Significance
Vibrio cholerae O1, the causative agent of the disease cholera, has two biotypes: namely classical and El Tor. The disease is the principal manifestation of a potent toxin known as cholera toxin, produced by the bacteria. Generation of a toxin producing V. cholerae strain arises by infection of a toxin gene-carrying phage (CTX phage) that becomes integrated into its genome. Experimental evidence has shown the replication and transmission of the type-specific phage in the El Tor strains, but this has not been experimentally demonstrated in the classical strains and the exact mechanism was not known. This study provides laboratory evidence that illustrates the genesis of toxigenic classical biotype strains.
Keywords: Vibrio cholerae, cholera toxin, CTX phage, biotype
Abstract
The toxigenic classical and El Tor biotype Vibrio cholerae serogroup O1 strains are generated by lysogenization of host-type–specific cholera toxin phages (CTX phages). Experimental evidence of the replication and transmission of an El Tor biotype-specific CTX phage, CTX-1, has explained the evolution of V. cholerae El Tor biotype strains. The generation of classical biotype strains has not been demonstrated in the laboratory, and the classical biotype-specific CTX phage, CTX-cla, is considered to be defective with regard to replication. However, the identification of atypical El Tor strains that contain CTX-cla–like phage, CTX-2, indicates that CTX-cla and CTX-2 replicate and can be transmitted to V. cholerae strains. The replication of CTX-cla and CTX-2 phages and the transduction of El Tor biotype strains by various CTX phages under laboratory conditions are demonstrated in this report. We have established a plasmid-based CTX phage replication system that supports the replication of CTX-1, CTX-cla, CTX-2, and CTX-O139. The replication of CTX-2 from the tandem repeat of lysogenic CTX-2 in Wave 2 El Tor strains is also presented. El Tor biotype strains can be transduced by CTX phages in vitro by introducing a point mutation in toxT, the transcriptional activator of the tcp (toxin coregulated pilus) gene cluster and the cholera toxin gene. This mutation also increases the expression of cholera toxin in El Tor strains in a sample single-phase culture. Our results thus constitute experimental evidence of the genetic mechanism of the evolution of V. cholerae.
Cholera is a severe diarrheal disease that is caused by toxigenic strains of Vibrio cholerae (1). Two serogroups of V. cholerae—O1, which is further classified into the classical and El Tor biotypes, and O139—are toxigenic, triggering toxin-mediated epidemic cholera (2). Toxigenic V. cholerae strains are generated by infection and lysogenization of a 6.9-kb single-stranded DNA filamentous phage, cholera toxin phage (CTX phage), which carries the cholera toxin gene (3). Three host-specific CTX phages that are discriminated by the rstR (repressor of CTX phage) type have been described: CTX-cla in classical biotype strains, CTX-1 in El Tor biotype strains, and CTX-O139 in serogroup O139 strains (4, 5). A model by which type-specific CTX phages are acquired by each biotype strain in the evolution of toxigenic V. cholerae strains has been developed (3).
The replication of lysogenic CTX-1 in El Tor biotype strains has been demonstrated, based on transmission of the CTX-1 phage by transduction to classical biotype strains, indicating that the infection and replication of CTX phage are not restricted by host bacterial biotype (3). The replication of CTX-O139 and transmission to a classical strain have also been recapitulated under laboratory conditions (4). However, the replication and transmission of CTX-cla remain unconfirmed (6).
Recently, atypical El Tor strains that harbor new types of CTX phages—genetic mosaics of CTX-cla and CTX-1—have increased their prevalence worldwide (7). These mosaic phages contain classical-type ctxB instead of El Tor-type ctxB. Among the new types of CTX phages in atypical El Tor strains is CTX-2, a genetic homolog of CTX-cla, because it contains classical-type rstR (rstRcla), whereas others (CTX-3 ∼ CTX-6) contain El Tor-type rstR (rstREl Tor) (8). According to variations in the bacterial genome and the CTX phages that they contain, the El Tor strains from the seventh cholera pandemic have been classified into several waves (9). Wave 1 strains, or prototype El Tor strains, contain CTX-1, whereas one of the main characteristics of Wave 2 strains is that they contain a tandem repeat of CTX-2 on chromosome 2. Wave 3 strains primarily harbor TLC:RS1:CTX (CTX3 ∼ CTX6).
The generation and replication of CTX-2 in Wave 2 strains have not been corroborated. A tandem repeat of lysogenic CTX-2 on chromosome 2 might support the replication of CTX-2, but the replication of CTX-2 has not been demonstrated experimentally (10). Alternative mechanisms of CTX phage acquisition by V. cholerae strains, generalized transduction of the CTX phage genome by the bacteriophage CP-T1, and uptake of classical CTX prophage by El Tor strains by chitin-induced competence are potential explanations of the dissemination of CTX-cla and CTX-2 (11, 12). However, the replication of CTX-cla and CTX-2 has been suspected to occur in nature, which we believe has not been proven because of the absence of a suitable strain that can function as the recipient of CTX-cla and CTX-2 (13).
Classical biotype strains already contain CTX-cla, and they are not expected to allow superinfection of the same type of CTX phage, a phenomenon called phage immunity (14). Although El Tor biotype strains can be transduced by CTX-1 in vivo at high frequencies, the transduction efficiency of El Tor strains by CTX-1 phage under laboratory conditions remains low, possibly because the conditions have been inadequate for transduction (15). We hypothesize that El Tor strains can be transduced by CTX phages under the appropriate laboratory conditions.
In this study, the replication of various CTX phages under laboratory conditions was verified by developing a plasmid-based CTX phage replication system and constructing El Tor strains that can be transduced by CTX phages. The replication and transmission of CTX-2 were confirmed in the plasmid-based system and from the tandem repeat of lysogenic CTX-2 in Wave 2 strains. We show that El Tor strains can be transduced by CTX phages under laboratory conditions by introducing a point mutation in toxT [the transcriptional activator of the toxin coregulated pilus (tcp) gene cluster, from which the CTX phage receptor is encoded]. The El Tor biotype strains that harbored the same point mutation in toxT produced elevated levels of cholera toxin under laboratory conditions.
Results
CTX-1 Phage Is Replicated from the Plasmid-Cloned CTX Phage Genome.
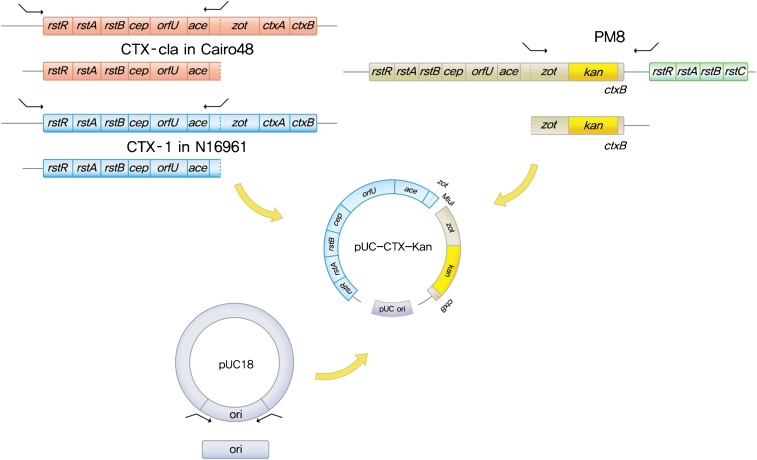
An Escherichia coli- and V. cholerae-compatible recombinant plasmid was constructed by linking the replication origin of pUC18 to the CTX-1 phage genome, in which ctxA and the first 126 nucleotides of ctxB were replaced by a kanamycin cassette (Fig. S1). V. cholerae strains that did not contain lysogenic CTX-1 were transformed with the recombinant plasmid, and CTX phage production from the plasmid-cloned CTX phage genome was monitored by transduction of recipient strains (V. cholerae strains are listed in Table S1). The three host strains for the recombinant plasmid were PM14, an N16961 derivative that has lost the CTX-1 prophage from chromosome 1, thus containing the TLC:RS1 array; O395, a classical biotype strain; and A213, a United States Gulf Coast strain that harbors only the toxin-linked cryptic (TLC) element on chromosome 1 (Table 1).
Fig. S1.
Genetic map and construction of pUC-CTX plasmids. The recombinant pUC-CTX plasmid consists of three DNA fragments: a replication origin fragment of pUC18; a DNA fragment spanning nucleotide 245 of zot to the termination codon of rstR of RS1 amplified from PM8; and a fragment spanning the att sequence of CTX phage or from nucleotide 119 nucleotide upstream of the termination codon of rstR to nucleotide 244 of zot in various CTX phages. Therefore, the zot-3′UTR fragment is common to all constructs, and the 5\x{2032}UTR-zot fragments are phage type-specific.
Table S1.
V. cholerae strains used in this study
| Strains | Genetic structure | Note | Genome sequence information and source | |
| Chromosome 1 CTX array | Chromosome 2 CTX array | |||
| V212-1 derivatives | ||||
| V212-1 | TLC:RS1:CTX-1:RS1 | CTX-2:CTX-2 | Wave 2 El Tor strain | ERS013132 (9) |
| PM8 | TLC:CTX-1kan:RS1 | CTX-2:CTX-2 | (16) | |
| PM9 | TLC:RS1:CTX-1:RS1 | CTX-2kan:CTX-2 | (16) | |
| PM10 | TLC:RS1:CTX-1:RS1 | CTX-2:CTX-2kan | (16) | |
| N16961 derivatives | ||||
| N16961 | TLC:CTX-1:RS1 | No element | Wave 1 El Tor strain | AE003852/AE003853 (31) |
| PM20 | TLC:CTX-1kan:RS1 | No element | Present study | |
| PM14 | TLC:RS1 | No element | (16) | |
| B33 derivatives | ||||
| B33 | No TLC, No element | CTX-2:CTX-2 | Wave 2 El Tor strain | ACHZ00000000 (10) |
| PM22 | No TLC, No element | CTX-2kan:CTX-2 | Present study | |
| IB4122 derivatives | ||||
| IB4122 | TLC:RS1:CTX-3 | No element | Wave 3 El Tor strain | ERS013264 (9, 18) |
| IB4122-toxTY139F | TLC:RS1:CTX-3 | No element | Present study | |
| MG116025 derivatives | ||||
| MG116025 | TLC:RS1:CTX-1:RS1 | No element | Wave 2 strain (Matlab type 3) | ERS013135 (9) |
| PM25 | TLC:CTX-1:RS1 | No element | Present study | |
| PM26 | TLC:RS1:RS1 | No element | Present study | |
| PM27 | TLC:RS1 | No element | Present study | |
| PM28 | TLC | No element | Present study | |
| PM29 | No TLC, No element | No element | Present study | |
| MG116025-toxTF139Y | TLC:RS1:CTX-1:RS1 | No element | Present study | |
| A213 derivatives | ||||
| A213 | TLC | No element | US Gulf Coast strain, att+ | ERS013191 (9) |
| A213-toxTY139F | TLC | No element | Present study | |
| O395 | TLC:TrunCTX-cla:CTX-cla | CTX-cla | Classical biotype | CP000626/CP000627 (9) |
| Cairo 48 | TLC:CTX-cla | (?)-CTX-cla | Classical biotype | ERS013171 (9) |
| AR196157 | (?):CTX-O139:(?) | (?) | O139 strain | (32) |
Table 1.
Transduction efficiency of selected V. cholerae strains and plasmid-based replication
| Strains | Description | Genetic structure | CTX phage produced | Transduction recipient | Transduction efficiency* | |
| Chromosome 1 | Chromosome 2 | |||||
| PM20 | N16961 derivative | TLC:CTX-1kan:RS1 | No element | CTX-1kan-C1† | O395 | 105 |
| MG116025 | 102 | |||||
| PM14-U1 | PM14 transformed with pUC-CTX-1kan | TLC:RS1 | No element | CTX-1kan-P‡ | O395 | 105 |
| PM14-U2 | PM14 transformed with pUC-CTX-2kan | TLC:RS1 | No element | CTX-2kan-P | MG116025 | 2 × 10 |
| PM14-U3 | PM14 transformed with pUC-CTX-clakan | TLC:RS1 | No element | CTX-clakan-P | MG116025 | 102 |
| PM14-U4 | PM14 transformed with pUC-CTX-O139kan | TLC:RS1 | No element | CTX-O139kan-P | O395 | 2 × 102 |
| MG116025 | 10 | |||||
| PM9 | V212-1 derivative | TLC:RS1:CTX-1:RS1 | CTX-2kan:CTX-2 | CTX-2kan-C2† | MG116025 | 103 |
| PM22 | B33 derivative | No TLC, No element | CTX-2kan:CTX-2 | CTX-2kan C2 | MG116025 | 5 × 103 |
| O395-U1 | O395 transformed with pUC-CTX-1kan | TLC:TrunCTX-cla:CTX-cla | CTX-cla | CTX-1kan-P | O395 | 5 × 104 |
| A213-U1 | A213 transformed with pUC-CTX-1kan | TLC | No element | CTX-1kan-P | O395 | 3 × 103 |
Transduction efficiency was calculated as the number of transductants per 6 × 108 recipient cells per 1 mL of culture supernatant of the donor strain. The data represent the average of at least three independent experiments.
C denotes CTX phage produced from lysogenic phage integrated in chromosome 1 or 2.
P denotes CTX phage produced from phage genome cloned in the recombinant plasmid.
Although there were variations between host strains, ∼105 transductants were obtained in 1 mL of donor strain supernatant when O395 was used as the recipient, which was comparable with the production of CTX-1 phage from the lysogenic CTX-1:RS1 array of N16961. This result confirms that the replication and dissemination of CTX phages do not depend on the host bacterial biotype. The 119 nucleotides upstream of rstR were sufficient to initiate replication of CTX-1 in this system, because a similar number of transductants were obtained from the plasmids that contained the entire intergenic region 1 (ig-1) sequence or only the last 119 nucleotides. Subsequently, CTX phages were produced from the recombinant plasmid that contained the last 119 nucleotides of ig-1.
The replication origin of pUC18 was no longer present in the transduced CTX-1kan phage genome, and the DNA sequence of CTX-1kan that was generated from the recombinant plasmid was identical to the CTX-1kan that was produced from the lysogenic CTX-1kan in PM20, indicating that authentic CTX phages were obtained by replication from the plasmid-cloned CTX phage genome.
Replication of CTX-cla, CTX-2, and CTX-O139 in the Plasmid-Based CTX Phage Replication System.
Based on the confirmation that CTX-1 could be replicated from a plasmid-cloned CTX phage genome in V. cholerae, we could predict the replication of CTX-2 and CTX-cla using the plasmid-based replication system. Recombinant plasmids that contained the CTX-clakan and CTX-2kan genomes were constructed (Materials and Methods) and used to transform PM14.
We did not expect the classical biotype strains to be transduced by CTX-clakan or CTX-2kan because of phage immunity; thus, various El Tor biotype strains were tested for transduction using the envisaged CTX-clakan and CTX-2kan phages that were produced from the pUC-CTX plasmids in PM14 (14). Recipient El Tor strains were prepared for transduction in the same manner as the classical strains (agglutinated; i.e., grown in LB, pH 6.5 at 30 °C).
No El Tor strains were transduced by CTX-clakan and CTX-2kan, except for a Wave 2 El Tor strain, MG116025; this strain was transduced by CTX-clakan and CTX-2kan phages that were replicated using the plasmid-based replication system. Approximately 100 and 20 transductants were obtained from the plasmid-cloned CTX-clakan and CTX-2kan genomes, respectively (Tables 1 and 2). The replication of CTX-clakan and CTX-2kan was further confirmed by secondary transduction (described below). Based on these results, the replication of CTX-2 and CTX-cla was verified with the appropriate recipient strain, and the plasmid-based CTX phage replication system was demonstrated to be working properly. Similarly, CTX-O139kan phages were also produced using the plasmid-based replication system.
Table 2.
Replication and transduction of CTX-cla and CTX-2
| Primary transduction | Secondary transduction | |||||
| Donor | pCTX produced from the donor | Transduction recipient strain | Transduction efficiency | Transductants (transduced pCTX) | Transduction recipient strain | Transduction efficiency |
| PM14-U2 | pCTX-2kan-P* | MG116025 | 2 × 10 | MG116025 (pCTX-2kan-P) | MG116025 | 2 × 104 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 3 × 103 | |||||
| IB4122 toxTY139F | 0 | |||||
| MG116025-toxTF139Y | 0 | |||||
| PM14-U3 | pCTX-clakan-P† | MG116025 | 1 × 102 | MG116025 (pCTX-clakan-P) | MG116025 | 3 × 104 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 2 × 104 | |||||
| IB4122- toxTY139F | 0 | |||||
| MG116025-toxTF139Y | 0 | |||||
| PM9 | pCTX-2kan-VL‡ | MG116025 | 103 | MG116025 (pCTX-2kan-VL) | MG116025 | 2 × 107 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 106 | |||||
| IB4122-toxTY139F | 1 × 103 | |||||
| MG116025-toxTF139Y | 0 | |||||
pCTX-2kan-P: pCTX-2kan produced from the plasmid pUC-CTX-2kan.
pCTX-clakan-P: pCTX-clakan produced from the plasmid pUC-CTX-clakan.
pCTX-2kan-VL: pCTX-2kan produced from lysogenic CTX-2 in PM9, which is a derivative of strain V212-1.
CTX-2 Replication from the Tandem Repeat of Lysogenic CTX-2 in Wave 2 El Tor Strains.
Replication of CTX-2 from a tandem repeat of lysogenic CTX-2 on chromosome 2 in Wave 2 El Tor strains was also substantiated. PM9 and PM22, derivatives of V212-1 and B33, respectively, were constructed as described previously (16). Approximately 103 transductants were obtained from the primary transduction of PM9 and PM22 (Table 1 and 2). Secondary transduction of CTX-cla and CTX-2 from the transductants that contained the replicative form pCTX-clakan and pCTX-2kan was further verified (described below). No classical strain that contains a tandem repeat of CTX-cla or the CTX-cla:RS1 array is available, preventing us from demonstrating the replication of lysogenic CTX-cla in such strains.
A Point Mutation in toxT Is Responsible for the CTX Phage Transduction-Competent Phenotype.
To identify genetic changes that facilitated the susceptibility of strain MG116025 to CTX phage, we have examined genetic changes in the tcp gene cluster of MG116025, because CTX phage infection is mediated by TCP. Two SNPs that are specific to MG116025 in the tcp gene cluster compared with other El Tor strains were identified from genomic sequencing data (9, 17). Ala56 (C269) in tcpA is changed to Asp (A269), and Tyr139 (A416) in toxT is replaced by Phe (T416) in MG116025. The transduction efficiency was unchanged or increased by up to 10-fold, depending on CTX phage, when Asp56 in tcpA of MG116025 was switched to Ala, indicating that this change was not critical for the transduction-competent phenotype of MG116025. When Phe139 in toxT of MG116025 was replaced by Tyr, the transduction competency was impaired, suggesting that this change in toxT mediates the ability of MG116025 to be transduced by CTX phages (Table 2 and Tables S2–S5).
Table S2.
Replication and transduction of CTX-1
| Primary transduction | Secondary transduction | |||||
| Donor | pCTX produced from the donor | Transduction recipient strain | Transduction efficiency | Transductants (transduced pCTX) | Transduction recipient strain | Transduction efficiency |
| PM14-U1 | pCTX-1kan-P* | O395 | 105 | O395 (pCTX-1kan-P) | O395 | 5 × 107 |
| MG116025 | 3 ×105 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 105 | |||||
| IB4122- toxTY139F | 5 × 10 | |||||
| MG116025 | 102 | MG116025 (pCTX-1kan-P) | O395 | 5 × 106 | ||
| MG116025 | 103 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 10 | |||||
| IB4122-toxTY139F | 0 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 104 | A213-toxTY139F (pCTX-1kan-P) | O395 | 5 × 104 | ||
| MG116025 | 2 × 10 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 2 × 102 | |||||
| IB4122- toxTY139F | 0 | |||||
| IB4122 toxTY139F | 102 | B4122 toxTY139F(pCTX-1kan-P) | ||||
| PM20 | pCTX-1kan-L† | O395 | 105 | O395 (pCTX-1kan-L) | O395 | 3 × 107 |
| MG116025 | 3 × 105 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 106 | |||||
| IB4122- toxTY139F | 103 | |||||
| MG116025 | 102 | MG116025 (pCTX-1kan-L) | O395 | 5 × 104 | ||
| MG116025 | 5 × 10 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 102 | |||||
| IB4122- toxTY139F | 10 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 104 | A213- toxTY139F (pCTX-1kan-L) | O395 | 2 × 105 | ||
| MG116025 | 10 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 102 | |||||
| IB4122- toxTY139F | 0 | |||||
| IB4122- toxTY139F | 10 | |||||
pCTX-1kan-P: pCTX-1kan produced from the plasmid pUC-CTX-1kan.
pCTX-1kan-L: pCTX-1kan produced from the lysogenic CTX prophage.
Table S5.
Replication and transduction of CTX-O139
| Primary transduction | Secondary transduction | |||||
| Donor | pCTX produced from the donor | Transduction recipient train | Transduction efficiency | Transductants (transduced pCTX) | Transduction recipient strain | Transduction efficiency |
| PM14-U4 | pCTX-O139kan-P* | O395 | 2 × 102 | O395 (pCTX-O139kan-P) | O395 | 106 |
| MG116025 | 5 × 103 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 106 | |||||
| IB4122- toxTY139F | 102 | |||||
| MG116025 | 10 | MG116025 (pCTX-O139kan-P) | O395 | 5 × 106 | ||
| MG116025 | 3 × 104 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 3 × 103 | |||||
| IB4122- toxTY139F | 4 × 102 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 30 | A213- toxTY139F (pCTX-O139kan-P) | ||||
| IB4122- toxTY139F | 20 | IB4122- toxTY139F (pCTX-O139kan-P) | ||||
pCTX-O139kan-P: pCTX-O139kan produced from the plasmid pUC-CTX-O139kan.
The toxT allele (Tyr139) of two El Tor strains that were not transduced by CTX phages—A213 and IB4122 (a Wave 3 El Tor strain, which contains RS1:CTX-3 on chromosome 1) (18)—was replaced by the toxT allele (Phe139) of MG116025, and the strains were examined for their ability to be transduced by CTX phages. A213-toxTY139F and IB4122-toxTY139F were transduced by CTX phages (Tables S2–S5) when grown in LB, pH 6.5 at 30 °C, whereas A213 and IB4122 were not transduced by CTX phages.
toxT 139Phe Allele Up-Regulates TCP.
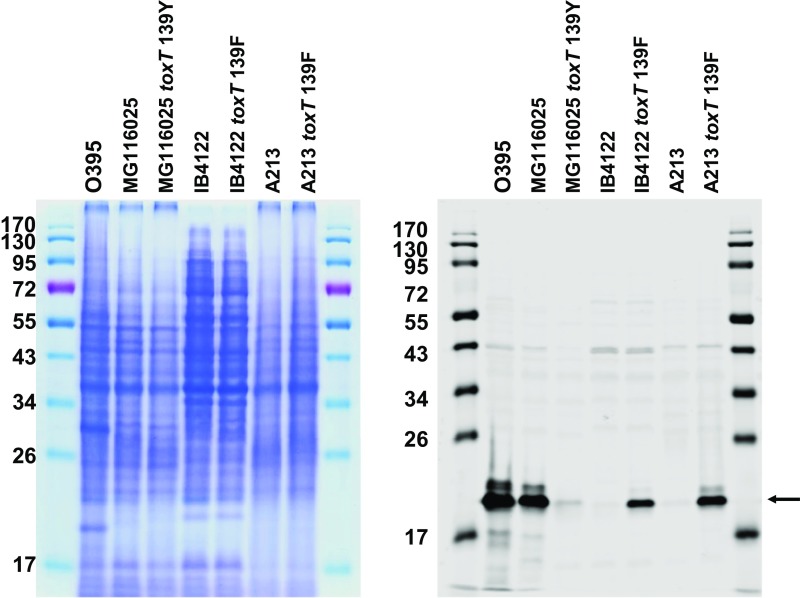
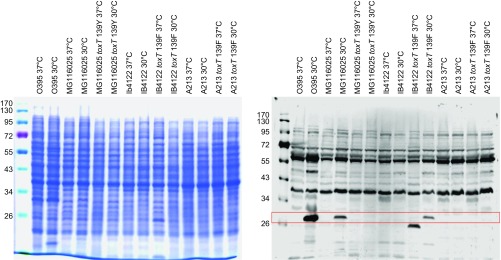
CTX phage infection in V. cholerae strains is mediated by TCP as the phage receptor. The transduction-competent phenotype of strains that harbor the toxT 139Phe allele is perhaps mediated through the elevated expression of TCP. TCP levels were monitored by examining the expression of TcpA by Western blot (Fig. 1 and Fig. S2). El Tor strains that contained the toxT 139Phe allele expressed more TcpA, whereas no TcpA was detected in the strains that contained the toxT 139Tyr allele. High TcpA expression in the O395 strain was observed, despite it harboring the toxT 139Tyr allele. These results indicate that the toxT 139Phe allele in El Tor strains up-regulates TCP, allowing the bacteria to become susceptible to CTX phage infection.
Fig. 1.
toxT 139Phe allele increases TCP expression in El Tor strains. Expression of TCP was assessed by Western blot using anti-TcpA. Approximately 5 × 107 cells grown in LB, pH 6.5 at 30 °C were loaded onto each lane. Lane 1: O395; lane 2: MG116025; lane 3: MG116025-toxTF139Y; lane 4: IB4122; lane 5: IB4122-toxTY139F; and lane 6: A213, 7: A213-toxTY139F.
Fig. S2.
Coomassie Brilliant blue staining of bacterial cells grown in LB pH 6.5 at 30 °C and full-size Western blot image of TcpA expression. Arrow indicates TcpA.
Replication and Transduction of CTX Phages.
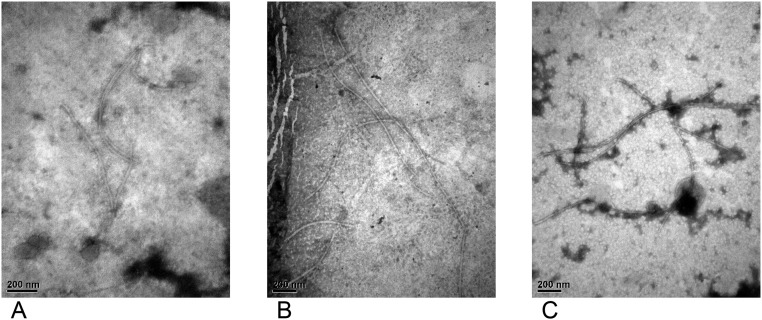
Primary and secondary transduction of various CTX phages was further evaluated with O395, MG116025, A213-toxTY139F, and IB4122-toxTY139F recipients (3). In brief, El Tor strains with the toxT-Phe139 allele can be transduced by CTX phages, although the efficiencies vary, depending on the strain. The production of CTX phage was ∼102- (CTX-1 and CTX-cla) to 104-fold (CTX-2 and CTX-O139) higher by the replicative form pCTXs (secondary transduction) compared with the lysogenic or plasmid-cloned CTX phage genome (primary transduction). Electron microscopy images of CTX-1kan, CTX-2kan, and CTX-O139kan were obtained when the viral titer exceeded 106 particles per milliliter (Fig. S3). In the transductants of various CTX phages, the integration of pCTXs into chromosome 1 or 2 was confirmed, as shown earlier (19).
Fig. S3.
Electron microscopy images of CTX phages. (A) CTX-1kan produced from O395 harboring pCTX-1kan. (B) CTX-2kan replicated from MG116025 transductant by pCTX-2kan. (C) CTX-O139kan replicated from O395 transduced with pCTX-O139kan. Filamentous CTX phages were observed; 100,000×.
CTX-1.
Approximately 105, 104, and 102 transductants were obtained from PM14-U1 (PM14 transformed with pUC-CTX-1kan) when O395, A213-toxTY139F, and MG116025 (and IB4122-toxTY139F) were the recipients (Table S2). Secondary transduction efficiency by pCTX-1kan in the O395 transductant increased up to 5 × 107 when O395 was the recipient, indicating that ∼10% of recipient cells were transduced by CTX-1kan. With the same titer of the CTX-1kan phage, 104 transductants were obtained when MG116025 was the recipient, perhaps because of phage immunity by lysogenic CTX-1 and RS1 (described below). The replication efficiency of pCTX-1kan also varied, depending on the host strain. Whereas 5 × 107 transductants were obtained from O395 (pCTX-1kan), fewer transductants (5 × 106 and 5 × 104, respectively) were generated from MG116025 (pCTX-1kan) and A213-toxTY139F (pCTX-1kan). The replication efficiencies of CTX-1kan phages that were produced from the lysogenic CTX-1 (strain PM20) were similar to those from the pUC-CTX-1kan plasmid. Electron microscopy images of CTX-1kan phages were taken from the culture supernatant of O395 (pCTX-1kan), as shown in Fig. S3A.
CTX-2.
pCTX-2kan, replicated from pUC-CTX-2kan plasmid, was transmitted to MG116025 and IB4122-toxTY139F at an efficiency of 20 and 10 transductants formed, respectively (Table S3). Approximately 2 × 104 transductants were obtained in the secondary transduction from the primary transductant, MG116025, which harbored pCTX-2kan-P. The replication efficiency of lysogenic CTX-2 varied, depending on the host strain: ∼5 × 103 transductants were obtained from PM22 (B33 derivative) and 103 transductants formed from PM9 (V212-1 derivative) with MG116025 as the recipient. No CTX phage was produced from the second CTX-2 prophage of V212-1 (PM10).
Table S3.
Replication and transduction of CTX-2
| Primary transduction | Secondary transduction | |||||
| Donor | pCTX produced from the donor | Transduction recipient strain | Transduction efficiency | Transductants (transduced pCTX) | Transduction recipient strain | Transduction efficiency |
| PM14-U2 | pCTX-2kan-P* | MG116025 | 2 × 10 | MG116025 (pCTX-2kan-P) | MG116025 | 2 × 104 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 3 × 103 | |||||
| IB4122 toxTY139F | 0 | |||||
| O395 | 0 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 10 | |||||
| IB4122 toxTY139F | 0 | |||||
| O395 | 0 | |||||
| PM22 | pCTX-2kan-BL† | MG116025 | 5 × 103 | MG116025 (pCTX-2kan-BL) | MG116025 | 107 |
| MG116025-toxTF139Y | 10 | |||||
| A213- toxTY139F | 4 × 106 | |||||
| IB4122-toxTY139F | 102 | |||||
| O395 | 300‡ | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 5 × 102 | |||||
| IB4122- toxTY139F | 10 | |||||
| O395 | 0 | |||||
| PM9 | pCTX-2kan-VL§ | MG116025 | 103 | MG116025 (pCTX-2kan-VL) | MG116025 | 2 × 107 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 106 | |||||
| IB4122-toxTY139F | 1 × 103 | |||||
| O395 | 200* | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 103 | A213- toxTY139F (pCTX-2kan-VL) | MG116025 | 2 × 103 | ||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 2 × 103 | |||||
| IB4122- toxTY139F | 0 | |||||
| O395 | 0 | |||||
| IB4122- toxTY139F | 10 | IB4122- toxTY139F (pCTX-2kan-VL) | ||||
| O395 | 5¶ | |||||
| PM10 | MG116025 | 0 | ||||
pCTX-2kan-P: pCTX-2kan produced from the plasmid pUC-CTX-2kan.
pCTX-2kan-BL: pCTX-2kan produced from lysogenic CTX-2 in PM22, which is a derivative of strain B33.
O395 transductants were transduced by CTX-2kan (one-third of transductants) or CTX phage (two-thirds of transductants) that contained rstREl Tor generated by interstrand recombination between CTX-2kan and lysogenic CTX-1 in MG116025.
pCTX02kan-VL: pCTX-2kan produced from lysogenic CTX-2 in PM9, which is a derivative of strain V212-1.
These five O395 transductants were transduced by CTX phage that contained rstREl Tor generated by interstrand recombination between CTX-2kan and lysogenic CTX-1 in V212-1 derivative PM9.
A213-toxTY139F was also transduced at a slightly lower efficiency than MG116025, whereas the transduction efficiency of IB4122-toxTY139F was substantially lower. Several O395 recipients were transduced by the supernatant of PM9, but the pCTX that was transmitted to O395 contained rstREl Tor, which was perhaps generated by interstrand recombination between CTX-2kan that was generated from the plasmid and CTX-1 on chromosome 1, as reported previously (16). O395 was not transduced by CTX-2kan at a low titer, perhaps because of the immunity that was conferred by rstRcla in lysogenic CTX-cla in O395.
The secondary transduction efficiencies of pCTX-2kan, produced from PM9 or PM22, exceeded 106 when MG116025 was the recipient. A213-toxTY139F and IB4122-toxTY139F were transduced by a high titer of CTX-2kan phage. Although O395 was not transduced by the low titer of CTX-2kan phage, as shown above, the classical strain was transduced by a high titer of CTX-2kan in the secondary transduction. Of several hundred transductants, approximately two-thirds of O395 transductants were transduced by CTX phage that contained rstREl Tor, and one-third was transduced by CTX-2. Electron microscopy images of CTX-2kan phages in the culture supernatant of MG116025 (pCTX-2kan) were taken (Fig. S3B).
CTX-cla.
The transduction efficiency of CTX-clakan from pUC-CTX-clakan to MG116025 was 102 transductants, and 104 transductants were formed in the secondary transduction (Table S4). A213-toxTY139F and IB4122-toxTY139F were also transduced by CTX-clakan in the primary and secondary transduction at a slightly lower efficiency than MG116025. Because of the low titer of CTX-clakan, electron microscopy images of CTX-clakan phages were not available.
Table S4.
Replication and transduction of CTX-cla
| Primary transduction | Secondary transduction | |||||
| Donor | pCTX produced from the donor | Transduction recipient strain | Transduction efficiency | Transductants (transduced pCTX) | Transduction recipient strain | Transduction efficiency |
| PM14-U3 | pCTX-clakan-P* | MG116025 | 1 × 102 | MG116025 (pCTX-clakan-P) | MG116025 | 3 × 104 |
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 2 × 104 | |||||
| IB4122- toxTY139F | 0 | |||||
| O395 | 0 | |||||
| MG116025-toxTF139Y | 0 | |||||
| A213- toxTY139F | 4 × 10 | A213- toxTY139F (pCTX-clakan-P) | ||||
| IB4122- toxTY139F | 3 × 10 | IB4122- toxTY139F (pCTX-clakan-P) | ||||
| O395 | 0 | |||||
pCTX-clakan-P: pCTX-clakan produced from the plasmid pUC-CTX-clakan.
CTX-O139.
Replication of CTX-O139 from the plasmid-cloned CTX-O139 is demonstrated in this study, as replication and production of infectious CTX-O139 from the lysogenic CTX-O139 genome have been reported (4). CTX-O139kan phages that were produced from PM14-U4 could be transmitted by transduction, with ∼200 and 10 transductants formed when O395 and MG116025 were the recipients, respectively (Table S5). The secondary transduction efficiency increased up to 106 transductants formed, and electron microscopy images of CTX-O139kan phages were taken (Fig. S3C). A213-toxTY139F and IB4122-toxTY139F were also transduced by a high titer of CTX-O139kan phages.
Phage Immunity.
To measure the degree of inhibition of superinfection of CTX-1 phage by lysogenic CTX-1 and RS1, a set of isogenic strains of MG116025 was constructed by stepwise removal of CTX-1 and RS1 (strains PM25 ∼ PM29). PM28 and PM29, which do not contain CTX-1 or RS1, were transduced by CTX-1 with ∼80% efficiency; 5 × 108 transductants were obtained when 6 × 108 recipient cells were used (Table S6), which was 10-fold higher compared with the classical strain O395. The transduction efficiency was nearly the same when PM27 that contained a single RS1 (thus, 1 rstREl Tor) was used as the recipient. The transduction efficiency was reduced by ∼10−4-fold in strains PM26 and PM25, as well as MG116025. These results indicate that the superinfection of CTX-1 is restricted by rstREl Tor of lysogenic CTX-1 or RS1. However, one rstR is not sufficient to repress the superinfection, and at least two rstR genes are necessary to repress the superinfection. Whereas the superinfection of CTX-1kan phage was restricted by the preexisting RS1 and CTX-1, the infection of CTX-2 kan that contained rstRcla was unaffected by rstREl Tor (heteroimmunity).
Table S6.
Repression of superinfection of CTX-1kan phage by resident rstREl Tor in lysogenic CTX-1 and RS1
| Recipient | No. of transductants by CTX-1kan | No. of transductants by CTX-2kan |
| MG116025 (TLC:RS1:CTX-1:RS1) | 3 × 105 | 5 × 107 |
| PM25 (TLC:CTX-1:RS1) | 3 × 105 | 4 × 107 |
| PM26 (TLC:RS1:RS1) | 5 × 105 | 5 × 107 |
| PM27 (TLC:RS1) | 5 × 108 | 6 × 107 |
| PM28 (TLC) | 5 × 108 | 5 × 107 |
| PM29 (No TLC, No element) | 5 × 108 | 5 × 107 |
toxT and Cholera Toxin Production.
ToxT is a 32-kDa AraC family transcriptional activator (20). ToxT contains a conserved C-terminal DNA-binding domain that comprises 100 amino acids, and its 176-amino acid N terminus is a dimerization domain (21). ToxT positively controls the expression of the tcp gene cluster and ctxAB directly. The toxT-F139 allele of El Tor strains up-regulated TCP at 30 °C, pH 6.5, enabling the bacteria to be transduced by CTX phages. This result indicates that the toxT-F139 allele also increases the expression of cholera toxin. The production of cholera toxin in V. cholerae strains was examined by Western blot (Fig. 2 and Fig. S4). No cholera toxin was produced in the 37 °C culture, but production was observed at 30 °C, pH 6.5 in O395, MG116025, and IB4122-toxTY139F. Cholera toxin expression was shut off in MG116025-toxTF139Y, demonstrating that the SNP in toxT mediates the elevated expression of TCP and cholera toxin of El Tor biotype strains under laboratory conditions.
Fig. 2.
Immunoblot analysis of CtxA production. Expression of CtxA was examined by immunoblot using anticholera toxin. Approximately 5 × 107 cells were loaded onto each lane. Lanes 1 and 2: O395; lanes 3 and 4: MG116025; lanes 5 and 6: MG116025-toxTF139Y; lanes 7 and 8: IB4122; lanes 9 and 10: IB4122-toxTY139F; lanes 11 and 12: A213; and lanes 13 and 14: A213-toxTY139F. Odd-numbered lanes are bacterial samples grown at 37 °C in LB media, and even-numbered lanes are bacterial samples grown at 30 °C in LB, pH 6.5. Arrow indicates CtxA.
Fig. S4.
Coomassie Brilliant blue staining of bacterial cells grown in LB at 37 °C or LB, pH 6.5 at 30 °C and full-size Western blot image of CtxA expression. Red box indicates CtxA.
Discussion
To study the evolution of toxigenic classical biotype strains and atypical El Tor strains, experimental evidence of replication of CTX-cla and CTX-2, which harbor rstRcla, must be generated. The development of atypical El Tor strains from prototype El Tor strains was proposed as follows (7): (i) infection and lysogenization of CTX-2 (although the replication and transmission of CTX-2 have remained unverified) in a prototype El Tor strain that contains TLC:RS1:CTX-:RS1 (the same array as MG116025) to generate an intermediary strain, such as V212-1; (ii) generation of Wave 2 strains from the intermediary strain by stepwise removal of CTX-1 and RS1; and (iii) recombination between CTX-2 and CTX-1/RS1 in the intermediary strain to generate CTX-3 ∼ CTX-6 and transmission of the recombinant CTX phages to other El Tor strains to give rise to Wave 3 strains.
The replication and dissemination of CTX-2 phage have been demonstrated under laboratory conditions in this report (3, 16). Because of the absence of classical biotype strains that contain a tandem repeat of lysogenic CTX-cla, the replication of lysogenic CTX-cla has not been provided in this report; however, the presence of a tandem repeat of CTX-2 in Wave 2 atypical El Tor strains suggests that classical strains that harbor a tandem repeat of CTX-cla existed. The primary genesis of CTX-2 can be recapitulated in a strain that contains a tandem repeat of CTX-cla and is transduced by pCTX-1. CTX-2 could have been generated in such a strain by interphage recombination between pCTX-1 and a tandem repeat of CTX-cla.
Our results on the replication of CTX-cla/CTX-2 and interstrand recombination between two types of CTX phage in a single host cell explain the mechanisms by which toxigenic V. cholerae classical biotype strains and atypical El Tor strains are generated.
The replication efficiencies of pCTX-cla and pCTX-2, which were replicated from pUC-CTX-cla and pUC-CTX-2 (both with ∼104 transductants formed), respectively, were lower than that of pCTX-2 that was replicated from the lysogenic CTX-2 of PM9 and PM22 (more than 106 transductants formed), perhaps because of variations in the ig-1 sequences between CTX-1 and CTX-cla (22). The lysogenic CTX-cla and thus the authentic CTX-cla phage contain the ig-1 sequence that is identical to that of CTX-2; however, the recombinant plasmid pUC-CTX-clakan contained the ig-1 sequence of CTX-1. Although we attempted repeatedly to construct an authentic pUC-CTX-cla or pUC-CTX-2 that contains the ig-1 sequence of CTX-cla, we failed and continue to examine this procedure.
Each biotype-specific RstR inhibits the superinfection of the same type of CTX phage (14). RstR suppresses the transcription of rstA, and the transcription is sufficiently repressed by a single rstR (14). Our results also indicate that rstREl Tor in lysogenic RS1 and CTX-1 can suppress the superinfection of the CTX-1 phage that contains the same type of rstR. However, at least two copies of residential rstREl Tor are necessary to inhibit the superinfection of CTX-1.
Amino acid 139 in ToxT resides in a loop between helices α2 and α3 (21). Nineteen residues, including several hydrophobic residues in the N-terminal domain of ToxT, are essential for its dimerization and transcriptional activation activity (23, 24). The replacement of Tyr with Phe in the toxT allele might increase its hydrophobicity, enhancing the function of ToxT as a transcriptional activator.
Induction of cholera toxin expression in El Tor strains under laboratory conditions has been challenging, whereas it has been established in classical biotype strains at 30 °C in LB, pH 6.5 (25, 26). AKI conditions (biphasic culture in a medium that contains 0.3% NaHCO3) and low-oxygen/high CO2 concentrations induce cholera toxin expression in El Tor strains (27, 28). Here, we showed that the toxT-Y139F allele enables El Tor strains to produce cholera toxin in single-phase cultures. Increased Tcp and cholera toxin expression might affect the colonization and virulence of V. cholerae. Potential changes in colonization in animal intestine models and the virulence of isogenic strains that contain various toxT alleles will need to be studied further.
In this report, we have described the replication and production of CTX phages in a plasmid-based replication system and the transduction of El Tor strains by various CTX phages by introducing an SNP into toxT. These results are experimental evidence of the replication of CTX-cla and CTX-2 phages, explaining the generation of toxigenic classical biotype strains and atypical El Tor strains (7, 8).
Materials and Methods
CTX Phage Plasmid Construction.
A 674-bp origin of replication DNA fragment of pUC18 was amplified by PCR with the primer pair pUC18-ori-MluIF (CCG CGC ACG CGT ATG TGA GCA AAA GGC) and pUC18-ori-KpnIR (CGC GCC GGT ACC CCC GTA GAA AAG ATC). A MluI restriction enzyme site (at nucleotide 245) in zot, which is common in various CTX phage genomes, was used for the plasmid construction (Fig. S1). A zot-3′UTR fragment that spans nucleotide 245 of zot to the termination codon of rstREl Tor of RS1 was amplified by PCR with the primer pair zot-MluIF (CGC GCG ACG CGT TTC TCT TTA TCG ATG) and 3′UTR-KpnIR (CCG GCC GGT ACC CAA GAC TCG CTA GCG) from the strain PM8, which contains TLC:CTX-1kan:RS1 on chromosome 1 (16). This fragment was common to all CTX phages that were constructed in this study. The zot genes of CTX-1 and CTX-cla differ by 14 SNPs; however, zot does not affect the morphogenesis of CTX phages (5). A recombinant plasmid that consisted of the pUC18 replication origin fragment and zot-3′UTR fragment was constructed first, and then the 5′ fragment was inserted into the MluI site of this plasmid. Two 5′UTR-zot fragments of each CTX phage were amplified using a common reverse primer, MluIR (CCG GCG ACG CGT CCT TTC TCG CCC AGT GCC), and the forward primer 5′UTR MluIF (CGC CCG ACG CGT TAG AGA CAA AAT GTT CCT, from the att site, so that the entire ig-1 sequence is included) or 5′-119MluIF (CGC CCG ACG CGT GCC TGT CCG CTG TGG, from nucleotide −119 upstream of the termination codon of rstR of the lysogenic CTX genome). The 5′ fragment of CTX-cla was amplified in a classical strain, Cairo48, and the 5′ fragment of CTX-O139 was amplified in an O139 strain, AR1961537. The forward primer B33Ch2MluF (CGC CCG ACG CGT ATG ATG TTT TTA TTC CAC), instead of 5′UTR MluIF, was used to amplify the 5′UTR-zot fragment of CTX-2 on chromosome 2 of strain B33.
CTX Phage Transduction.
Transduction of CTX phages from a lysogenic CTX phage genome, a recombinant plasmid in a V. cholerae strain that does not contain a CTX prophage, or strains that contained the replicative form CTX phage was conducted, as described previously (3, 16). Briefly, 0.5 mL of the culture supernatant of the donor strain, which was grown overnight in the presence of 20 ng/mL mitomycin C, was mixed with 3 × 108 CFU of agglutinated (grown in 30 °C, LB pH 6.5) recipient strains (11). The transduction efficiencies of the recipient strains were calculated by the number of transductants per 6 × 108 CFU recipient cells per 1 mL of the supernatant of the donor strains. The transduction efficiency of each recipient strain does not reflect the actual titer of the CTX phage that is produced from the donor strain, because the transduction efficiency differs between recipient strains. Transduction efficiencies were presented as the average of at least three independent experiments.
toxT Allele Exchange.
DNA fragments from 50 nucleotides upstream of the ATG start codon to nucleotide 793 of toxT were amplified with the primers toxT-XbaIF: CCG GCC TCT AGA TAC GTG GAT GGC TCT CTG CG and toxTSacIR: CCG GCC GAG CTC CAC TTG GTG CTA CAT TCA by PCR from the strains MG116025 and N16961 and inserted into a suicide plasmid, pCVD442. The SNP position at nucleotide 416 (A416 of N16961 and T416 of MG116025) lies in the center of this fragment. The MG116025 toxT 139Phe allele was replaced by N16961 toxT 139Tyr allele by allelic exchange method, and similarly, IB4122 and A213 toxT 139Tyr allele was replaced with MG116025 toxT 139Phe allele (29).
Western Blot.
Bacterial strains were grown at 30 °C overnight in LB medium and subcultured at a dilution of 1:100 in LB for the 37 °C culture or in LB (pH 6.5) for the 30 °C culture and grown overnight again. Cells were pelleted, resuspended in 1× sample buffer, and boiled for 5 min. Approximately 5 × 107 cells were loaded onto each lane in an SDS-polyacrylamide gel. Proteins were transferred to Protran nitrocellulose membrane (GE Healthcare) and probed with rabbit anti-CT (Sigma) or rabbit anti-TcpA (a generous gift from W. F. Wade, Dartmouth University, Hanover, NH) (30).
Construction of MG116025 Derivatives.
A set of isogenic strains of MG116025, PM25 (TLC:CTX-1:RS1), PM26 (TLC:RS1:RS1), PM27 (TLC:RS1), PM28 (TLC), and PM29 (no TLC, no element) was constructed by stepwise excision of CTX-1 and RS1 from chromosome 1, as described previously (16). pCVDrstRET that contained a DNA fragment that harbored rstREl Tor (339 bp) and the first 226 bp of rstA was inserted into rstREl Tor of CTX-1 and RS1s of MG116025 individually (29). The excision of CTX-1 and RS1 was verified by analyzing the genetic structure of strains that were selected on LB plates with 15% (wt/vol) sucrose.
CTX-1 Phage Immunity.
Inhibition of superinfection of CTX-1 by the lysogenic CTX-1 or RS1 was monitored by measuring the transduction efficiency of isogenic strains of MG116025. MG116025 and PM25 ∼ PM29 were transduced by pCTX-1kan phages that were produced from the O395 transductant by pCTX-1kan. Immunity against CTX-2 was tested in the same set of strains with pCTX-2kan phages that were produced from the MG116025 transductant by pCTX-2kan.
Acknowledgments
We thank Dr. Claude Parsot and Lisa Wawrynchuk for critical review of the manuscript. This work was supported by Grants NRF-2015R1A2A2A01007297 and NRF-2015M3C9A2054024 from the National Research Foundation of Korea.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701335114/-/DCSupplemental.
References
- 1.Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004;363(9404):223–233. doi: 10.1016/s0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8(1):48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 4.Davis BM, Kimsey HH, Chang W, Waldor MK. The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor. J Bacteriol. 1999;181(21):6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim EJ, Lee D, Moon SH, Lee CH, Kim DW. CTX prophages in Vibrio cholerae O1 strains. J Microbiol Biotechnol. 2014;24(6):725–731. doi: 10.4014/jmb.1403.03063. [DOI] [PubMed] [Google Scholar]
- 6.Boyd EF, Heilpern AJ, Waldor MK. Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(phi)s by toxigenic Vibrio cholerae. J Bacteriol. 2000;182(19):5530–5538. doi: 10.1128/jb.182.19.5530-5538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim EJ, Lee CH, Nair GB, Kim DW. Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol. 2015;23(8):479–489. doi: 10.1016/j.tim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18(1):46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque SM, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci USA. 2007;104(12):5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd EF, Waldor MK. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: Generalized transduction of CTXPhi by bacteriophage CP-T1. Infect Immun. 1999;67(11):5898–5905. doi: 10.1128/iai.67.11.5898-5905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udden SM, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci USA. 2008;105(33):11951–11956. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: Functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;182(24):6992–6998. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimsey HH, Waldor MK. CTXphi immunity: Application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95(12):7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruque SM, et al. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXPhi: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66(12):5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim EJ, et al. Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog. 2014;10(9):e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose KE. Regulation of virulence in Vibrio cholerae. Int J Med Microbiol. 2001;291(2):81–88. doi: 10.1078/1438-4221-00104. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen BM, et al. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47(5):1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467(7318):982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowden MJ, et al. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci USA. 2010;107(7):2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Childers BM, et al. Identification of residues critical for the function of the Vibrio cholerae virulence regulator ToxT by scanning alanine mutagenesis. J Mol Biol. 2007;367(5):1413–1430. doi: 10.1016/j.jmb.2007.01.061. [DOI] [PubMed] [Google Scholar]
- 22.Kim EJ, Yu HJ, Kim DW. Sequence variations in the non-coding sequence of CTX phages in Vibrio cholerae. J Microbiol Biotechnol. 2016;26(8):1473–1480. doi: 10.4014/jmb.1604.04022. [DOI] [PubMed] [Google Scholar]
- 23.Childers BM, et al. N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J Biol Chem. 2011;286(32):28644–28655. doi: 10.1074/jbc.M111.258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plecha SC, Withey JH. Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J Bacteriol. 2015;197(10):1716–1725. doi: 10.1128/JB.02409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez J, Medina G, Buhse T, Holmgren J, Soberón-Chavez G. Expression of cholera toxin under non-AKI conditions in Vibrio cholerae El Tor induced by increasing the exposed surface of cultures. J Bacteriol. 2004;186(5):1355–1361. doi: 10.1128/JB.186.5.1355-1361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobaxin M, et al. Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb Pathog. 2014;66(1):5–13. doi: 10.1016/j.micpath.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Iwanaga M, et al. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30(11):1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, et al. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci USA. 2011;108(2):810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor RK, Kirn TJ, Meeks MD, Wade TK, Wade WF. A Vibrio cholerae classical TcpA amino acid sequence induces protective antibody that binds an area hypothesized to be important for toxin-coregulated pilus structure. Infect Immun. 2004;72(10):6050–6060. doi: 10.1128/IAI.72.10.6050-6060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, et al. Mozambique Cholera Vaccine Demonstration Project Coordination Group Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J Med Microbiol. 2006;55(Pt 2):165–170. doi: 10.1099/jmm.0.46287-0. [DOI] [PubMed] [Google Scholar]