Significance
Vitamin D has been suggested to be associated with beneficial immunomodulation in autoimmune diseases. We demonstrate that the protective effect of vitamin D in an animal model of multiple sclerosis (MS) is linked to multiple signaling and metabolic pathways critical for T-cell activation and differentiation into pathogenic T helper (Th) 1 and Th17 subsets in vivo. This effect is mediated by epigenetic mechanisms as reflected by genome-wide reduction of DNA methylation and upregulation of microRNAs, with concomitant downregulation of their protein-coding target genes. Our data support the role of vitamin D in modulating risk for human disease, because orthologues of nearly 50% of MS candidate risk genes changed their expression in vivo in CD4+ T cells upon vitamin D supplementation.
Keywords: vitamin D, experimental autoimmune encephalomyelitis, multiple sclerosis, epigenetics, DNA methylation
Abstract
Vitamin D exerts multiple immunomodulatory functions and has been implicated in the etiology and treatment of several autoimmune diseases, including multiple sclerosis (MS). We have previously reported that in juvenile/adolescent rats, vitamin D supplementation protects from experimental autoimmune encephalomyelitis (EAE), a model of MS. Here we demonstrate that this protective effect associates with decreased proliferation of CD4+ T cells and lower frequency of pathogenic T helper (Th) 17 cells. Using transcriptome, methylome, and pathway analyses in CD4+ T cells, we show that vitamin D affects multiple signaling and metabolic pathways critical for T-cell activation and differentiation into Th1 and Th17 subsets in vivo. Namely, Jak/Stat, Erk/Mapk, and Pi3K/Akt/mTor signaling pathway genes were down-regulated upon vitamin D supplementation. The protective effect associated with epigenetic mechanisms, such as (i) changed levels of enzymes involved in establishment and maintenance of epigenetic marks, i.e., DNA methylation and histone modifications; (ii) genome-wide reduction of DNA methylation, and (iii) up-regulation of noncoding RNAs, including microRNAs, with concomitant down-regulation of their protein-coding target RNAs involved in T-cell activation and differentiation. We further demonstrate that treatment of myelin-specific T cells with vitamin D reduces frequency of Th1 and Th17 cells, down-regulates genes in key signaling pathways and epigenetic machinery, and impairs their ability to transfer EAE. Finally, orthologs of nearly 50% of candidate MS risk genes and 40% of signature genes of myelin-reactive T cells in MS changed their expression in vivo in EAE upon supplementation, supporting the hypothesis that vitamin D may modulate risk for developing MS.
Vitamin D has been recognized not only for its functions in homeostasis of calcium and phosphate but also for its important role in regulating cellular growth and the immune system. The active form of vitamin D, 1,25(OH)2D3, binds to the vitamin D receptor (VDR), which has wide tissue distribution, including immune cells such as dendritic cells, macrophages, and activated T and B cells, and exerts multiple immunomodulatory functions (1, 2). Epidemiological studies have linked poor vitamin D status with the increased prevalence of multiple chronic inflammatory diseases, including systemic and organ-specific autoimmune diseases (3), and several genes in the vitamin D pathway have been associated with the increased risk of developing autoimmune diseases (www.ebi.ac.uk/gwas) (3, 4).
Vitamin D deficiency is one of the most consistently reported environmental factor in the etiology of multiple sclerosis (MS) (5), a chronic inflammatory disease of the CNS characterized by autoimmune destruction of myelin, axonal loss, and brain atrophy (6). Increased risk of developing MS has been described in carriers of rare and common variants of the CYP27B gene (7, 8), which encodes the enzyme that catalyzes the last step in converting vitamin D to its active form, from 25(OH)D3 to 1,25(OH)2D3. These studies imply a causal role of low vitamin D in MS, which has recently been further supported by Mendelian randomization studies in two large cohorts demonstrating that three genetic variants that associate with serum 25(OH)D3 levels also associate with the risk of developing MS (9). However, high levels of vitamin D have been associated not only with the reduced risk of developing MS (10, 11) but also with the reduced risk for relapses, new brain lesions, and subsequent disability (12, 13). Moreover, it has been described that increased levels of vitamin D can reduce serum levels of IL-17 in MS patients (14). Most of what is known about the immunological mechanisms of vitamin D in MS comes from the studies in its animal model, experimental autoimmune encephalomyelitis (EAE). Vitamin D has been shown to impact both myeloid cells and T cells in EAE. This protective effect has been associated with reduced development of pathogenic T helper (Th) 1 (15, 16) and Th17 (17, 18) subsets, as well as with differentiation into regulatory T cells (Tregs) (19).
The cellular mechanisms of 1,25(OH)2D3 are mediated by the transcription factor VDR, which belongs to the steroid superfamily of nuclear receptors. Ligand-bound VDR forms a heterodimer with retinoid X receptor (RXR), which becomes translocated to the nucleus where it exerts its functions on gene regulation. The effects of vitamin D are cell type-specific because they depend on VDR/RXR binding, which is influenced by the cellular chromatin state and the availability of interacting DNA-binding protein partners (20). Similar to other nuclear receptors, VDR/RXR interacts with a variety of coactivators and corepressors, resulting in local epigenetic changes that have either permissive or repressive effects on gene expression. The cellular epigenetic state comprises highly interconnected mechanisms such as DNA methylation, histone modifications, and expression of noncoding RNAs (ncRNAs), which is critical for cell survival and its physiological function. Although the impact of vitamin D on histone modifications is well documented, because of VDR/RXR associations with histone acetyltransferases, deacetylases, and histone methyltransferases, its impact on DNA methylation is just beginning to emerge (21, 22). Additionally, recent studies in cancer suggest that ncRNAs, including long ncRNAs and microRNAs (miRNA), may be involved in mediating VDR signaling (22).
We have previously reported the protective effect of dietary vitamin D supplementation in myelin oligodendrocyte glycoprotein (MOG)-induced EAE in Dark Agouti (DA) rats (23), a well-established model of MS that shares numerous features with the human disease (24). This effect was associated with down-regulation of Th1/Th17-associated cytokines and transcription factors and a reduced amount of MOG-specific T cells (23). Several studies demonstrated that VDR expression is necessary for its suppressive activity in EAE, suggesting that vitamin D impacts gene regulation on the genomic level via VDR/RXR (17, 25, 26). Specifically, Mayne et al. (26) described the necessity of VDR expression in CD4+ T cells to ameliorate EAE, because vitamin D failed to inhibit EAE in mice with selective VDR gene deletion in CD4+ T cells. Our present study uses functional genomics to characterize effect of vitamin D supplementation in vivo on CD4+ T cells in actively induced EAE and shows that acquired changes due to vitamin D treatment in vitro impact T-cell capacity to induce disease in an adoptive transfer EAE model.
Results
Vitamin D Supplementation Affects CD4+ T Cells in MOG–EAE.
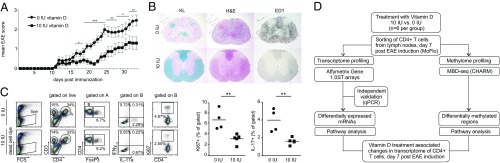
We initially reproduced our previous findings (23) demonstrating efficacy of the dietary vitamin D supplementation in ameliorating MOG-induced EAE in juvenile/adolescent rats (Fig. 1 A and B). We then extended our analyses toward characterization of the T-cell responses in the local draining lymph nodes 7 d postimmunization (p.i.) using flow cytometry (Fig. 1C). At this stage, DA rats develop an intense immune response leading to infiltration of pathogenic MOG-specific T cells into the CNS and subsequent neurological disabilities (27). Vitamin D supplementation did not affect the relative proportions of CD4+ and CD8+ T cells or Foxp3-expressing cells within each of these compartments (Fig. S1), but significantly decreased proliferation of CD4+ T cells (Fig. 1C) after recall with MOG. Additionally, vitamin D supplemented animals had significantly lower frequency of IL-17–producing CD4+ T cells (Fig. 1C). This finding is in line with our previous report on the vitamin D supplementation-mediated decrease of Rorc mRNA expression in the lymph nodes (23), which encodes the master transcription factor that drives IL-17–producing Th17 cells. To characterize observed differences in CD4+ T cells on the functional genomic level, we analyzed transcriptome and DNA methylome of CD4+ T cells. The experimental design is summarized in Fig. 1D.
Fig. 1.
Protective effect of vitamin D supplementation associates with changes in CD4+ T cells. (A) Supplementation with 10 IU vitamin D ameliorates clinical symptoms of EAE during entire disease course (n = 14 for 0 IU and n = 15 for 10 IU). (B) Histopathological and IHC analyses performed in the rat CNS harvested on day 34 p.i. Rats subjected to the vitamin D-supplemented diet displayed less severe neuroinflammation and myelin loss than the vitamin D-deprived group (n = 5 for each diet group). Group representative images of Kluever (KL), H&E, and ED1 staining in the rat spinal cord are shown. (C) Flow cytometry analysis of cells isolated from lymph nodes 7 d p.i. and restimulated for 48 h in vitro with MOG shows a significant decrease in frequency of proliferating CD4+ T cells, as shown by Ki67 staining. Additionally, vitamin D supplementation led to a significant decrease in frequency of IL-17–producing CD4+ T cells (n = 4 for 0 IU and n = 5 for 10 IU). (D) Schematic illustration of study design for further functional genomics analysis. Error bars represent SEM. For clinical EAE scores and FACS data, statistical analyses were performed using Mann–Whitney and t test, respectively (*P < 0.05; **P < 0.01; ***P < 0.001).
Fig. S1.
Flow cytometry analysis of cells isolated from lymph nodes harvested on day 7 p.i from rats subjected to the vitamin D-supplemented and -deprived diet (n = 5 for 10 IU and n = 4 for 0 IU), and restimulated for 48 h in vitro with MOG. Vitamin D supplementation did not affect the frequency of CD3− cells, CD4+, and CD8+ T cells as well as FoxP3-expressing cells within the CD4+ and CD8+ compartment. Additionally, vitamin D supplementation did not induce changes in CD8+ T cells, whereas there was a significant decrease in frequency of proliferating CD4+ T cells (as shown by Ki67 staining), both FoxP3+ and FoxP3−, in vitamin D-supplemented rats. Additionally, vitamin D supplementation led to a significant decrease in frequency of IL-17–producing CD4+ T cells and a tendency for a decrease of IFN-γ–producing CD4+ T cells. Statistical analyses were performed using the Student t test (*P < 0.05; **P < 0.01).
Vitamin D Supplementation Induces Marked Changes in the Transcriptome of CD4+ T Cells and Down-Regulates Multiple Signaling and Metabolic Pathways.
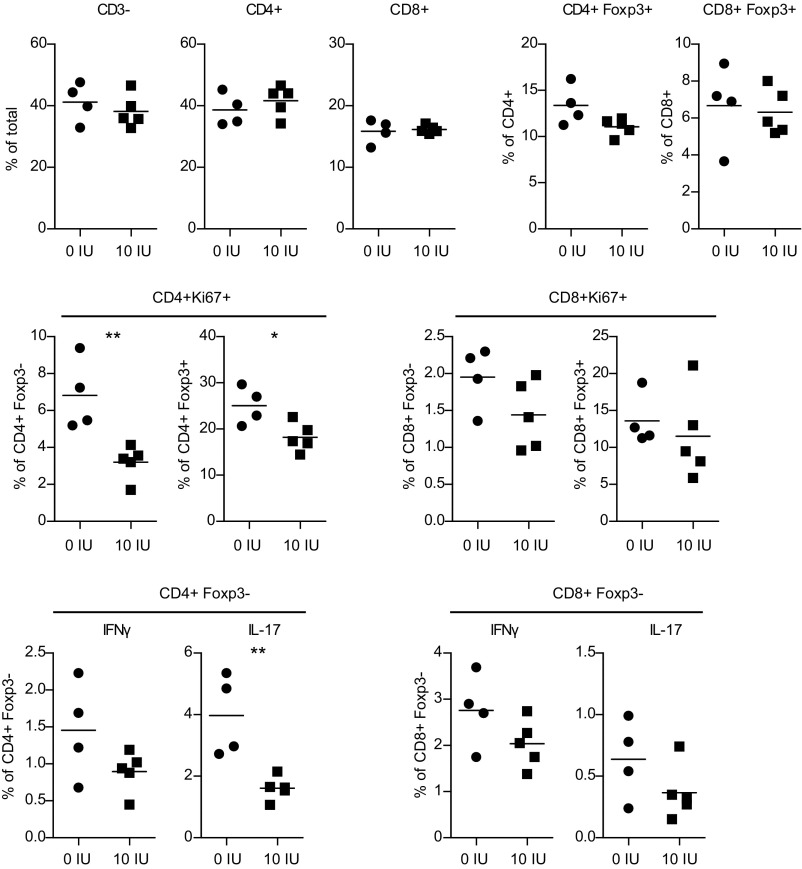
Transcriptome analysis was performed on CD4+ T cells isolated from the draining lymph nodes 7 d p.i. from rats fed vitamin D-supplemented [10 international units (IU) of vitamin D], vitamin D-deprived (0 IU of vitamin D), and a regular rodent diet (2 IU vitamin D) using Affymetrix microarrays (GeneChip Rat Gene 1.0 ST Array; Dataset S1). Principal component analysis (PCA) demonstrated that animals on different vitamin D dietary regimens formed distinct clusters (Fig. 2A). Animals subjected to the vitamin D-supplemented diet displayed marked changes in gene expression compared with animals subjected to a regular and vitamin D-deprived diet (Fig. 2B). Using 1% false discovery rate (FDR), 3,460 probes (3,400 Ensembl genes) were differentially expressed between animals fed with the vitamin D-supplemented and vitamin D-deprived diet (Dataset S2). We observed 1,617 probes (1,598 Ensembl genes) to be differentially expressed between animals fed with a regular and the vitamin D-supplemented diet (Dataset S3), whereas only six probes (six Ensembl genes) showed expression differences between animals fed with a regular and the vitamin D-deprived diet (Dataset S4). Hence, vitamin D supplementation and not deprivation was responsible for transcriptomic changes in CD4+ T cells during EAE. Therefore, we continued the analyses, focusing exclusively on comparison between vitamin D-supplemented and vitamin D-deprived group.
Fig. 2.
Gene signatures of CD4+ T cells upon vitamin D supplementation. Total RNA from CD4+ T cells isolated from inguinal lymph nodes 7 d p.i. was subjected to transcriptomics analysis using GeneChip Gene 1.0 ST Arrays (n = 5 for 0 IU, n = 6 for 10 IU, and n = 6 for 2 IU). (A) PCA demonstrates that the expression profile of CD4+ T cells is different between the vitamin D treatment groups. (B) A heat map diagram of differentially expressed genes shows that the expression profile of CD4+ T cells from rats subjected to vitamin D supplementation is significantly different from that from CD4+ T cells from rats fed with regular diet or diet lacking vitamin D. (C) IPA demonstrates down-regulation of disease-relevant pathways upon vitamin D supplementation. The numbers on the right side of the panel represent the number of molecules associated with the respective pathway. We used FDR 1% and no fold-change cutoff for differentially expressed genes. For pathway analysis, significance was determined with the right-tailed Fisher's exact test and adjusted using the Benjamini-Hochberg correction depicted by –log(B-H P value).
To identify biological functions that are regulated between the vitamin D-supplemented and -deprived groups, we performed functional ingenuity pathway analysis (IPA) on differentially expressed transcripts. IPA revealed that vitamin D had impact on functions such as cell death and survival, cell growth and proliferation, energy production, protein synthesis and trafficking, gene expression, and posttranscriptional modifications, as well as DNA replication, recombination, and repair (Dataset S5). Notably, IPA canonical pathway analysis revealed that multiple molecules involved in activation and differentiation of T cells were down-regulated upon vitamin D supplementation (Fig. 2C, Table S1, and Dataset S6). Multiple transcripts in the TCR, CD28, RhoA, and Erk/Mapk signaling pathways, which are critical for activation and proliferation of T cells, were down-regulated. For example Cd4, Cd3e/Cd3d/Cd3g, Lck, Fyn, and Vav1/Vav3, important members of the TCR signaling pathway, and Shp2, Grb2, Ras, and Mek1, important members of the Erk/Mapk signaling pathway, displayed lower expression in vitamin D-supplemented animals. In addition, multiple members of the Pi3K/Akt/mTor pathway, a signaling cascade crucial for cell proliferation, growth, and metabolism, which acts downstream of TCR, CD28, and IL-2R, were also down-regulated upon vitamin D supplementation. Transcripts of multiple catalytic and regulatory subunits of Pi3K, Akt2/Akt3, and Mtor displayed lower expression in vitamin D-supplemented animals as well as one of the key downstream transcription factors, Hif1a. Interestingly, many molecules involved in the TCA cycle and in glycolysis, which provide not only energy but also building blocks during proliferation, were also down-regulated by vitamin D supplementation. All three key regulatory enzymes of the TCA, Cs, Idh, and Sdh, as well as two members of glycolysis Bpgm and Pgk1, were down-regulated upon vitamin D supplementation, indicating decreased energy consumption of CD4+ T cells in animals supplemented with vitamin D. Notably, multiple transcripts in the Jak/Stat pathway, a signaling cascade engaged by cytokines critical for differentiation into distinct T-helper types, such as IL-2, IL-12, IFN-γ, IL-6, IL-21, IL-23, and GM-CSF (28), were down-regulated upon vitamin D supplementation. Thus, Jak1/Jak2 as well as Stat1/Stat4 and Stat3, which are important for Th1 and Th17 differentiation (29, 30), displayed lower levels in vitamin D-supplemented animals. Additional transcription factors such as NfkB1, Fos, and Jun, all activated by the interaction of Jak/Stat and Erk/Mapk pathways, were down-regulated upon vitamin D supplementation. In contrast, transcripts of genes associated with antiinflammatory properties, such as Il13, Il19, and Il24, were up-regulated upon vitamin D-supplemented diet.
Table S1.
Up- and down-regulated immunologically relevant transcripts in EAE in the supplemented (10 IU) compared with the vitamin D-deprived group (0 IU)
| Gene | Entrez gene name | P value | Fold change, (10 vs. 0) | Probe ID |
| TCR signaling | ||||
| Cd4 | CD4 molecule | 8.53E-04 | −1,47 | 10865585 |
| Cd3d | CD3d molecule | 1.93E-06 | −1,27 | 10909583 |
| Cd3e* | CD3e molecule | 2,76E-02 | −1,15 | 10916955 |
| Cd3g | CD3g molecule | 8.28E-04 | −1,09 | 10916946 |
| Fyn* | Proto-oncogene tyrosine-protein kinase Fyn | 3,51E-03 | −1,32 | 10830382 |
| Lck | LCK proto-oncogene, Src family tyrosine kinase | 1.39E-04 | −1,30 | 10880033 |
| Tec | Tec protein tyrosine kinase | 1.04E-03 | −1,43 | 10772408 |
| Vav1 | Vav guanine nucleotide exchange factor 1 | 9.75E-06 | −1,38 | 10931196 |
| Vav3 | Vav guanine nucleotide exchange factor 3 | 3.66E-04 | −1,22 | 10818454 |
| Jak/Stat signaling | ||||
| Ptpn6 | Protein tyrosine phosphatase, nonreceptor type 6 | 5.91E-06 | −1,35 | 10865463 |
| Ptpn11 | Protein tyrosine phosphatase, nonreceptor type 11 | 5.09E-04 | −1,34 | 10758727 |
| Stat1 | Signal transducer and activator of transcription 1 | 3.79E-04 | −1,18 | 10927842 |
| Stat3 | Signal transducer and activator of transcription 3 | 5.31E-04 | −1,42 | 10747506 |
| Stat4 * | Signal transducer and activator of transcription 4 | 3,79E-04 | −1,18 | 10927875 |
| Stat5b | Signal transducer and activator of transcription 5B | 1.13E-03 | −1,26 | 10747494 |
| Jak1 | Janus kinase 1 | 4.39E-06 | −1,31 | 10878286 |
| Jak2 | Janus kinase 2 | 4.00E-06 | −1,58 | 10714667 |
| Erk/Mapk signaling | ||||
| Grb2 | Growth factor receptor bound protein 2 | 9.95E-04 | −1,36 | 10749005 |
| Kras | Kirsten rat sarcoma viral oncogene homolog | 5.76E-05 | −1,28 | 10866850 |
| Map2k1 | Mitogen-activated protein kinase kinase 1 | 7.49E-08 | −1,37 | 10918209 |
| Sos1 | SOS Ras/Rac guanine nucleotide exchange factor 1 | 2.01E-07 | −1,55 | 10888021 |
| Dusp1 | Dual specificity phosphatase 1 | 3,53E-04 | −2,16 | 10732652 |
| Prkca* | Protein kinase c, alpha | 4,43E-03 | −1,23 | 10748473 |
| Prkcb* | Protein kinase c, beta | 1,95E-03 | −1,22 | 10710647 |
| Prkci | Protein kinase c, iota | 5,90E-04 | −1,40 | 10822644 |
| Pi3K/Akt/mTor signaling | ||||
| Akt2 | V-Akt murine thymoma viral oncogene homolog 2 | 1.11E-03 | −1,34 | 10705394 |
| Akt3 | V-Akt murine thymoma viral oncogene homolog 3 | 3.21E-04 | −1,41 | 10770197 |
| Pik3c3 | Phosphatidylinositol 3-kinase catalytic subunit type 3 | 1.48E-03 | −1,19 | 10800603 |
| Pik3r3 | Phosphoinositide-3-kinase regulatory subunit 3 | 7.47E-04 | −1,43 | 10871229 |
| Pik3r6 | Phosphoinositide-3-kinase regulatory subunit 6 | 1.92E-04 | 1,20 | 10734740 |
| Hif1a | Hypoxia inducible factor 1, alpha subunit | 3.99E-05 | −1,26 | 10885251 |
| Ppp2r1a | Protein phosphatase 2 regulatory subunit A, alpha | 4,56E-05 | −1,46 | 10703479 |
| Ppp2r2a | Protein phosphatase 2 regulatory subunit B, alpha | 1,21E-04 | −1,12 | 10784702 |
| Ppp2r2c | Protein phosphatase 2 regulatory subunit B, gamma | 8,21E-04 | 1,23 | 10777422 |
| Ppp2r5a | Protein phosphatase 2 regulatory subunit B′, alpha | 8,71E-05 | −1,24 | 10770721 |
| TCA cycle | ||||
| Cs | Citrate synthase | 2.94E-04 | −1,39 | 10893127 |
| Dld | Dihydrolipoamide dehydrogenase | 5.64E-05 | −1,35 | 10889522 |
| Dlst | Dihydrolipoamide S-succinyltransferase | 1.05E-03 | −1,25 | 10885998 |
| Idh3a | Isocitrate dehydrogenase 3 (NAD+) alpha | 1.65E-05 | −1,26 | 10910071 |
| Idh3B | Isocitrate dehydrogenase 3 (NAD+) beta | 6.10E-05 | −1,62 | 10849875 |
| Mdh1 | Malate dehydrogenase 1 | 5.85E-06 | −1,41 | 10778661 |
| Mdh2 | Malate dehydrogenase 2 | 1.52E-03 | −1,23 | 10761152 |
| Sdha | Succinate dehydrogenase complex subunit a | 5.19E-05 | −1,32 | 10702467 |
| Sdhb | Succinate dehydrogenase complex subunit b | 1.40E-05 | −1,36 | 10873597 |
| Sdhc | Succinate dehydrogenase complex subunit c | 6.29E-04 | −1,37 | 10769807 |
| Sdhd | Succinate dehydrogenase complex subunit d | 8.74E-05 | −1,43 | 10917236 |
| Sucla2 | Succinate-CoA ligase, ADP-forming, beta subunit | 2.03E-05 | −1,41 | 10781451 |
| Glycolysis/gluconeogenesis | ||||
| Bpgm | Bisphosphoglycerate mutase | 1.09E-04 | −1.497 | 10854437 |
| Fbp1 | Fructose-bisphosphatase 1 | 1.35E-03 | 1.278 | 10793579 |
| Pgk1 | Phosphoglycerate kinase 1 | 8.43E-07 | −2.280 | 10934610 |
| Transcription factors | ||||
| Nfkb1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 6.72E-04 | −1,27 | 10826918 |
| Jun | Jun protooncogene | 1.40E-04 | −3,65 | 10878112 |
| Fos | Fos protooncogene | 3,87E-03 | −1,88 | 10886031 |
| Interleukins and chemokines with antiinflammatory properties in T cells | ||||
| Il-13 | Interleukin 13 | 2.46E-05 | 1,56 | 10742612 |
| Il-19 | Interleukin 19 | 6.46E-04 | 1,23 | 10767482 |
| Il-24 | Interleukin 24 | 4.95E-04 | 1,29 | 10767465 |
| Il-33 | Interleukin 33 | 4.10E-04 | 1,40 | 10714745 |
| Cd52 | CD52 antigen | 1.66E-03 | 1,22 | 10880429 |
| miRNAs implicated in T-cell function and development | ||||
| Mir125a | microRNA 125a | 2.78E-05 | 1,27 | 10703430 |
| Mir210 | microRNA 210 | 7,03E-05 | 1,28 | 10726756 |
| Mir181b2 | microRNA 181a-1 | 3.70E-04 | 1,58 | 10835987 |
| Mir483 | microRNA 483 | 1,23E-03 | 1,32 | 10727006 |
| Mir30c-1 | microRNA 30c-1 | 9,98E-05 | 1,33 | 10879442 |
| Mir449a | microRNA 449a | 2,55E-04 | 1,28 | 10813101 |
| Mir134 | microRNA 134 | 2,16E-04 | 1,28 | 10887092 |
| Mir23a | microRNA 23a | 9,17E-04 | 1,37 | 10810152 |
| Mir377 | microRNA 377 | 1,93E-04 | 1,24 | 10887100 |
| Mir7a-1 | microRNA let-7a-1 | 7.69E-04 | 1,69 | 10793838 |
| miRNAs implicated in MS/EAE | ||||
| Mir125a | microRNA 125a | 2.78E-05 | 1,27 | 10703430 |
| Mir210 | microRNA 210 | 7,03E-05 | 1,28 | 10726756 |
| Mir760 | microRNA 760 | 6.80E-06 | 1,35 | 10826500 |
| Mir186 | microRNA 186 | 1.59E-05 | 3,31 | 10819903 |
| Mir181c * | microRNA 181c | 3,18E-03 | 1,15 | 10806724 |
| Mir181b2 | microRNA181b2 | 3,70E-04 | 1,58 | 10835987 |
| Mir30c-1 | microRNA 30c-1 | 9,98E-05 | 1,33 | 10879442 |
| Mir23a | microRNA 23a | 9.17E-04 | 1,37 | 10810152 |
| Mir328 | microRNA 328 | 2.17E-04 | 1,25 | 10810583 |
| Mir346 | microRNA 346 | 4.86E-04 | 1,30 | 10787029 |
| Mir7a-1 | microRNA 7a-1 | 7.69E-04 | 1,69 | 10793838 |
| Mir23a | microRNA 23a | 9,17E-04 | 1,37 | 10810152 |
Selected based on T-cell function. FDR 1%, and no fold-change cutoff was used.
Significantly up-regulated but with FDR 5%.
Taken together, changes in the transcriptome of CD4+ T cells implicate that vitamin D supplementation down-modulates T-cell metabolism and signaling pathways that are critical for T-cell activation and differentiation into Th1 and Th17 cells.
Vitamin D Supplementation Increases Expression of ncRNAs, Including miRNA Genes.
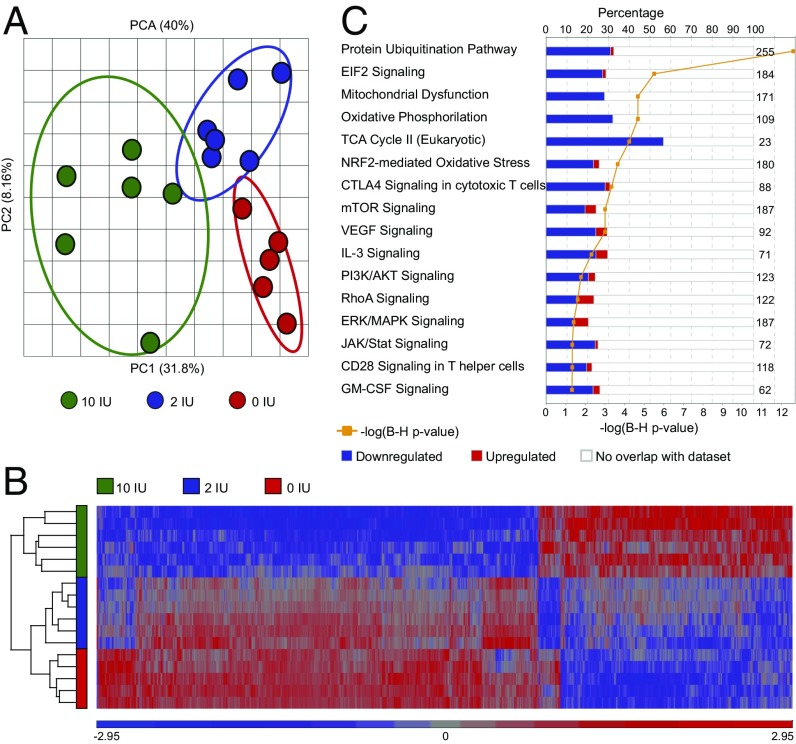
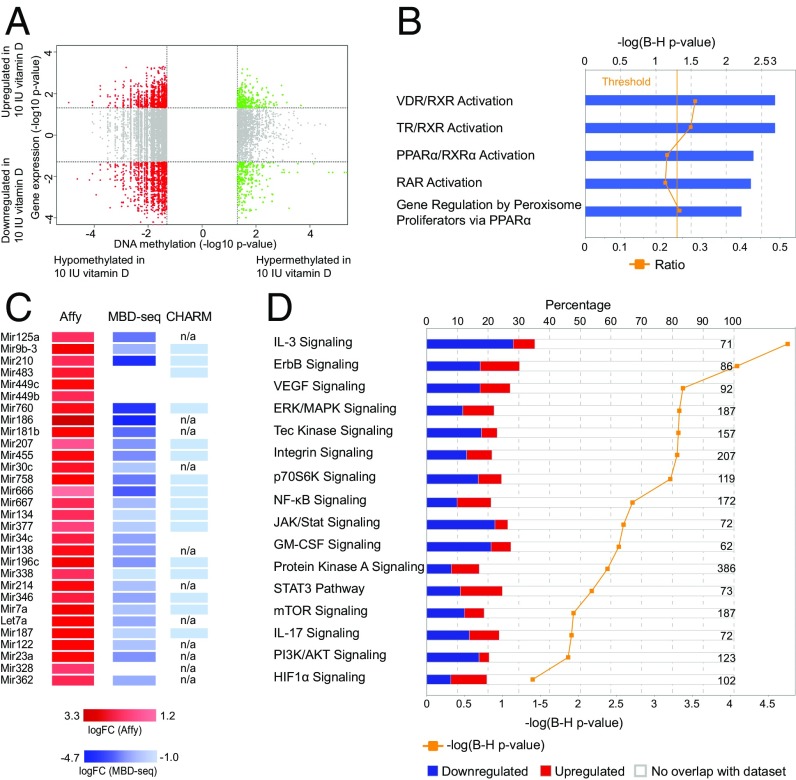
Examining the type of differentially expressed genes, we observed that although protein-coding genes preferentially showed lower expression (62%, enrichment P < 2 × 10−3), noncoding genes displayed preferential higher expression (70%, enrichment P < 2 × 10−24) in the vitamin D-supplemented group (Fig. 3A). In particular, snRNA, miRNA, and ribosomal RNA genes showed predominant higher expression in vitamin D-supplemented animals (with 96%, 89%, and 100% up-regulated probes, respectively).
Fig. 3.
Vitamin D supplementation influences miRNA gene expression. (A) Representation of the proportion of up- and down-regulated classes of RNAs differentially expressed in CD4+ T cells upon vitamin D supplementation. Statistical analysis was done using the χ2 test. (B) A heat map diagram of well-annotated differentially expressed probes with FDR 1% uniquely mapping to miRNA genes in CD4+ T cells upon vitamin D supplementation. (C) IPA of the predicted miRNA targets, using conserved targets predicted by TargetScan, shows that many of the pathways between predicted miRNA targets and differential expressed genes following vitamin D supplementation were overlapping. Based on the significance of predicted target genes, we classified the pathways according to their dependence on miRNAs. Significance was determined with the right-tailed Fisher's exact test and adjusted using the Benjamini-Hochberg correction depicted by –log(B-H P value).
All 30 well-annotated differentially expressed miRNA probes at 1% FDR demonstrated higher levels in the vitamin D-supplemented group (Fig. 3B) and 92 of 100 differentially expressed miRNA probes were up-regulated at nominal significance (P < 0.05). Because the best-described function of miRNAs is to reduce the amount of the target mRNAs on the posttranscriptional level, we speculated that the increased miRNAs can be responsible for lower levels of protein-coding genes. Indeed, TargetScan predicted target genes of the up-regulated miRNAs were enriched among genes that were down-regulated in the vitamin D-supplemented group (P < 1 × 10−3). In addition, IPA identified multiple miRNAs as activated upstream regulators based on the observed expression changes of IPA predicted (experimentally validated) miRNA targets in our dataset (Dataset S7). By analyzing pathways of the predicted targets of 30 detected up-regulated miRNAs, we observed that many of the pathways overlap with the pathways affected by all differentially expressed genes (Dataset S8). Based on their significance, we classified these pathways according to their dependence on miRNAs (Fig. 3C). Notably, the pathways of general importance for cell survival, such as mitochondrial functions, protein synthesis, telomere extension, and repair mechanisms, did not seem to be regulated by miRNAs. However, pathways important for T-cell activation and differentiation, such as Pi3k/Akt/mTor, Erk/Mapk, and Jak/Stat pathways, seemed to be at least in part mediated by miRNAs, whereas the TCR and IL-2 signaling appear to be heavily dependent on regulation by miRNAs. Notably, genes critical for T-cell activation and signaling are predicted targets of multiple up-regulated miRNAs (e.g., Kras is a target of miR-30c, -134, -181b, and -483; Vav3 is a target of miR-9, -30c, -449c, and let-7a; and Pik3r3 is a target of miR-9, -23a, -181b, and -377).
Our data indicate that miRNAs may, at least in part, mediate the effect of vitamin D supplementation on signaling pathways in CD4+ T cells in MOG–EAE.
Vitamin D Supplementation Reduces DNA Methylation Genome Wide in CD4+ T Cells.
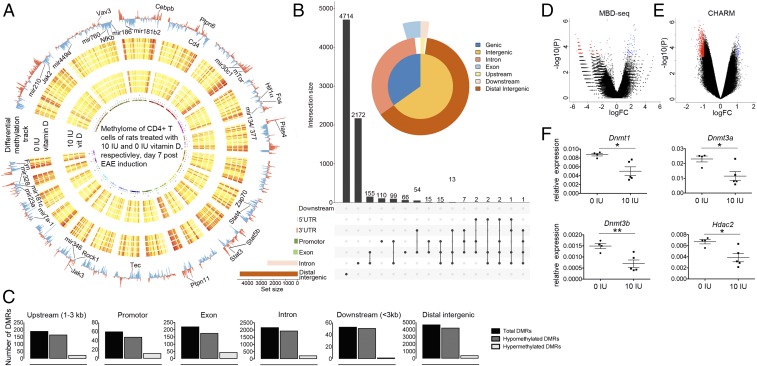
To address the impact of vitamin D on DNA methylation, an important epigenetic mark that can actively impact gene regulation on the transcriptional level or be a marker of the genome activity (31), we used methyl-CpG binding domain sequencing (MBD-seq). This method investigated differentially methylated regions (DMRs) between the vitamin D-supplemented and -deprived groups in the entire genome (Fig. 4A and Dataset S9). The methylation changes predominantly affected distal intergenic regions, whereas changes in genic regions primarily affected introns (Fig. 4B). For example, there were 491 DMRs, which associated with 413 Ensembl genes, with a P value lower than 0.001. Of these DMRs, 14 mapped to promoter (<3 kb) regions, 11 to exons, 153 to introns, 4 to downstream (<3 kb) regions, and 309 to distal intergenic regions. Interestingly, the vast majority of DMRs, irrespective of the genomic location and significance level, displayed lower methylation in the vitamin D-supplemented group (Fig. 4 C and D). This preferential lower methylation in the vitamin D-supplemented group was confirmed with a different method to measure methylation genome wide, the comprehensive high-throughput arrays for relative methylation method (CHARM) (Fig. 4E); consistent with this, CD4+ T cells from vitamin D-supplemented animals displayed lower expression of all three active DNA methyltransferases, Dnmt1, Dnmt3a, and Dnmt3b, as well as many other members of the cellular epigenetic machinery—for example, Cbx1, Hdac1, and Hdac2 (Dataset S2). Significantly lower expression of the enzymes involved in establishing and maintaining DNA methylation marks, which could explain preferential lower methylation upon vitamin D supplementation, was validated in independent samples using qPCR (Fig. 4F). Thus, vitamin D supplementation reduces the levels of enzymes involved in establishing and maintaining DNA methylation marks, subsequently contributing to widespread reduction of DNA methylation.
Fig. 4.
Widespread DNA methylation changes in CD4+ T cells upon vitamin D supplementation. gDNA from CD4+ T cells isolated from inguinal lymph nodes 7 d p.i. was subjected to DNA methylome analysis using MBD-seq (0 IU and 10 IU, n = 4 for each diet group). (A) The genome-wide map of all autosomal DMRs (P < 0.01) is shown as a circular ideogram, composed of concentric circles depicting the entire autosome complement, with chromosomal location annotated in a clockwise manner. In the Circos plot, 0 IU DMRs are visualized as a blue histogram plot and 10 IU DMRs as a red histogram plot. Each sample separately is shown as a heat map with hypomethylation in yellow and hypermethylation in red. Selected genes of important pathways evaluated in this study are indicated. (B) Graphic representation of DMRs (P < 0.01) at different genomic locations. (C) Numbers of hypo, hyper, and total DMRs (P < 0.01) at different genomic locations. Volcano plots of DMRs identified using (D) MBD-seq and (E) CHARM shows that more DMRs are hypomethylated upon vitamin D supplementation. The x and y axis show log (fold change) and −log10 (P value), respectively. Red dots correspond to DMRs that are significantly hypomethylated (P < 0.001) and blue dots correspond to DMRs that are significantly hypermethylated (P < 0.001). (F) qPCR analyses in independent samples confirms that transcripts necessary for active DNA methylation and histone acetylation, Dnmt1, Dnmt3a, 3b, and Hdac2, respectively, are down-regulated upon vitamin D supplementation (n = 4 for 0 IU and n = 5 for 10 IU). Error bars represent SEM, and statistical analysis was performed using the Student t test (*P < 0.05; **P < 0.01). Details of MBD-seq and CHARM analyses are provided in Materials and Methods.
Vitamin D Modulates Signaling Pathways in CD4+ T Cells on Both Epigenetic and Transcriptional Level and Impacts Function and Encephalitogenic Potential of CD4+ T Cells.
Finally, we wanted to investigate T-cell pathways that are affected by changes on both epigenetic and transcriptional levels upon vitamin D supplementation. To that end, we investigated genes that displayed changes in expression profile and methylation levels (Fig. 5A). We detected 2,562 Ensembl genes associated with 6,418 DMRs exhibiting changes in methylation and expression with nominal significance (Dataset S10). We observed negative and positive correlation of methylation with gene expression in the promoter (<1 kb) and exon regions, respectively. There was a statistically significant correlation between hypomethylated DMRs in promoter regions and increased gene expression (P < 2 × 10−5). There were too few hypermethylated DMRs in promoters to test the inverse correlation. In contrast, in exons we observed a significant correlation between hypomethylated DMRs and lower gene expression (P < 2 × 10−19) and hypermethylated DMRs and higher gene expression (P < 3 × 10−6).
Fig. 5.
Analysis of genes that display changes both in expression and methylation in CD4+ T cells upon vitamin D supplementation. (A) Quadrant plot of DMRs and expression of associated genes. On the x axis the −log10 (P value) for DMRs is shown, and on the y axis the −log10 (P value) of differential expression for associated genes is shown. Vertical dashed lines indicate a threshold of P < 0.05 and horizontal dashed lines indicate a threshold corresponding to P < 0.05. The four quadrants shown are (i) hypermethylated and up-regulated in 10 IU (green, Upper), (ii) hypermethylated and down-regulated in 10 IU (green, Lower), (iii) hypomethylated and up-regulated in 10 IU (red, Upper), and (iv) hypomethylated and down-regulated in 10 IU (red, Lower). (B) Topmost significant compounds identified by Ingenuity Toxicogenomic analysis on those genes that show both differential expression and DMRs (P < 0.05). (C) Up-regulated microRNA genes display evidence of hypomethylation with MBD-seq and CHARM (the exact fold-change values are provided for DMRs identified with MBD-seq, whereas for CHARM only the direction of the change is indicated). (D) Pathway analysis in Ingenuity of those genes that show both differential expression and DMRs (P < 0.05). Significance was determined with the right-tailed Fisher's exact test and adjusted using the Benjamini-Hochberg correction depicted by –log(B-H P value). The numbers on the right side of the panel represent the number of molecules associated with the respective pathway.
IPA toxicogenomics analysis on transcripts that displayed changes in both expression and methylation revealed VDR/RXR activation among the topmost significant compound, strongly suggesting that genes with both methylation and expression changes are more proximal mediators of the VDR signaling (Fig. 5B). Significant upstream regulator analysis predominantly implicated miRNAs as upstream regulators based on the observed expression changes of genes affected by both methylation and expression, which suggests that vitamin D supplementation may affect DNA methylation of miRNA genes that regulates expression levels of these miRNAs, which in turn down-regulates levels of protein-coding genes. Indeed, there was evidence of lower methylation in the regions encoding 30 miRNAs detected to be up-regulated in the vitamin D-supplemented group (Fig. 5C).
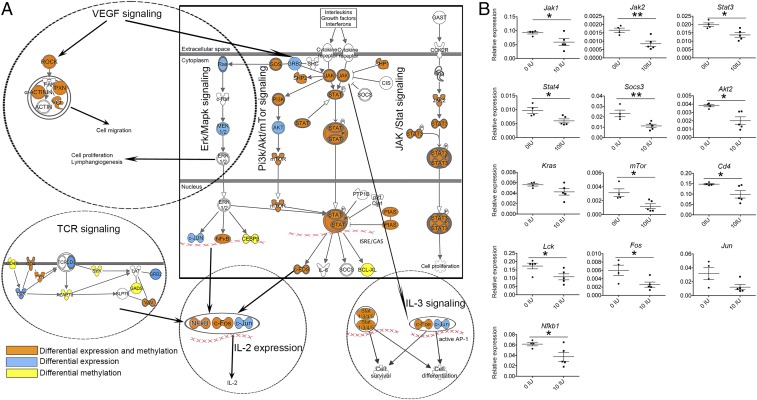
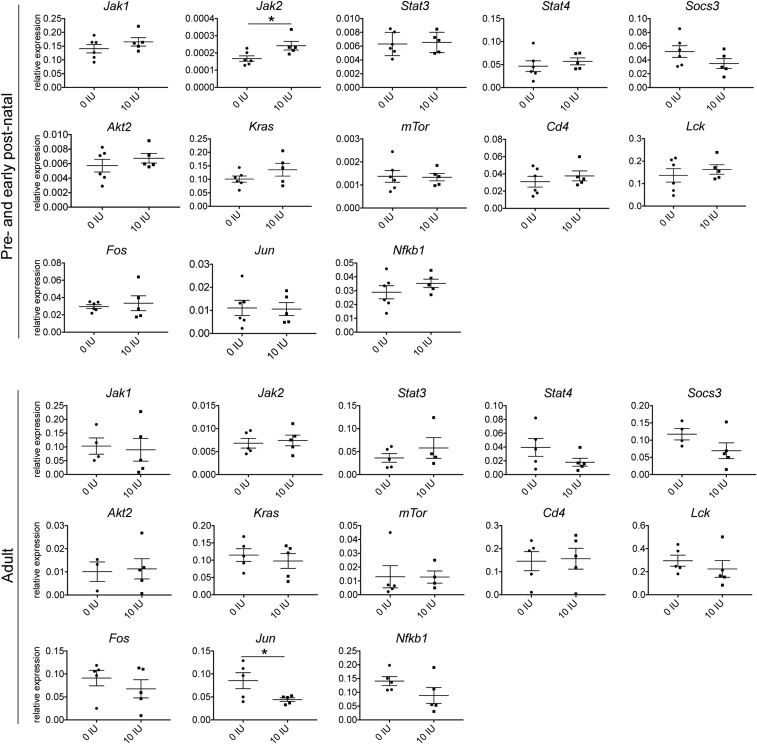
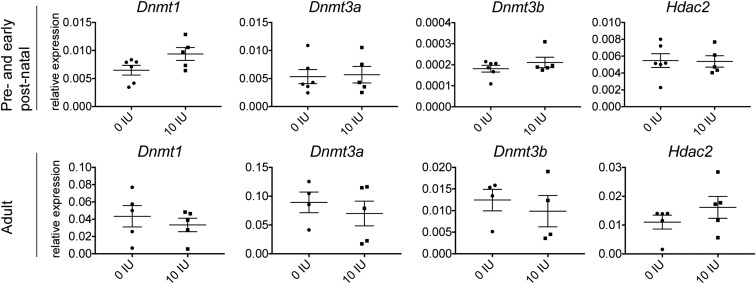
Canonical IPA pathway analysis revealed that members of multiple pathways that were down-regulated showed also changes in methylation levels (Fig. 5D and Dataset S11). Multiple members of TCR and Erk/Mapk pathways important for activation and differentiation of T cells were affected both by expression and methylation (Fig. 6A). For example, Sos and Nfkb1 showed methylation and expression changes, whereas Grb2, Ras, Mek1/2, and Jun showed expression changes only. In addition, T-cell coreceptors Cd4 and Fyn, which are important for activation of the TCR, showed both differential expression and methylation; Cd3 and Lck showed differential expression and Cd45, Zap70, Lat, and Syk showed differential methylation. In addition, members of the Pi3k/Akt/mTor pathway, crucial for cell proliferation, growth, and metabolism and the Jak/Stat pathway crucial for differentiation of T cells into distinct subsets, showed differential expression and methylation (Fig. 6A). For instance, downstream mediators such as Stat3, Stat4, Jak2, Pi3k family members, Mtor, and Fos exhibited differential expression and methylation changes, whereas Akt showed differential expression only. Moreover, the Vegf pathway important for cell migration and cell proliferation was down-regulated and differentially methylated upon vitamin D supplementation. Differences in expression of key genes of the pathways were confirmed in independent samples from rats treated during juvenile/adolescent age, by real-time qPCR analysis (Fig. 6B). Interestingly, the same genes were not affected in CD4+ T cells from rats treated during either adult or pre- and early postnatal age, i.e., the treatment regimens that we have previously shown not to be efficient in ameliorating EAE (23). In contrast to juvenile/adolescent rats, significant changes with regard to the signaling pathway genes were reduced to down-regulation of Jun in adult rats and up-regulation of Jak2 in pre- and early postnatally treated rats (Fig. S2). Similarly, in contrast to changes in epigenetic enzymes observed in juvenile/adolescent rats, we could not detect any differences in Dnmt1, Dnmt3a, Dnmt3b, and Hdac2 expression in the two other age groups (Fig. S3).
Fig. 6.
Vitamin D supplementation down-regulates multiple disease-driving pathways in CD4+ T cells. (A) Schematic representation of Jak/Stat, Erk/Mapk, Pi3k/Akt, Vegf, TCR, and IL-3 signaling that are affected by vitamin D. Orange indicates genes that are differentially expressed and methylated. Blue indicates genes that are differentially expressed only, and yellow indicates genes that are differentially methylated only. (B) qPCR confirmation in independent samples of key players of the pathways depicted in A. Error bars represent SEM, and statistical analysis was performed using the Student t test (*P < 0.05; **P < 0.01).
Fig. S2.
Key transcripts of the Jak/Stat, Erk/Mapk, and Pi3k/Akt/mTor pathways are not differentially expressed upon vitamin D treatment in CD4+ T cells from pre- and early postnatally treated and adult rats. Graphs presenting mRNA levels of indicated targets in CD4+ T cells isolated from inguinal lymph nodes harvested on day 7 p.i. from pre- and early postnatally treated and adult rats, respectively, subjected to different amounts of vitamin D through the diets; analysis performed by qPCR (n = 5–6 for pre- and early postnatally treated rats; n = 4–5 for adult rats). Pre- and early postnatally treated rats did not show any significant differences for the tested transcripts of the Jak/Stat, Erk/Mapk, and Pi3k/Akt/mTor pathways, except for Jak2, which was up-regulated in rats supplemented with vitamin D. In addition, adult rats did not show any significant differences for the transcripts tested except for Jun, which was down-regulated in rats supplemented with vitamin D. Statistical analysis was performed using the Student t test (*P < 0.05).
Fig. S3.
Enzymes important for establishing and maintaining DNA methylation marks are not differentially expressed upon vitamin D treatment in CD4+ T cells from pre- and early postnatally treated and adult rats. Graphs presenting mRNA levels of indicated targets in CD4+ T cells isolated from inguinal lymph nodes harvested on day 7 p.i. from pre- and early postnatally treated and adult rats, respectively, subjected to different amounts of vitamin D through the diets; analysis performed by qPCR (n = 5–6 for pre- and early postnatally treated rats; n = 4–5 for adult rats).
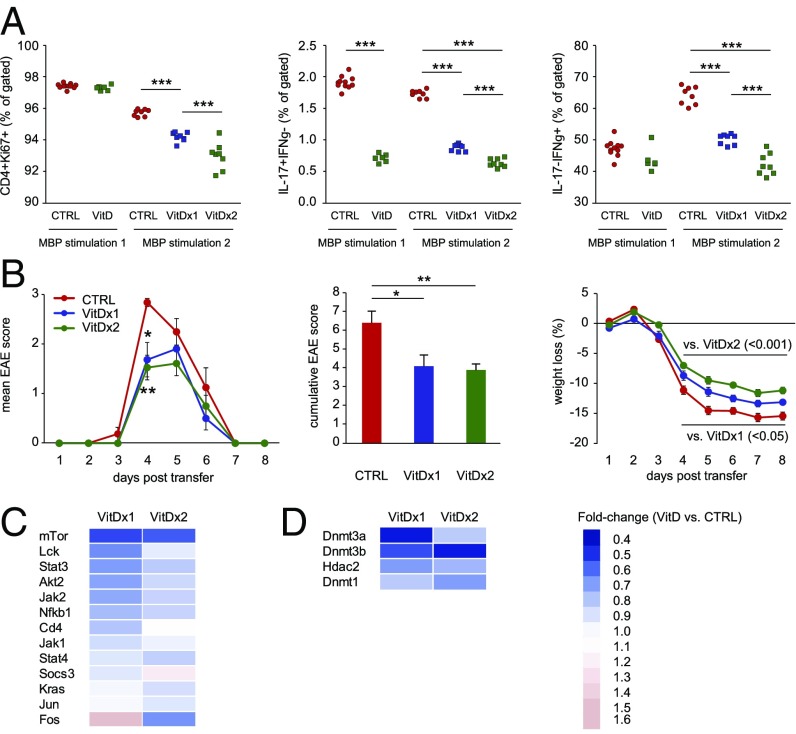
To assess the impact of vitamin D on CD4+ T-cell function and their encephalitogenic potential, we conducted adoptive transfer experiments in DA rats with myelin basic protein (MBP)63–88-specific T-cell lines treated with calcitriol (1,25(OH)2D3). Similar to our findings in CD4+ T cells from actively induced MOG–EAE, vitamin D-treated MBP63–88-specific T cells displayed significantly lower proliferation of CD4+ T cells and frequency of Th1 and Th17 cells (Fig. 7A). This effect was dependent on the number of exposures to vitamin D, i.e., T cells treated during two rounds of stimulation with MBP63–88 exerted a more prominent effect compared with cells treated only during the last stimulation round. Moreover, upon transfer to naïve DA rats, MBP63–88-specific T cells treated with vitamin D induced milder EAE with significantly lower cumulative disease score and weight loss compared with untreated cells (Fig. 7B). In addition, the majority of the key genes of Jak/Stat, Erk/Mapk, and Pi3k/Akt/mTor pathways (Fig. 7C) and enzymes important for establishing and maintaining DNA methylation marks (Fig. 7D) were down-regulated in vitamin D-treated MBP63–88-specific T cells.
Fig. 7.
Vitamin D treatment of MBP63–88-specific T-cell lines reduces their encephalitogenic potential and associates it with changes in CD4+ T-cell function and down-regulation of signaling pathway and epigenetic machinery genes. (A) Flow cytometry analysis of MBP63–88-specific T-cell lines treated with 10 nM 1,25(OH)2D3 (calcitriol) during two rounds of stimulation with MBP63–88 (VitD×2) and during the last stimulation round only (VitD×1) shows a significant decrease in frequency of proliferating CD4+ T cells, as shown by Ki67 staining. Additionally, 1,25(OH)2D3 treatment led to a significant decrease in frequency of IL-17– and IFN-γ–producing CD4+ T cells in an exposure-dependent manner. (B) Transfer of 1,25(OH)2D3-treated MBP63–88-specific T-cell lines (VitD×2 and VitD×1) to naïve rats induces milder EAE with significantly lower cumulative score and weight loss compared with untreated MBP63–88-specific T-cell lines (CTRL). (C) qPCR analysis demonstrates that 1,25(OH)2D3 treatment of MBP63–88-specific T-cell lines induces down-modulation of multiple key transcripts of the Jak/Stat, Erk/Mapk, and Pi3k/Akt/mTor pathways, as depicted by differences in fold change. (D) qPCR analysis demonstrates that 1,25(OH)2D3 treatment of MBP63–88-specific T-cell lines induces down-regulation of key enzymes important for establishing and maintaining DNA methylation marks, as depicted by differences in fold change. Error bars represent SEM. Statistical analysis was performed using ANOVA with Bonferroni correction for multiple testing for FACS data and weight loss, and Kruskal–Wallis test with Dunn’s correction for multiple testing for clinical EAE scores and cumulative score (*P < 0.05; **P < 0.01; ***P < 0.001).
These data demonstrate a link among vitamin D, CD4+ T-cell function, and their encephalitogenic potential and associate them with changes in signaling pathways and epigenetic machinery enzymes.
Discussion
We used transcriptome, methylome, and pathway analyses to elucidate biological processes in CD4+ T cells that mediate the in vivo protective effect of vitamin D on autoimmunity. Moreover, we show that these processes associate with changes in T cells treated with vitamin D in vitro and their capacity to induce disease. Vitamin D down-regulated multiple signaling and metabolic pathways that are critical for T-cell activation and differentiation into pathogenic Th1 and Th17 subsets. This effect was associated with epigenetic mechanisms and involved global reduction in DNA methylation and up-regulation of several classes of ncRNAs, including miRNAs.
We observed striking changes in the transcriptome of CD4+ T cells with 3,400 transcripts displaying differential expression between the vitamin D-supplemented and -deprived group. Vitamin D has the potential to affect a large number of genes, because VDR binds thousands of genomic sites in immune cell lines stimulated with 1,25(OH)2D3 (20, 32, 33). There is, moreover, a positive correlation between vitamin D levels and the number of VDR binding sites in the genome (32, 34, 35). For example, although in vitamin D-deficient individuals VDR binds to 601 sites in primary CD4+ T cells, this number increases to 4,518 (7.5-fold) in vitamin D-sufficient individuals (35). Thus, a constant dietary vitamin D supplementation in rats, which we have previously shown to significantly increase levels of 25(OH)D, the major circulating form of vitamin D (23), may engage thousands of additional VDR binding sites, leading to marked changes in gene expression.
Our data additionally demonstrate a widespread effect of vitamin D on DNA methylation of CD4+ T using two methods that use different principles to quantify DNA methylation. CHARM enriches for unmethylated DNA using the McrBC restriction enzyme, followed by identification of digested DNA by hybridization to preselected loci on the array (36). However, MBD-seq combines precipitation of methylated DNA by recombinant methyl-CpG binding domain of the MBD2 protein and identification, across the entire genome, of the isolated DNA using parallel sequencing (37). Thus, we could confirm that vitamin D supplementation induces a decrease in DNA methylation at numerous regions in the genome, although the sample size was too small to reliably establish all DMRs. The impact of vitamin D on DNA methylation is rather unique, with several studies in cancer starting to reveal this link, although the underlying mechanisms are still unknown (21, 22). Changes in leukocyte DNA methylation between vitamin D-deficient and -sufficient healthy adolescent males of African-American origin have recently been reported using 27,000 CpG methylation arrays (38). To date, epigenetic effects by vitamin D have primarily been associated with histone modifications, and it is well known that binding of vitamin D to VDR/RXR induces conformational changes that favor release of corepressors and interaction with coactivators and histone acetyltransferases (21, 22). Indeed, a recent study using FAIRE-seq in a monocytic cell line demonstrated that vitamin D affects chromatin accessibility at nearly 9,000 sites in the genome (39). The most pronounced effect was observed early after VDR/RXR engagement, causing opening of the chromatin with CCCTC-binding factor (CTCF) being likely involved in this early reprograming. Because transcriptionally active regulatory regions are devoid of DNA methylation, it is likely that in our model of constant exposure to vitamin D, a large fraction of detected hypomethylated regions is a consequence of VDR/RXR binding to these loci. VDR/RXR can either induce demethylation at bound loci or protect them from methylation through the recruitment of CTCF and other interacting partners. This hypothesis is further supported by unbiased IPA analysis that identified VDR/RXR as the most significant upstream compound when genes that displayed changes in both methylation and expression were analyzed.
Furthermore, our data support an additional mechanism whereby vitamin D can induce global hypomethylation through the reduced levels of DNA methyltransferases. We demonstrated lower expression of all three active DNA methyltransferases, Dnmt1, Dnmt3a, and Dnmt3b, in CD4+ T cells from the vitamin D-supplemented group as well as in MBP63–88-specific T-cell lines treated with vitamin D (calcitriol). A similar mechanism has been suggested in cancer, where it has been shown that vitamin D can induce hypomethylation and reactivation of tumor suppressor genes through regulation of transcriptional regulators of Dnmt1 (40). Described transcriptional regulators of Dnmt1, Fos and Jun (comprising AP-1), Stat3, Sp1, and Nfkb1 (41–43), were all down-regulated in CD4+ T cells upon vitamin D supplementation. Besides affecting the DNA methylation machinery, vitamin D supplementation associated with down-regulation of several histone modifiers, which generally associate with gene repression, including Hdac1, Hdac2, Kdm1b, Kdm2a, and Kdm5a. Vitamin D, thus, likely uses several mechanisms to impact the epigenome of CD4+ T cells. The methylation changes were particularly abundant in intergenic and intronic regions, suggesting that vitamin D supplementation may target those regions that have recently been shown to be important for differentiation into Th subsets (44). Indeed, a number of signaling pathways important for Th differentiation—for example, Stat3 signaling crucial for Th17 differentiation—emerged when genes that displayed changes in both methylation and expression were analyzed. These changes may explain the observed significant reduction in frequency of highly pathogenic Th17 cells, observed both in actively induced MOG–EAE and in MBP63–88-specific T cells treated in vitro, because it has been shown that DNA methylation controls the high plasticity of Th17 cells (45). Interestingly, the transcription profile induced by vitamin D supplementation resembled that induced by valproic acid, which is a class I and II histone deacetylase (HDAC) inhibitor that also causes genome-wide DNA demethylation (46) and proteosomal degradation of HDAC2 (47) and has been shown by us and others to ameliorate EAE and affect Th17 cells (48, 49). This finding suggests that vitamin D might share protective mechanisms with other epigenetic drugs. Notably, vitamin D has been shown to act in synergy with several epigenetic drugs (50), providing interesting prospects for future combined therapies, especially in the light of dynamic development of HDAC inhibitors for clinical use in various cancers.
The type of VDR/RXR-interacting proteins suggests that vitamin D can induce both gene activation and repression (21). Although early response to vitamin D in a monocytic cell line is characterized by activation of gene expression, late response is characterized by the closing of the affected chromatin and similar proportion of up- and down-regulated genes (39). Equal proportion of up- and down-regulated genes has also been described in primary T cells after 10 d of exposure to a vitamin D analog (51). We observed similar distribution with 59% (2,050/3,460) of affected probes being down-regulated. The mechanisms that lead to vitamin D-induced gene repression seem to be more diverse, and new modes of VDR actions have been suggested, including ncRNAs (21). Indeed, we observed a predominant up-regulation of several classes of ncRNAs upon vitamin D supplementation. Additionally, although VDR/RXR emerged as the upstream regulator when genes that displayed changes in methylation and expression were analyzed, this was not the case when only differentially expressed genes were analyzed, suggesting involvement of RNA mediators. Particularly attractive candidates are miRNAs, ∼22-nucleotide-long ncRNAs that bind to complementary sequences on the corresponding mRNA and cause their degradation. Interestingly, the Affymetrix probes specific for primary miRNAs that displayed differential expression were primarily up-regulated in the vitamin D-supplemented group, and their predicted targets were significantly enriched among down-regulated genes. The involvement of miRNAs was further functionally supported by unbiased IPA prediction of multiple miRNAs as the most significant activated upstream regulators that can explain the observed pattern of differential gene expression. In the context of cancer, studies are emerging that suggest that miRNAs constitute an integral part of VDR signaling and participate in anticancer actions of vitamin D (52, 53). IPA analysis of conserved targets of multiple up-regulated miRNAs suggests that they can target signaling pathways critical for T-cell activation and differentiation. Some of the up-regulated miRNAs have well-documented roles that support this hypothesis. Notably, miR-125a suppresses several effector T-cell factors such as Stat3 and Ifng and stabilizes the commitment and immunomodulatory capacity of Tregs during EAE (54). Moreover, most of the up-regulated miRNAs analyzed here were also hypomethylated, which suggests that VDR/RXR binding may directly activate miRNAs, and they in turn mediate down-regulation of protein-coding genes. Based on our observations and the published data, miRNA are likely an important mediator of a protective vitamin D effect in CD4+ T cells in the context of autoimmunity.
The most pronounced effect of vitamin D supplementation in our study was reduced proliferation of CD4+ T cells. This effect of vitamin D has previously been demonstrated in vitro and in vivo in different species and conditions, including murine EAE (15, 16, 18). In addition to a decrease in expression of cyclins that control cell cycle progression, our functional genomics data suggest that the effect is mediated through down-regulation of pathways critical for TCR signaling. We observed down-regulation of several coreceptors and Src family kinases, which have a critical role in proximal TCR signal transduction. Multiple members of distal TCR signaling pathways, such as Erk/Mapk and Pi3K/Akt, which are also engaged downstream IL-2R, were down-regulated in the vitamin D-supplemented group, including components of AP-1 transcription factor. It has been shown in vitro that VDR/RXR can repress IL-2 expression by direct inhibition of NFATp/AP-1 formation (55). Altogether, our data indicate that vitamin D supplementation most likely influences composition of the immunologic synapse and propagation of signaling events, which ultimately contributes to reduced CD4+ T-cell proliferation. We also observed down-regulation of key enzymes involved in glycolysis, TCA cycle, and oxidative phosphorylation, the metabolic pathways that get activated to meet increased biosynthetic demands during polyclonal T-cell expansion and effector functions (56). We observed effect of vitamin D on Th17 cells, which has also been shown in murine EAE (17, 18). Notably, the Pi3K/Akt/mTor pathway, including the key metabolic regulator Mtor and transcription factors Myc and Hifa (57, 58), were down-regulated upon vitamin D supplementation. These changes may explain the observed impact on Th17 cells, because expression of Hif1α is mTorc1 dependent, and treatment with the mTorc1 inhibitor rapamycin was shown to impair differentiation of Th17 subset (59). Further impact on Th17 cells is likely caused by down-regulation of the Jak/Stat pathway—in particular, Stat3, which is critical for Th17 differentiation (29). We also observed impact on Stat1 and Stat4, which are critical for Th1 differentiation (30). The effect of vitamin D has previously been associated with inhibition of IL-12/IFN-γ axis and Th1 development (15), partly through modulation of JAK/STAT signaling in T cells and myeloid cells (16). However, Stat4 also induces secretion of GM-CSF in both Th1 and Th17 cells (60), and GM-CSF has recently been shown to be essential for induction of EAE (61). In contrast to the observed changes in the signaling and metabolic pathways in juvenile/adolescent rats, no consistent changes were detected in CD4+ T cells from either adult or pre- and early postnatally treated rats, which are not protected by vitamin D supplementation (23), suggesting that these pathways in CD4+ T cells are important in mediating protective vitamin D effect in EAE. Moreover, down-regulation of Jaks, Stats, and Mtor and decreased proliferation and frequency of Th1 and Th17 cells were also confirmed when T cells were treated in vitro with vitamin D, and were associated with decreased encephalitogenic potential of T cells to transfer disease. Thus, although additional mechanisms may be involved in vitamin D protection in vivo, our data demonstrate that one important mechanism involves direct impact on signaling and metabolic pathways in CD4+ T cells.
Patterns of VDR binding (20, 35) and gene expression (39, 51) in cell lines and healthy subjects have suggested an effect of vitamin D on metabolic and signaling pathways crucial for cell survival, growth, and proliferation. Our data confirm that these pathways are also affected in vivo in CD4+ T cells, mediating the protective effect of the vitamin D supplementation in experimental autoimmune disease. VDR binding has been found enriched near autoimmune risk genes (32, 35), and together with epidemiological data, this finding strongly indicates that vitamin D modulates risk to autoimmune diseases. We found that nearly 50% of the rat orthologs of established candidate MS risk genes that bind VDR in primary human CD4+ T cells (62) and ∼40% of the signature genes of myelin-reactive T cells in MS (63) changed their expression in vivo upon vitamin D supplementation in EAE (Table S2). Remarkably, nearly 80% of the latter reverted their expression profile toward physiological upon vitamin D supplementation. Hence, our in vivo data from the animal model of MS support the role of vitamin D in modulating genes important for human disease. In addition, our study highlights significance of vitamin D supplementation for prevention or treatment of autoimmune diseases in general because CD4+ T cells are driving target organ destruction in autoimmune diseases (64) and because many of the autoimmune loci are shared by multiple autoimmune diseases (65).
Table S2.
Comparison of genes affected with vitamin D supplementation in EAE with candidate MS genes
| Genes enriched in myelin-reactive T cells from MS (ref. 63, figure S12), % | Pathway genes enriched in myelin-reactive T cells in MS (ref. 63, figure S12), % | MS risk genes (ref. 62, tables S1 and S2), % | |||||||
| IL-23 | AP1 | NFAT | ATF2 | GPCR | All | VDRE | VDR | ||
| Differentially expressed | 42 | 57 | 33 | 30 | 57 | 28 | 48 | 48 | 54 |
| Up-regulated | 19 | 8 | 33 | 18 | 24 | 55 | 38 | 38 | 30 |
| Down-regulated | 81 | 92 | 67 | 82 | 76 | 45 | 62 | 62 | 70 |
| Effect reversed by vitamin D | 81 | 92 | 73 | 55 | 57 | 36 | |||
This table shows comparison of genes affected by vitamin D supplementation in CD4+ T cells from MOG–EAE with the genes found enriched in myelin-reactive T cells from MS patients (ref. 63, figure S14) and the current candidate MS risk genes (62), which comprise VDRE (as defined by ref. 62, tables S1 and S2) and/or display VDR binding in primary human CD4+ T cells (as defined by ref. 62, table S2). Genes that display consistent vitamin D effect and/or P < 0.05 in minimum of two comparisons (i.e., dose, 10 IU vs. 0 IU, 10 IU vs. 2 IU, or 2 IU vs. 0 IU) in CD4+ T cells from MOG–EAE were considered for analysis. Vitamin D supplementation was considered to reverse an effect on a gene if vitamin D induced down-regulation (or up-regulation) of a gene found up-regulated (or down-regulated) in myelin-reactive T cells from MS. AP1, activator protein 1; ATF2, activating transcription factor 2; GPCR, G protein coupled receptor; IL-23, interleukin-23; NFAT, nuclear factor of activated T cells.
Despite numerous studies suggesting a beneficial effect of vitamin D in MS, there is still a controversy whether the supplementation can be used therapeutically (66). Based on the current state of knowledge and our data, vitamin D supplementation may be considered as a preventative measure for decreasing the risk for developing autoimmune diseases and potentially as adjunctive therapy. Moreover, we here show that the protective effect of vitamin D involves epigenetic mechanisms—in particular, DNA methylation, which may provide a molecular basis for cellular memory that mediates long-term effects (67–69) and suggests potential for future combined therapies (50).
Materials and Methods
Additional experimental details are provided in SI Materials and Methods.
Animals, Diet Regimen, and EAE Induction.
Inbred DA rats were housed in the animal facility at Karolinska University Hospital. Experimental setting and diet regime based on different contents of vitamin D3 (cholecalciferol; referred to as vitamin D) is described in detail elsewhere (23). MOG (amino acids 1–125 from the N terminus) used for active EAE induction was expressed in Escherichia coli and purified to homogeneity by chelate chromatography (70). Passive EAE was induced by transfer of MBP63–88-specific T-cell lines.
FACS Analysis and Sorting.
Lymph node cells day 7 p.i. were washed with cold PBS and resuspended in 100 μL of PBS. Cells were stained and visualized on a FACSCalibur (BD). CD3+, CD4+, CD45RA−, and CD8− cells were sorted and constituted the pure CD4+ T-cell population (MoFlo, >99% purity) which was used for further extraction of mRNA and genomic DNA (gDNA).
Histopathological and Immunohistochemical Analyses.
For histopathology and immunohistochemistry (IHC), the animals were euthanized 34 d p.i. As previously described (23), paraffin-embedded brain and spinal cord cross-sections (3–5 μm thick) were stained with H&E and Luxol fast blue (Kluever) to assess inflammation and demyelination, respectivley. Lysosomes of activated macrophages and microglia cells were targeted using an anti-rat ED1 antibody.
mRNA Extraction and Quantitative Real-Time PCR.
RNA was extracted from sorted CD4+ T cells from lymph nodes 7 d p.i. using the RNeasy kit (Qiagen) and the QIAcube (Qiagen). qPCR was performed using a Bio-Rad iQ5 iCycler Detection System. The primers used in this study are listed in Table S3.
Table S3.
Primers used for real-time quantitative PCR analyses
| Gene symbol | Primers for expression validation |
| Hdac2 | CCTAACTGTCAAAGGTCACGC |
| TTCCGGATTGTGTAGCCACC | |
| mTOR | TCTGGCCAAAAGACAGGTGG |
| AGCACTTCAAGCAGAGTGGG | |
| Jak2 | CTCCACAGAAGAAGAGGCCC |
| TTCAGAACATCGGCCTTCCC | |
| Stat3 | ATCCTAAGCACAAAGCCCCC |
| GGGTCTTGCCACTGATGTCC | |
| Stat4 | CATGGCTGAAAACATCCCCG |
| CGGTCTTGAAACTTCGCACG | |
| Jak1 | AGGCAAGAGTGCATAGAGCG |
| TGGGATCTCGCCATTGTAGC | |
| Akt2 | GAATACCAGGCACCCCTTCC |
| GATCCTCCGTGAAGACTCGC | |
| Fos | GAGGGAGCTGACAGATACGC |
| CAATCTCGGTCTGCAACGC | |
| Jun | CATCACCACTACACCGACCC |
| TATGCAGTTCAGCTAGGGCG | |
| Kras | TAGACACGAAACAGGCTCAGG |
| TAGAAGGCATCGTCAACACCC | |
| Nfkb1 | CCGTGTTTGTTCAGCTTCGG |
| AACTGTCGGAGAAGTTGGGC | |
| Socs3 | CTACTGGAGTGCCGTAACCG |
| ATGCGTAGGTTCTTGGTCCC | |
| Lck | AATCTGAGCCGTAAGGACGC |
| CTTCTCCCTGGTTCTGGTCG | |
| L19 | CCTGTGACTGTCCATTCCCG |
| GCATCCAGGTCACCTTCTCG | |
| Cd4 | AAGGACTGGCCAGAGACTCAGAT |
| ACGACTATACAGCTCAAGTGAACC | |
| Dnmt1 | GGAGGACAAAGAGAACACCATGA |
| TCCCACACTCAGGCTGTTGA | |
| Dnmt3a | CGCCAGAAGTGCCGAAAC |
| GGTGCTCCAGGGTAACATTGA | |
| Dnmt3b | GATGCCAGGACTCCCTCTGA |
| CCAGGCTGGAGATACTGTTGCT |
Expression Array Hybridization and Data Processing.
Array hybridization was done on GeneChip ST Arrays (GeneChip Gene 1.0 ST Array) by the Bioinformatics and Expression Analysis (BEA) core facility (Huddinge, Sweden). The data were deposited on the NCBI Gene Expression Omnibus database (accession no. GSE92680).
MBD-seq.
MBD based methylation sequencing was done by NXT-Dx (Ghent, Belgium). For sequencing, an Illumina Hi-Seq 2000 with 2 × 51 + 7 (index) sequencing cycles was used. The raw data can be provided upon request. Analysis was done using edgeR.
CHARM.
A total of 1 µg DNA per sample was sheared, McrBC digested, and gel fractionated before labeling and hybridization onto arrays containing 2.1 M probes. For a detailed protocol, see ref. 71 and SI Materials and Methods. The raw data can be provided upon request.
Pathway Analysis.
Molecules from the dataset with a cutoff of FDR of 1% and no cutoff for fold change were uploaded to the Ingenuity Pathways Analysis platform (Ingenuity Systems).
Ethics Statement.
All experiments in this study were approved and performed in accordance with the guidelines from the Swedish National Board for Laboratory Animals, which was approved by the North Stockholm Animal Ethics Committee. Rats were tested according to a health monitoring program at the National Veterinary Institute in Uppsala, Sweden.
SI Materials and Methods
Animals, Diet Regimen, and EAE Induction.
Inbred Dark Agouti rats were housed in the animal facility at Karolinska University Hospital in a specific pathogen-free and climate-controlled environment in polystyrene cages containing aspen wood shavings with free access to rodent chow and water with regulated 12-h light/dark cycles free of UV radiation.
Experimental setting and diet regime based on different contents of vitamin D3 (cholecalciferol; referred to as vitamin D) is described in detail elsewhere (23). Briefly, age-matched inbred females were subjected to one of the following: (i) supplemented diet containing fivefold increased amount of vitamin D than the regular rat diet (10 IU/g); (ii) regular rat diet containing 2 IU/g of vitamin D; or (iii) vitamin D-deprived diet (0 IU/g). The cholecalciferol-supplemented and -deprived diet formulations were obtained from TestDiet Limited.
MOG (amino acids 1–125 from the N terminus) used for active EAE induction was expressed in E. coli and purified to homogeneity by chelate chromatography (70). The purified protein, dissolved in 6 M urea, was dialyzed against PBS to obtain a physiological preparation that was stored at −70 °C. To induce EAE, rats were anesthetized with isoflurane [2-chloro-2-(difluoromethoxy)-1,1,1-trifluoro-ethane] (Forene; Abbott Laboratories) and injected s.c. in the tail base with a 200-µL inoculum containing 15 µg MOG in PBS, emulsified 1:1 with incomplete Freund’s adjuvant (Sigma-Aldrich).
Passive EAE was induced by transfer of MBP63–88-specific T-cell lines. For generation of T-cell lines, DA rats were injected s.c. in the tail base with a 200-µL inoculum containing 100 μg gpMBP63–88 peptide (EZBiolab) emulsified 1:1 with Freund's adjuvant containing 200 μg Mycobacterium tuberculosis (strain H37 RA; Difco Laboratories). Single-cell suspension was prepared from inguinal lymph nodes 10 d p.i. and cells were cultured 3 d in DMEM (Sigma-Aldrich) containing 10% (vol/vol) FBS and 20 μg/mL gpMBP63–88 peptide. Lymphoblasts were isolated using Ficoll (GE Healthcare Sciences) density gradient and expanded with IL-2 containing supernatant from mouse lymphoma assay cell cultures for 5 d. Two subsequent rounds of T-cell stimulation were done with 20 μg/mL gpMBP63–88 and a mix of irradiated thymocytes and splenocytes as antigen-presenting cells, with or without 10 nM 1,25(OH)2D3 (calcitriol; Sigma-Aldrich), each followed by IL-2 expansion. After separation on the Ficoll density gradient, cells were resuspended in saline and 300 μL containing 10 × 106 T cells was injected i.v. into 8- to 10-wk-old age-matched naïve rats.
Phenotyping.
Rats were weighed and monitored daily for clinical signs of EAE. The clinical scoring was as follows: 0, no clinical signs of EAE; 1, tail weakness or tail paralysis; 2, hind leg paraparesis or hemiparesis; 3, hind leg paralysis or hemiparalysis and 4, tetraplegy or moribund. The following clinical parameters were used: daily clinical scores; cumulative EAE score, the sum of all daily clinical scores; and weight loss, represented as a percentage of weight loss in relation to the weight at immunization.
Tissue Collection.
Animals were killed using CO2 7 or 34 d post-EAE induction. Draining inguinal lymph nodes were collected on day 7 p.i. and placed in DMEM (Gibco-BRL) before being mechanically separated by passage through a 100-μm mesh screen followed by a 40-μm mesh screen (Sigma) with the bolus of a syringe. Cells were spun at 300 × g and resuspended in PBS. Aliquots of the single-cell suspension from the lymph nodes were used for sorting of CD4+ T cells or for FACS analysis.
For histopathology and IHC, animals were euthanized 34 d p.i. and perfused with PBS. The spinal cords were carefully dissected and postfixed overnight in 4% (vol/vol) paraformaldehyde in PBS. Subsequently, the spinal cords were paraffin-embedded and serial sections were cut on a microtome.
FACS Analysis and Sorting.
Lymph node cells day 7 p.i. were washed with cold PBS and resuspended in a further 100 μL of PBS. Cells were stained for 5 min at room temperature (RT) and 20 min at 4 °C with the following antibodies: CD3-APC, CD4-PE, CD8a-PE, and CD45RA-PE:Cy5 (all from BD Biosciences).
Staining was visualized on a FACSCalibur (BD) with Cell Quest (version 3.2.1f1; BD) and analyzed using Kaluza (Beckman Coulter). CD3+, CD4+, CD45RA−, and CD8− cells were sorted and constituted the pure CD4+ T-cell population (MoFlo; >99% purity), which was used for further extraction of mRNA and gDNA.
Cytokine Flow Cytometry and Proliferation Assay.
Cells were acquired as described above. Following mechanical separation, cells were counted and 0.4 × 106 cells were placed in 96-well U-bottom plates and stimulated with either 20 μg/mL recombinant rat MOG or 1 μg/mL Con A, as a positive control for proliferation, in 200 μL of complete medium at 37 °C. To assess cytokine production, after 48 h, cells were stimulated with phorbol 12-myristate 13-acetate (PMA) (50 ng/mL), ionomycin (1 µg/mL), and Golgi Plug (1 µL/mL) in complete medium for 4 h at 37 °C followed by surface staining in PBS with CD3- and CD4-specific antibodies (both from BD Biosciences), as well as LIVE/DEAD fixable dead-cell exclusion dye (Life Technologies). After fixation/permeabilization for intranuclear antigens (all reagents from eBioscience), cells were stained with antibodies to IFN-γ and Ki67 (both from BD Biosciences) as well as IL-17A and Foxp3 (both from eBiosciences). Cells were acquired in a Gallios flow cytometer and analyzed with Kaluza software (both from Beckman Coulter). A minimum of 105 events per lymph node was acquired.
Histopathological and Immunohistochemical Analyses.
Sections were stained with H&E and Luxol fast blue (Kluever) to assess tissue inflammation and demyelination, respectively. All images were captured using a Leica Polyvar 2 microscope, and representatives of the respective stainings are shown.
For IHC analysis, paraffin-embedded brain and spinal cord cross-sections (3–5 μm thick) were treated as previously described (23). After deparaffinization in xylol, sections were transferred to 90% (vol/vol) ethanol. Endogenous peroxidase was blocked by incubation in methanol with 0.02% H2O2 for 30 min at RT and rehydration to distilled water followed via a 90% (vol/vol), 70% (vol/vol), and 50% (vol/vol) ethanol series. Antigen retrieval was performed with ethylenediamine tetraacetic acid buffer, pH 8.5, or citrate buffer, pH 6.0, by heating for 1 h in a steamer device at 98 °C (Braun). Sections were incubated in 10% (vol/vol) FCS in PBS 30 min before incubation with primary antibody on 4 °C, overnight. After washing in PBS, sections were incubated with biotinylated secondary antibody (1:200 biotinylated anti-mouse or rabbit; Amersham Pharmacia Biotech) for 1 h at RT. Biotin-avidin peroxidase was used as detection system, and 3,3′diaminobenzidine-tetrahydrochloride (Sigma) for visualization. Lysosomes of activated macrophages and microglia (ED1, mouse anti rat monoclonal antibody, AbD Serotec; 1–1,000) were stained to detect macrophages/ monocytes.
mRNA Extraction and qPCR.
RNA was extracted from sorted CD4+ T cells from lymph nodes 7 d p.i. using the RNeasy Kit (Qiagen) and the QIAcube (Qiagen), including on-column DNA digestion for fully automated sample preparation. RNA concentration and purity was determined through measurement of A260/A280 ratios with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). Next, 100 ng RNA was used to prepare cDNA using the iScript Kit (Bio-Rad). qPCR was performed using a Bio-Rad iQ5 iCycler Detection System with a three-step PCR protocol (95 °C for 10 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s and 40 cycles of melt curve analysis), using SYBR Green (Bio-Rad) as the fluorophore and 50 ng cDNA in each qPCR reaction. As a control, we used ddH2O and instead of –RT control we analyzed the melt curves. Relative expression levels, corrected for amplification efficiency, were analyzed using iQ5 v2.0 software (Bio-Rad). Relative expression was calculated as the ratio between the target and ribosomal protein L19 (Rpl19), which is a frequently used housekeeping gene (72). The primers used in this study are listed in Table S3.
Expression Array Hybridization and Data Processing.
Array hybridization was done by the BEA core facility (Huddinge, Sweden). RNA concentration and purity was determined through measurement of A260/A280 ratios with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). Confirmation of RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples were immediately frozen and stored at −80 °C.
A total of 250 ng of total RNA from each sample was used to generate amplified and biotinylated sense-strand cDNA from the entire expressed genome according to the Ambion WT Expression Kit (P/N 4425209 Rev C 09/2009) and Affymetrix GeneChip WT Terminal Labeling and Hybridization User Manual (P/N 702808 Rev. 6; Affymetrix Inc.). GeneChip ST Arrays (GeneChip Gene 1.0 ST Array) were hybridized for 16 h in a 45 °C incubator, rotated at 60 rpm on a mutliplatform shaker (Fisher Scientific). According to the GeneChip Expression Wash, Stain and Scan Manual (PN 702731 Rev 3; Affymetrix), the arrays were then washed and stained using the Fluidics Station 450 and finally scanned using the GeneChip Scanner 3000 7G. The data were deposited in the NCBI Gene Expression Omnibus database (accession no. GSE92680).
The raw data were normalized using robust multiarray average method (73, 74). Subsequent analysis of the gene expression, including PCA and hierarchical clustering, was carried out using Partek Genomics Suite v6.0 (Partek Inc.) together with ANOVA model with a filter to select transcripts that show significant differential expression determined at a 1% FDR (75).
Targets of miRNA were predicted using TargetScan v6.2, and only conserved targets were considered. Enrichment of predicted miRNA targets among differentially expressed genes was calculated using a χ2 test.
Methylated Sequence Capturing Using a MBD-Based Enrichment Followed by MBD-seq and Data Processing.
gDNA was isolated from CD4+ T cells from lymph nodes 7 d p.i. using a standard protocol. MBD-based methylation sequencing was done by NXT-Dx. For sequencing, an Illumina Hi-Seq 2000 with 2 × 51 + 7 (index) sequencing cycles was used. The raw data are available upon request.
edgeR, which performs differential abundance analysis for predefined genomic features, was used to perform a differential methylation analysis. The package implements exact statistical methods for multigroup experiments developed by Robinson and Smyth (76, 77); it also implements statistical methods based on generalized linear models (glms), suitable for multifactor experiments of any complexity, developed by McCarthy et al. (78) and Lund et al. (79). A particular feature of edgeR functionality, both classic and glms, are empirical Bayes methods that permit the estimation of gene-specific biological variation, even for experiments with minimal levels of biological replication.
The significance of enrichment of genes that displayed hypomethylation up-regulation, hypomethylation down-regulation, hypermethylation up-regulation, and hypermethylation down-regulation (using P < 0.05) in different regions of the genome (promoter, exon, intron, and distal intergenic) was calculated using Fisher’s exact Gene Overlap package in R.
CHARM.
One microgram DNA per sample was sheared, McrBC digested, and gel fractionated before labeling and hybridization onto arrays containing 2.1 M probes. For detailed protocol, see ref. 71. The raw data are available upon request. To estimate methylation percentages from signal intensities, the methp function (CHARM Bioconductor package) (80) was used. A three-step methodology was used: (i) within-sample normalization using locally estimated scatterplot smoothing (LOESS); (ii) between-sample normalization by subset quantile normalization; and, finally, (iii) the percentage methylation estimation. Differential methylation analysis per probe was computed using the limma package (81) over the logit transformation of the beta values. None of the DMRs in CHARM did survive genome-wide correction, but there was correlation between MBD-seq and CHARM for P < 0.001, which is why we selected this cutoff for reporting methylation findings.
Pathway Analysis.
Molecules from the dataset with a cutoff of FDR of 1% and no cutoff for fold change were uploaded to the IPA platform (Ingenuity Systems). The molecules in this dataset were grouped in biological functions and/or diseases or were associated with a canonical pathway in Ingenuity’s knowledge base. Right-tailed Fisher’s exact test was used to calculate a P value, determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. The significance of the association between the data set and the canonical pathway was measured in two ways: (i) a ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed; (ii) Benjamini-Hochberg multiple testing was used to calculate P values, determining the probability that the association between the genes in the dataset and canonical pathway is by chance alone.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Research Council (M.J. and J.N.T.); the Swedish Association for Persons with Neurological Disabilities (M.J.); the Swedish Brain Foundation (M.J. and J.N.T.); the Swedish Medical Society (M.J.); the Petrus and Augusta Hedlunds Foundation (M.J.); Karolinska Institutet funds (to M.J. and S.R.); AFA Insurance (T.J.E. and J.N.T.); Wenner-Gren Foundations Grant (to M.Z.); and Biogen Idec Grant (to M.Z.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE92680).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615783114/-/DCSupplemental.
References
- 1.Plum LA, DeLuca HF. Vitamin D, disease and therapeutic opportunities. Nat Rev Drug Discov. 2010;9(12):941–955. doi: 10.1038/nrd3318. [DOI] [PubMed] [Google Scholar]
- 2.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: Vitamins A and D take centre stage. Nat Rev Immunol. 2008;8(9):685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agmon-Levin N, Theodor E, Segal RM, Shoenfeld Y. Vitamin D in systemic and organ-specific autoimmune diseases. Clin Rev Allergy Immunol. 2013;45(2):256–266. doi: 10.1007/s12016-012-8342-y. [DOI] [PubMed] [Google Scholar]
- 4.Kriegel MA, Manson JE, Costenbader KH. Does vitamin D affect risk of developing autoimmune disease? A systematic review. Semin Arthritis Rheum. 2011;40(6):512–531. doi: 10.1016/j.semarthrit.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ascherio A, Munger KL, Simon KC. Vitamin D and multiple sclerosis. Lancet Neurol. 2010;9(6):599–612. doi: 10.1016/S1474-4422(10)70086-7. [DOI] [PubMed] [Google Scholar]
- 6.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 7.Ramagopalan SV, et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann Neurol. 2011;70(6):881–886. doi: 10.1002/ana.22678. [DOI] [PubMed] [Google Scholar]
- 8.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 9.Rhead B, et al. Mendelian randomization shows a causal effect of low vitamin D on multiple sclerosis risk. Neurol Genet. 2016;2(5):e97. doi: 10.1212/NXG.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296(23):2832–2838. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- 11.Salzer J, et al. Vitamin D as a protective factor in multiple sclerosis. Neurology. 2012;79(21):2140–2145. doi: 10.1212/WNL.0b013e3182752ea8. [DOI] [PubMed] [Google Scholar]
- 12.Simpson S, Jr, et al. Higher 25-hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Ann Neurol. 2010;68(2):193–203. doi: 10.1002/ana.22043. [DOI] [PubMed] [Google Scholar]
- 13.Mowry EM, et al. Vitamin D status predicts new brain magnetic resonance imaging activity in multiple sclerosis. Ann Neurol. 2012;72(2):234–240. doi: 10.1002/ana.23591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toghianifar N, Ashtari F, Zarkesh-Esfahani SH, Mansourian M. Effect of high dose vitamin D intake on interleukin-17 levels in multiple sclerosis: A randomized, double-blind, placebo-controlled clinical trial. J Neuroimmunol. 2015;285:125–128. doi: 10.1016/j.jneuroim.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Mattner F, et al. Inhibition of Th1 development and treatment of chronic-relapsing experimental allergic encephalomyelitis by a non-hypercalcemic analogue of 1,25-dihydroxyvitamin D(3) Eur J Immunol. 2000;30(2):498–508. doi: 10.1002/1521-4141(200002)30:2<498::AID-IMMU498>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Muthian G, Raikwar HP, Rajasingh J, Bright JJ. 1,25 Dihydroxyvitamin-D3 modulates JAK-STAT pathway in IL-12/IFNgamma axis leading to Th1 response in experimental allergic encephalomyelitis. J Neurosci Res. 2006;83(7):1299–1309. doi: 10.1002/jnr.20826. [DOI] [PubMed] [Google Scholar]
- 17.Chang JH, Cha HR, Lee DS, Seo KY, Kweon MN. 1,25-Dihydroxyvitamin D3 inhibits the differentiation and migration of T(H)17 cells to protect against experimental autoimmune encephalomyelitis. PLoS One. 2010;5(9):e12925. doi: 10.1371/journal.pone.0012925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi S, et al. 1,25-dihydroxyvitamin D(3) ameliorates Th17 autoimmunity via transcriptional modulation of interleukin-17A. Mol Cell Biol. 2011;31(17):3653–3669. doi: 10.1128/MCB.05020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spanier JA, Nashold FE, Mayne CG, Nelson CD, Hayes CE. Vitamin D and estrogen synergy in Vdr-expressing CD4(+) T cells is essential to induce Helios(+)FoxP3(+) T cells and prevent autoimmune demyelinating disease. J Neuroimmunol. 2015;286:48–58. doi: 10.1016/j.jneuroim.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Tuoresmäki P, Väisänen S, Neme A, Heikkinen S, Carlberg C. Patterns of genome-wide VDR locations. PLoS One. 2014;9(4):e96105. doi: 10.1371/journal.pone.0096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell MJ. Vitamin D and the RNA transcriptome: More than mRNA regulation. Front Physiol. 2014;5:181. doi: 10.3389/fphys.2014.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fetahu IS, Höbaus J, Kállay E. Vitamin D and the epigenome. Front Physiol. 2014;5:164. doi: 10.3389/fphys.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzemovic MZ, Zeitelhofer M, Hochmeister S, Gustafsson SA, Jagodic M. Efficacy of vitamin D in treating multiple sclerosis-like neuroinflammation depends on developmental stage. Exp Neurol. 2013;249:39–48. doi: 10.1016/j.expneurol.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Storch MK, et al. Autoimmunity to myelin oligodendrocyte glycoprotein in rats mimics the spectrum of multiple sclerosis pathology. Brain Pathol. 1998;8(4):681–694. doi: 10.1111/j.1750-3639.1998.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meehan TF, DeLuca HF. The vitamin D receptor is necessary for 1alpha,25-dihydroxyvitamin D(3) to suppress experimental autoimmune encephalomyelitis in mice. Arch Biochem Biophys. 2002;408(2):200–204. doi: 10.1016/s0003-9861(02)00580-5. [DOI] [PubMed] [Google Scholar]
- 26.Mayne CG, Spanier JA, Relland LM, Williams CB, Hayes CE. 1,25-Dihydroxyvitamin D3 acts directly on the T lymphocyte vitamin D receptor to inhibit experimental autoimmune encephalomyelitis. Eur J Immunol. 2011;41(3):822–832. doi: 10.1002/eji.201040632. [DOI] [PubMed] [Google Scholar]
- 27.Thessen Hedreul M, Gillett A, Olsson T, Jagodic M, Harris RA. Characterization of multiple sclerosis candidate gene expression kinetics in rat experimental autoimmune encephalomyelitis. J Neuroimmunol. 2009;210(1-2):30–39. doi: 10.1016/j.jneuroim.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 29.Yang XO, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282(13):9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 30.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35(6):919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 32.Ramagopalan SV, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikkinen S, et al. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011;39(21):9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26(1):37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handel AE, et al. Vitamin D receptor ChIP-seq in primary CD4+ cells: Relationship to serum 25-hydroxyvitamin D levels and autoimmune disease. BMC Med. 2013;11:163. doi: 10.1186/1741-7015-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu H, et al. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J Pediatr. 2013;162(5):1004–1009 e1001. doi: 10.1016/j.jpeds.2012.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seuter S, Neme A, Carlberg C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016;44(9):4090–4104. doi: 10.1093/nar/gkv1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stefanska B, Karlic H, Varga F, Fabianowska-Majewska K, Haslberger A. Epigenetic mechanisms in anti-cancer actions of bioactive food components--the implications in cancer prevention. Br J Pharmacol. 2012;167(2):279–297. doi: 10.1111/j.1476-5381.2012.02002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene. 2000;242(1-2):407–418. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, et al. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108(3):1058–1064. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111(4):2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto S, et al. Coordinated changes in DNA methylation in antigen-specific memory CD4 T cells. J Immunol. 2013;190(8):4076–4091. doi: 10.4049/jimmunol.1202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang BH, et al. Development of a unique epigenetic signature during in vivo Th17 differentiation. Nucleic Acids Res. 2015;43(3):1537–1548. doi: 10.1093/nar/gkv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278(30):27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 47.Krämer OH, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22(13):3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castelo-Branco G, et al. Acute treatment with valproic acid and l-thyroxine ameliorates clinical signs of experimental autoimmune encephalomyelitis and prevents brain pathology in DA rats. Neurobiol Dis. 2014;71:220–233. doi: 10.1016/j.nbd.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Lv J, et al. The antiepileptic drug valproic acid restores T cell homeostasis and ameliorates pathogenesis of experimental autoimmune encephalomyelitis. J Biol Chem. 2012;287(34):28656–28665. doi: 10.1074/jbc.M112.356584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan L, et al. Vitamin D stimulates apoptosis in gastric cancer cells in synergy with trichostatin A/sodium butyrate-induced and 5-aza-2′-deoxycytidine-induced PTEN upregulation. FEBS J. 2010;277(4):989–999. doi: 10.1111/j.1742-4658.2009.07542.x. [DOI] [PubMed] [Google Scholar]
- 51.Baeke F, et al. The vitamin D analog, TX527, promotes a human CD4+CD25highCD127low regulatory T cell profile and induces a migratory signature specific for homing to sites of inflammation. J Immunol. 2011;186(1):132–142. doi: 10.4049/jimmunol.1000695. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez-Díaz S, et al. MicroRNA-22 is induced by vitamin D and contributes to its antiproliferative, antimigratory and gene regulatory effects in colon cancer cells. Hum Mol Genet. 2012;21(10):2157–2165. doi: 10.1093/hmg/dds031. [DOI] [PubMed] [Google Scholar]
- 53.Padi SK, Zhang Q, Rustum YM, Morrison C, Guo B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology. 2013;145(2):437–446. doi: 10.1053/j.gastro.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan W, et al. MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat Commun. 2015;6:7096. doi: 10.1038/ncomms8096. [DOI] [PubMed] [Google Scholar]
- 55.Alroy I, Towers TL, Freedman LP. Transcriptional repression of the interleukin-2 gene by vitamin D3: Direct inhibition of NFATp/AP-1 complex formation by a nuclear hormone receptor. Mol Cell Biol. 1995;15(10):5789–5799. doi: 10.1128/mcb.15.10.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13(10):907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 59.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McWilliams IL, Rajbhandari R, Nozell S, Benveniste E, Harrington LE. STAT4 controls GM-CSF production by both Th1 and Th17 cells during EAE. J Neuroinflammation. 2015;12:128. doi: 10.1186/s12974-015-0351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Codarri L, et al. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 62.Berge T, et al. The multiple sclerosis susceptibility genes TAGAP and IL2RA are regulated by vitamin D in CD4+ T cells. Genes Immun. 2016;17(2):118–127. doi: 10.1038/gene.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao Y, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Transl Med. 2015;7(287):287ra74. doi: 10.1126/scitranslmed.aaa8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wucherpfennig KW, Sethi D. T cell receptor recognition of self and foreign antigens in the induction of autoimmunity. Semin Immunol. 2011;23(2):84–91. doi: 10.1016/j.smim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4(1):6. doi: 10.1186/gm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papeix C, Lubetzki C. If I had a clinically isolated syndrome with MRI diagnostic of MS, I would take vitamin D 10,000 IU daily: No. Mult Scler. 2013;19(2):140–142. doi: 10.1177/1352458512474092. [DOI] [PubMed] [Google Scholar]
- 67.Munger KL, et al. Dietary intake of vitamin D during adolescence and risk of multiple sclerosis. J Neurol. 2011;258(3):479–485. doi: 10.1007/s00415-010-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mowry EM, et al. Vitamin D status is associated with relapse rate in pediatric-onset multiple sclerosis. Ann Neurol. 2010;67(5):618–624. doi: 10.1002/ana.21972. [DOI] [PubMed] [Google Scholar]
- 69.Munger KL, et al. Vitamin D in the Finnish status during pregnancy and risk of multiple sclerosis in offspring of women maternity cohort. JAMA Neurol. 2016;73(5):515–519. doi: 10.1001/jamaneurol.2015.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amor S, et al. Identification of epitopes of myelin oligodendrocyte glycoprotein for the induction of experimental allergic encephalomyelitis in SJL and Biozzi AB/H mice. J Immunol. 1994;153(10):4349–4356. [PubMed] [Google Scholar]
- 71.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Curr Protoc Hum Genet. 2010 doi: 10.1002/0471142905.hg2001s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lucas S, Ghilardi N, Li J, de Sauvage FJ. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2003;100(25):15047–15052. doi: 10.1073/pnas.2536517100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 74.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 76.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 77.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 78.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lund SP, Nettleton D, McCarthy DJ, Smyth GK. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol. 2012 doi: 10.1515/1544-6115.1826. [DOI] [PubMed] [Google Scholar]
- 80.Aryee MJ, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics. 2011;12(2):197–210. doi: 10.1093/biostatistics/kxq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.