Significance
Enzymes find utility as therapeutics and for the production of specialty chemicals. Changing the amino acid sequence of an enzyme can increase solubility, but many such mutations disrupt catalytic activity. To evaluate this trade-off, we developed an experimental system to evaluate the relative solubility for nearly all possible single point mutants for two model enzymes. We find that the tendency for a given solubility-enhancing mutation to disrupt catalytic activity depends, among other factors, on how far the position is from the catalytic active site and whether that mutation has been sampled during evolution. We develop predictive models to identify mutations that enhance solubility without disrupting activity with an accuracy of 90%. These results have biotechnological applications.
Keywords: protein solubility, high-throughput screening, fitness landscapes, yeast surface display, deep mutational scanning
Abstract
Proteins are marginally stable, and an understanding of the sequence determinants for improved protein solubility is highly desired. For enzymes, it is well known that many mutations that increase protein solubility decrease catalytic activity. These competing effects frustrate efforts to design and engineer stable, active enzymes without laborious high-throughput activity screens. To address the trade-off between enzyme solubility and activity, we performed deep mutational scanning using two different screens/selections that purport to gauge protein solubility for two full-length enzymes. We assayed a TEM-1 beta-lactamase variant and levoglucosan kinase (LGK) using yeast surface display (YSD) screening and a twin-arginine translocation pathway selection. We then compared these scans with published experimental fitness landscapes. Results from the YSD screen could explain 37% of the variance in the fitness landscapes for one enzyme. Five percent to 10% of all single missense mutations improve solubility, matching theoretical predictions of global protein stability. For a given solubility-enhancing mutation, the probability that it would retain wild-type fitness was correlated with evolutionary conservation and distance to active site, and anticorrelated with contact number. Hybrid classification models were developed that could predict solubility-enhancing mutations that maintain wild-type fitness with an accuracy of 90%. The downside of using such classification models is the removal of rare mutations that improve both fitness and solubility. To reveal the biophysical basis of enhanced protein solubility and function, we determined the crystallographic structure of one such LGK mutant. Beyond fundamental insights into trade-offs between stability and activity, these results have potential biotechnological applications.
Solubility is a fundamental biophysical property of proteins. In this work, we use the term solubility to refer to the probability that a protein is properly folded upon translation. Soluble expression is known to be a function of protein aggregation propensity, thermodynamic stability, and folding rate, among other factors. Understanding the distribution of solubility-modulating mutations can sharpen biophysical models undergirding evolutionary theories combining molecular evolution and population genetics. There are also a number of biotechnological applications: Improving protein solubility can enhance the total turnover number for biocatalysts, increase expression yield of enzymes needed in biomanufacturing, or bolster formulation stability of therapeutic proteins. From this applied perspective, general approaches are desired to identify mutations that improve the solubility of a protein while maintaining function.
Computational approaches have been used to evaluate and design thermodynamic stability (1) and aggregation propensity (2, 3). There also exist several high-throughput experimental screens that can be used to increase soluble protein expression and protein solubility (4–7). In the specific case of enzymes, a major challenge for the above approaches is that solubility-enhancing mutations often have lower specific activity (8, 9). Additionally, because the stabilizing effect of most beneficial mutations is modest (3), many mutations from the starting sequence are typically needed to increase solubility over wild type. Together, these facts necessitate running a secondary screen for activity for positive hits from the solubility screen (10), increasing time and effort.
Comprehensive evaluation of the trade-off between enzyme activity and solubility could identify classifiers used to predict whether a given solubility-enhancing mutation is deleterious for enzyme activity. For example, earlier directed evolution experiments have shown that solubility-enhancing mutations are enriched at or near active site residues; most such mutations are deleterious for activity (11, 12). Additionally, a powerful enzyme engineering approach is to choose only those mutations that have been oversampled in the evolutionary history of the protein family (13–15). This “back-to-consensus” strategy rests on the supposition that mutations to the consensus sequence of the protein family maintain enzyme function and improve stability. Other classifiers beyond the above two may exist.
In this work, we used deep mutational scanning (16, 17) to assess the sequence determinants of solubility for two different full-length enzymes. We chose enzymes with known fitness landscapes using experimentally derived functional selections (8, 17), allowing a direct comparison between protein fitness and solubility. We assessed two existing complementary in vivo high-throughput solubility screens/selections for the ability to identify mutations that confer solubility. For one enzyme, the fraction of solubility-enhancing mutations was between 4% and 5%, which is in line with theoretical predictions (18). We also identified several limitations in commonly used in vivo screens, which should help limit false-positive and false-negative results in high-throughput datasets. Mutations that improve solubility without impacting fitness can be identified with an accuracy of 90% using classifiers that do not require a high-resolution protein structure or homology model. We also solved the structure of a Pareto optimal enzyme variant to show the biophysical basis of enhanced solubility and function. Together, these results provide experimental illumination of the trade-off between enzyme solubility and function, as well as a means by which active, stable mutants can be uncovered without a high-throughput activity screen.
Results
Deep Mutational Scanning for Solubility.
We performed deep mutational scanning on two full-length enzymes: a 263-residue TEM-1 beta-lactamase (BLA) variant and a 439-residue levoglucosan kinase (LGK) using different in vivo high-throughput solubility screens/selections (Fig. 1). For TEM-1 BLA, we abolished catalytic activity by mutating the active site residue Ser70 to Ala because one selection involves growth on beta-lactam antibiotics. We also introduced the destabilizing mutation D179G (12, 19) because TEM-1 is stable at the selection and screening temperature of 30–37 °C. We refer to this resulting construct (TEM-1 S70A, D179G) as TEM-1.1 in the remainder of this work. In vitro, His-tagged TEM-1.1 had an apparent melting temperature (Tm) of 41.8 ± 0.3 °C, which is less stable than TEM-1 [Tm = 51.5 °C (12)] as expected. Comprehensive single-site saturation mutagenesis libraries were constructed in all genetic backgrounds using nicking mutagenesis (20), and libraries were harvested, prepared, and deep-sequenced in a standardized pipeline (21) (SI Appendix, Fig. S1).
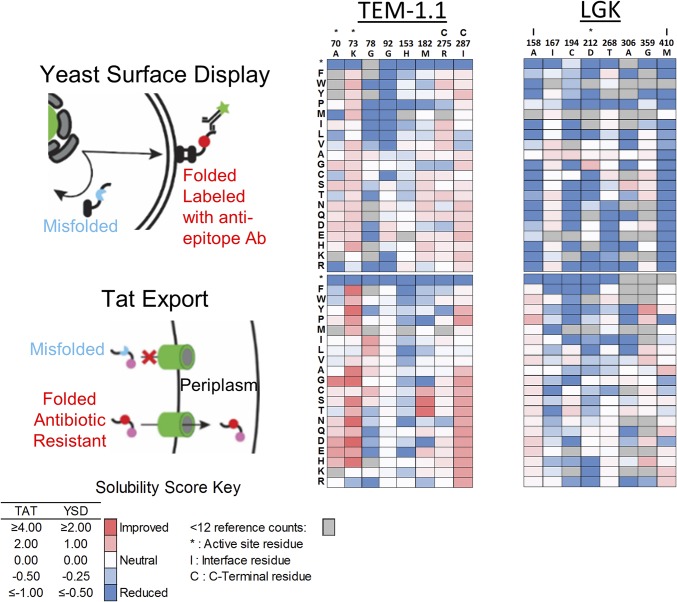
Fig. 1.
Overview of solubility deep mutational scans for TEM-1.1 and LGK. (Left) Screens used in the present work. In YSD, the protein is exported to the surface and labeled by a fluorescent antibody that is specific for a C-terminal epitope tag. The top 5% of cells by fluorescence intensity are collected by FACS. For Tat export, a protein is fused to a C-terminal beta-lactamase that requires periplasm localization for activity. Variants are selected on plates containing high antibiotic concentrations. (Center and Right) Heat maps of solubility scores for selected residues of TEM-1.1 and LGK. Residues in the active site are indicated by (*), interface by (I), and proximal to the C terminus by (C).
We tested three previously developed screens/selections that purport to gauge protein solubility (Fig. 1). In yeast surface display (YSD) (22), proteins are fused in-frame with a C-terminal epitope tag and an N-terminal Aga2p domain that localizes the fusion protein to the outer cell surface. Before successful display on the yeast surface, nascent proteins are subject to the endoplasmic reticulum quality-control system. Misfolded proteins are marked for degradation and subsequently destroyed by proteasomes (6). Binding with a fluorescently conjugated antiepitope antibody allows discrimination of variants that express on the cell surface from ones that cannot. We used fluorescence-activated cell sorting (FACS) to collect a reference population of all yeast and the top 5% of displaying population determined by fluorescence intensity. Sort statistics are given in SI Appendix, Table S1.
We also performed a genetic selection for solubility based on twin-arginine translocation (Tat)–selective export of “folded” proteins into the bacterial periplasm (23). A protein of interest is fused in-frame with an N-terminal ssTorA Tat periplasmic export signal peptide and a C-terminal TEM-1 BLA with a deleted Sec export signal sequence (23). Escherichia coli expressing this fusion protein will survive in the presence of beta-lactam antibiotics if the fusion protein is present in the periplasm, because TEM-1 BLA activity is dependent on the formation of a disulfide bond. Therefore, cells producing folded soluble proteins will permit growth on ampicillin plates. This selection has been applied to enzymes (10), amyloid beta (23), and multiple other proteins (7). We prepared a selection plasmid using a codon-swapped C-terminal Δ4–25 TEM-1 designed to minimize recombination during experiments with TEM-1.1.
There was, on average, 93.2% [4,929 of 5,260 (TEM-1.1) and 7,945 of 8,560 (LGK)] coverage of single nonsynonymous mutations identified across all libraries. A total of 5,466 (67.2%) single amino acid mutations were present in both LGK screens, whereas 3,690 (73.8%) single amino acid substitutions were shared in both TEM-1 screens. Enrichment ratios calculated from deep sequencing were converted to a solubility score centered about a wild-type score of 0. The per-position scores are visualized in heat maps shown in Fig. 1 and SI Appendix, Figs. S2–S5. Detailed statistics for each deep mutational scan are provided in SI Appendix, Tables S2 and S3.
Validation of Solubility Datasets.
We performed a number of checks on the quality of the resulting solubility datasets. A major source of error in deep mutational scanning experiments is in the frequency calculation of each member of the library (8, 21, 24). This calculation involves counting members of a population a discrete number of times, with larger errors found for those variants underrepresented in the population either through low abundance in the unselected population or through depletion during selection. To evaluate errors in our deep mutational scanning experiments, we performed experimental replicates for a subset of the full datasets for each screen and enzyme combination (SI Appendix, Fig. S6). Consistent with the above error predictions, the correlation coefficient between replicates was above 0.85 for all data and above 0.87 for variants represented above 100 times in the unselected population (SI Appendix, Fig. S6). Another means to evaluate error is by comparing the solubility score of synonymous mutations with the wild-type sequence, operating under the assumption that mutations at the nucleic acid level do not impact solubility (17). We note that this simplification is an overestimate for the error in the method, because replicates of synonymous mutations show that up to 20% of the variance cannot be explained by noise (SI Appendix, Fig. S7). Nevertheless, the distributions of synonymous mutations can be fit with Gaussian distributions, with a mean centered on a solubility score of 0, with an SD ranging from 0.18 for TEM-1.1 YSD to 0.43 for the LGK Tat genetic selection dataset (SI Appendix, Fig. S8 and Table S4). Furthermore, for all datasets, the SD decreased with increasing depth of coverage, as expected (SI Appendix, Fig. S8).
Next, we evaluated the ability of the screens to select for higher solubility variants and to deplete low-solubility mutants in several ways. First, all five solubility datasets showed statistically significant reductions in nonsense compared with missense mutations (P < 0.0001; Fig. 2A and SI Appendix, Fig. S9). Second, the fraction of residues tolerated at each position was negatively correlated with contact number (average number of neighboring residues, which is a measure of packing density) (25) for LGK and a subset of the TEM-1.1 datasets [positions 61–215 using the Ambler sequence convention (justification is provided below)] (Fig. 2B and SI Appendix, Fig. S10).
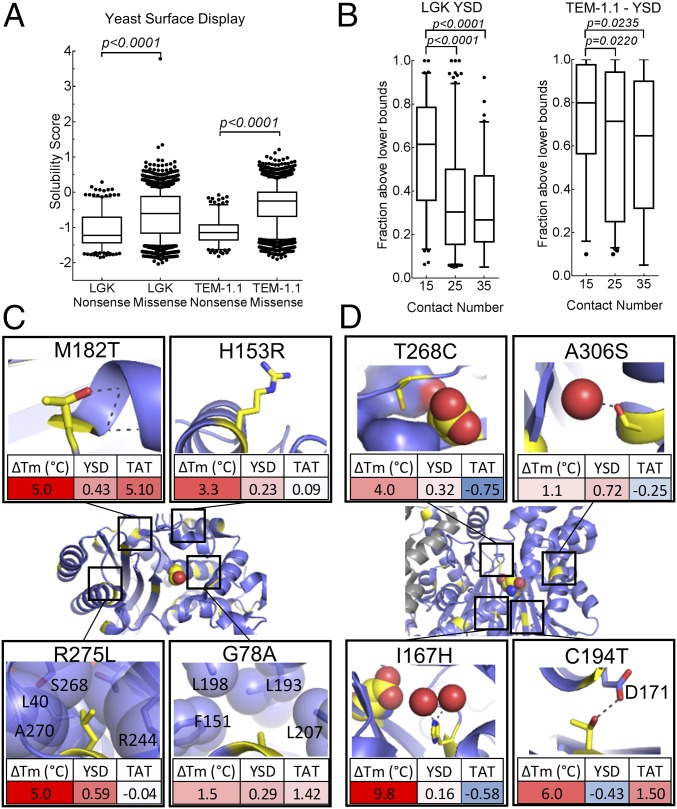
Fig. 2.
Validation of solubility datasets. (A) Nonsense vs. missense solubility scores for YSD (LGK, TEM-1.1). (B) Fraction of beneficial mutations above the lower bounds versus contact number for LGK and TEM-1.1 (residues 61–215). Known stabilizing mutations (yellow) are mapped onto TEM-1.1 (C; PDB ID code 1M40) and LGK (D; PDB ID code 4ZLU). (Insets) Structural basis of the stabilizing mutations, shown as yellow sticks, along with the corresponding solubility scores identified by deep sequencing.
We also reasoned that there would be correlation between these solubility scores and existing deep mutational scanning fitness datasets for LGK (8) and TEM-1 (17), because enzyme fitness is subject to the biophysical constraints of folding. The correlations between solubility and fitness datasets were statistically significant (P < 10−17 in all cases), with correlation coefficients ranging from 0.61 (LGK) and 0.45 (positions 61–215 for TEM-1.1) for YSD datasets down to 0.22 (positions 61–215 for TEM-1.1) for the Tat genetic selection (SI Appendix, Figs. S11 and S12 and Table S5).
We evaluated the ability of the solubility deep mutational scans to identify known stabilizing mutations in TEM-1 (Fig. 2C and SI Appendix, Table S6) and LGK (Fig. 2D and SI Appendix, Table S7) that are located at the surface and core (and at the homodimer interface for LGK). These mutations were previously shown to rescue enzyme solubility in the context of other destabilizing mutations, and all had an in vitro-characterized change in melting temperature (ΔTm) ≥1 °C in the parental background. We identified a mutation as solubility-enhancing if its solubility score was above 0.15; for screens using FACS, this value corresponds to a mean fluorescence intensity of 10% above the wild-type sequence. For TEM-1, 15 of 19 mutations were recorded as solubility-enhancing in the YSD dataset (Fisher’s exact test, P = 2.9 × 10−10). For the LGK datasets, YSD identified six of 11 solubility-enhancing mutations (P = 3.2 × 10−6). Changing the threshold for identifying solubility-enhancing mutations did not alter the significance of the results, except for the most stringent threshold for the YSD LGK dataset (SI Appendix, Table S8). Five of six of the false-negative results from YSD were just below the 0.15 cutoff used. The notable exception was LGK C194T, which had a very low YSD solubility score (Fig. 2D). There is an ASN at position 192 that is surface-exposed but in the catalytic active site, introduction of THR194 introduces a potential N-linked glycosylation site. We speculate that this aberrant glycosylation at the active site results in misfolded protein that would be retained in the endoplasmic reticulum. We conclude that YSD and GFP fusion solubility screens are able to identify gain of thermodynamic stability variants.
By contrast, in our hands, the Tat selection identifies five of 17 (P = 0.36) and one of 12 (P = 0.20) known stabilizing mutations for TEM-1 and LGK, respectively, although we note that the very stabilizing mutations TEM-1 M182T (ΔTm = 5 °C) and LGK C194T (ΔTm = 6 °C) were strongly enriched. The inability of the Tat screen to enrich known stability-enhancing mutations may reflect that the selection criteria for Tat export are not dominated by the effect of thermal stability on protein solubility.
Distribution of Solubility Scores.
What is the distribution of solubility scores for the two enzymes? Here, we restricted our analysis to YSD because of the number of false-negative results observed for the Tat pathway selection. The distributions for LGK and TEM-1.1 are multimodal, with the mean value below the wild-type solubility score of 0 (Fig. 3A). For the LGK dataset, 4.5% of possible single missense mutations were above a solubility score of 0.15. However, 14.5% of mutations were identified in the TEM-1.1 dataset using the same criteria. The numbers reported above may overestimate the percentage of solubility-enhancing mutations if the number of false-positive results outweighs the number of false-negative results, and vice versa.
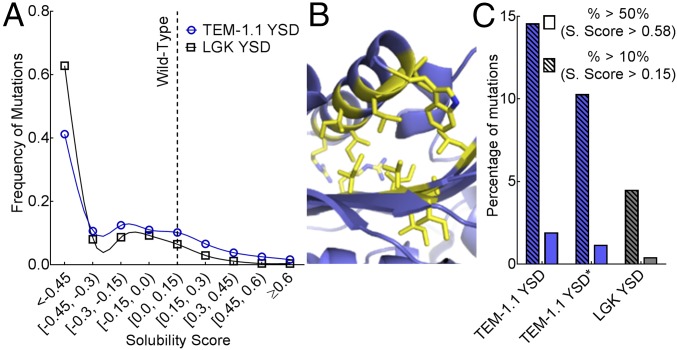
Fig. 3.
Distribution of solubility-enhancing mutations. (A) Frequency of mutations for TEM-1.1 YSD (blue) and LGK YSD (black) found at each solubility score. Each dataset is fit with a cubic spline to help guide the eye. (B) Positions with more than 10 beneficial mutations in the TEM-1.1 YSD dataset are shown as yellow sticks. These false-positive results are predicted to disrupt the C-terminal helix, presumably to promote accessibility of the c-myc epitope tag. (C) Percentage of mutations with solubility scores above a 10% (hatched fill) and 50% (solid fill) increase in function for TEM-1.1 YSD (blue) and LGK YSD (gray). TEM-1.1 YSD* covers residues 61–215 to remove the section with false-positive results indicated in B.
Because we expected to find a similar distribution of solubility-enhancing mutations between proteins, we diagnosed potential problems with the screens. In YSD, most positions that allow 10 or more of these substitutions map to the C terminus (Fig. 3B). Because the N and C termini are so close to one another, and we did not include a linker region between the C terminus and the myc epitope tag, we speculate that mutations enriched from this screen destabilized the helix positioning to avoid steric clashes between the anti-myc antibody and either the N-terminal Aga2p or TEM-1.1. Thus, the YSD screen rewards mutations that enhance antibody binding at the expense of core destabilization. Restricting analysis to portions of the protein not affected by this set of false-positive results (Ambler positions 61–215) results in 10.3% of solubility-enhancing mutations, which is closer to the solubility score distributions found in the LGK dataset (Fig. 3C).
Restricting mutational search space to variants represented in the evolutionary history of the enzyme family is a proven stabilization strategy in protein engineering (back-to-consensus) (13–15). We asked for the proportion of solubility and fitness-maintaining hits that could be uncovered by back-to-consensus using previously published near-comprehensive experimental fitness landscapes for TEM-1 (17) and a thermally stabilized LGK (8). Under the selection conditions used in the above experiments, the fitness (W) can be restated as a product of the catalytic activity or function level of enzyme (f) and solubility or amount of active enzyme in the cell ([E]):
| [1] |
In both cases, experimental fitness landscapes were determined for enzyme variants stable in their genetic background; that is, although there are mutations that can further stabilize an enzyme, such mutations would not increase the amount of active enzyme in the cell (“solubility”). Thus, most neutral or beneficial mutations maintain similar catalytic efficiencies to wild type. We first classified the experimental fitness values into neutral (≥80% of wild type), slightly deleterious (>50% and <80%), and deleterious (<50%) bins. It is important to note that the “neutral” bin also includes those mutations that improve fitness. The tolerance for a mutation at a given position as determined by evolutionary history was evaluated using a position-specific scoring matrix (PSSM). Whereas 32% of all TEM-1 mutations are neutral, 69% of mutations with a PSSM score ≥3 were neutral (SI Appendix, Table S9). For LGK, 28% of all mutations were neutral but 57% of conserved (PSSM ≥ 3) mutations were neutral (SI Appendix, Table S9). Using a less restrictive cutoff (PSSM ≥ 0) does not appreciably change the findings. Although these results suggest that including evolutionary history increases the probability of a hit, we also note that the probability of an evolutionary sampled deleterious or moderately deleterious mutation is 31% (TEM-1) to 43% (LGK) (SI Appendix, Table S9). Thus, choosing mutations solely through evolutionary conservation is insufficient to engineer stable, active enzymes without a secondary activity screen.
Mutations That Enhance Solubility and Fitness Are Rare.
It is well known that mutations that enhance catalytic activity are, on average, destabilizing. We asked what is the percentage of mutations that enhance solubility in addition to fitness (here taken as correlative with catalytic activity). Of 33 LGK mutations with a statistically significant increase in fitness, only seven have an YSD solubility score above 0.15. Similarly, of 28 TEM-1 beneficial mutations, eight have an YSD solubility score greater than 0.15. Thus, the odds that a given mutation enhances stability and fitness is between 0.05% and 0.15%.
Classification Methods Improve Chances of Finding Soluble, Active Enzyme Variants.
Many solubility-enhancing mutations decrease enzyme-specific activity. For example, it is well known that catalytically active residues are poorly optimized for solubility (8, 11, 12). Additionally, false-positive results like the results observed in the TEM-1.1 datasets are often deleterious for fitness. Thus, additional metrics are needed to identify mutations that impart solubility and do not decrease activity.
For this analysis, we used published near-comprehensive experimental fitness landscapes for TEM-1 (17) and a thermally stabilized LGK (8) using the same classification bins (neutral, slightly deleterious, and deleterious) as above. For the datasets developed in this work, the probability of finding a deleterious mutation among the list of solubility-enriched variants ranges between 15% (LGK-YSD) and 55% (TEM-1.1–YSD) (Fig. 4 A and B). To improve our chances of finding solubility-enhancing mutations of neutral fitness, we assessed mutations according to size, chemical type, contact number of the original residue, distance to active site as determined by Cα distance to the nearest active site ligand, and evolutionary conservation as quantified by PSSM.
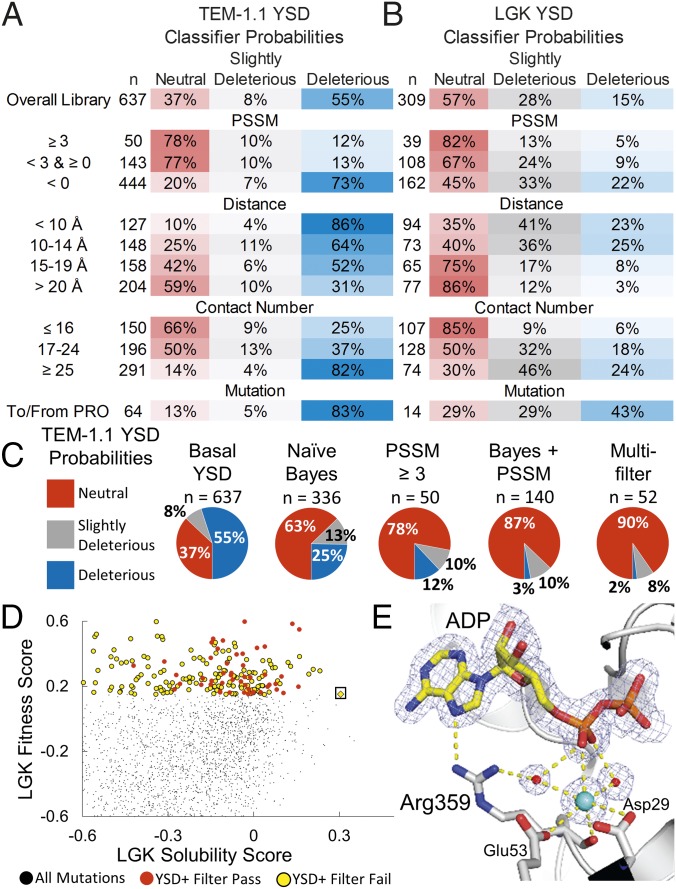
Fig. 4.
Classification methods improve probabilities of selecting mutations conferring solubility and activity but remove rare, globally optimal mutations. Classifier probabilities for YSD deep mutational scan for TEM-1.1 (A) and LGK (B). The total number of mutations found in a given bin (n) is provided, and the PSSM represents the site-specific preferences found in the evolutionary history of the enzyme. (C) Classification methods improve probabilities of selecting neutral mutations. (D) LGK fitness versus the LGK solubility score of individual mutations. Beneficial mutations from the YSD screen are shown as circles colored by whether they pass (red) or fail (yellow) the multiple-filter classification method. The Pareto optimal mutation G359R (boxed) fails the filtering due to its close distance to the active site, low evolutionary conservation, and high contact number. (E) Crystal structure of LGK G359R (PDB ID code 5TKR). G359R makes direct and water-mediated hydrogen bonds with ADP near the active site. A potassium ion also appears to be coordinated in this region, possibly contributing to the stability of the enzyme. Carbon atoms are shown in gray and yellow for the protein and ligand atoms, respectively. Nitrogen, oxygen, and phosphorous atoms are shown in blue, red, and orange, respectively. Waters and the potassium are shown as red and cyan spheres, respectively. The 2mFo-DFc electron density map is contoured to 1σ. For clarity, the magnesiums in the active site have been omitted from the figure.
We first addressed whether PSSM improves the identification of active mutations. Solubility-enhancing mutations with a PSSM score ≥3 are more likely to maintain fitness and are less likely to be deleterious (Fig. 4 A and B). For example, only 12% of solubility-enhancing mutations observed from the TEM-1 YSD datasets are deleterious with regard to fitness. In contrast, nonconserved solubility-enhancing mutations are likely to be deleterious for fitness, with probabilities ranging from 22% for LGK-YSD to 73% for TEM-1.1–YSD.
Other classifiers yielded similar results. For example, distance to active site was correlated with increasing the probability of finding a neutral mutation, whereas contact number was anticorrelated (Fig. 4 A and B). However, mutations sorted by size and chemical type did not show improvements in classifications across all selections (SI Appendix, Table S10), with the exception of mutations to or from proline, which are generally disfavored.
Next, we tested different classification methods to improve the chances of finding solubility-enhancing mutations conferring neutral fitness. We looked at filtering, naive Bayes classification, and a hybrid method combining filtering on PSSM score (≥3) with Bayes analysis on the remaining classifiers (Fig. 4C and SI Appendix, Table S11). One filter was strictly on PSSM score, whereas multiple filtering included PSSM score (≥0), distance to active site (≥15 Å), contact number (≤16), and no mutations involving a proline. The multiple-filter classification performed best in all three datasets: For the YSD datasets, the probability of finding a neutral mutation is 90%, with only a 2% (TEM-1.1) or 3% (LGK) chance of uncovering a deleterious mutation. The hybrid Bayes method was next, with a 77–87% chance of finding a neutral mutation and a 3–4% chance of a deleterious mutation for the YSD datasets. However, increased accuracy using multiple filtering is at the expense of number of mutations identified, because the hybrid Bayes method identified approximately threefold more mutations than the multiple-filtering method (mean of 130 versus 43 for filtering).
The trade-off with strict filtering is the removal of rare, globally beneficial mutations. This balance can best be visualized by plotting the fitness values of individual variants versus their respective solubility score (Fig. 4D). In this case, the multiple-filtering method removes the Pareto optimal mutation (26), G359R, that is enriched in all four datasets (Fig. 4D). In vitro, LGK G359R shows an increased ΔTm of 1.1 °C and improves the kcat over wild type by ∼60% (8). G359R fails the filtering because it is not evolutionarily conserved, it is close to the ATP binding cleft, and Gly359 is in a relatively packed portion of the protein. To evaluate the structural basis for this mutation, we solved the structure of this mutant in the presence of ADP and magnesium (Fig. 4E) (Protein Data Bank ID code 5TKR). The additional hydrogen-bonding interactions from the Arg359 side chain to the nucleotide in the active site may lead to stronger binding of ATP during the catalytic reaction and possibly have polarizing effects that enhance phosphate transfer. From this regard it is also interesting to note that a strong electron density peak near Arg359 and ADP, modeled as a potassium ion, may affect electrostatic interactions of the required reactants and promote catalysis.
Discussion
Deep mutational scanning has previously been used to evaluate enzyme function on a massive scale (8, 17, 27). In the present work, we combine deep mutational scanning with two high-throughput techniques purporting to screen for solubility for two different full-length enzymes. The addition of mutational solubility data produces a more complete picture of the fitness landscape for an enzyme. In the present work, we find that at least 40% of given solubility-enhancing mutations result in enzymes with impaired fitness. Although this trade-off has generally been modeled as solubility-reducing residues in the active site of the enzyme (11), a significant number of mutations occur at distances up to 15 Å away from any active site residues for all datasets.
As with other deep mutational scanning experiments, caution must be exercised when interpreting results. First, analysis of replicates of the Tat genetic selection and synonymous mutations to wild type showed that error in solubility score increased with lower depth of coverage. Screening and sequencing at a higher depth of coverage in the population will increase the accuracy. Second, the solubility measured in YSD is of a protein fusion partner, not the individual protein. Fusion partners can drastically affect folding ability of target proteins. Additionally, environmental effects like temperature or the expression host (secretory pathway in Saccharomyces cerevisiae, cytoplasm of E. coli) itself can make significant differences in the fraction of successfully folded variants. For example, TEM-1 is known to be a client of E. coli GroEL/ES chaperones that are not present in the YSD system. Finally, the YSD readout is the per-cell number of epitope tags labeled by a fluorescently conjugated antibody. The relationship between fluorescence and fraction of protein surface displayed breaks down when destabilization can increase accessibility of the epitope tag, as seen for a subset of the TEM-1.1 dataset.
The current theoretical framework based on thermodynamically stabilizing mutations (18) is likely missing key solubility determinants like folding rate and aggregation propensity. The datasets presented in this work could potentially be used to refine theory, although the low dynamic range (approximately fivefold) inherent in the present deep-sequencing–based measurements is a limitation. To that end, future experiments should focus on increasing the dynamic range of the measurements, along with the number of enzymes tested.
Applying simple classification methods to the deep mutational scanning screens increases the probability of selecting soluble, active mutants. Filtering on multiple classifiers gives a probability of selecting a deleterious mutation of 2–3%; this error rate is low enough for 10–20 mutations to be combined additively and still maintain activity. From a protein engineering and design perspective, the next step is to test the generality of the approach to find active solubility-enhancing mutations to improve the solubility of difficult proteins involved in biomanufacturing and metabolic engineering. There are a number of considerations that make this deep mutational scanning approach attractive. First, there exist multiple screens shown to identify gain of stability mutations. Second, enzymes stable in their genetic background can be scanned using the recursive destabilizing approach developed by Bradbury and coworkers (19). In fact, we used this approach in the present work by destabilizing the robust TEM-1 using the known D179G mutation. Finally, the classifiers used were designed so that high-resolution structures are not necessary. PSSM and type of mutation do not require structural information, whereas contact number and distance to active site can be approximated even with crude homology models. The remaining step is to automate a design process to combine many solubility-enhancing mutations simultaneously. To that end, excellent results have already been demonstrated on the LGK system (8) and in the presence of a solved structure using the PROSS (Protein Repair One-Stop Shop) method (3). Because the mutations are already known, we anticipate that all-atom design using Rosetta or a similar software package will prove successful even using crude homology models.
Materials and Methods
Library Construction.
Comprehensive single-site mutagenesis was performed on the gene-encoding sequences for LGK and TEM-1 using nicking mutagenesis (20). Full details of the preparation of cell libraries are given in SI Appendix, SI Text.
Screening Procedures.
For YSD screening, yeast cells were suspended to a concentration of 2 × 106 cells per milliliter of PBSF (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4 with 1 g/L BSA), and labeled with 1 μL of anti–c-myc-FITC (Miltenyi Biotec) per 2 × 105 cells. Cell sorting and collection were done on a BD Influx cell sorter (Becton Dickinson). Tat genetic selection was performed according to Fisher et al. (28). The full protocol and postscreening DNA extraction conditions are given in SI Appendix, SI Text.
Data Analysis and Deep Sequencing.
The PSSM analysis was performed following the method of Goldenzweig et al. (3). The yeast display and GFP fusion solubility score for a variant i (ζi) is defined as:
| [2] |
where Fi is the mean fluorescence of variant i and Fwt is the mean fluorescence of the wild-type sequence. This solubility score is calculated using experimental observables in the deep-sequencing experimental pipeline according to:
| [3] |
where εi is the enrichment ratio of the variant, εwt is the enrichment ratio of the starting sequence, σ is the SD of the population, and ϕ is the percentage of cells collected of the gating population (21).
The Tat solubility score is defined as:
| [4] |
where εi is the enrichment ratio of the variant and εwt is the enrichment ratio of the starting sequence.
Protein Expression, Purification, Crystallization, Data Collection, and Structure Determination.
Expression, purification, and crystallization of LGK-G359R were performed closely following the method of Klesmith et al. (8). Specific details are given for crystallization methods in SI Appendix, SI Text.
Supplementary Material
Acknowledgments
We thank B. Hackel for helpful comments, M. Ostermeier for helpful suggestions on an earlier version of this manuscript and the gift of the TEM-1 BLA mutagenic primer set, and S. Thorwall and V. Kelly for their help in the laboratory. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (including Grant P41GM103393). This work was supported by US Department of Agriculture National Institute of Food and Agriculture (NIFA) Award 2016-67011-24701 (to J.R.K.) and US National Science Foundation Career Award 1254238 CBET (to T.A.W.). J.-P.B. was partially funded through the Protein Crystallography Station from the DOE Office of Biological and Environmental Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5TKR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614437114/-/DCSupplemental.
References
- 1.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79(3):830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sormanni P, Aprile FA, Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427(2):478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Goldenzweig A, et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol Cell. 2016;63(2):337–346. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldo GS. Genetic screens and directed evolution for protein solubility. Curr Opin Chem Biol. 2003;7(1):33–38. doi: 10.1016/s1367-5931(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 5.Park S, et al. Limitations of yeast surface display in engineering proteins of high thermostability. Protein Eng Des Sel. 2006;19(5):211–217. doi: 10.1093/protein/gzl003. [DOI] [PubMed] [Google Scholar]
- 6.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 7.Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol. 2006;60(1):373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klesmith JR, Bacik JP, Michalczyk R, Whitehead TA. Comprehensive sequence-flux mapping of a levoglucosan utilization pathway in E. coli. ACS Synth Biol. 2015;4(11):1235–1243. doi: 10.1021/acssynbio.5b00131. [DOI] [PubMed] [Google Scholar]
- 9.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Boock JT, et al. Repurposing a bacterial quality control mechanism to enhance enzyme production in living cells. J Mol Biol. 2015;427(6 Pt B):1451–1463. doi: 10.1016/j.jmb.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLOS Comput Biol. 2008;4(2):e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320(1):85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 13.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 2008;379(5):1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann M, et al. From DNA sequence to improved functionality: Using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 2000;13(1):49–57. doi: 10.1093/protein/13.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Steipe B, Schiller B, Plückthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J Mol Biol. 1994;240(3):188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 16.Araya CL, Fowler DM. Deep mutational scanning: Assessing protein function on a massive scale. Trends Biotechnol. 2011;29(9):435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 2014;31(6):1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369(5):1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 19.Kiss C, Temirov J, Chasteen L, Waldo GS, Bradbury ARM. Directed evolution of an extremely stable fluorescent protein. Protein Eng Des Sel. 2009;22(5):313–323. doi: 10.1093/protein/gzp006. [DOI] [PubMed] [Google Scholar]
- 20.Wrenbeck EE, et al. Plasmid-based one-pot saturation mutagenesis. Nat Methods. 2016;13(11):928–930. doi: 10.1038/nmeth.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalsky CA, et al. High-resolution sequence-function mapping of full-length proteins. PLoS One. 2015;10(3):e0118193. doi: 10.1371/journal.pone.0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1(2):755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 23.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15(3):449–458. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloom JD. Software for the analysis and visualization of deep mutational scanning data. BMC Bioinformatics. 2015;16(1):168. doi: 10.1186/s12859-015-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleishman SJ, et al. Community-wide assessment of protein-interface modeling suggests improvements to design methodology. J Mol Biol. 2011;414(2):289–302. doi: 10.1016/j.jmb.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klesmith JR, Whitehead TA. High-throughput evaluation of synthetic metabolic pathways. Technology (Singap World Sci) 2016;4(1):9–14. doi: 10.1142/S233954781640001X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014;42(14):e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher AC, Rocco MA, DeLisa MP. Genetic selection of solubility-enhanced proteins using the twin-arginine translocation system. In: Evans TC, Xu M-Q, editors. Heterologous Gene Expression in E.coli: Methods and Protocols. Humana Press; Totowa, NJ: 2011. pp. 53–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.