Significance
The population dynamics of species arise from individual-level inter- and intraspecies interactions, driven by genetic and neurobehavioral factors. However, linking ecological and evolutionary dynamics to underlying mechanisms represents a major challenge, largely due to experimental intractability. Here, we study the population dynamics of a predator–prey system comprising the nematode worm Caenorhabditis elegans and bacteria Escherichia coli. We find that the worms engage in a form of primitive agriculture, driven by their foraging behavior, by redistributing their bacterial food source, which subsequently grows. Our findings have ecoevolutionary consequences that are broadly applicable not only to worm–bacterial dynamics but also for diverse situations such as the spread of epidemics, foraging behavior, seed dispersal, and the organisms’ engineering of their habitat.
Keywords: foraging behavior, public goods, predator–prey, population dynamics, farming
Abstract
The ecological and evolutionary dynamics of populations are shaped by the strategies they use to produce and use resources. However, our understanding of the interplay between the genetic, behavioral, and environmental factors driving these strategies is limited. Here, we report on a Caenorhabditis elegans–Escherichia coli (worm–bacteria) experimental system in which the worm-foraging behavior leads to a redistribution of the bacterial food source, resulting in a growth advantage for both organisms, similar to that achieved via farming. We show experimentally and theoretically that the increased resource growth represents a public good that can benefit all other consumers, regardless of whether or not they are producers. Mutant worms that cannot farm bacteria benefit from farming by other worms in direct proportion to the fraction of farmers in the worm population. The farming behavior can therefore be exploited if it is associated with either energetic or survival costs. However, when the individuals compete for resources with their own type, these costs can result in an increased population density. Altogether, our findings reveal a previously unrecognized mechanism of public good production resulting from the foraging behavior of C. elegans, which has important population-level consequences. This powerful system may provide broad insight into exploration–exploitation tradeoffs, the resultant ecoevolutionary dynamics, and the underlying genetic and neurobehavioral driving forces of multispecies interactions.
The fitness of an organism is affected by its strategies to produce, explore, and exploit resources (1, 2). These strategies are influenced, in large part, by interdependencies among organisms, such as competition, predation (3, 4), mutualism (5–7), or the production of public good resources (8–10). Despite the wide prevalence of such interactions in nature as well as numerous theoretical and empirical studies, we are still limited in our mechanistic understanding of the interplay between different survival strategies, the resultant evolutionary dynamics and the underlying genetic, neurobehavioral, and ecological driving forces. In this pursuit, model systems in the laboratory have served as a useful bridge between the complexity of nature and the simplifications inherent in theoretical investigations. Such model systems have predominantly been either microbial (11, 12) or higher organisms, such as primates and humans (13–16). Microbial systems are very convenient due to their genetic tractability and short generation times but are limited in the space of behavioral traits they exhibit. At the other extreme, higher organisms exhibit rich neurobehavioral and genetic traits, but they are difficult to experimentally manipulate and generation times are very long.
Recently, organisms such as the nematode worm Caenorhabditis elegans and the fruit fly Drosophila melanogaster have been increasingly used in evolutionary and behavioral studies (17–19). These organisms demonstrate complex behavior and yet retain experimental tractability due to their extensive development as model systems in neurobiology and genetics. The C. elegans worms, in particular, are amenable to experimental tracking of large populations and multiple generations at high resolution (20–23), which has made them one of the most widely used model organisms in behavioral, genetic, and neurobiological studies.
The ecological and evolutionary backgrounds of C. elegans, however, have remained unclear for a long time, and only recently have insights into the organism’s natural habitat begun to be uncovered. Contrary to the common perception that it is a soil nematode, C. elegans is primarily a colonizer of microbe-rich habitats including decaying organic matter where resources are finite and are quickly depleted (24, 25). C. elegans populations are characterized by a rich set of ecological dynamics: (i) a boom and bust population dynamics due to ephemeral resources (26), self-fertilization (27), and dauer developmental stages (28); (ii) dispersal and migration by various means (26); (iii) competition (26, 29, 30); and even (iv) host–microbe interactions (24, 25, 30, 31). Such a lifestyle is naturally tied with the movement patterns of the worms through the complex environments where they dwell. In the laboratory, the foraging strategies of C. elegans, which are a key determinant of their fitness, are influenced both by the distribution and quality of resources (32) and by the presence of competitors (33), whether they are of the same or different genotypes, a scenario that likely results from local genetic diversity induced by worm movement (25). Therefore, C. elegans is an ideal model organism to study the interplay between ecology (resource distribution, inter and intraspecies interactions) and behavior (e.g., foraging strategies, public goods production) and to explore the genetic and neural circuits responsible for integrating ecological information. However, the potential for exploring these areas using C. elegans populations remains largely untapped.
Here, we use the C. elegans–bacteria (E. coli) system to study the emergent population dynamics of each species. C. elegans feed on bacteria and persistently forage for new bacterial food sources. Using both experimental and theoretical approaches, we uncover a relationship between foraging and a hitherto unrecognized mechanism of public goods production. This production of public goods leads to a long-term fitness advantage for both the worms and the bacteria, but one that can easily be exploited by nonproducing types.
Results and Discussion
Our experimental setup comprises a homogeneous surface containing nutrients for bacterial (E. coli) growth; C. elegans can dwell and feed on the growing bacteria or they can forage about the surface by crawling, their predominant form of motility. At the start of an experiment, an individual N2 (laboratory “wild type”) worm is placed near a circular patch of bacteria growing at the center of a nutrient agar-filled Petri dish. As the experiment progresses, new bacterial colonies appear, both as discrete new patches or as continuous trails leading away from the initial patch (Fig. 1A). Because the bacterial strain we use is nonmotile, the worm’s movement within and outside of the initial patch of bacteria (34, 35) is responsible for redistributing the bacterial resource.
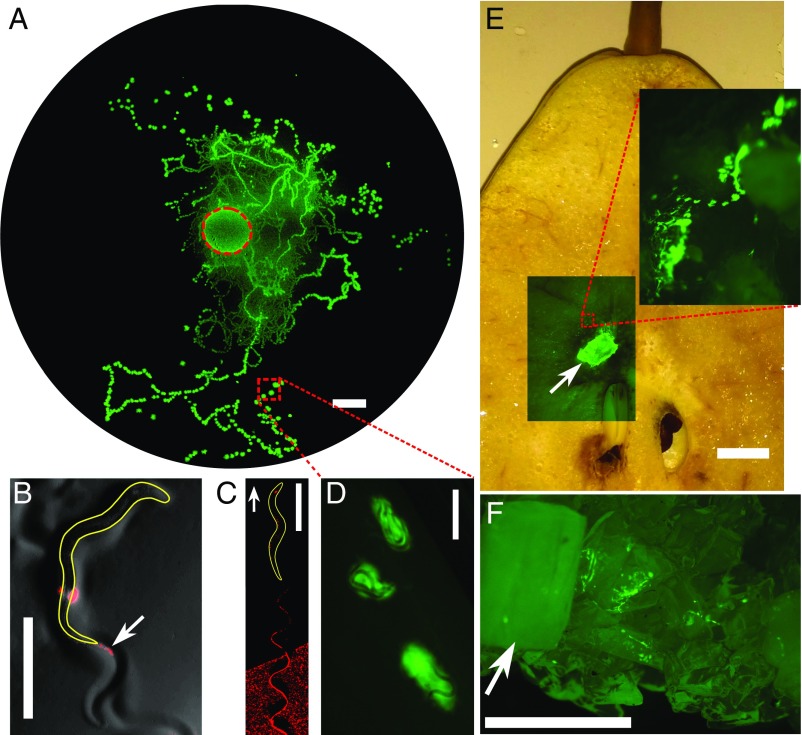
Fig. 1.
Redistribution and growth of E. coli bacteria due to locomotion of C. elegans. (A) Trails and patches of bacteria are found away from an initial circular patch (red dashed circle) of bacterial inoculation in which a single worm is placed. The image is taken 4 d after seeding of a bacterial patch with a worm. The bacterial redistribution is due to two main mechanisms: defecation of ingested bacteria (white arrow points to red “feces”) by the worm (B) and entrainment of the bacteria (red) in the wake of the locomoting worm (white arrow shows direction of motion) (C). (D) Worms revisit and colonize the redistributed, growing bacterial patches. Worms disperse bacteria in natural settings such as rotting fruit (E) and soil-like porous medium (F). Arrows indicate initial chunk of worms and bacteria. (Scale bars: A, E, and F, 1 cm; B–D, 500 m.)
In our experiment, the entire homogeneous agar surface is available for the worms to explore. Worms that have been feeding on fluorescent bacteria reveal two mechanisms for the formation of new bacterial colonies. Bacterial food is propelled through the worm digestive system, and undigested material is excreted (36). Thus, while locomoting, worms may defecate undigested bacteria as illustrated in Fig. 1B (also see Movie S1), which can eventually grow into larger bacterial colonies. A second and much more prevalent source of bacterial dispersal arises from adhesion of the bacteria to the surface of the worm body. When the worm moves out of a dense bacterial colony, some bacteria hitchhike on the worm’s surface and are sloughed off through fluid entrainment as the worm crawls around its arena (Fig. 1C; also see Movie S2); these bacteria then also grow into colonies. Redistributed colonies represent a new food resource, which can be revisited and used by the worms (Fig. 1D). The dynamics are not unique to flat Petri-dish surfaces but also occur in native environments such as rotting fruit (Fig. 1E) and 3D porous soil (Fig. 1F).
Farming of Bacteria by the C. elegans Worms.
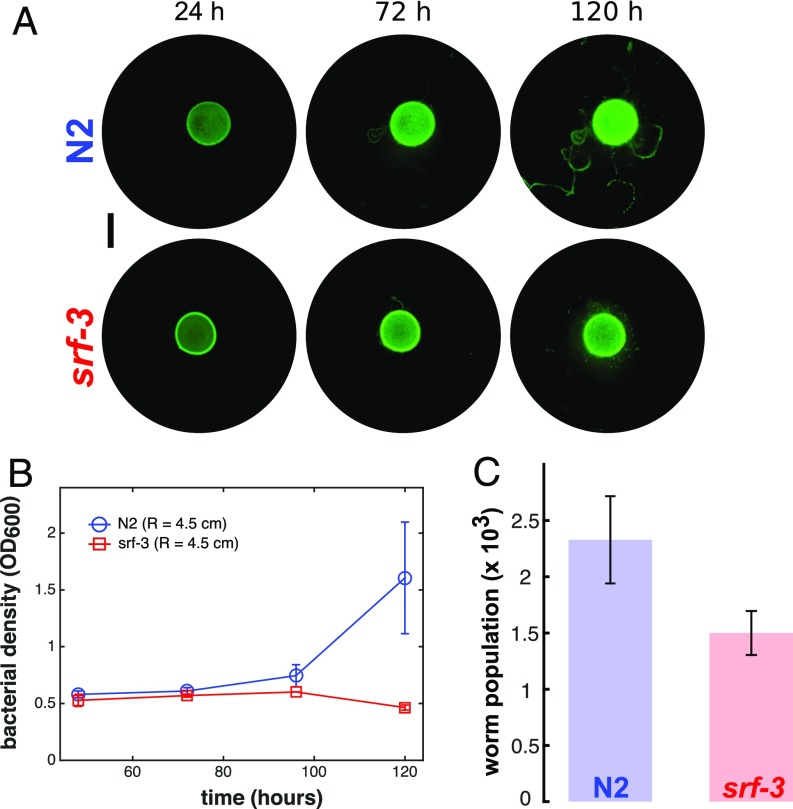
The bacterial redistribution has consequences for the population dynamics of both bacteria and worms, which we explore via experiments and mathematical modeling. We first sought to examine how the population dynamics are impacted when we limit the ability of the worms to redistribute bacteria. We take advantage of the genetic tractability of this system by using worms with a mutation in the srf-3 gene, which causes altered body surface properties that reduce surface adhesion by bacteria (37, 38). The exploratory behavior of srf-3 mutants is comparable to that of the N2 worms (SI Appendix, Fig. S1), but they do not cause significant bacterial dispersal (Fig. 2A). By dispersing bacteria from a single large patch into several smaller patches, N2 worms decrease the density of the bacterial colonies. Smaller, less dense patches of bacteria grow faster than large, denser patches due to an increased availability of local nutrients on the agar surface (SI Appendix, Fig. S2). Thus, bacterial dispersal by N2 worms, but not by the srf-3 mutant, results in an increase in the overall population growth rate of the bacteria (Fig. 2B). This difference in their ability to disperse their bacterial resource has a significant impact on worm population dynamics; under identical initial conditions, the N2 population grew larger than that of the srf-3 worms (Fig. 2C; brood sizes of both worm types are comparable as shown in SI Appendix, Fig. S3).
Fig. 2.
Farming confers a population growth advantage. (A) N2 worms redistribute bacteria, whereas srf-3 mutants do not. (Scale bar: 1 cm.) (B) Bacterial density on plates (R = 4.5 cm) with N2 (blue circles, n = 4) and srf-3 (red squares, n = 4). (C) The population sizes of N2 worms and the mutant type srf-3 worms 144 h after the start of the experiment for the same initial conditions.
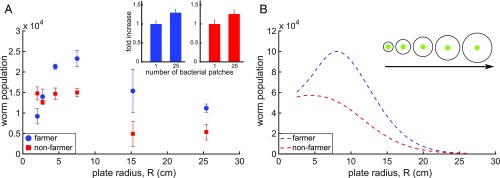
This behavior appears to reflect a type of “bacterial farming,” whereby the consumer (worm) benefits from increasing its food resource (bacteria) by facilitating the bacteria’s access to a third resource, the agar nutrients. To test this idea, we asked whether increasing the Petri dish size—and thus increasing the local availability of agar nutrients—would increase worm proliferation. Keeping the size of the initial bacterial seed colony constant and with saturated concentrations of bacterial nutrients in the agar (SI Appendix, Fig. S4), increasing dish size up to a critical radius of 7.5 cm had little effect on the srf-3 population, while having a dramatic positive effect—more than twofold increase with the available exploration area—on the N2 population (Fig. 3). This difference between N2 and srf-3 worms occurs despite the fact that both worm types engage in similar space exploration strategies (SI Appendix, Fig. S1). Moreover, upon artificial dispersal of bacteria into multiple patches, both worms are found to exhibit a similar up-regulation of their reproduction in response to the increased food availability (Fig. 3A, Inset). These results suggest that the N2 behavior seen in Fig. 3 does indeed represent a type of farming. Beyond the critical plate size, although N2 populations remained larger than srf-3 ones, both srf-3 and N2 population sizes decreased with plate size, which may reflect a decreased birth rate associated with long-distance exploration.
Fig. 3.
Effect of available farming area on the worm population. Worm population as a function of the available dispersal area for the farmer (N2, blue) and nonfarmer (srf-3 mutant, red) worms. (A) Experiments. (B) Numerical results. (A, Inset) Experiments indicating the fold increase in population size of the worms grown on artificially distributed bacterial patches. The data are normalized to a onefold increase in the case of one bacterial patch. Measurements were made 72 h after initialization. The initial amount of bacteria is the same but the patches are distributed either as 1 large patch or the 25 smaller patches formed in a 5 × 5 grid; n = 2–5 for each plate size. (B, Inset) Schematic of the experiment. The bacterial patch is kept constant, whereas the dish size is increased.
Mathematical Model.
To understand the farming behavior, we constructed a theoretical model that allowed us to test the proposed farming mechanism but also to explore further scenarios of interest and make testable predictions. To retain the simplicity and versatility of the model, we sacrificed complex and little-understood details related to spatial movement, such as the use of memory, and instead captured the relevant qualitative behavior implicitly via physically and experimentally motivated assumptions. The model consists of age-structured, spatially implicit ordinary differential equations that capture the population dynamics of the worms and the impact of their foraging behaviors on bacterial dynamics (the text in SI Appendix and SI Appendix, Figs. S5–S9). Because the feeding and farming behaviors of the worms affect the area and density of bacteria in ways that cannot be captured by the total number of bacteria alone, we characterize the bacteria via their spatial distribution (captured by area, ) and via their density , assumed to be homogeneous but time-dependent. In the absence of worms, both area and density grow logistically. The worm population is age-structured into four stages: eggs, sexually immature, mature, and infertile worms. Eggs hatch at a fixed rate to become immature worms, which then progress through the three stages of their adult life cycle at a rate proportional to the amount of bacteria consumed. As the bacteria get depleted, their density decreases, whereas their area is preserved, an assumption supported visually by experiments (Fig. 2A). Worms feed according to a Holling’s type II form, , featuring an encounter rate between worms and bacteria. To capture this behavior in a spatially implicit way we let the encounter rate, , be a function of bacterial area and plate radius, : the more bacteria relative to the size of the plate, the higher the encounter rate:
| [1] |
| [2] |
The parameter controls how quickly the feeding rate increases in response to increased bacteria, whereas the parameter controls how quickly the encounter rate declines with plate size. In reality, the encounter rate between worms and bacteria depends on a range of C. elegans characteristics, including their exploration–exploitation strategies, foraging behavior, and memory. In our simplified model, we propose the phenomenological form above for to capture the experimental observation that the mean feeding/reproductive rate decreases with large plate sizes (Fig. 3).
Farmers increase the spatial distribution of the bacteria, thus simultaneously decreasing its density. The rate of bacterial spreading is proportional to the amount of free space on the plate, the density of the bacteria, and the encounter rate:
| [3] |
Nonfarmers do not spread bacteria and, hence, . We are able to derive experimentally the majority of our parameters (the text in SI Appendix and SI Appendix, Table S1). The only free parameters pertain to the worm spatial feeding behavior ( and ) and, in the case of the farmer, the spreading behavior, . Despite its simplicity and low-dimensionality, our model robustly recapitulates qualitatively (with fourfold quantitative difference) the complex population dynamics of the worms and bacteria (Fig. 3, SI Appendix, Figs. S5–S6, and Movies S3 and S4). A further simplified version of this model that ignores the age structure of the worm population also recapitulates the dynamics qualitatively, showing the robustness of our physically motivated assumptions, but does so at the expense of having more free parameters and a worse quantitative fit (see SI Appendix and Movie S5 for details).
Redistributed Bacteria Is a Potentially Costly Public Good.
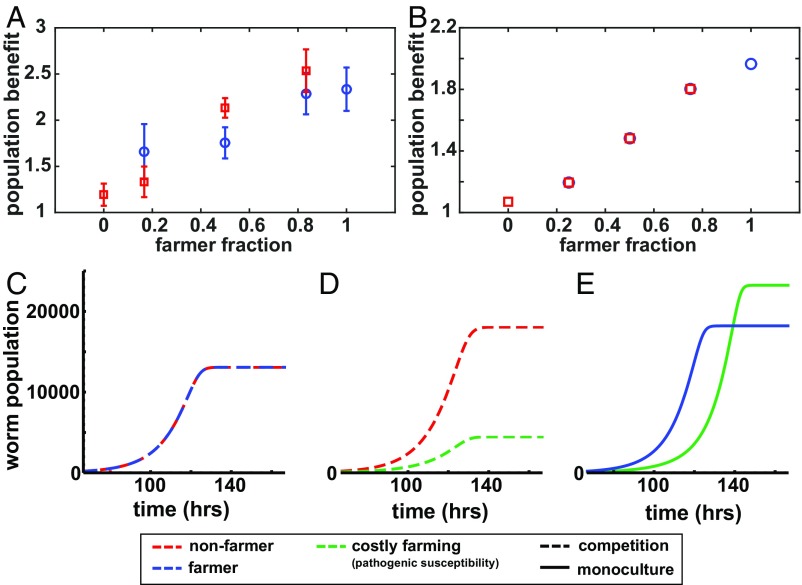
Despite the inability of srf-3 worms to farm, if a mixed population includes N2 worms that farm and therefore increase the bacterial resource, all worms may be able to take advantage of it. To experimentally test this hypothesis, we competed mixed populations containing both worm types, starting with varying initial ratios of farmer (N2) to nonfarmer (srf-3). We found that both phenotypes equally share the farming-increased resource, and the benefits scale linearly with the proportion of farmers (Fig. 4A). This result shows that the farming of bacteria by N2 worms is a public good and that on agar plates there is no significant spatial clustering that would cause the public good to be mostly shared with kin. Given the small difference in the benefit gained by the farming and nonfarming populations, this public good appears to be generated with negligible additional metabolic cost to the farmers.
Fig. 4.
Redistributed bacteria is a potentially costly public good. (A) Normalized worm population from competition experiments between N2 worms and srf-3 mutant for two different plate sizes (R = 7.5 cm and R = 2.75 cm). The population increase is the ratio of the worm population on the R = 7.5 cm plate to the population on the R = 2.75 cm plate. n = 4 for each experiment. (B) Corresponding data of the competition between farmers and nonfarmers from mathematical model (see SI Appendix for details). (C–E) Mathematical model trajectories of worm population sizes in competitive (solid curves) or clonal (dashed curves) growth conditions. Worm counts are normalized by the initial numbers of that phenotype in the simulation. Competitive dynamics of farmers and nonfarmers (red) when farming inflicts no mortality cost (blue) (C) and some mortality cost (green) (D). (E) Clonal dynamics of farmers with and without mortality cost. The plate size is R = 7.5 cm. All parameters are as in SI Appendix, Table S1.
Although we are unable to detect significant fitness costs incurred by N2 farmers compared with the srf-3 nonfarmers, costs associated with public goods production are likely to occur in nature and can strongly impact the evolutionary dynamics of mixed populations of producers and nonproducers. In our system, there are at least two ways in which costs may arise. First, although prolonged exposure to E. coli does not appear to be harmful for the worms, other types of bacteria commonly found in nature are highly pathogenic to the worm (37, 38). Consequently, bacterial entrainment arising from worm stickiness could increase this pathogenicity, whereas the nonstickiness of the srf-3 worms could confer resistance to pathogenic bacteria (37, 38). Second, metabolic costs associated with foraging are known to occur and mutants adopting different foraging strategies will necessarily incur different costs.
We use our mathematical model to explore the public good production and the effects of possible costs on the worm population dynamics. If there is no cost to farming, farmers and nonfarmers perform equally well in mixed cultures, consistent with our experimental findings (Fig. 4 A–C). However, we find significant difference in performance in two competition scenarios: between farmers and nonfarmers when the farmers pay an increased mortality cost due to bacterial pathogenicity (scenario 1) and between two farmers with different foraging behaviors (scenario 2); a more efficient forager that can find bacteria quickly but at a higher cost to its reproduction and a slower forager that incurs a lower reproductive cost. As expected, in scenario 1, we find that farmers that pay a mortality cost are worse off, whereas nonfarmers are better off (Fig. 4D and SI Appendix, Fig. S10). In scenario 2, depending on the magnitude of the cost, better-foraging farmers can perform worse in competition with the poorer foragers (SI Appendix, Figs. S11 and S12). These outcomes may change however in a spatially structured environment where worm movement is limited (e.g., soil), and the public good is mainly available to related individuals (6, 39). Interestingly, we found that when the composition of the population is homogeneous (i.e., comprising only a single phenotype), farmers that pay a cost either to mortality (Fig. 4E) or to reproduction (SI Appendix, Figs. S10 and S12) reach higher population densities than farmers that pay lower or no cost. This counterintuitive result stems from the fact that increased worm mortality or lowered worm reproduction can reduce the pressure on the bacterial resource and in turn lead to higher worm growth in the long term. Thus, although a cost makes farmers vulnerable to exploitation in mixed cultures, it leads to higher population densities if interactions are clonal, which reinforces the importance of spatially structured environments with limited dispersal in shaping C. elegans behavior.
Conclusions
We have shown that C. elegans worms engage in a primitive form of farming of the bacterial resource that they feed on. The farming is brought about by the redistribution of bacteria by foraging worms, resulting in an increased amount of bacteria, which can be exploited by nonproducers. This form of public goods production, which may be incidental to the foraging behavior of the worms, is qualitatively different from situations in which the good production is associated only with the explicit metabolic cost of chemical synthesis of the good, a mechanism often at play in microbial systems (6, 8–10), which lack complex behaviors. In contrast, the mechanism of public goods production that we describe here could be associated with neurobehavioral traits, such as exploration–exploitation strategies (18, 19, 29, 40–42) or the use of spatial memory (42, 43), in addition to potential metabolic costs associated with carrying the bacteria (37, 38). Moreover, C. elegans also appear to be capable of dispersing Dictyostelium discoideum spores (44), another food source; given that D. discoideum themselves farm bacteria (7), we anticipate a rich set of multitrophic level dynamics and niche partitioning to emerge in multispecies interactions involving the kind of effects that we have uncovered here. More specifically, these previously unobserved effects of worm-foraging behavior are likely to have significant consequences for experimental work involving C. elegans populations; even the most routine aspects of worm maintenance in the laboratory are likely to be affected by these dynamics.
The dynamics in our system have a striking similarity to a range of spreading processes in nature such as the dispersal of seeds or the carrying of commensal infectious agents by mobile vectors (14, 45, 46). Empirical data in these cases are limited, and even when available, the data are observational rather than experimental. Moreover, in processes such as the dispersal of seeds (46), the benefit to the disperser likely occurs on a much longer time scale compared with the benefit accrued by the dispersed. In contrast, the impact of the bacterial redistribution reported here occurs on a fast time scale, with effects similar to those of farming in other organisms (7, 47–51). This characteristic allows for experimental and theoretical investigations into the role of farming in driving and shaping the evolutionary dynamics of foraging. In addition, the microbial populations on which the worms feed are redistributed through the ecological landscape, which affects the composition of microbial communities and their relationships and interactions. Altogether, these effects will shape the local microbial and worm ecologies in ways that significantly affect their dynamics. Although further investigations are needed to determine the impact of such dynamics in the wild, this incidental dropping of “resource seeds” is remarkably similar to the early stages of human agriculture during which “… people who gathered [wheat] grains carried them back to their temporary campsites for processing…some of them inevitably fell on the way to the campsite and were lost. Over time, more and more wheat grew along favorite human trails and near campsites” (52).
Materials and Methods
C. elegans Strains and Culture.
N2 Bristol (laboratory wild type) and AT10 (srf-3 (yj10)) (mutant type) were obtained from the Caenorhabditis Genetics Center (CGC) and maintained on standard nematode growth medium (NGM) plates supplemented with ampicillin and seeded with OP50-GFP E. coli (GFP plasmid pFVP25.1 with ampicillin resistance) also obtained from the CGC. For competition experiments, CPB089 (), with the same brood size as N2 worms, generated in house by CRISPR technology, was used as a substitute. For all experiments, 20 of bacteria at per worm were seeded on NGM plates of the appropriate size. Worms were age-synchronized by bleaching and individual larval stage 4 (L4) worms were placed onto dishes of the appropriate size. Brood size was quantified by counting the number of embryos laid in 24-h intervals by age-synchronized worms on standard NGM plates, at which time worms were moved to a fresh dish. OD600 shown in Fig. 2 was measured using a NanoDrop (ThermoScientific) by washing each plate with the same volume of M9 buffer.
Imaging.
To image entire Petri dish surfaces such as in Fig. 1A, we used a desktop flatbed scanner (Epson V700) custom-fitted with a blue light LED strip to excite fluorescence emission in the OP50-GFP E. coli and a corresponding photographic emission filter (Kodak) to record the image.
Flow Cytometry.
Individual plates were carefully washed with M9 buffer and inspected to collect all worms. Worm samples were washed to remove bacteria and then transferred to a Complex Object Parametric Analyzer and Sorter Biosort (Union Biometrica) sample cup at a dilution of approximately one nematode per microliter in M9 buffer. To distinguish N2 and mutant worms, fluorescent gates were determined by running fluorescent worms and nonfluorescent worms separately. All data are shown as means F0B1 SEM.
Supplementary Material
Acknowledgments
We thank Christina DeCoste (Princeton Flow Cytometry Resource Facility) for invaluable assistance with the COPAS Biosort. We are grateful to Paulina Orillac for help with the initial setup of the experiments. We thank Mochi Liu for help with worm tracking and Bindu Madhav U for help with worm counting. Worm and bacterial strains were obtained from the CGC, which is funded by NIH Office of Research Infrastructure Programs Grant P40 OD010440. S.T. acknowledges the Human Frontier Science Program (Cross Disciplinary Fellowship) for funding. C.P.B. and S.U. acknowledge support from NIH Director’s New Innovator Award 1DP2GM105437-01 and Searle Scholars Program Grant 12-SSP-217. S.L. was supported by Simons Foundation Grant 395890.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608961114/-/DCSupplemental.
References
- 1.Charnov EL. Optimal foraging, the marginal value theorem. Theor Popul Biol. 1976;9(2):129–136. doi: 10.1016/0040-5809(76)90040-x. [DOI] [PubMed] [Google Scholar]
- 2.MacArthur RH. Geographical Ecology: Patterns in the Distribution of Species. Harper and Row; New York: 1972. [Google Scholar]
- 3.Brooks JL, Dodson SI. Predation, body size, and composition of plankton. Science. 1965;150(3692):28–35. doi: 10.1126/science.150.3692.28. [DOI] [PubMed] [Google Scholar]
- 4.Oksanen L. Evolution of exploitation ecosystems I. Predation, foraging ecology and population dynamics in herbivores. Evol Ecol. 1992;6(1):15–33. [Google Scholar]
- 5.Grossart HP, Dziallas C, Leunert F, Tang KW. Bacteria dispersal by hitchhiking on zooplankton. Proc Natl Acad Sci USA. 2010;107(26):11959–11964. doi: 10.1073/pnas.1000668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24(1):50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469(7330):393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 8.Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404(6778):598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 9.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 10.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459(7244):253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jessup CM, Forde SE, Bohannan BJM. Microbial experimental systems in ecology. Adv Ecol Res. 2005;37(04):273–307. [Google Scholar]
- 12.Kawecki TJ, et al. Experimental evolution. Trends Ecol Evol. 2012;27(10):547–560. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Vishwanath G, da Luz M, Raposo EP, Stanley E. The Physics of Foraging. Cambridge Univ Press; Cambridge, UK: 2011. [Google Scholar]
- 14.Brockmann D, Hufnagel L, Geisel T. The scaling laws of human travel. Nature. 2006;439(7075):462–465. doi: 10.1038/nature04292. [DOI] [PubMed] [Google Scholar]
- 15.Ledyard JO. Public goods: A survey of experimental research. In: Kagel J, Roth AE, editors. Handbook of Experimental Economics. Princeton Univ Press; Princeton: 1995. pp. 111–251. [Google Scholar]
- 16.Grujic J, Fosco C, Araujo L, Cuesta JA, Sanchez A. Social experiments in the mesoscale: Humans playing a spatial prisoner’s dilemma. PLoS One. 2010;5(11):e13749. doi: 10.1371/journal.pone.0013749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad NG, Joshi A. What have two decades of laboratory life-history evolution studies on Drosophila melanogaster taught us? J Genet. 2003;82(1-2):45–76. doi: 10.1007/BF02715881. [DOI] [PubMed] [Google Scholar]
- 18.Gray JC, Cutter AD. Mainstreaming Caenorhabditis elegans in experimental evolution. Proc Biol Sci. 2014;281(1778):20133055. doi: 10.1098/rspb.2013.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol. 2008;4(4):e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen JP, et al. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc Natl Acad Sci USA. 2016;113(8) doi: 10.1073/pnas.1507110112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatachalam V, et al. Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc Natl Acad Sci USA. 2015;113(8):1082–1088. doi: 10.1073/pnas.1507109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroustrup N, et al. The Caenorhabditis elegans lifespan machine. Nat Methods. 2013;10(7):665–70. doi: 10.1038/nmeth.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yemini E, Jucikas T, Grundy LJ, Brown AE, Schafer WR. A database of Caenorhabditis elegans behavioral phenotypes. Nat Methods. 2013;10(9):877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Félix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20(22):R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 25.Frézal L, Félix MA. C. elegans outside the Petri dish. Elife. 2015;4:1–14. doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10(1):59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutter AD. Sperm-limited fecundity in nematodes: How many sperm are enough? Evolution. 2004;58(3):651–655. [PubMed] [Google Scholar]
- 28.Green JWM, Snoek LB, Kammenga JE, Harvey SC. Genetic mapping of variation in dauer larvae development in growing populations of Caenorhabditis elegans. Heredity. 2013;111(4):306–313. doi: 10.1038/hdy.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gloria-Soria A, Azevedo RBR. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18(21):1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 30.Cutter AD. Caenorhabditis evolution in the wild. Bioessays. 2015;37(9):983–995. doi: 10.1002/bies.201500053. [DOI] [PubMed] [Google Scholar]
- 31.Dirksen P, et al. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol. 2016;14(1):1–16. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209(Pt 1):89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greene JS, et al. Balancing selection shapes density-dependent foraging behaviour. Nature. 2016;539(7628):254–258. doi: 10.1038/nature19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milward K, Emanuel K, Joseph R, Bono MD, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108(51):20672–20677. doi: 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature. 2011;472(7343):313–318. doi: 10.1038/nature09821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGhee J. The C. elegans intestine. WormBook. 2007;Mar 27:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Höflich J, et al. Loss of srf-3-encoded nucleotide sugar transporter activity in Caenorhabditis elegans alters surface antigenicity and prevents bacterial adherence. J Biol Chem. 2004;279(29):30440–30448. doi: 10.1074/jbc.M402429200. [DOI] [PubMed] [Google Scholar]
- 38.Cipollo JF, Awad AM, Costello CE, Hirschberg CB. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J Biol Chem. 2004;279(51):52893–52903. doi: 10.1074/jbc.M409557200. [DOI] [PubMed] [Google Scholar]
- 39.Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314(5805):1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croll N. Behavioural analysis of nematode movement. Adv Parasitol. 1975;13:71–122. doi: 10.1016/s0065-308x(08)60319-x. [DOI] [PubMed] [Google Scholar]
- 41.Salvador LCM, Bartumeus F, Levin SA, Ryu WS. Mechanistic analysis of the search behaviour of Caenorhabditis elegans. J R Soc Interface. 2014;11(92):20131092. doi: 10.1098/rsif.2013.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calhoun AJ, Chalasani SH, Sharpee TO. Maximally informative foraging by Caenorhabditis elegans. Elife. 2014;3:1–13. doi: 10.7554/eLife.04220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calhoun AJ, et al. Neural mechanisms for evaluating environmental variability in Caenorhabditis elegans. Neuron. 2015;86(2):428–441. doi: 10.1016/j.neuron.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kessin RH, Gundersen GG, Zaydfudim V, Grimson M. How cellular slime molds evade nematodes. Proc Natl Acad Sci USA. 1996;93(10):4857–4861. doi: 10.1073/pnas.93.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallatschek O, Fisher DS. The acceleration of evolutionary spread by long-range dispersal. Proc Natl Acad Sci USA. 2014;111(46):E4911–E4919. doi: 10.1073/pnas.1404663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howe HE, Smallwood J. Ecology of seed dispersal. Annu Rev Ecol Syst. 2013;13(1982):201–228. [Google Scholar]
- 47.Rindos D. The Origins of Agriculture. Academic; Orlando, FL: 1984. [Google Scholar]
- 48.Hata H, Kato M. A novel obligate cultivation mutualism between damselfish and Polysiphonia algae. Biol Lett. 2006;2:593–596. doi: 10.1098/rsbl.2006.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller UG, Gerardo NM, Aanen DK, Six DL, Schultz TR. The evolution of agriculture in insects. Annu Rev Ecol Evol Syst. 2005;36:563–595. [Google Scholar]
- 50.Silliman BR, Newell SY. Fungal farming in a snail. Proc Natl Acad Sci USA. 2003;100(26):15643–15648. doi: 10.1073/pnas.2535227100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu D, Huang L, Lin S. Cryptophyte farming by symbiotic ciliate host detected in situ. Proc Natl Acad Sci USA. 2016;113(43):12208–12213. doi: 10.1073/pnas.1612483113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harari YN. Sapiens - A Brief History of Humankind. Harper; New York: 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.