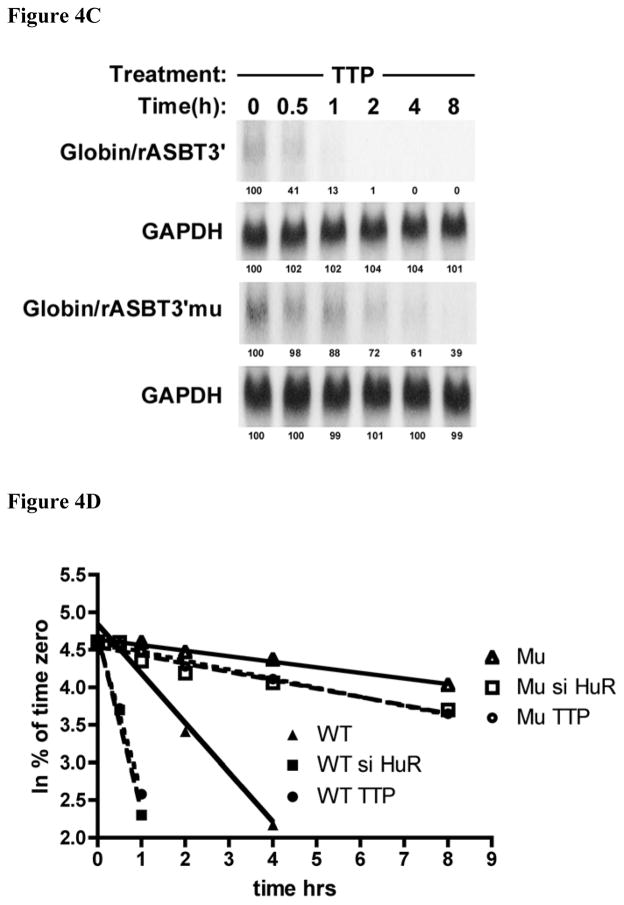
The luciferase reporter system used in the aforementioned experiments can be influenced by effects on both mRNA stability and translation. Therefore kinetic analyses of mRNA half-life of globin:ASBT 3′UTR mRNA hybrid constructs were utilized to test for a direct effect on mRNA stability. The same 0.3 kb rat ASBT 3′UTR fragments (wild type and MTA1 mutant) were incorporated into Globin/rASBT3′ constructs and mRNA half-life was measured by a transcriptional-pulsing method based on the cfos-serum inducible promoter system [24]. mRNA half-life was assessed for the wild type 0.3 kb Globin/rASBT3′ construct and the MTA1 mutant Globin/rASBT3′ hybrid construct in the presence or absence of siHuR or TTP wild type expression vector (Figures 4A – 4D). The basal half-life of the wild type construct was 2.6±0.2hrs (Figure 4A and 4D) and decreased to 0.60±0.04hrs when HuR was silenced (77% reduction) (Figure 4B and 4D) and to 0.80±0.06hrs when TTP was overexpressed (69% reduction) (Figure 4C and 4D). In contrast, compared to the wild type construct, the Globin/rASBT3′-muMTA1 had a markedly longer half-life of 21.7±1.6hrs (Figure 4A and 4D). As in the luciferase-based studies, this finding indicates that in IEC-6 cells an intact MTA1 site imparts relative instability to mRNA (Figure 5). Silencing HuR or overexpressing TTP led to an attenuated change in mRNA decay rate compared to the wild type with a half-life of 14.4±2.1hrs (34% reduction) (Figure 4B and 4D) and 13.3±0.7hrs (39% reduction) (Figure 4C and 4D), respectively. This attenuated but not absent response may be explained by previous studies demonstrating that the β-globin construct itself may be affected by HuR as the basal construct had a half-life of 15.8±4 hrs, and with si HuR treatment the β-globin construct had a half-life of 13.6±2.1 hrs (14% reduction) [17]. These findings strongly implicate the MTA1 site in mediating changes in ASBT mRNA stability in response to both HuR and TTP (Figure 5).
Figure 4. An intact MTA-1 site is required for full changes in mRNA decay in response to modulation of HuR and TTP expression.
The wild type Globin/rASBT3′ or the Globin/rASBT3′ MTA1 mutant (Globin/rASBT3′mu) were positioned downstream of a globin c-DNA driven by a c-fos promoter. A timed pulse of transcriptional activity in IEC6 cells was induced by serum withdrawal, restitution and withdrawal. Time 0 coincides with the second withdrawal of serum from the media. At the specified time points RNA was extracted and analyzed by northern blot for globin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Signal intensity as measured with a phosphorimager is indicated below each band and has been normalized to 100% for time 0 for each probe. Linear regression analysis was used for derivation of mRNA half-life. (A) mRNA half-life analysis of the Globin/rASBT3′ construct. The stability of the Globin/rASBT3′mu construct is greater than the wild type construct. (B) mRNA half-life analysis of siHuR treated Globin/rASBT3′ constructs. The stability of the Globin/rASBT3′mu construct is greater than the wild type construct. (C) mRNA half-life analysis of TTP overexpression treated Globin/rASBT3′ constructs. The stability of the Globin/rASBT3′mu construct is greater than the wild type construct. Overall, mRNA half-life is diminished with either HuR silencing or TTP overexpression. The magnitude of the change in half-life is reduced when the MTA1 site is mutated. (D) Data from Figures 4A – C are analyzed by linear regression of the ln of the % of transcript remaining at various time points relative to time 0. The best-fit line for each experiment is shown. Analyses of wild type are indicated by the closed symbols and MTA1 mutant by open symbols. The triangles represent untreated IEC6 cells, the squares siHuR treated cells and circles for cells treated with a TTP expression construct.
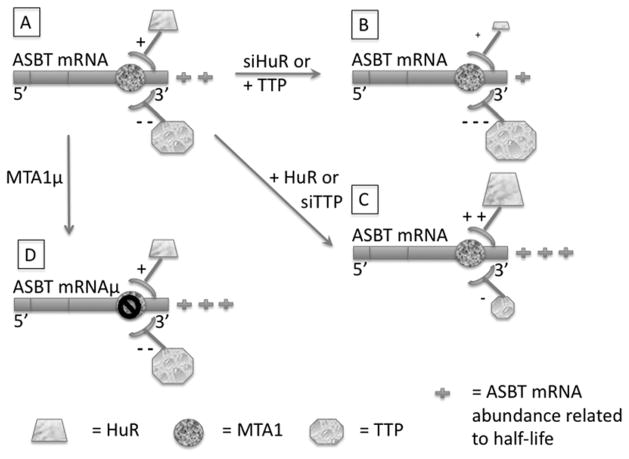
Figure 5. Model of the effects of HuR and TTP on ASBT mRNA abundance.
ASBT mRNA is depicted by the horizontal bar with the site of the MTA1 cis-element indicated by the circle (panel A). In the native state, ASBT mRNA abundance (2+) is dually impacted by HuR (trapezoid) and TTP (octagon), with stabilization and positive regulation (+) by HuR and destabilization and negative (−) regulation by TTP. In the setting of siHuR or TTP overexpression (panel B), ASBT mRNA abundance is diminished (1+) due to either diminished stabilization by HuR or enhanced destabilization by TTP. In the setting of siTTP or overexpression of HuR (panel C) ASBT mRNA abundance in increased (3+) due to the opposite regulation of either diminished destabilization by TTP or enhanced stabilization by HuR. When the MTA1 cis-element is mutated (panel D), the net effect in IEC-6 cells is an increase in ASBT mRNA abundance (3+). Presumably this is the result of the overall dominant effect of TTP on destabilizing ASBT mRNA through the MTA1 cis-element.
ASBT expression is controlled by a complex regulatory process, which may include regulated changes in mRNA stability. This is most strongly suggested on the basis of measurements of ASBT mRNA abundance and transcription in developing rat ileum and kidney [15, 16]. During normal development in the ileum there is a greater than 100-fold increase in ASBT mRNA abundance, while there is only a 10-fold increase in transcription as measured by nuclear run-on analysis. Prior to weaning transcription rates in ileum and kidney are similar, yet ASBT mRNA abundance in the ileum is one tenth that in the kidney. These findings suggest that ASBT mRNA is relatively unstable in the preweaning ileum. Coordinate changes in the ontogenic expression of HuR and TTP in rat ileum and kidney imply a novel role for the RNA-binding proteins HuR and TTP in intestinal development [17]. HuR is a protein that belongs to the Hu/ELAV family. It is relatively ubiquitously expressed and was originally identified as binding to AU-rich elements (AREs) in the 3′untranslated regions of different cytokines and proto-oncogenes [25]. TTP is a member of the mammalian family of CCCH tandem zinc-finger proteins. It suppresses inflammation by accelerating the degradation of cytokine mRNAs [26]. HuR stabilizes ASBT mRNA leading to enhanced expression, whereas TTP induces the opposite effect [17]. During development in the ileum HuR goes from barely detectable to strongly expressed after weaning. The opposite is observed for TTP. Therefore, relative instability of ASBT mRNA in the preweaning rat ileum is induced by the coordinate down-regulation of HuR and up-regulation of TTP.
Counter-regulation of mRNA abundance/stability is a relatively common mechanism for control of gene expression. Fine-tuning by simultaneous up- and down-regulation may yield more stable transitions in gene expression. Intestinal expression of cyclooxygenase-2 (COX-2) is also controlled by counter-regulation by HuR and TTP. This may be relevant for colorectal tumorigenesis. In normal colonic tissue, low levels of nuclear HuR and higher TTP levels have been observed. In contrast, HuR overexpression and enhancement of its cytoplasmic localization has been observed in adenomas and adenocarcinomas, with the loss of TTP expression in both. In both adenomas and adenocarcinomas, cyclooxygenase (COX-2) overexpression was associated with HuR overexpression and TTP repression [18]. Similar counter-regulatory effects on interleukin-3 mRNA stability have been described [27]. Translation of tumor necrosis factor-alpha mRNA is counter-regulated by HuR and TTP, with enhancement of translation by HuR and potential displacement of HuR from relevant cis-elements by TTP [28]. Counter-regulation by HuR and TTP may also be relevant for gliomas via regulation of vascular endothelial growth factor and interleukin-8 [29].
Counter-regulation by RNA binding proteins is not limited to HuR and TTP. Typically HuR enhances expression, with counter-regulation (i.e. repression) by the other RNA binding protein. The CUG-binding protein 1 represses translation of occludin mRNA [30]. The KH-type splicing regulatory protein (KSRP) represses expression of human inducible nitric oxide [31]. The AU-rich element RNA-binding protein 1 (AUF1) is a negative regulator of the androgen receptor, c-fos, cyclin D1 and p21 [32–34]. These interactions become more complex in light of the descriptions of the binding of TTP by either KSRP or AUF1 [31, 35].
There is limited information as to the specific RNA sequence elements that mediate regulated changes in RNA stability. HuR and AUF1 may compete for binding to elements in the 3′UTR of p21 and cyclin D1, although the specific elements for which they may compete are not known [34]. RNase footprint analysis reveals a similar binding sequence for HuR and AUF1 in the androgen receptor 3′ UTR [32]. TTP and HuR may compete for binding to the same AU rich element in the 3′UTR of tumor necrosis factor RNA with implications for translational regulation of that gene [28]. KSRP and HuR compete for binding to a 90 base pair fragment of the 3′ UTR of the inducible nitric oxide synthase mRNA [31]. The findings from this study are the first to identify specific nucleotides in a predicted binding site that alter both binding by and functional consequences of two distinct RNA binding proteins. The mutated nucleotides in the ASBT MTA-1 element are highly conserved across species (Figure 1A). The mutations that were introduced are predicted to have profound consequences on an alternative predicted secondary structure for this region of RNA (http:\\rna.tbi.univie.ac.at) (Figures 1B and 1C). The predicted secondary structure for this region is lost in the face of the mutations used in these studies (data not shown) [36].
In summary, our data suggest that ASBT mRNA stabilization and destabilization occurs via post-transcriptional regulation by HuR and TTP, and these effects are mediated at least in part by the MTA1 sequence in the 3′UTR ASBT mRNA (Figure 5). Further investigations will be required to assess the functional role of other predicted RNA binding protein sites in the rat ASBT 3′UTR. The MTA1 element is sufficient to mediate effects of both HuR and TTP. The molecular mechanism by which this occurs needs to be explored. It is not known if this is a function of competition between HuR and TTP for the same binding sequence or whether it is the result of direct interactions between HuR and TTP. In light of the description of a similar phenomenon in the regulation of Cox-2 and the remarkable changes in gene expression during weaning in the intestine, the counter-regulatory effects of HuR and TTP may have much broader biologic significance.
Summary Statement.
A single cis-element in the 3′UTR of the rat apical sodium dependent bile acid transporter mediates both mRNA stabilization by Hu antigen R and destabilization by tristetraprolin.
Acknowledgments
We thank Drs. Ann-Bin Shyu and Paul Dawson for their advice and critical reading of the manuscript.
Funding: This work was supported by NIH NIDDK grant DK054165 to BLS.
Abbreviations
- ASBT
apical sodium dependent bile acid transporter
- HuR
Hu antigen R
- MTA
metastasis associated gene
- TTP
tristetrapolin
- UTR
untranslated region
Footnotes
Author Contribution: Dellys Soler, Frank Chen, Ayantika Ghosh and Benjamin Shneider contributed to the conceptualization of these studies. Delly Soler, Frank Chen and Ayantika Ghosh contributed to the conduct of the experimentation. All authors participated in the analysis of the data and the writing of the manuscript.
References
- 1.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent biel acid transporter gene (SLC10A2) J Clin Invest. 1997;99:1880–1887. doi: 10.1172/JCI119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duane W, Hartich L, Bartman A, Ho S. Diminished gene expression of ileal apical sodium bile acid transporter explains impaired absorption of bile acid in patients with hypertriglyceridemia. J Lipid Res. 2000;41:1384–1389. [PubMed] [Google Scholar]
- 4.Holzer A, Harsch S, Renner O, Strohmeyer A, Schimmel S, Wehkamp J, Fritz P, Stange EF. Diminished expression of apical sodium-dependent bile acid transporter in gallstone disease is independent of ileal inflammation. Digestion. 2008;78:52–59. doi: 10.1159/000159379. [DOI] [PubMed] [Google Scholar]
- 5.Jung D, Fantin AC, Scheurer U, Fried M, Kullak-Ublick GA. Human ileal bile acid transporter gene ASBT (SLC10A2) is transactivated by the glucocorticoid receptor. Gut. 2004;53:78–84. doi: 10.1136/gut.53.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halpern MD, Weitkamp JH, Mount Patrick SK, Dobrenen HJ, Khailova L, Correa H, Dvorak B. Apical sodium-dependent bile acid transporter upregulation is associated with necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G623–631. doi: 10.1152/ajpgi.00242.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, Ma L, Al-Ansari N, Shneider B. The role of AP-1 in the transcriptional regulation of the rat apical sodium-dependent bile acid transporter. J Biol Chem. 2001;276:38703–38714. doi: 10.1074/jbc.M104511200. [DOI] [PubMed] [Google Scholar]
- 9.Shih D, Bussen M, Sehayek E, Ananthanarayanan M, Shneider B, Suchy F, Shefer S, Bollileni J, Gonzalez F, Breslow J, Stoffel M. Hepatocyte nuclear factor-1a is an essential regulator of bile acid and plasma cholesterol metabolism. Nature Genetics. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 10.Jung D, Fried M, Kullak-Ublick G. Human apical sodium-dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator-activated receptor a. J Biol Chem. 2002;277:30559–30566. doi: 10.1074/jbc.M203511200. [DOI] [PubMed] [Google Scholar]
- 11.Chen F, Ma L, Dawson P, Sinal C, Sehayek E, Gonzalez F, Breslow J, Ananthanarayanan M, Shneider B. Liver receptor homologue-1 mediated species- and cell line-specific bile acid dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chemistry. 2003;278:19909–19916. doi: 10.1074/jbc.M207903200. [DOI] [PubMed] [Google Scholar]
- 12.Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–156. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- 13.Thomas C, Landrier JF, Gaillard D, Grober J, Monnot MC, Athias A, Besnard P. Cholesterol dependent downregulation of mouse and human apical sodium dependent bile acid transporter (ASBT) gene expression: molecular mechanism and physiological consequences. Gut. 2006;55:1321–1331. doi: 10.1136/gut.2005.085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Chen F, Liu S, Glaeser H, Dawson PA, Hofmann AF, Kim RB, Shneider BL, Pang KS. Transactivation of rat apical sodium-dependent bile acid transporter and increased bile acid transport by 1alpha,25-dihydroxyvitamin D3 via the vitamin D receptor. Mol Pharmacol. 2006;69:1913–1923. doi: 10.1124/mol.105.020792. [DOI] [PubMed] [Google Scholar]
- 15.Shneider BL, Dawson PA, Christie DM, Hardikar W, Wong MH, Suchy FJ. Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J Clin Invest. 1995;95:745–754. doi: 10.1172/JCI117722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie DM, Dawson PA, Thevananther S, Shneider BL. Comparative analysis of the ontogeny of a sodium-dependent bile acid transporter in rat kidney and ileum. Am J Physiol. 1996;271:G377–G385. doi: 10.1152/ajpgi.1996.271.2.G377. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, Shyu AB, Shneider BL. Hu antigen R and tristetraprolin: counter-regulators of rat apical sodium-dependent bile acid transporter by way of effects on messenger RNA stability. Hepatology. 2011;54:1371–1378. doi: 10.1002/hep.24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CY, Ezzeddine N, Shyu AB. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 2008;448:335–357. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 22.Toh Y, Oki E, Oda S, Tokunaga E, Ohno S, Maehara Y, Nicolson GL, Sugimachi K. Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int J Cancer. 1997;74:459–463. doi: 10.1002/(sici)1097-0215(19970822)74:4<459::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis. 2009;26:215–227. doi: 10.1007/s10585-008-9233-8. [DOI] [PubMed] [Google Scholar]
- 24.Shyu A, Greenberg M, Belasco J. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 25.Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo. 2006;20:17–23. [PubMed] [Google Scholar]
- 26.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 27.Ming XF, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. The role of tristetraprolin in cancer and inflammation. Front Biosci (Landmark Ed) 2012;17:174–188. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J Biochem. 2012;151:423–437. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- 33.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kedar VP, Zucconi BE, Wilson GM, Blackshear PJ. Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J Biol Chem. 2012;287:5459–5471. doi: 10.1074/jbc.M111.312652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]