Significance
The tumor suppressor Trp53 (p53) is a gene that regulates the expression of many genes. However, the role of p53 in the heart has not been well characterized. This work documents the important role for p53 in the heart as a master regulator of the cardiac transcriptome. The contribution of p53 to the maintenance of cardiac tissue homeostasis is complex under physiological conditions.
Keywords: heart failure, tumor suppressor, cardiac hypertrophy, transcriptome, cardiomyopathy
Abstract
The tumor suppressor Trp53 (p53) inhibits cell growth after acute stress by regulating gene transcription. The mammalian genome contains hundreds of p53-binding sites. However, whether p53 participates in the regulation of cardiac tissue homeostasis under normal conditions is not known. To examine the physiologic role of p53 in adult cardiomyocytes in vivo, Cre-loxP–mediated conditional gene targeting in adult mice was used. Genome-wide transcriptome analyses of conditional heart-specific p53 knockout mice were performed. Genome-wide annotation and pathway analyses of >5,000 differentially expressed transcripts identified many p53-regulated gene clusters. Correlative analyses identified >20 gene sets containing more than 1,000 genes relevant to cardiac architecture and function. These transcriptomic changes orchestrate cardiac architecture, excitation-contraction coupling, mitochondrial biogenesis, and oxidative phosphorylation capacity. Interestingly, the gene expression signature in p53-deficient hearts confers resistance to acute biomechanical stress. The data presented here demonstrate a role for p53, a previously unrecognized master regulator of the cardiac transcriptome. The complex contributions of p53 define a biological paradigm for the p53 regulator network in the heart under physiological conditions.
The tumor suppressor Trp53 (p53) is a transcription factor that translates growth and survival signals into specific gene expression patterns, regulating tumor-free survival of an organism (1). In normal cells, p53 expression is kept at low levels by the E3 ubiquitin ligase Mdm2, which targets p53 for proteasomal degradation (2). In response to acute stress, Mdm2 is inactivated and increased p53 levels block cell division and induce apoptosis (3). Conversely, p53 can activate Mdm2 transcription, thereby forming a negative feedback loop that curtails p53 activity (4). Cells with mutated p53 proliferate aberrantly and generate outgrowths of genetically unstable cells, leading to tumorigenesis as demonstrated by the early cancer predisposition of p53 knockout mice (p53KO) (5). Loss of Mdm2 in mice leads to death in embryogenesis through p53-induced apoptosis, which is prevented by p53 codeletion (6). These findings provide genetic evidence that Mdm2 exerts a physiologically critical role in regulating p53 in vivo.
Adult mammalian cardiomyocytes (CM) are differentiated postmitotic cells that lack significant proliferative potential through their inability to reactivate the cell cycle (7). This is caused by the lack of G1 cyclin/cyclin-dependent kinases (Cdk), crucial positive cell-cycle modulators, and high levels of cell-cycle inhibitors (8), such as the retinoblastoma proteins pRb/p130 (9) and the Cdk inhibitors (Cdki) p21/p27 (10). The limited mitotic capacity of mature CM (11) renders the adult mammalian heart functionally unable to repair itself after ischemic injury (12). Instead, surviving CM undergo hypertrophy to compensate for the ensuing hemodynamic stress manifested as cell enlargement, myofibrillar disarray, and re-expression of fetal genes (13). This process becomes maladaptive with time, leading to the development of heart failure (HF) with significant morbidity and mortality (14). The molecular mechanisms underlying HF remain poorly understood. As such, identifying the factors that effectively maintain cardiac tissue homeostasis is of great scientific and clinical importance.
Elevated p53 levels correlate with CM apoptosis and hypertrophy in end-stage human HF (15). The heart, as an obligate aerobic organ, has the largest mitochondrial (mt) content to meet its high energy demand (16). Reactive oxygen species (ROS), normal by-products of aerobic respiration, induce DNA damage (17), thereby activating cellular defense systems against ROS through stabilization of p53 (18). Therefore, we hypothesized that impairment of p53 will have deleterious consequences on the heart. Here we report that p53 forms a critical hub in a comprehensive transcriptional network. Our study suggests that p53 acts as a pleiotropic regulator of cardiac structure and function.
Results
Cardiac-Specific Ablation of p53 Induces Age-Dependent Cardiac Hypertrophy and HF.
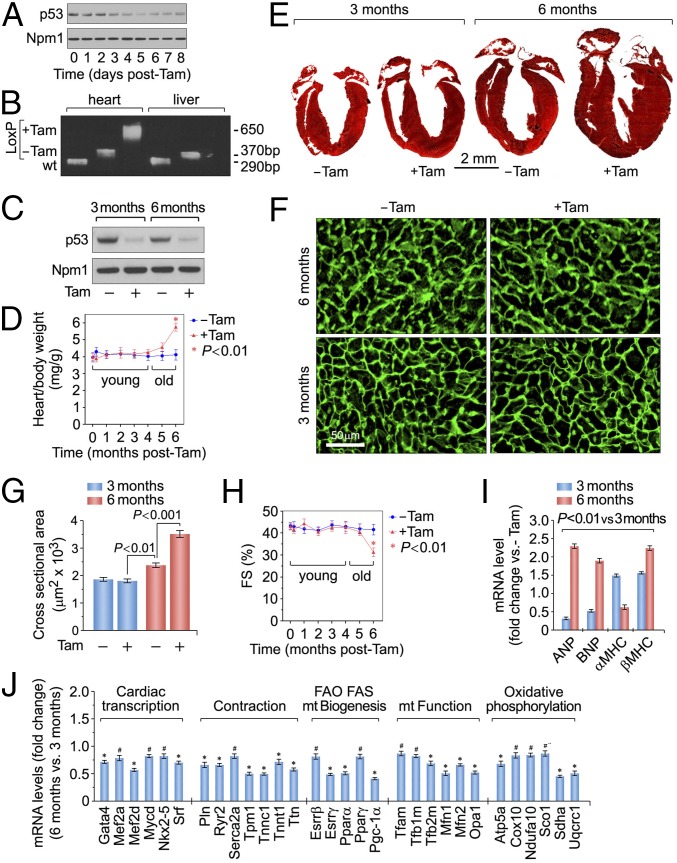
We crossed transgenic mice expressing Cre recombinase flanked by mutated estrogen receptors (MerCreMer; mcm) with mice carrying loxP-flanked alleles of p53 to obtain p53fl/fl;mcm animals. The day of the last injection was arbitrarily set to 0. Four consecutive daily i.p. Tamoxifen (Tam) injections induced genetic ablation of p53 with high recombination efficiency (Fig. 1 A–C). To determine the physiological consequence of p53 ablation, cardiac morphology and function in p53fl/fl;mcm mice were assessed in the presence and absence of Tam.
Fig. 1.
Heart-specific ablation of p53 triggers age-dependent concentric hypertrophy with cardiac dysfunction. (A) Immunoblot analysis of LV extracts (60 μg total protein/lane) of p53fl/fl;mcm mice at various time points after Tam using anti-p53 antibodies. Animals were 13 wk old. For normalization, Western blots were probed with anti-nucleophosmin (Npm1). Immunoblots were repeated once with similar results. (B) PCR of DNA isolated from LV and liver samples of wild-type, vehicle-injected control p53fl/fl;mcm mice 7 d post-Tam. Animals were 13 wk old. (C) Immunoblot analysis of p53 expression in LV extracts at the indicated time points after Tam administration (Tam) (D) Heart/body weight ratios of Tam- and vehicle-injected p53fl/fl;mcm mice at various time points post-treatment. Data are means ± SEM; n = 6. *P < 0.01 vs. −Tam; mg, milligram; g, gram. (E) Masson staining of longitudinal cardiac sections. (F) Immunofluorescence microscopy of wheat germ agglutinin (WGA; green)-stained LV sections. (G) Quantification of cross-sectional area of adult cardiomyocytes. n = 6–8. (H) Fractional shortening (FS) determined by echocardiography at various time points post-Tam or vehicle. n = 4. (I) Levels of hypertrophic marker genes as analyzed by RT-qPCR at 14 d post-Tam. n = 4. *P < 0.05 vs. 3 mo post-Tam. Atrial natriuretic factor, ANF; brain natriuretic factor, BNP; myosin heavy chain, MHC. (J) Cardiac-specific gene expression in Tam-treated p53fl/fl;mcm animals at 6 mo post-Tam vs. 3 mo post-Tam as analyzed by RT-qPCR. n = 4. *P < 0.01; #P < 0.05.
Although p53fl/fl;mcm mice were initially normal post-Tam, they developed concentric hypertrophy by 6 mo. These mice showed significant increases in heart weight/body weight (HBW) ratios (Fig. 1D) (P < 0.001) and wall thickening (Fig. 1E). In contrast, p53fl/fl;mcm mice developed normally in the absence of Tam (Fig. 1 D and E). The cross-sectional area of cardiomyocytes was 1.5-fold greater in p53fl/fl;mcm mice post-Tam (46 ± 3.4%; P < 0.001) compared with vehicle-injected controls (Fig. 1 F and G). The analysis of cardiac performance by echocardiography on older p53fl/fl;mcm mice post-Tam revealed significant decreases in fractional shortening (31 ± 4.5%) in comparison with vehicle-injected controls (44 ± 5.1%; P < 0.001) (Fig. 1H). As expected, transcripts for atrial/brain natriuretic factors (ANF, BNP) and β-myosin heavy chain (β-MHC), canonical markers of cardiac hypertrophy and heart failure, were up-regulated in p53-deficient hearts compared with controls (P < 0.05) (Fig. 1I). Interestingly, no effect on life span was observed in p53fl/fl;mcm mice at 14 mo post-Tam (n = 14; P > 0.05). Taken together, acute loss of p53 induces spontaneous hypertrophy in the heart with loss of function.
In the younger p53fl/fl;mcm mice there was an apparent cardioprotective effect from the ablation of p53 that was lost over time. To support this, a significant down-regulation of the core set of heart-specific transcription factors (Gata4, Mef2a/d, Myocd, Nkx-2.5, Srf) was observed in p53fl/fl;mcm at 6 mo post-Tam compared with 3 mo (Fig. 1J). We also found significantly lower levels of expression for genes encoding important regulators of cardiac contractility (Pln, Ryc2, Serca2a) in older p53fl/fl;mcm mice post-Tam (Fig. 1J). Finally, sarcomere organization profoundly influences cardiac function with abnormalities in this structure commonly observed in human HF. Key contractile proteins (Tpm1, Tnnc1, Tnnt1, Ttn) were down-regulated in older p53fl/fl;mcm mice post-Tam (Fig. 1J), demonstrating that p53 is important for proper expression of the contractile apparatus. Beyond these genes, we also found key regulators of fatty acid metabolism and mitochondrial respiration to be transcriptionally down-regulated in older p53KO, including Esrrβ/γ, Pparα/γ, Pgc-1a, Tfam, and Tfb1/2m (Fig. 1J). These data strongly suggest that p53 acts in concert with other transcriptional components that are necessary for the regulation of physiological hypertrophy.
p53-Deficient Hearts Are Resistant to Pressure Overload.
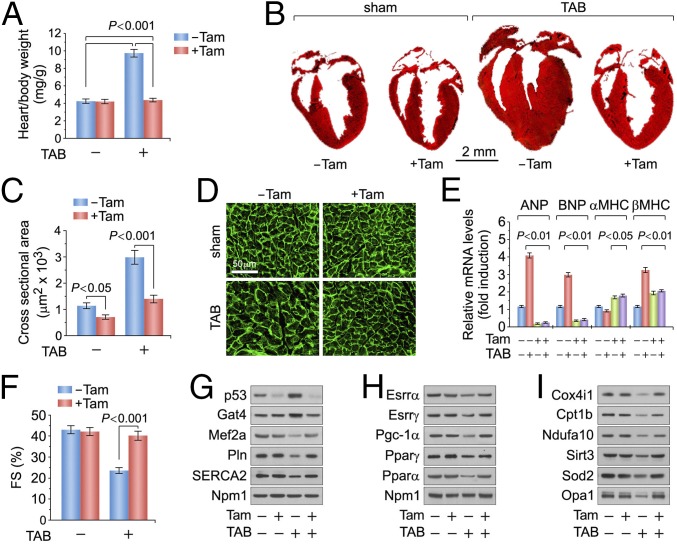
We then examined the responses of p53 loss to mechanical stress in younger mice by subjecting 10-wk-old Tam-treated p53fl/fl;mcm mice to transaortic banding (TAB). After 3 wk of TAB, HBW in vehicle-injected p53fl/fl;mcm mice increased by 2.3-fold, compared with Tam-treated sham p53fl/fl;mcm animals (P < 0.001) (Fig. 2A). Vehicle-treated hearts subjected to TAB exhibited significantly greater dilatation and increases in left ventricular wall thickness than Tam-injected p53fl/fl;mcm mice (Fig. 2B). To confirm that higher HBW in control hearts after TAB represents cardiomyocyte hypertrophy, we determined that the width of cardiomyocytes from hearts of vehicle-treated mice was 2.1-fold greater than in p53fl/fl;mcm mice injected with Tam post-TAB (P < 0.001) (Fig. 2 C and D). This was accompanied with significantly higher levels of ANP, BNP, and β-MHC compared with p53-deficient hearts (Fig. 2E). Importantly, TAB treatment reduced fractional shortening in vehicle-injected p53fl/fl;mcm mice, as compared with p53fl/fl;mcm animals in the presence of Tam (P < 0.001) (Fig. 2F).
Fig. 2.
Ablation of p53 confers protection against pressure overload-induced cardiac dysfunction. (A) Heart/body weight ratios after 3 wk post-transaortic banding performed at 10 wk of age. n = 6. (B) Masson stain of longitudinal cardiac sections. (C) Quantification of cross-sectional area of adult cardiomyocytes. n = 4. (D) Immunofluorescence microscopy of WGA (green)-stained LV sections. (E) Levels of hypertrophic marker genes as analyzed by RT-qPCR. n = 4. (F) FS determined by echocardiography. n = 4. (G–I) Immunoblot analysis of LV extracts (60 μg total protein/lane) from p53fl/fl;mcm mice at 6 mo post-Tam using antibodies as indicated (left). Data are means ± SEM.
Next, we investigated the insensitivity of p53fl/fl;mcm mice to TAB by immunoblotting of left ventricular lysates (Fig. 2 G–I). Mef2a, Pln, and Serca2 were markedly down-regulated in vehicle-treated p53KO, but not in Tam-treated p53KO (Fig. 2G). Administration of Tam to p53fl/fl;mcm mice also markedly maintained the protein levels of Esrrβ/γ, Pparα/γ, and Pgc-1a after TAB (Fig. 2H), which was not observed in vehicle-injected mice subjected to TAB. Notably, we observed significant decreases in the expression of crucial components of respiratory chain complexes in vehicle-treated littermates post-TAB, including Cox4i (CIV) and Ndufa10 (CI), and of the key antioxidant factors Sod2 and Sirt3, which play important roles in redox homeostasis and ROS detoxification (Fig. 2I). All these effects were not observed in Tam-treated p53fl/fl;mcm mice subjected to TAB (Fig. 2I). Collectively, these results suggest that p53 is important in the regulation of pressure overload-induced cardiac hypertrophy and deterioration of cardiac function.
p53 Leads to Genome-Wide, Specific Transcriptional Changes.
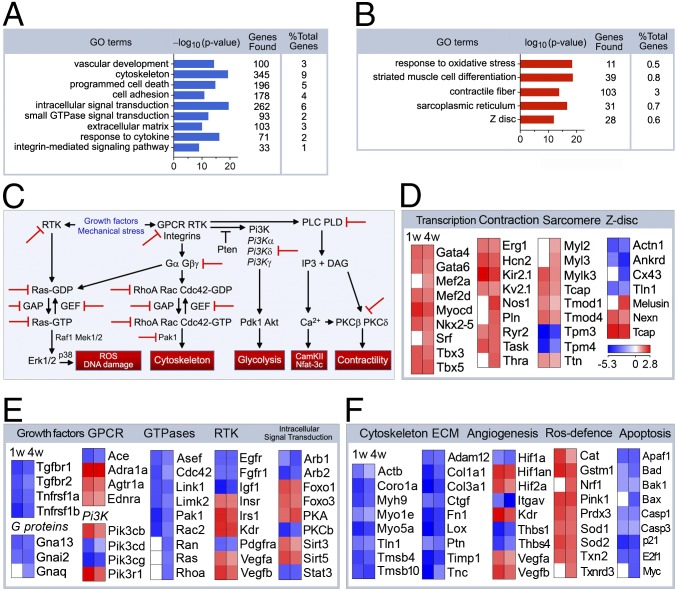
To elucidate mechanisms underlying the phenotypes described above, we performed genome-wide messenger RNA (mRNA) microarray profiling. Unsupervised hierarchical cluster analysis at a high confidence threshold revealed that 5,221 (7 d post-Tam) and 5,172 (4 wk post-Tam) of 26,166 individual transcripts changed significantly relative to vehicle-injected controls (Fig. 3A). We observed that p53 inactivation causes widespread alterations in gene expression profiles, indicative of a high degree of complexity in the regulation of the transcriptome by p53. Heat-map analysis of 87 canonical p53-target genes (19) (Fig. 3B and SI Appendix, Fig. S1) was performed. Genes in this test set regulate diverse classes of biological processes such as antioxidants (Gpx1), apoptosis (Apaf1), autophagy (Atg7), cell cycle (PCNA), DNA damage (Dram1/Dnmt1), growth arrest (pRb/p21), metabolism (Vdr), oncogene activation (Pml), and signal transduction (Egfr/Ptprn). The majority of these factors was already down-regulated in p53f/f;mcm at 7 d post-Tam (Fig. 3B; SI Appendix, Fig. S1). We also observed several repressed genes related to autophagy (Atg10), DNA damage (Gadd45), and glycolysis (Tigar), which may reflect p53’s ability to interfere with the basal transcription machinery (1) or hindrance of p53’s transactivation domain by Mdm2. This pattern reveals a previously unrecognized role for p53 whereby it may act in concert with a subset of p53-related transcription factors (TFs) rather than simply reflecting the binding of p53 to target promoters. This model is supported by the known ability of Mdm2 to physically interact with pleiotropic TFs, such as E2f1 (20), or transcriptional repressors, including pRb (21).
Fig. 3.
Genome-wide transcriptional changes in the p53-deficient myocardium. (A) Heat-map examination of genes enriched in the hearts of p53fl/fl;mcm mice 7 d and 4 wk post-Tam (columns) relative to vehicle-injected controls. Values (log2 expression) are shown by color and intensity of shading. Blue, repressed; red, induced. n = 3 biological replicates. P < 0.01. Fold change ± 1.3. (B) Heat map examining the impact of genomic modifications in p53fl/fl;mcm post-Tam of p53-target genes. log2 expression values. n = 3 biological replicates. P < 0.01. Fold change >1.3. (C) GSEA of different biological processes assessed by overrepresentation of GSEA terms for the biological function of each transcript in p53fl/fl;mcm at 4 wk post-Tam. NES, normalized enrichment scores.
To identify gene clusters with similar biological functions in all mutant transcriptomes, we performed Gene Set Enrichment Analysis (GSEA) at 4 wk post-Tam. G-protein signaling, oxidative phosphorylation (OxPhos), tricarboxylic acid (TCA), cell cycle, and apoptosis were among the highest ranked GSEA terms (Fig. 3C). GSEA hits were further categorized by gene ontology (GO) analysis (Fig. 3C), illustrating the presence of a comprehensive transcriptional network regulating >1,000 genes in >20 biological processes. We conclude from this data that p53 is functionally active in the adult heart under physiologic conditions.
p53 Broadly Regulates the Transcriptome to Maintain Cardiac Architecture and Function.
Although p53 loss protects the heart against biomechanical stress (22), the basis for this was not clearly identified. The cardioprotective mechanism may be explained by the detailed GO term analysis delineating the down-regulated (Fig. 4A) and up-regulated biological processes (Fig. 4B) at 4 wk post-Tam. The gene sets identified are involved in the regulation of programmed cell death, small GTPase signal transduction, extracellular matrix composition (Fig. 4A), or striated muscle-cell differentiation and response to oxidative stress (Fig. 4B). Fig. 4C identifies regulators and targets of potential pathways that were obtained by Natural Language Processing (NLP) examination onto a known interaction network for these enriched GO gene sets displayed in Fig. 4 A and B. Intriguingly, GO analysis identified a gene signature with induction of cardiac transcription factors (Gata4, Mef2a, Nkx-2.5) accompanied by repression of genes regulating hypertrophic signaling, for example, G proteins, G-protein–coupled receptors, phosphatidylinositol 3-kinase signaling, receptor tyrosine kinases, and intracellular signal transduction (Fig. 4 C and E and SI Appendix, Fig. S2A). In contrast, unfavorable changes observed in Z disk (Actn1, Cx43, Melusin), sarcomere (Mylk3, Tnnc1, Ttn), extracellular matrix (Col1a1, Col3a1, Fn1), and angiogenesis (Kdr, Thbs1, Vegfa) components may well have induced HF in older p53f/f;mcm after administration of Tam (Fig. 4 D and E; SI Appendix, Fig. S2 A–C). All these changes in the cardiac transcriptome were already detectable as early as 7 d post-Tam. Thus, many alterations in mRNA expression appear to be a primary consequence of the experimental genetic ablation of p53. These changes are sufficient to produce the cardiac phenotypes at baseline and the in vivo response to biomechanical stress.
Fig. 4.
p53 regulates gene sets involved in cardiac tissue architecture and function. GO term enrichment representing down-regulated (A) and up-regulated (B) biological processes involved in cardiac structure and function in p53fl/fl;mcm mice post-Tam. (C) Visualization of regulators and targets of potential pathways that were obtained by NLP onto a known interaction network for enriched GO gene sets in Fig. 3 A and B. (D) Heat maps displaying GO gene sets for cardiac differentiation. Shown are subsets of the most abundantly enriched or down-regulated genes (rows), comparing p53 mutant hearts (columns) post-Tam vs. vehicle control tissue samples. log2 expression values. n = 3 biological replicates. P < 0.01. Fold change ±1.3. On the right are listed the selected genes significantly induced (red) and repressed (blue) upon genetic modification of p53. (E) Heat maps examining major signal transduction pathways as determined by GO analysis. (F) Heat map examining gene transcripts important for the regulation of the organization of the extracellular matrix and the cellular defense mechanisms against ROS.
p53 Regulates Mitochondrial Biogenesis and Bioenergetics.
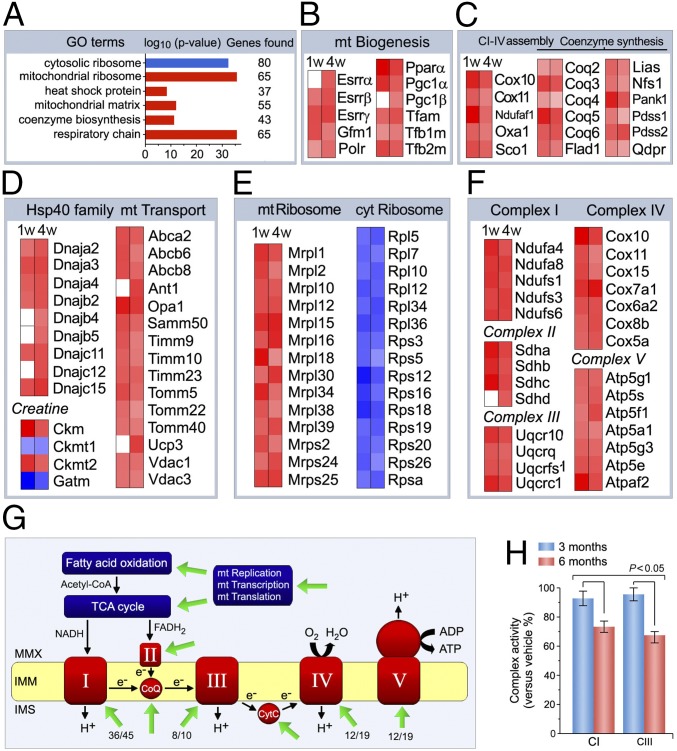
GO analysis of the p53-regulated transcriptome identified “mitochondrion organization” and “mitochondrial matrix” as enriched GO terms (Fig. 5A). With the exception of “cytosolic ribosome,” all of the gene sets contained in these GO terms were up-regulated in p53fl/fl;mcm post-Tam. Given the importance of mt ATP production for cardiac contractility, we analyzed gene expression datasets relating to mt biogenesis and capacity. Heatmap examination revealed that transcripts of mt tRNA synthesis (for example, Fars2), translation (Gfm2), transcription, and replication (EndoG, Polrmt, Tfam) were increased in p53-deficient hearts (Fig. 5B). Moreover, increased expression of the estrogen-related receptors Esrr α/β/γ, Pparα, and Pgc-1α/β were also observed in this strain. The p53-dependent regulation of these factors was unexpected as previous work has shown that mt biogenesis is controlled by the interaction of Esrr/Ppar and Nrf/Tfam with Pgc-1α, a known downstream target of Mef2a (23, 24).
Fig. 5.
p53 regulates mitochondrial biogenesis and bioenergetics. (A) GO enrichment analysis of differentially expressed transcripts encoding proteins involved in mt bioenergetics and function in p53fl/fl;mcm mice post-Tam. Blue, repressed; red, induced. (B–F) Heat maps showing enrichment of induced (red) and repressed (blue) genes involved in the regulation of mt biogenesis (B), mt electron complex assembly and coenzyme Q/C synthesis (C), mt Hsp40 homologs and mt transporters (D), ribosomal subunits (E), and respiratory chain complexes and ATP synthase subunits (F) as determined by GO analysis in A. n = 3. P < 0.01 vs. −Tam. Fold change ±1.3. (G) Potential p53 targets that map onto known interaction networks of mt biogenesis, bioenergetics, and respiratory chain function as obtained by NLP pathway examination for enriched GO gene sets in A. Green arrows indicate significantly up-regulated genes in p53KO mice post-Tam. Numbers refer to the number of genes with altered expression vs. total number of genes in the complex. CoQ, coenzyme Q10; CytC, cytochrome c; IMM, inner mt membrane; IMS, inner membrane space; MMX, mt matrix; ROS, reactive oxygen species. (H) Activity of complexes I and III in the electron transport chain. n = 4. Data are means ± SEM.
In addition, we observed increases in gene expression for complex I–V assembly, creatine kinase system, CoQ/CycC synthesis, Hsp40 proteins, mt transport, ribosome subunit composition, and subunits of complexes I–V (Fig. 5 B–F). Fig. 5G illustrates these potential p53 targets in the context of a known interaction network for regulation of mt biogenesis, bioenergetics, and respiratory chain function that was generated by NLP pathway examination for the enriched GO gene sets shown in Fig. 5A. Notably, deletion of p53 significantly reduced activities of the mt electron transfer complexes CI and CIII (Fig. 5H) post-Tam. This suggests that mt dysfunction due to impaired complex I and complex III activities is potentially a direct cause of the heart failure observed in older p53fl/fl;mcm mice post-Tam.
p53 Is Involved in the Regulation of Glucose and Fatty Acid Metabolism.
GO annotation of various mutant transcriptomes identified metabolic signatures for fatty acid (FA) and glucose utilization (Fig. 6A). Significantly increased Lkb1/Ampk1 mRNA levels, metabolic sensors promoting insulin sensitivity, and FA oxidation (FAO) (25) in p53-deficient hearts indicate an essential role for p53 in metabolic fuel sensing (Fig. 6A). Genes essential for FAO, FA transport (CD36/Lpl/Cpt1b), and FA synthesis (ACC2) were up-regulated in the absence of p53, as were the related mt pathways including pyruvate dehydrogenase complex (PDH) and TCA (26) (Fig. 6A). Glucose transporter Glut4 and 9 of the 11 steps to glycolysis were up-regulated after p53 ablation. Intriguingly, Key factors in the pentose phosphate pathway (PPP) were significantly reduced in p53fl/fl;mcm mice post-Tam versus vehicle-injected controls. Taken together, central metabolic pathways were deregulated as a consequence of perturbed p53 regulation during development of the mutant cardiac phenotype.
Fig. 6.
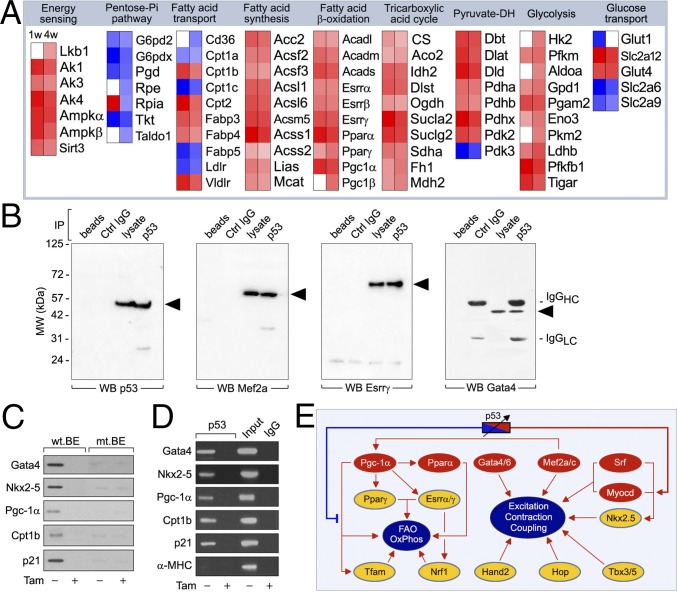
p53 forms a major transcription factor hub in a metabolic network regulating fatty acid and glucose utilization. (A) Heat maps to identify p53 target genes associated with regulation of FA and glucose metabolism. Depicted are regulation of normalized signals of enriched genes (rows) in mutant strains (columns). PPP, FAO, FA transport and storage, FAS, PDH), tricarboxylic acid cycle (TCA = citric acid cycle). n = 3. P < 0.01. Fold change ± 1.3. The numbers in the PPP, TCA and glycolysis represent the different enzymatic steps. (B) Immunoprecipitation of LV derived from 14-wk-old wild-type mice with antibodies specific to p53 and immunoblotting with either anti-Mef2a, Esrrγ, and Gata4 or antibodies. IP, immunoprecipitation. WB, Western blot. One result of two independent experiments is shown. (C) Binding of endogenous p53 to biotinylated double-stranded wild-type binding element (wt.BE) oligonucleotide probes derived from promoters as indicated (left), which is diminished by specific mutations (mt) in mt.BE. Determination of the presence of p53 in the precipitate was analyzed by immunoblotting with antibodies to p53. p53fl/fl;mcm mice were used. (D) ChIP analysis was performed with p53-specific antibodies (Top). PCRs with primers specific to promoter regions harboring the p53-binding element as indicated (left). Cptb1 (carnitine palmitoyltransferase 1B, muscle) and p21 primers were used in positive control reactions. Primers specific for the α-MHC gene promoter served as an unresponsive control. (E) p53 exerts a rheostat-like function to adjust global gene expression in the cardiac transcriptome in this network prediction. Red represents top-level TFs; yellow represents midlevel TFs.
p53 Regulates Process-Specific Transcription Factors.
Loss of p53 also resulted in activation of process-specific TFs (Mef2a, Myocd, Pgc-1α, Tfam). TFs bind in a combinatorial fashion to specify the on-and-off states of genes (27). In determining whether p53 can exert such control, we performed coimmunoprecipitation of adult cardiac extracts (Fig. 6B). The presence of Esrrγ, Gata4, and Mef2a in p53 immunoprecipitates indicates complex formation between p53 and process-specific TFs. In oligonucleotide precipitation experiments and PCR chromatin immunoprecipitation (ChIP) assays, endogenous p53 protein specifically bound to these promoters, in addition to Cpt1b (mt FA transporter) and p21 (Fig. 6 C and D).
The promoter regions of Gata4, Nkx-2.5, and Pgc-1α contain consensus DNA-binding sites for p53. Therefore, we investigated the connection between p53 expression and transrepression of Gata4, Nkx-2.5, and Pgc-1α gene promoters by oligonucleotide precipitation using left ventricular (LV) extracts prepared from p53fl/fl;mcm mice (Fig. 6C). As positive controls, we included the Cpt1b and p21 gene promoters in this analysis. The p53-binding element (BE) in the Pgc-1α, Cpt1b, and p21 gene promoters is conserved in the human and mouse promoters. As synthetic oligonucleotides, their p53-binding regions bound to endogenous p53 protein in the absence of Tam. Importantly, Tam treatment eliminated the induction of p53 binding to all probes (Fig. 6C). This effect was specific because mutation of the p53 site abrogated p53 binding to Pgc-1α, Cpt1b, and p21 promoter constructs (Fig. 6C).
Next, we used ChIP assays to analyze Pgc-1α, Cpt1b, and p21 gene-promoter occupancy by p53. We observed that p53 specifically bound to contiguous sites in these promoters in samples prepared from hearts of vehicle-treated p53fl/fl;mcm mice, reflecting p53’s repressive impact in vivo (Fig. 6D). In contrast, such an effect was not observed in extracts from Tam-treated p53fl/fl;mcm littermates, confirming that this process requires p53. The specificity of our ChIP assay was confirmed with primers annealing to the α-myosin heavy chain promoter because transcription of this gene is not thought to be under the control of p53 (Fig. 6D). Thus, significant levels of α-myosin heavy chain were never amplified. We infer from these results that cardiac p53 is recruited to the Pgc-1α, Cpt1b, and p21 gene promoters and that this cellular response is dependent on Tam. A model consistent with our findings is depicted in Fig. 6E.
Discussion
Fundamental to biological systems is the presence of TF networks with sequence specificity (27). In this paper, we demonstrate that p53 functions as a master regulator of such an intricate network to maintain cardiac tissue homeostasis. In the classical view, the default position for the p53 network is “off” whereby p53 is inactive until induced by acute genotoxic or oncogenic stress (3). Activated p53 promotes apoptosis, senescence, and DNA repair, activities critical for tumor suppression. Here, we investigated the role of the p53 circuitry in the regulation of cardiac hypertrophy. Our results demonstrate that deletion of p53 was sufficient to trigger the development of spontaneous pathological hypertrophy in older mice (Fig. 1). Intriguingly, ablation of p53 provided protection against biochemical stress (Fig. 2). These remarkable effects were tightly coupled to the activation of beneficial gene sets, including proteins involved in excitation-contraction coupling, energy metabolism, and the oxidative stress response with the inhibition of hypertrophic signaling and apoptosis (Figs. 4–6). Drugs that transiently suppress cardiac p53 and modulate these pathways would be of clinical importance. Our analysis clearly demonstrates that p53 operates within a complementary network that allows other major cardiac transcription factors to have opposite and even independent transcriptional effects that ultimately determine the observed phenotype. Our data also support the role of other p53-related factors, specifically p63/p73, in the regulation of the transcriptional network, and this should be analyzed further.
Recent evidence suggests that p53 may also regulate certain aspects of cell metabolism, such as oxidative phosphorylation and glycolysis, in a cell- and context-specific manner. These include the transcriptional activation of Sco2, COXII, p52R2, GLUT1/4, Hk2, Pgm, and Tigar (28). In contrast, based on the data presented in our study, most of the genes encoding main regulators and key enzymes of metabolic processes [OxPhos, FAO, FA synthesis (FAS), glycolysis, PPP] are extremely sensitive to alterations in p53 protein levels. Overall, these findings indicate that the p53 network is able to dynamically balance between energy demand, fuel uptake, and metabolism, thus providing an explanation for the critical importance of stable cardiac function in various hemodynamic settings.
The most exciting observation from our study is the high degree of connectivity of the p53 network hub with defined nodes (Mef2a/Myocd/Esrr/Pgc-1α). Studies in animal models have linked several master TFs (Gata4, c-Myc, Nfat3, NF-κB) to the induction of pathological gene sets that leads to the development of HF (14, 29–31). Moreover, physical exercise also highly influences cardiac function through increases in expression of these specific gene sets (32). Based on these findings, we envision a rheostat-like role for p53 whereby environmental cues induce small, but highly significant, changes in the p53/Mdm2 circuitry that are transformed to broad, pleiotropic output signals that maintain the overall temporal stability of cardiac performance (Fig. 6E).
Cardiovascular disease is likely a multifactorial phenomenon and should not be studied in a monogenic fashion through the analysis of individual factors in isolation. Although single-gene or single-factor–based approaches have uncovered many of the individual components of biological systems, a systems-based approach is required to integrate all factors relevant to the underlying pathology in an effort to understand the dynamic nature of a system with all of the individual parts studied in context with each other (33). A systems biology approach has begun to successfully complement the single-gene–focused biology. From this type of analysis, it is clear that gene expression is regulated by specific sets of transcription factors. Specifically, normal adult heart function is maintained by a core set of transcription factors (Gata, Mef2, Nfat, Nkx2.5) that control expression of cardiac-specific genes, known as a transcriptional signature (34, 35). The molecular mechanism coordinating these transcriptional profiles is not well understood in heart disease. Therefore, understanding the biological network underlying transcriptional alterations in heart disease will enable us to better define the interdependency of molecular mechanisms that coordinate these gene expression patterns (36).
In human and mouse, it is thought that more than 2,000 TFs modulate the mRNA profiles corresponding to 23,000 genes (33). Major insights have been gained into the regulation of the transcription process by DNA-binding TFs (37–39). However, there is a lack of data analyzing the interaction between TFs at the level of combined transcriptional regulation, which is the main driving force of maintenance of tissue homeostasis. In line with this, we observed a panel of cardiac- or process-specific TFs that regulate gene expression together with p53, rather than p53 functioning in isolation. Our genetic data suggest that there is a high degree of interdependency between TFs at different levels of cellular organization. In addition, our data also underline the high potency of compensatory regulation between TFs and clearly demonstrate that primarily unrelated TFs share common targets in the adult heart. We identified different nodes of the regulatory p53 network that exhibited a high degree of interdependency. Thus, these TFs all have the potential to modulate each other and should therefore be viewed in a tissue-dependent context. Further experimentation will provide a deeper mechanistic insight into the evaluation of the regulatory circuits involved in this biological process. It will be of interest to study how the interdependency of different TF factors stabilizes the overall function of given networks, a role of p53 that has only just begun to reveal itself. Another emerging question is how would the activity of p53 protect against external stressors in addition to providing a potential avenue for novel therapeutic drug development. Our study defines a physiologic role for the p53 pathway in differentiated CM that appears to be equally important to its well-accepted tumor suppressive function.
Materials and Methods
All animal use in this study was in accordance with approved institutional animal care guidelines of the University Health Network (AUP 1815/1379, Canadian Council in Animal Care). Age-matched syngeneic adult male mice (12–13 wk old; 22–27 g body weight) were used in this study. All experiments used vehicle-injected controls of matched age and sex. Functional analysis was done at various time points and included echocardiographic, biochemical, histological, and immunofluorescent assessment. Details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by grants awarded by the Canadian Institute of Health Research (to F.B.). F.B. is the recipient of the Canadian Institute of Health Research Phase II Clinician-Scientist Award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621436114/-/DCSupplemental.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: Alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8(15):1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 5.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 6.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 7.Rubart M, Field LJ. Cardiac regeneration: Repopulating the heart. Annu Rev Physiol. 2006;68:29–49. doi: 10.1146/annurev.physiol.68.040104.124530. [DOI] [PubMed] [Google Scholar]
- 8.MacLellan WR, Schneider MD. Genetic dissection of cardiac growth control pathways. Annu Rev Physiol. 2000;62:289–319. doi: 10.1146/annurev.physiol.62.1.289. [DOI] [PubMed] [Google Scholar]
- 9.MacLellan WR, et al. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol. 2005;25(6):2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poolman RA, Li JM, Durand B, Brooks G. Altered expression of cell cycle proteins and prolonged duration of cardiac myocyte hyperplasia in p27KIP1 knockout mice. Circ Res. 1999;85(2):117–127. doi: 10.1161/01.res.85.2.117. [DOI] [PubMed] [Google Scholar]
- 11.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271(5 Pt 2):H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 12.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246(1):14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 13.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. N Engl J Med. 1999;341(17):1276–1283. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 14.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: Pathophysiology and therapy. Circulation. 2000;101(25):2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 15.Song H, Conte JV, Jr, Foster AH, McLaughlin JS, Wei C. Increased p53 protein expression in human failing myocardium. J Heart Lung Transplant. 1999;18(8):744–749. doi: 10.1016/s1053-2498(98)00039-4. [DOI] [PubMed] [Google Scholar]
- 16.Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest. 2005;115(3):500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puente BN, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157(3):565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11(12):1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 20.Martin K, et al. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375(6533):691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 21.Xiao ZX, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375(6533):694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 22.Sano M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446(7134):444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 23.Arany Z, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1(4):259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Naya FJ, et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8(11):1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 25.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85(3):1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 27.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2(4):a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Berlo JH, Elrod JW, Aronow BJ, Pu WT, Molkentin JD. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc Natl Acad Sci USA. 2011;108(30):12331–12336. doi: 10.1073/pnas.1104499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong W, et al. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25(16):3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier HJ, et al. Cardiomyocyte-specific IκB kinase (IKK)/NF-κB activation induces reversible inflammatory cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2012;109(29):11794–11799. doi: 10.1073/pnas.1116584109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boström P, et al. C/EBPβ controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143(7):1072–1083. doi: 10.1016/j.cell.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer S, et al. Translational regulation shapes the molecular landscape of complex disease phenotypes. Nat Commun. 2015;6:7200. doi: 10.1038/ncomms8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger J, et al. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toenjes M, et al. Prediction of cardiac transcription networks based on molecular data and complex clinical phenotypes. Mol Biosyst. 2008;4(6):589–598. doi: 10.1039/b800207j. [DOI] [PubMed] [Google Scholar]
- 36.Herrer I, et al. Gene expression network analysis reveals new transcriptional regulators as novel factors in human ischemic cardiomyopathy. BMC Med Genomics. 2015;8:14. doi: 10.1186/s12920-015-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birney E, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10(9):605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.