Significance
Since the discovery of oosporein more than 70 years ago, there have been conflicting reports on its potential antimicrobial and insecticidal activities. Our results indicate that oosporein is unlikely to function as an insect toxin or to be involved in early to mid-infection processes, including penetration and immune evasion. Instead, oosporein most likely functions after death of the host to thwart bacterial competition on a host cadaver, allowing the fungus to maximally use host nutrients and complete its life cycle. Our data also reveal that oosporein production is regulated by a cascade of transcription factors, with BbSmr1 acting as an upstream negative regulator, targeting the expression of OpS3, which in turn acts as a positive regulator of the oosporein biosynthetic gene cluster.
Keywords: Beauveria bassiana, oosporein, biological role, transcription factor, fungal–bacterial competition
Abstract
The regulatory network and biological functions of the fungal secondary metabolite oosporein have remained obscure. Beauveria bassiana has evolved the ability to parasitize insects and outcompete microbial challengers for assimilation of host nutrients. A novel zinc finger transcription factor, BbSmr1 (B. bassiana secondary metabolite regulator 1), was identified in a screen for oosporein overproduction. Deletion of Bbsmr1 resulted in up-regulation of the oosporein biosynthetic gene cluster (OpS genes) and constitutive oosporein production. Oosporein production was abolished in double mutants of Bbsmr1 and a second transcription factor, OpS3, within the oosporein gene cluster (ΔBbsmr1ΔOpS3), indicating that BbSmr1 acts as a negative regulator of OpS3 expression. Real-time quantitative PCR and a GFP promoter fusion construct of OpS1, the oosporein polyketide synthase, indicated that OpS1 is expressed mainly in insect cadavers at 24–48 h after death. Bacterial colony analysis in B. bassiana-infected insect hosts revealed increasing counts until host death, with a dramatic decrease (∼90%) after death that correlated with oosporein production. In vitro studies verified the inhibitory activity of oosporein against bacteria derived from insect cadavers. These results suggest that oosporein acts as an antimicrobial compound to limit microbial competition on B. bassiana-killed hosts, allowing the fungus to maximally use host nutrients to grow and sporulate on infected cadavers.
Fungi produce a multitude of low molecular weight molecules of diverse chemical structures collectively termed secondary metabolites. These compounds are involved in many different biological processes, including fungal development, intercellular communication, and interaction with other organisms in complex niches (1). Fungal secondary metabolites are of significant biotechnological and biopharmaceutical interest because they include antibiotics and other molecules/drugs relevant to human health (2). The biosynthesis of many secondary metabolites often involves large gene clusters with obscure regulatory networks and cryptic induction parameters (1, 3). Indeed, significant effort has been expended in determining the appropriate conditions for the production of many fungal secondary metabolites, and, equally importantly, the biological functions of many of these compounds remain unknown (4–6).
Oosporein is a red dibenzoquinone pigment initially reported in Oospora colorans (7) and subsequently found in a number of soil and endophytic fungi, as well as in the entomopathogenic Beauveria genus (8–11). The physiochemical properties of oosporein have been explored, and its chemical synthesis has been reported (12–14). Anecdotal evidence seen in our laboratory and reported by others has indicated that B. bassiana can sometimes produce oosporein during growth in various artificial media, although no consistent results have been reported. In one study, 5 of 16 B. bassiana strains freshly isolated from insect hosts were found to be capable of producing oosporein under the conditions tested; however, the oosporein-producing strains lost their ability to synthesize the red pigment after serial passage on artificial media (15). Nonetheless, it has been noted that insect cadavers killed by B. bassiana appear reddish immediately at death (16). Several studies have indicated that oosporein may possess some insecticidal activity. Limited mortality (15–20%) was seen when partially purified oosporein was topically applied onto silverleaf whitefly (Bemisia tabaci) nymphs, with a synergistic effect (>90% mortality) seen when oosporein was combined with B. bassiana spores compared with fungal spores alone, in which only 60% host mortality was noted (17). Oosporein also has been documented to exert antimicrobial, antioxidant, and cytotoxic activities (18). At high concentrations, oosporein reportedly exhibits toxicity to poultry, and various techniques for monitoring and detecting oosporein with high sensitivity have been reported (9, 19, 20).
The availability of the B. bassiana genome has led to the identification of at least 45 different putative secondary metabolite biosynthetic gene clusters, including the prediction of one cluster potentially responsible for oosporein production (21, 22). The latter gene cluster was later verified experimentally as being responsible for oosporein synthesis via both genetic and biochemical characterization (23). The oosporein biosynthetic cluster comprises at least seven ORFs, including a nonreducing polyketide synthase [oosporein synthase 1 (OpS1)], a membrane transporter (OpS2), a transcription factor (TF) (OpS3), and four additional enzymes involved in oosporein biosynthesis (OpS4–OpS7). Disruption of the OpS3 TF was found to abolish oosporein production, indicating that it acts as a positive regulator of the system (23). The mechanism behind OpS3 activation is unknown; however, it was previously shown that the B. bassiana ortholog of the Msn2 stress response TF acts as a pH-dependent negative regulator of oosporein biosynthesis (24). Based on characterization of the various OpS gene deletion mutants, a putative role for oosporein in hyphal body development and immune evasion has been suggested (23).
In the present study, we initiated a genetic screen for B. bassiana T-DNA insertion mutants that overproduce oosporein. One such mutant was mapped to an ORF coding for a zinc finger TF designated B. bassiana secondary metabolite regulator 1 (Bbsmr1) owing to its regulation of secondary metabolite production. Targeted gene deletion of Bbsmr1 resulted in constitutive oosporein production and derepressed expression of the OpS genes, including OpS3, a regulatory gene found within the oosporein gene cluster. Double ΔBbsmr1::ΔBbOpS3 mutants did not produce oosporein. Studies of gene expression coupled to GFP promoter indicated that oosporein production was induced during late stages of infection, but not in early stages, including attachment, penetration, proliferation, and immune evasion during hyphal body growth in the host hemocoel. Oosporein itself was not detected until after host death, with levels increasing at 24–48 h post death (hpd). Oosporein was shown to display strong antimicrobial activity toward host bacterial flora, inhibiting the ability of bacteria to proliferate on cadavers. These results suggest that the biological function of oosporein is to act as a late-stage antimicrobial compound, decreasing bacterial competition and allowing the fungus to complete its life cycle and sporulate on the host cadaver.
Results
Screening of B. bassiana T-DNA Insertion Library for Derepression of Oosporein Production and Identification of the BbSmr1 TF.
We screened a collection of 5,000 clones derived from a B. bassiana T-DNA insertion mutant library (25) for red pigment production during growth in 0.5× Sabouraud dextrose broth (SDB) medium. One mutant (designated T120) was identified based on morphological assessment of the production of a deep red pigment in 0.5× SDB medium, conditions under which the wild type (WT) colony was yellow-white. The pigmented compound was partially purified, and comparison with an oosporein standard using HPLC-MS confirmed its identity to oosporein.
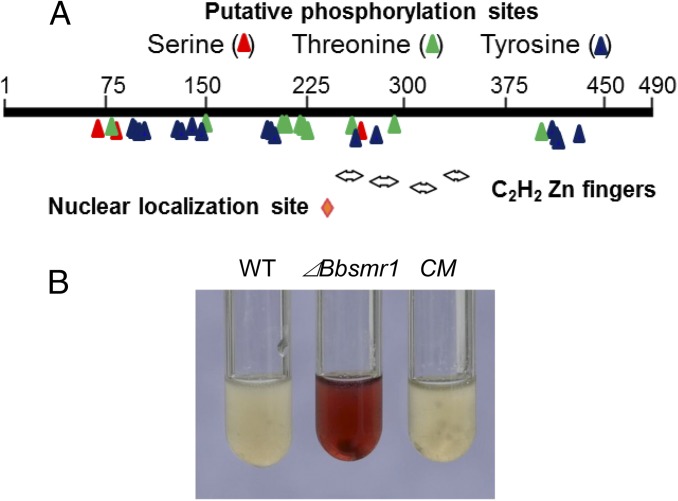
The T-DNA insertion site in strain T120 was mapped to an asparagine-rich C2H2-type zinc finger protein (GenBank accession no. EJP66373.1) annotated as Bbazf1 (21) and renamed Bbsmr1. The full-length genomic sequence of Bbsmr1, including 5′ and 3′ flanking sequences (3.9 kb), was obtained via chromosome walking as detailed in Materials and Methods. (The genome of B. bassiana was not available at that time.) Analysis of the sequence indicated that the ORF of Bbsmr1 was 1,473 bp long, with no introns, and encoding a highly basic protein of 490 amino acids with a molecular mass of 55 kDa and an isoelectric point of 9.4. Phylogenetic analysis revealed Smr1 orthologs broadly distributed in filamentous fungi (SI Appendix, Fig. S1), with a potential ortholog identified in yeast (32% identity to Saccharomyces cerevisiae; GenBank accession no.AJU08930.1). To date, the function of Smr1/Azf1 remains uncharacterized. The predicted BbSmr1 protein showed the best matches to predicted proteins found in Cordyceps militaris protein (GenBank accession no. EGX92404.1; 86% identity), with ∼60% identity to known proteins of Metarhizium anisopliae (GenBank accession no. EFY97004.1), Trichoderma reesei (GenBank accession no. EGR53090.1), and Claviceps purpurea (GenBank accession no. CCE27792.1) (SI Appendix, Fig. S1). Protein domain analyses of BbSmr1 indicated the presence of four C2H2-zinc finger consensus motifs at Cys246-His268, Cys276-His298, Cys306-His326, and Cys334-His357; a nuclear localization signal, PKKKWV, at amino acids 240–245; and several putative phosphorylation sites (Fig. 1A).
Fig. 1.
BbSmr1 protein structure and its negative regulation of oosporein production. (A) Schematic of BbSmr1 protein features indicating the presence of a nuclear localization signal (red diamond), zinc finger motifs (double arrows), and putative phosphorylation sites as indicated. (B) BbSmr1 negatively regulates oosporein production in B. bassiana. Shown are images of culture supernatants derived from B. bassiana WT strain (WT), Bbsmr1-targeted gene knockout (ΔBbsmr1), and complementation mutant (CM), as detailed in Materials and Methods.
BbSmr1 Negatively Regulates Expression of the Oosporein Gene Cluster.
To confirm and further characterize the mutant phenotype, we constructed a targeted gene knockout mutant of Bbsmr1 as described in Materials and Methods. The targeted genomic insertion event and loss of Bbsmr1 expression were confirmed by Southern blot analysis and real-time quantitative PCR (qPCR), respectively (SI Appendix, Fig. S2). A complementation mutant (CM) was also constructed via ectopic integration of the entire Bbsmr1 ORF, as well as 1,966- and 481-bps 5′ and 3′ flanking sequences, respectively, into the ΔBbsmr1 mutant strain.
To confirm the original selection phenotype, we inoculated the ΔBbsmr1, CM, and WT type parental strains into 0.5× SDB. After 3 d of growth at 26 °C, the culture supernatant derived from the ΔBbsmr1 strain produced copious amounts of oosporein, whereas no oosporein was detected in either the WT or CM strain (Fig. 1B). The identity of the red pigmented compound as oosporein was confirmed via extraction and HPLC-MS analyses and comparison with an oosporein standard (SI Appendix, Fig. S3).
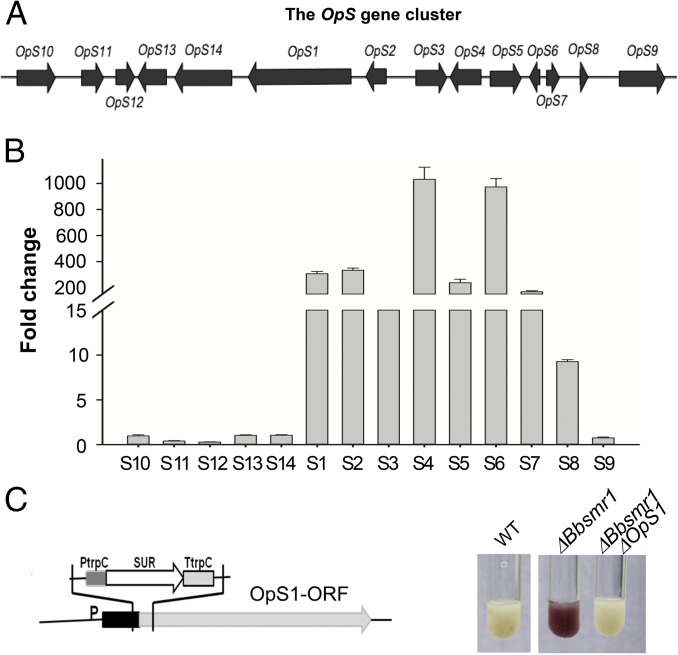
The oosporein biosynthetic gene cluster was partially characterized recently (22, 23). The cluster contains 14 putative ORFs (GenBank accession nos. EJP62787–EJP62800, antiSMASH predicted) and includes a gene for a (type I) nonreducing polyketide synthase gene (OpS1; GenBank accession no. EJP62792) that produces the orsellinic acid precursor to oosporein (23) (Fig. 2A). Gene expression analyses revealed that loss of the BbSmr1 TF resulted in derepression of ORFs corresponding to OpS1–OpS7 (following the nomenclature used in ref. 23), as well as one additional 3′-flanking ORF, designated OpS8 (Fig. 2B). The expression of five additional ORFs, designated OpS10–OpS14, located on the 5′-flanking side of OpS1, was examined as well. Of these, OpS11 expression decreased by ∼2.5-fold and OpS12 expression decreased by ∼3.3-fold, whereas little to no change in expression was seen for ORFs OpS9, OpS10, OpS13, and OpS14.
Fig. 2.
Regulation of the OpS gene cluster by Bbsmr1. (A) Schematic of the OpS gene cluster. (B) Fold change in gene expression of the OpS1-14 genes. Real-time qPCR expression analysis of the OpS gene cluster in the ΔBbsmr1 deletion mutant normalized to WT expression levels. (C) Schematic of OpS1 gene knockout in the Bbsmr1 deletion mutant and its effect on the oosporein production.
To provide further confirmation that oosporein overproduction in the ΔBbsmr1 strain was related to activity of the OpS1 polyketide synthase (PKS) gene, we constructed a double-mutant strain, ΔBbsmr1ΔΟpS1. Culture supernatants of the ΔBbsmr1ΔOpS1 strain were devoid of oosporein production (Fig. 2C).
The OpS3 TF Functions Downstream of the BbSmr1 TF.
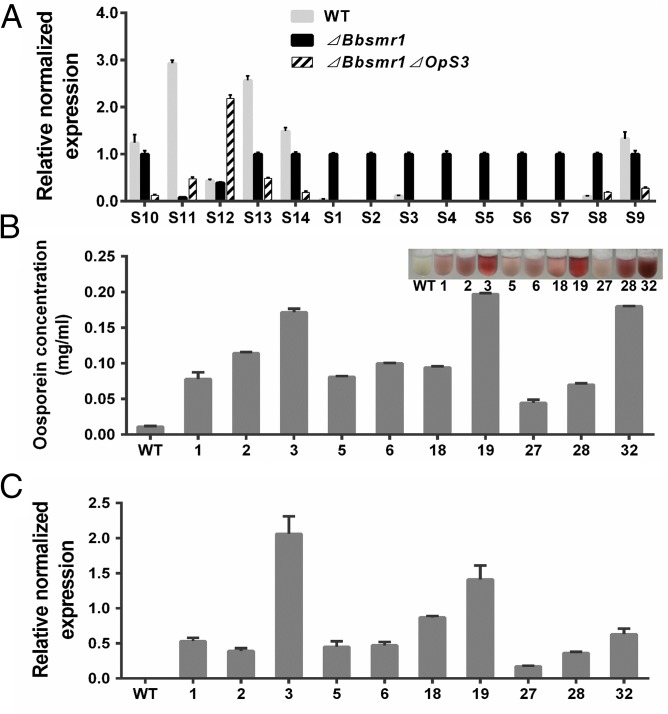
OpS3 (GenBank accession no. EJP62794) has been shown to code for a TF regulating the oosporein biosynthetic cluster (23). To examine whether OpS3 functions downstream of BbSmr1, we constructed a double-knockout strain, ΔBbsmr1ΔOpS3. Compared with the derepression of ORFs OpS1–OpS8 seen in the ΔBbsmr1 strain, we found little to no expression of these genes in the ΔBbsmr1ΔOpS3 double mutant (Fig. 3A). To further verify the function of OpS3 on the production of oosporein, we overexpressed the gene in WT B. bassiana by placing it under control of a constitutive promoter, PBbgpdA (26). Transformation and ectopic integration of the overexpression vector into the WT strain resulted in clones producing various amounts of oosporein, ranging from 0.05 to 0.2 mg/mL, in culture supernatants, compared with WT levels of <0.01 mg/mL (Fig. 3B). Oosporein production correlated with OpS3 expression in the various overexpression clones ranging from 5- to 20-fold higher than that in WT (Fig. 3C).
Fig. 3.
Regulation of OpS3 on the expression of OpS gene cluster and oosporein production. (A) RT-qPCR analysis of the expression of the OpS gene cluster in WT, ΔBbsmr1, and ΔBbsmr1ΔOpS3 strains. Gene expression data were normalized to gpd, actin, and cypA as detailed in Materials and Methods. (B) Oosporein production in OpS3 overexpression strains. Oosporein production in WT and 10 different OpS3 overexpression transformants was quantified. (Inset) The colors of the various culture supernatants. (C) Real-time qPCR analysis of OpS3 expression in the various B. bassiana overexpression transformants as in B. bassiana.
Oosporein Is Not Directly Related to Fungal Virulence.
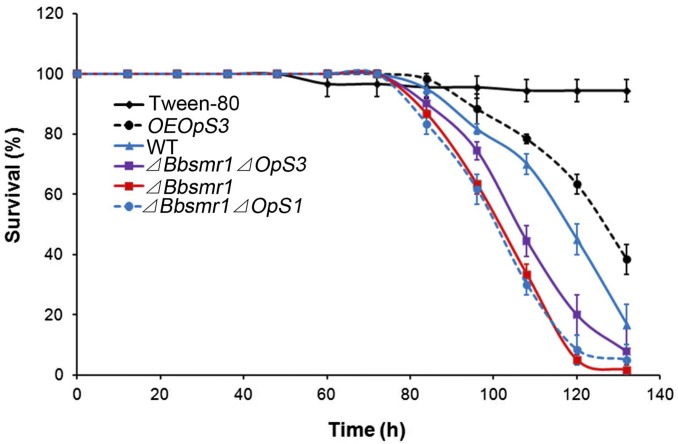
We examined the virulence of WT, gene deletion mutants, and OpS3 overexpression strains using the greater waxmoth, Galleria mellonella, as the insect host. Deletion of Bbsmr1 resulted in a small increase in host mortality (∼20% decrease in median lethal time [LT50]) compared with the WT strain (Fig. 4 and Table 1). Deletion of OpS1 or OpS3 in the ΔBbsmr1 background did not significantly affect virulence compared with the ΔBbsmr1 parent. Finally, the OpS3 overexpression strain (WT background) exhibited a ∼12% decrease in virulence compared with the WT strain. Both the ΔBbsmr1 and OpS3 overexpression strains, which produced oosporein in SDB and potato dextrose broth (PDB) media, exhibited reduced growth compared with WT and other strains examined; however, in Czapek–Dox broth (CZB) medium, all of the tested strains grew poorly, and no consistent results were obtained (SI Appendix, Fig. S4).
Fig. 4.
Insects bioassays. Survival of G. mellonella larvae infected with conidial suspensions (1 × 107 conidia/mL) of WT (solid blue line), ΔBbsmr1 (solid red line), OpS3 overexpression strain (OEOpS3; dashed black line), ΔBbsmr1ΔOpS3 (solid purple line), and ΔBbsmr1ΔOpS1 (dashed blue line).
Table 1.
Calculated LT50 values of B. bassiana WT, ΔBbsmr1, ΔBbsmr1Δ OpS1, ΔBbsmr1ΔOps3, and OEOpS3 strains against G. mellonella
| Strain | Lethal time50, h |
| WT B. bassiana | 116.9 ± 2.4a |
| ΔBbsmr1 | 100.8 ± 1.9b |
| ΔBbsmr1ΔOpS1 | 100.9 ± 2.0b |
| ΔBbsmr1ΔOpS3 | 106.7 ± 2.0b |
| OEOpS3 | 125.7 ± 3.2c |
Spore concentration, 1 x 107 conidia/mL. Different lowercase letters indicate significant differences between columns (P < 0.05, Tukey’s test).
Oosporein Is Produced After Host Death.
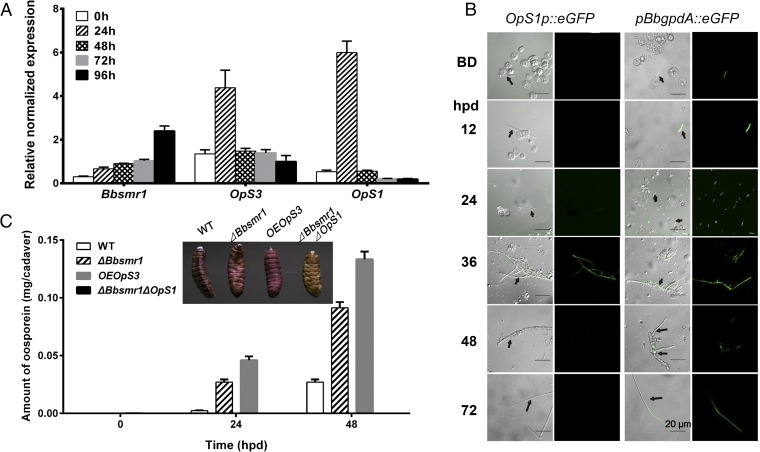
We investigated the expression pattern of Bbsmr1, OpS3, and OpS1 over a time course of 0–96 hpd of the host (G. mellonella larvae) due to B. bassiana mycosis (Fig. 5A). These data indicated basal expression of Bbsmr1, with a 2- to 2.5-fold induction at the 96-h time point. The expression of both OpS3 and OpS1 appeared to rise rapidly and to peak by 24 hpd, before returning to basal levels throughout the rest of the growth process.
Fig. 5.
Expression of Bbsmr1, OpS1, and OpS3 after host death. (A) Real-time qPCR analysis of Bbsmr1, OpS1, and OpS3 in G. mellonella cadavers killed by WT B. bassiana after topical infection. The time course represents time after death. (B) Analysis of OpS1 expression using an eGFP promoter fusion construct as detailed in Materials and Methods. The time course includes before host death (BD) and 12–72 hpd. Arrows indicate B. bassiana hyphal bodies (BD or 12 hpd) or hyphae (throughout) of B. bassiana. (C) Quantification of oosporein production on host cadavers killed by indicated strains, from 0 to 48 hpd. (Inset) The color of insect cadavers killed by the indicated strains at 48 hpd.
To further investigate the expression of OpS1 during the fungal infection process, we transformed an eGFP reporter construct in which the eGFP gene was placed under control of the OpS1 promoter (OpS1P::eGFP) into the B. bassiana WT strain, as described in Materials and Methods. The OpS1P::eGFP strain was used to infect G. mellonella larvae, and fungal cells were visualized by fluorescent microscopy at various stages of infection (Fig. 5B; images of control B. bassiana WT are shown in SI Appendix, Fig. S5). No GFP fluorescent signal was seen in the control WT strain (SI Appendix, Fig. S5), and no signal was detected in the OpS1p::eGFP strain when grown in vitro, i.e., in CZB, PDB, or SDB (SI Appendix, Fig. S6). No fluorescent signal was seen in the OpS1p::eGFP strain before host death and during the initial phases after death (i.e., 12–24 hpd), with a signal detected at 36 hpd that was reduced at 48 hpd and not detectable at 72 hpd (Fig. 5B). Of note, no GFP signal was detected during fungal proliferation in the hemocoel or in the hemocoel-produced fungal hyphal bodies. A positive control strain in which eGFP expression was driven by the B. bassiana pBbgpdA promoter showed constitutive fluorescent signals during in vitro growth irrespective of the infection stage (Fig. 5B).
Attempts at detecting oosporein after host infection (i.e., extraction and analyses of larvae infected by B. bassiana at 24–72 h postinfection) were unsuccessful. However, at 48 hpd, G. mellonella cadavers killed by B. bassiana turned dark red (Fig. 5C, Inset). Extraction and quantification of oosporein production revealed little to no oosporein immediately after death of the host, but showed a gradual increase to ∼0.02 mg per cadaver within 48 h in WT killed insects (Fig. 5C). Oosporein production followed a similar timeline of production in the ΔBbsmr1 and OpS3 overexpression strains, although levels of oosporein were 3- to 10-fold higher in the latter strains compared with WT at 24 and 48 hpd. No oosporein was detected in insects killed by the ΔBbsmr1ΔOpS1 strain.
B. bassiana–Produced Oosporein Inhibits Bacterial Growth on Insect Cadavers.
A negative correlation was seen between fungal growth and oosporein production, with strains producing oosporein (i.e., the ΔBbsmr1 and OpS3 overexpression strains) producing significantly less biomass compared with the WT and ΔBbsmr1ΔOpS1 strains (P < 0.05; SI Appendix, Fig. S4A), although the exogenous addition of oosporein to fungal culture medium had no significant effect on fungal growth (SI Appendix, Fig. S4D). Given the reported antimicrobial activities of oosporein and the expression data obtained earlier, we hypothesized that B. bassiana produces oosporein to limit the growth of other microorganisms on insect cadavers, allowing the fungus to complete its life cycle.
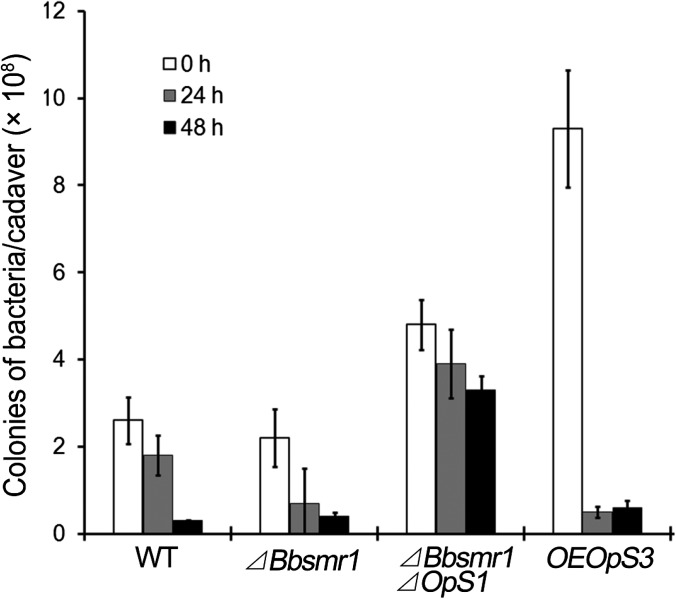
We first confirmed the in vitro antibacterial activity of oosporein using both Gram-positive and Gram-negative test strains, including Staphylococcus aureus N315, Escherichia coli, and Bacillus thuringiensis (SI Appendix, Figs. S7 and S8). We then analyzed total bacterial counts and sought to provide an estimate of the bacterial species found on B. bassiana-infected cadavers. Infection by WT B. bassiana resulted in a gradual decrease in total bacterial counts to <10% of the starting concentration (∼2.8 × 108/cadaver to <0.3 × 108/cadaver) within 48 hpd (Fig. 6 and SI Appendix, Fig. S9). Infection of G. mellonella larvae by the ΔBbsmr1 strain resulted in an earlier onset (at 24 hpd) of the decrease in bacterial counts (P < 0.01), although a large variation was noted. Infection by the non–oosporein-producing ΔBbsmr1ΔBbOpS1 resulted in higher overall bacterial counts throughout the experimental time course, with only a small decrease noted at the last time point (48 hpd). Infection by the oosporein-deregulated OpS3 overexpression strain resulted in a dramatic increase in initial bacterial count that was suppressed within the first 24 hpd.
Fig. 6.
Total culturable bacterial counts on G. mellonella cadavers killed by the indicated B. bassiana strains at 0, 24, and 48 hpd.
Five major bacterial colony morphotypes were seen on the bacterial enumeration plates. Selected colonies were analyzed by sequencing of 16S rDNA fragments, as described in Materials and Methods. Comparison of resultant sequences with the National Center for Biotechnology Information (NCBI) database using BLASTN revealed near-identity of the five different morphotypes to Enterococcus faecalis, Stenotrophomonas sp., Pantoea sp., Acinetobacter calcoceticus, and Staphylococcus sp., respectively (Table 2). The relative abundance of the bacterial species at the initial time point of infection followed the order E. faecalis (48%) > Stenotrophomonas (35%) > Pantoea (14%) > A. calcoaceticus (3.4%) > Staphylococcus (<1%), whereas at the 48-h time point, the order of abundance was E. faecalis (81%) > Stenotrophomonas (13%) > Staphylococcus (6%), with almost no Pantoea or A. calcoaceticus detected (Table 2). The minimum inhibitory concentration (MIC50, for 50% inhibition of growth) for oosporein tested against Pantoea, Staphylococcus, Stenotrophomonas, Acinetobacter, and Enterococcus was 3, 5, 10, 30, and 100 µg/mL, respectively (Table 2). The concentration required for ≥90% growth inhibition (MIC90) was ∼100 µg/mL for all of the bacteria tested except for Enterococcus, which required >200 µg/mL (Table 2).
Table 2.
Bacterial communities in host cadavers infected by B. bassiana strains by 16S rDNA sequencing and effect of oosporein on growth
| NCBI accession no. | Bacterial species (identity, %) | Percentage* | MIC50 of oosporein, µg/mL | MIC90 of oosporein, µg/mL | |
| 0 hpd | 48 hpd | ||||
| KX648537 | Enterococcus faecalis (99) | 48.3 | 81.3 | 100 | >200 |
| KX648538 | Stenotrophomonas sp. (99) | 34.5 | 12.5 | 10 | 100 |
| KX648539 | Pantoea sp. (99) | 13.8 | — | 3 | 100 |
| KX648541 | Acinetobacter calcoaceticus (99) | 3.4 | — | 30 | 100 |
| KX648542 | Staphylococcus sp. (99) | — | 6.3 | 5 | 100 |
Bacterial colonies were isolated at random from 0 hpd and 48 hpd cadavers.
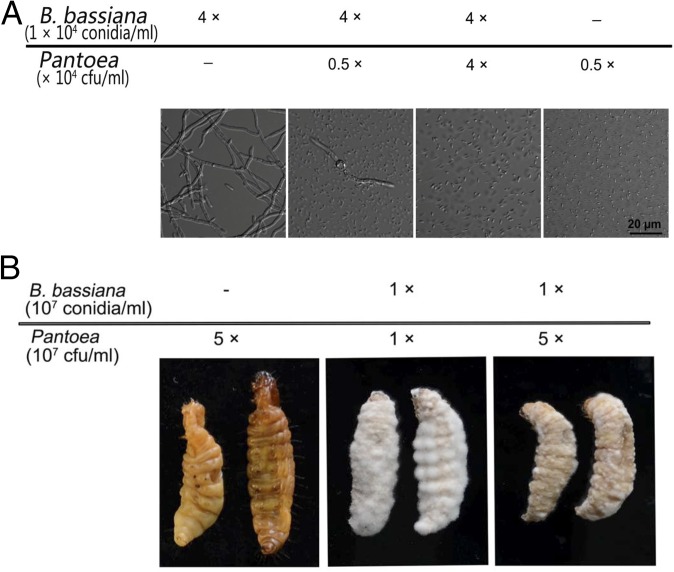
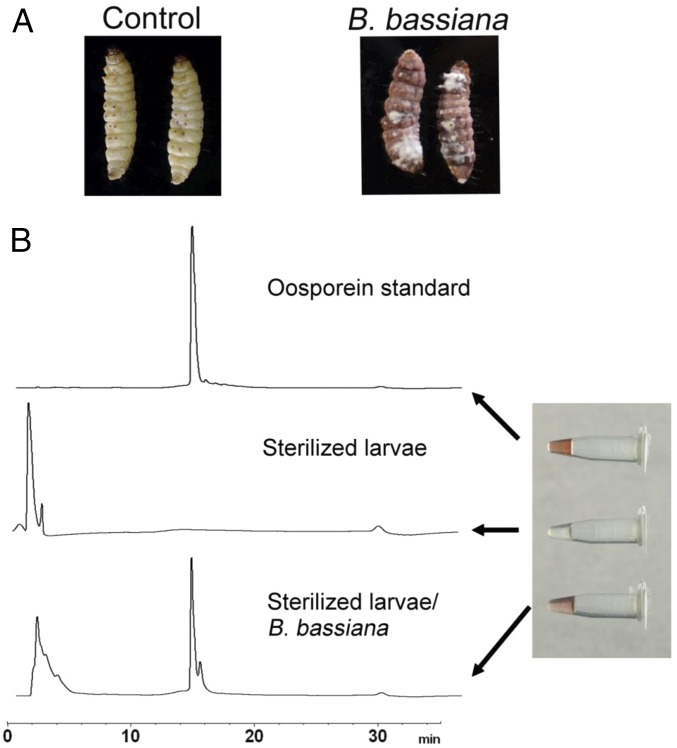
Because Pantoea were essentially eliminated from the host (from 14% to 0% within 48 h) and in vitro studies showed high sensitivity to oosporein (SI Appendix, Fig. S10), this isolate was chosen for competition and coinoculation studies using sterilized G. mellonella cadavers. Coinoculation of B. bassiana with Pantoea in SDB showed rapid proliferation of the bacteria that were easily able to outcompete the fungus (Fig. 7A). Cotopical infection of Pantoea with B. bassiana spores onto autoclaved insect cadavers (topical application) resulted in various degrees of competition depending on the ratio of fungal:bacterial cells used; however, the fungus successfully sporulated on cadavers (Fig. 7B). Coinjection of the bacteria with the fungi typically resulted in bacterial growth that overwhelmed the fungus, however (SI Appendix, Fig. S11). Oosporein was produced during B. bassiana growth on host cadavers in the absence of any competing bacteria, i.e., after injection of B. bassiana into autoclaved larvae (Fig. 8); however, no oosporein was produced when B. bassiana was cogrown with Pantoea in vitro, i.e., in 0.5× SDB (SI Appendix, Fig. S12). In addition, the ΔBbsmr1ΔOpS1 mutant showed slower growth and reduced/delayed sporulation on host cadavers compared with the WT strain (SI Appendix, Fig. S13).
Fig. 7.
B. bassiana and Pantoea competition. (A) In vitro growth in 0.5× SDB medium inoculated by indicated concentrations of B. bassiana conidia and Pantoea cells. (B) Fungal growth on sterilized G. mellonella cadavers inoculated with indicated concentrations of B. bassiana and Pantoea cells.
Fig. 8.
Oosporein produced in sterilized G. mellonella cadavers. (A) G. mellonella larvae were autoclaved and inoculated with B. bassiana conidia (by injection). (B) After treatment, cadavers were placed at 26 °C for 4 d before extraction of oosporein, as detailed in Materials and Methods. Shown are the results of HPLC analysis of the oosporein standard, untreated cadavers, and B. bassiana WT treated cadavers.
Discussion
Although there is significant interest in fungal secondary metabolites, several obstacles have hindered their characterization. Because many of these compounds are not produced in standard mycological media and growth conditions, one impediment to their characterization has been the often-cryptic nature of secondary metabolite expression in fungi. The lack of information about the conditions under which fungal secondary metabolites are produced extends to the genetic networks that regulate their synthesis. An additional factor that has complicated this research is the difficulty in establishing clear biological roles for many secondary metabolites. Various approaches have been used to address these issues (5, 27). Successful production of a number of secondary metabolites has been achieved via overexpression of pathway-specific regulators, genetic manipulation of signal transduction cascades, and microbial induction of secondary metabolite production via cocultivation (5). The availability of genomes, coupled to in silico bioinformatic predictions and in some cases further facilitated by metabolomics studies, has led to the discovery of an increasing number of compounds in a wide range of fungi (28). Additional strategies have included removal (via gene knockout) of repressors, epigenetic manipulation, characterization of “global regulators” of secondary metabolite production (e.g., LaeA), and screening for secondary metabolite hyperproducers (27, 29, 30).
Oosporein was identified more than 70 y ago as a pigment in the ascomycete Oospora colorans (7), and has since been detected in various fungi, including Beauveria sp. Although a number of properties have been attributed to this compound, including antimicrobial, antiviral, antiproliferative, and various cytotoxic effects, its biological role and the genetic pathway for its synthesis has remained elusive. The availability of the B. bassiana genome has allowed for the analysis of secondary metabolite gene clusters and probing of oosporein biosynthesis (22, 23). Oosporein biosynthesis in B. bassiana proceeds via a PKS (OpS1 gene product) pathway involving at least seven genes that exert hydroxylase, dioxygenase, and catalase activities (23). The gene cluster also encodes a positive transcriptional regulator, OpS3, whose overexpression stimulated expression of the OpS gene cluster. Here, in a screen for constitutive oosporein production, we isolated a T-DNA mutant based on a red colony phenotype. The T-DNA mutation was mapped to an ORF outside of the OpS gene cluster and provisionally annotated as coding for a zinc finger TF of unknown function, termed Bbsmr1 based on its regulatory role in mediating secondary metabolite production. Analyses of OpS gene expression profiles, coupled with quantification of oosporein production in a ΔBbsmr1-targeted gene knockout strain, indicated that Bbsmr1 acts as a negative regulator of the system, likely via regulation of OpS3, which in turn (positively) regulates the rest of the OpS gene cluster. This conclusion is supported by the phenotypes of the ΔBbsmr1ΔBbOpS1 and ΔBbsmr1ΔBbOpS3, well as the OpS3 overexpression strains, in which for the former two strains oosporein production is eliminated and the OpS1 or OpS gene cluster is transcriptionally silent, whereas in the latter strain the system is turned on and oosporein is produced.
Our data show that from 24 to 96 hpd, the expression of Bbsmr1 gradually increased, whereas the expression of both OpS3 and OpS1 gradually decreased. This pattern would support a negative role for BbSmr1 in expression of the OpS gene cluster; however, at the 0 hpd time point, Bbsmr1 expression was low, as were the expression levels of OpS3 and OpS1, an inconsistency with the rest of our results. There are several possible explanations for these data. First, at the immediate time point of host death, fungal growth is significantly lower than at the other time points, potentially resulting in poor/low recovery of fungal RNA and/or more (than at the other time points) significant amounts of host (insect) RNA in samples that could potentially interfere with the expression results. However, it is also possible that other (negative) regulatory inputs, including the requirement for as-yet uncharacterized induction factors, are not yet active at the early time points. These results highlight that host death/post-host death is likely a discrete stage of the infection that merits further genetic and biochemical dissection.
Despite advances in identification of secondary metabolite gene clusters and characterization of the chemical products produced, the biological roles of many fungal secondary metabolites remain unclear, with antibiotics and metabolites involved in pathogenic processes the best characterized to date. The discovery of antibiotics in Penicillium crysogenum is well known, and fungi—particularly those occupying unique ecological niches—continue to be a source for their discovery (31, 32). Iron-binding siderophores, synthesized via nonribosomal peptide synthase-mediated pathways, are involved in iron scavenging, uptake, and sequestration, and have been shown to act as virulence factors in various pathogenic fungi (33). Some secondary metabolites act as “toxins,” affecting/disrupting host tissues and processes; for example, the Cochliobolus carbonum HC toxin targets host histone deacetylases and is required for infection of maize cultivars that harbor the Hm resistance gene, but not of cultivars that lack Hm (34). In B. bassiana, a number of secondary metabolites, some implicated in virulence, have already been characterized. Although the cyclic depsipeptide beauvericin was originally thought to be uninvolved in entomopathogenicity, later genetic characterization of its synthesis indicated that it does contribute to virulence (35, 36). Manipulation of the beauvericin pathway also has been used to produce novel beauvericin-based compounds (37, 38). Similarly, the cyclic depsipeptide bassianolide also has been shown to contribute to B. bassiana virulence (39). In contrast, genetic analyses of the biosynthetic pathway of the 2-pyridone tenellin revealed that this compound does not appear to contribute to B. bassiana virulence (40, 41). This system also has been exploited for novel compound discovery, with expression of the tenellin nonribosomal peptide synthase in Aspergillus oryzae leading to the production of several new compounds and the use of chemical epigenetic modifiers in B. bassiana altering the selectivity of tenellin polyketide synthase to produce different products (42, 43).
Loss of oosporein reportedly results in delayed sporulation on host cadavers and has been linked to B. bassiana virulence and immune evasion (23). We also observed delayed growth and sporulation on host cadavers for the double-deletion mutant ΔBbsmr1ΔOpS1, which does not produce oosporein. Regarding the process of immune evasion, during the infection process, B. bassiana is known to undergo a dimorphic transition as it penetrates the host integument, producing free-floating in vivo hyphal bodies (44–46). Targeting of the OpS gene cluster reportedly decreases the number of hyphal bodies in infected hosts (23). Our data suggest that oosporein production is a late-stage event. Transcription of key OpS genes and production of oosporein itself was not detected until after death of the host. Once the host has died, a rapid increase in oosporein production occurs, with >20 μg of oosporein per larva detected in WT infections.
Because oosporein has been shown to exhibit antimicrobial activity, we investigated whether such activity is relevant to suppressing host microbes once the host dies, a critical stage at which competing microbes, especially faster growing bacteria, can potentially overtake the growth of the fungus. Analyses of G. mellonella bacteria revealed that after death of the host, ∼95% of the culturable bacteria were members of the Enterococcus, Stenotrophomonas, and Pantoea genera. Within 48 h after death, the vast majority of bacteria were Enterococcus (>80%), with Pantoea absent, and the proportion of Staphylococcus, which was not present at the early time point, rising to ∼6% of the culturable bacteria identified.
In a somewhat counterintuitive finding, our data indicate that bacterial counts in hosts during infection by the oosporein-overproducing strain were higher than those seen for WT infections at the initial time point. These results can be explained if oosporein has some mild toxicity, particularly with respect to suppression of the host immune system, as has been suggested by others (23). Thus, during the intial phases of infection, suppression of the immune system by misregulation and overproduction of oosporein would result in increased bacterial growth, which would not occur during WT infections. At the time of death, it is possible (as our data indicate) that overall insect bacterial counts can be higher than those found during WT infections, potentially requiring higher levels of oosporein for control. Eventually, oosporein levels are sufficient in the overproducing strains to suppress the bacterial population in the dead host.
The MIC50 for oosporein tested against the major bacterial isolates identified (i.e., Pantoea, Staphylococcus, Stenotrophomonas, and Acinetobacter) ranged from 3 to 30 µg/mL, with an MIC50 of 100 µg/mL for Enterococcus. Achieving >90% growth inhibition required ∼100 µg/mL of oosporein for the former group of bacteria and >200 µg/mL for Enterococcus. Our data indicated that is ≥20 μg per cadaver of oosporein is produced in G. mellonella larvae at 48 hpd. Because one larva has a volume of ∼0.4 mL, this would mean an oosporein concentration of ≥50 µg/mL. Thus, the amount of oosporein produced would be adequate for ≥50% inhibition of growth of most of the bacterial isolates examined, with the exception of Enterococcus. These results (and prediction) are consistent with the decreases seen in experiments examining the bacterial community changes after host death, i.e., significant losses of Pantoea and Acinetobacter, but significant levels of Enterococcus retained. These effects were further verified and expanded in coinoculation experiments showing that B. bassiana is capable of successfully competing with Pantoea when applied to the surface of G. mellonella, but not when grown in vitro in standard medium or when injected into the insect hemolymph. Both of the latter conditions represent relatively nutrient-rich conditions, and the bacteria are capable of quickly overtaking the growth of the fungus in these environments. Nonetheless, these data show that B. bassiana is exquisitely adapted to minimizing competitors during its “normal” infection process, i.e., via cuticle penetration.
Our data indicate that the biological function of oosporein is linked to the temporal program of infection, and that it acts to minimize bacterial competition once the host is dead. These data support the idea that the last stages of infection after host death represent a discrete event involving the induction of distinct (i.e., from the other stages of infection) fungal gene pathways. Thus, oosporein, and likely other late-stage processes that contribute to the ability of the fungus to complete its lifecycle on the host, allow for maximal assimilation of host nutrients and subsequent sporulation on the cadaver. The selective pressure for such an adaptation would be particularly strong, given that the conidia produced on the dead host represent the “progeny” of the fungus.
Materials and Methods
Microbial Strains and Media.
B. bassiana strain (CGMCC7.34; China General Microbiological Culture Collection Center) was isolated from a cadaver of Pieris rapae and conserved as a mixture of dry conidia at −80 °C. WT and mutant fungal strains were routinely grown on PDB/potato dextrose agar, SDB/Sabouraud dextrose agar, and/or CDB/Czapek–Dox agar. E. coli DH5α was used for plasmid propagation. E. coli strains were cultured in LB medium supplemented with ampicillin (100 μg/mL) or kanamycin (50 μg/mL) based on the plasmid selection markers used. The oosporein standard was kindly provided by Brian Love, East Carolina University, Greenville, NC.
Mutant Screening and Gene Manipulations.
T-DNA insertion library screen and gene identification.
Approximately 5,000 colonies derived from a B. bassiana T-DNA insertion library (25) were cultured in microtiter plates with SDB medium. Colonies that appeared visibly red after 4–6 d of growth were rescreened under the same conditions and single-spore purified. Mapping of the T-DNA integration site of the mutant was performed using the SiteFinding PCR protocol (47) with the primers listed in SI Appendix, Table S1. Zinc finger motifs, nuclear localization signals, and phosphorylation sites were predicted in NCBI, WoLF PSORT, and KinasePhos sites, respectively.
Construction of targeted gene knock and complementation strains.
For construction of the Bbsmr1 gene deletion vector, the phosphinothricin actetyltransferase gene (bar) cassette (∼1.6 kb) was used to replace a 400-bp gene fragment within the Bbsmr1 ORF. The upstream (BbsmrL) and downstream (BbsmrR) fragments of the construct were amplified via PCR with primer pairs PsmrL1/PsmrL2 and PsmrR1/PsmrR2, respectively, using B. bassiana genomic DNA as the template (SI Appendix, Table S1), and then cloned into the plasmid vector pK2-bar (48). The integrity of the resulting construct, pK2-BbsmrL-bar-BbsmrR, was verified by sequencing (Invitrogen), and the plasmid was transformed into Agrobacterium tumefaciens AGL-1. A. tumefaciens-mediated transformation of B. bassiana was performed as described previously (25), and putative homologous recombination transformants were screened with primers PsmrT1 and PsmrT2 for the correct integration event as described previously (49).
To construct the Bbsmr1 complementation vector, a fragment containing the entire ORF, along with 1,966-bp upstream and 481-bp downstream flanking sequences, was amplified by PCR using primers PsmrC1 and PsmrC2. The resultant fragment was digested with EcoRI and XbaI (unique sites engineered in the primers) and cloned into pCB1536 containing the sulfonylurea resistance gene (sur) cassette. The complementation vector was ectopically transformed into the ΔBbsmr1 strain via an LiCl-PEG–mediated blastospore transformation protocol as described previously (50). The integrity of the transformants was verified by PCR and Southern blot analysis. For Southern blotting, genomic DNA was digested with PstI and separated in 1.0% agarose gel. After electrophoresis, DNA was subsequently transferred onto a nylon membrane and probed with a 313-bp PCR product corresponding to genomic DNA, which was amplified with Psmr-sb1 and Psmr-sb2 and labeled with biotin (Roche) following the manufacturer’s instructions.
Construction of double-targeted gene knock mutant strains.
The polyketide synthetase genes EJP62792 (OpS1) and a TF factor encoding gene EJP62794 (OpS3) in oosporein cluster were deleted in ΔBbsmr1 via replacement of a ∼600-bp gene fragment within the ORF with the sur cassette (∼3.0 kb). The upstream fragments of these genes were amplified by primers XLB1/XLB2 (where X indicates various genes, listed in SI Appendix, Table S1) and cloned into pK2-Sur using the XbaI (SpeI)/HindIII sites to obtain pK2-XLB-Sur. The downstream fragments were amplified by XRB1/XRB2 (SI Appendix, Table S1) and cloned into pK2-XLB-Sur using the SpeI/EcoRI sites. The pK2-XLB-Sur-XRB constructions thus obtained were transformed into A. tumefaciens AGL-1. B. bassiana transformation (in ΔBbsmr1) and transformant screening were performed as described above.
Construction of OpS3 constitutive expression strain.
A ∼2.2-kb fragment containing the entire OpS3 (EJP62794) ORF was amplified by PCR using the primer pair POpS3-O1/POpS3-O2 and B. bassiana genomic DNA as the template. The PCR product was cloned into the XbaI site downstream of the constitutive PBbgpdA promoter (26). Clones containing the correct orientation of the ORF relative to the promoter were selected by PCR screening using primers Pgpda-t/POpS3-O2 (SI Appendix, Table S1).
Gene Expression and eGFP-Promoter Reporter Analyses.
B. bassiana WT and gene deletion strains were grown in 0.5× SDB medium for 3 d, after which cells were harvested by centrifugation and total RNA was extracted using the Aurum Total RNA Mini Kit (Bio-Rad). cDNA was synthesized using the RevertAid First-Strand cDNA Synthesis Kit (MBI Fermentas). Real-time qPCR was performed using the iCycler iQ Multicolor Real-Time PCR Detection System with SYBR Green (Bio-Rad). Reaction mixtures contained 5 μL of iQ SYBR Green Supermix (Bio-Rad), 0.5 μL (10 μM) of each primer pair for the indicated genes (SI Appendix, Table S1), and 4 μL of 1:10 diluted cDNA template. Reactions were incubated with a 5-min denaturation step at 95 °C, followed by 40 cycles of 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 30 s. The relative expression levels of specific genes were normalized to actin (GenBank accession no. HQ232398), gpd (GenBank accession no. AY679162), or cypA (GenBank accession no. HQ610831) as reference genes using iQ5 optical system software, version 2 (Bio-Rad) (51).
B. bassiana gene expression during growth on insect hosts was examined as follows. B. bassiana conidial suspensions (1 × 107 conidia/mL in 0.05% Tween-80) were topically inoculated on third instar Galleria mellonella larvae and incubated at 26 °C for 3–5 d. Immediately after larval death, cadavers were collected and placed in a 90-mm Petri dish with a wet cotton ball and incubated at 26 °C. Subsequently, at 0, 24, 48, 72, and 96 hpd, samples were ground in liquid nitrogen, RNA was extracted, and real-time qPCR was performed as described above.
Analysis of OpS1 expression.
An enhanced green fluorescent protein (eGFP) promoter reporter construct to monitor the expression of OpS1 was synthesized as follows. Primers Ppops1-1 and Ppops1-2 were used to amplify ∼1 kb of 5′ flanking sequences immediately upstream of the OpS1 ORF using B. bassiana genomic DNA as the template. The PCR fragment was cloned upstream of a promoterless eGFP ORF in the pUC-bar vector containing the phosphinothricin resistance cassette. The integrity of the construct was verified by sequencing and transformed into the B. bassiana WT strain as described previously (50). B. bassiana spores harboring the OpS1p::eGFP construct ectopically integrated into the fungal genome were topically inoculated on G. mellonella larvae and examined over a time course of infection.
For evaluation of the expression of OpS1p::eGFP during the infection process, hemolymph was collected at 12-h intervals starting at 48 h after topical inoculation as described previously (52). After larvae were killed by fungal infection, tissues containing fungal hyphae were isolated from the middle part of the cadavers and crushed in 100 µL of sterilized water. Samples were mounted on microscope slides, and the eGFP signal was measured using an Olympus FV1000 confocal microscope with a 488-nm filter.
Extraction and Detection of Oosporein.
Oosporein was extracted as described previously (9) with minor modifications. In brief, the B. bassiana WT and mutant strains were cultured in 0.5× SDB for 3 d, after which culture filtrates were collected and adjusted to pH 2.0 with 37% (wt/vol) HCl. Oosporein was extracted with an equal volume of ethyl acetate three times. The pooled ethyl acetate extracts were dried using a rotary evaporator, and the extract was resuspended in methanol. Samples were analyzed with a high-performance liquid chromatograph connected to a mass spectrometer in the negative ion mode (M-H) using a reverse-phase column (ZORBAX SB-C18; Agilent). The column was pre-equilibrated in 95:5 of solution A (0.05% formic acid) to solution B (100% acetonitrile), and after injection of the sample (5 µL), the column was run at 95:5 A:B for 3 min, followed by an increasing gradient to 100% B from 3 to 18 min, 100% B for a further 21 min at 100% B, and finally re-equilibration to 95:5 A:B. A column flow rate of 0.3 mL/min was used, and elution was monitored at a detection wavelength of 287 nm.
For metabolite extraction during infection, B. bassiana-infected G. mellonella larvae were cut into ∼0.3-cm × 0.3-cm pieces and immersed in methanol for 12 h. This process was repeated three times, after which the supernatants from the extractions were combined and centrifuged for 10 min at 13,800 × g to remove any particulate material, and then dried using a rotary evaporator. The extract was resuspended in 0.5 mL of methanol and analyzed by HPLC-MS as described above. Uninfected larvae served as controls, and synthetic oosporein was used as the standard. Each sample involved eight larvae, with three technical replicates, and the entire experiment was performed three times with independent batches of larvae and fungal conidia.
Insect Bioassays.
Fungal virulence bioassays were performed using G. mellonella larvae as the host. For each experimental condition, 3 × 30 larvae were treated topically by immersion in suspensions of 1 × 107 conidia/mL (in 0.05% Tween-80) for 3–5 s, after which excess liquid on the insect bodies was removed with dry paper towels. Control larvae were treated with 0.05% Tween-80. Treated larvae were placed in 150-mm Petri dishes and incubated at 26 °C. The number of dead insects was recorded daily. All bioassays were performed with at least three independent batches of larvae and conidia. Data were analyzed by PROC MIXED in SAS using a linear mixed model. The least significant difference test was used for comparisons between treatments.
Determination and Enumeration of Bacterial Species on Insect Cadavers.
B. bassiana conidial suspensions were prepared in 0.05% Tween-80 and topically inoculated on G. mellonella larvae as described above. Postmortem insects were collected and placed in 150-mm Petri dishes containing a wet cotton ball to maintain a high relative humidity. At 0, 24, and 48 hpd, eight cadavers per time point were homogenized in 5 mL of 50 mM phosphate buffer (pH 6.0) using a sterile mortar and pestle. The resultant suspensions were serially diluted (10-, 100-, 1,000-, and 10,000-fold), after which aliquots (100 µL) were spread onto LB plates (90 mm) and cultured at 37 °C for 24–48 h, and the total number of bacterial colonies was quantified. Living larvae treated with 0.05% Tween-80 served as controls for comparison. Each treatment was performed in triplicate, and the entire experiment was repeated three times with independent batches of larvae and conidia. Selected bacterial isolates were single-colony purified, and their 16S rDNA sequences were determined via PCR amplification using primers 338F and 806R (SI Appendix, Table S1) and subsequent sequencing of the resultant fragments. Sequences were analyzed using BLAST to determine the bacterial species.
Effects of Bacteria on B. bassiana Growth and Oosporein Production on Host Cadavers.
Coculturing assays using bacterial isolate identified as belonging to the genus Pantoea were performed as follows. B. bassiana conidia (final concentration, 1 × 104 conidia/mL) were inoculated into 5 mL of 0.5× SDB alone or with Pantoea (final concentration, 0.5∼4 × 104 cfu/mL in LB). After incubation at 26 °C for 2–3 d, the samples were examined microscopically. To examine the effects of bacterial-fungal coculturing on oosporein production, bacterial (106 ∼108 cfu/mL in LB) and fungal (107 conidia/mL) suspensions were injected (5 μL/injection) into autoclaved (15 min at 121 °C) G. mellonella at three different sites per larvae. Bacteria-fungus mixtures (fungus, 107 conidia/mL; bacterium, 0.1–10 × 107 cfu/mL) were also topically inoculated on autoclaved larvae. The treated larvae were placed in 90-mm Petri dishes with a wet cotton ball and then incubated at 26 °C for 3–5 d. Cadavers were extracted and analyzed for oosporein content as described above.
Induction of oosporein production in vitro was examined using Pantoea and a bacterial mixture from cadavers, with the addition of this population to a growing B. bassiana culture. In brief, 10 µL of B. bassiana (107 conidia/mL) was inoculated into 5 mL of 0.5× SDB and cultured for 2 d at 26 °C. Then 10 µL of bacteria, including Pantoea (108 cfu/mL), autoclaved Pantoea, and bacterial mixture (OD600 = 0.1) from host cadavers, were added into the fungal culture, followed by culturing for an additional 24∼48 h to measure oosporein production.
Supplementary Material
Acknowledgments
We thank Brian Love (East Carolina University) for providing the synthetic oosporein. Research was supported by the Initial Special Research for 973 Program (Grant 2012CB126304), the National Natural Sciences Foundation of China (Grants 31270092 and 31570137), the Program for Innovation Research Team of Chongqing (Grant CXTDX201601012), Fundamental Research Funds for the Central Universities (Grant XDJK2016A002), the Chongqing Foundation for Leaders of Disciplines in Science (Grant cstc2014kjcxljrc005), and the US National Science Foundation (Grant IOS-1557704, to N.O.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616543114/-/DCSupplemental.
References
- 1.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11(1):21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 2.Keller NP. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol. 2015;11(9):671–677. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin W, Keller NP. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49(3):329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiemann P, Keller NP. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41(2):301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 5.Nützmann H-W, Schroeckh V, Brakhage AA. Regulatory cross talk and microbial induction of fungal secondary metabolite gene clusters. In: David AH, editor. Methods in Enzymology. Vol 517. Academic; New York: 2012. pp. 325–341. [DOI] [PubMed] [Google Scholar]
- 6.Aghcheh RK, Kubicek CP. Epigenetics as an emerging tool for improvement of fungal strains used in biotechnology. Appl Microbiol Biotechnol. 2015;99(15):6167–6181. doi: 10.1007/s00253-015-6763-2. [DOI] [PubMed] [Google Scholar]
- 7.Kogl F, van Wessem GC. Analysis concerning pigments of fungi XIV: Concerning oosporein, the pigment of Oospora colorans van Beyma. Recl Trav Chim Pays Bas. 1944;63:5–24. [Google Scholar]
- 8.Eyal J, et al. Assessment of Beauveria bassiana Nov Eo-1 strain, which produces a red pigment for microbial control. Appl Biochem Biotechnol. 1994;44(1):65–80. [Google Scholar]
- 9.Strasser H, Abendstein D, Stuppner H, Butt TM. Monitoring the distribution of secondary metabolites produced by the entomogenous fungus Beauveria brongniartii with particular reference to oosporein. Mycol Res. 2000;104:1227–1233. [Google Scholar]
- 10.Abendstein D, Pernfuss B, Strasser H. Evaluation of Beauveria brongniartii and its metabolite oosporein regarding phytotoxicity on seed potatoes. Biocontrol Sci Technol. 2000;10(6):789–796. [Google Scholar]
- 11.Souza PN, et al. Production and chemical characterization of pigments in filamentous fungi. Microbiology. 2016;162(1):12–22. doi: 10.1099/mic.0.000168. [DOI] [PubMed] [Google Scholar]
- 12.Kalamar J, Steiner E, Charollais E, Posternak T. [The biochemistry of lower fungi, VIII: Chemical synthesis of diquinonic pigments.] Helv Chim Acta. 1974;57(8):2368–2376. doi: 10.1002/hlca.19740570809. French. [DOI] [PubMed] [Google Scholar]
- 13.Dallacker F, Löhnert G. [Derivatives of methylenedioxybenzene. 35. A novel synthesis of 3,6 dihydroxy-2-ethyl-1,4-benzoquinone, embelin, vilangin, rapanone, dihydromaesaquinone, bhogatin, spinulosin, and oosporein.] Chem Ber. 1972;105(2):614–624. doi: 10.1002/cber.19721050227. German. [DOI] [PubMed] [Google Scholar]
- 14.Love BE, Bonner-Stewart J, Forrest LA. An efficient synthesis of oosporein. Tetrahedron Lett. 2009;50(35):5050–5052. [Google Scholar]
- 15.Basyouni SH, Brewer D, Vining LC. Pigments of genus Beauveria. Can J Bot. 1968;46(4):441–448. [Google Scholar]
- 16.Zimmermann G. Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci Technol. 2007;17(5-6):553–596. [Google Scholar]
- 17.Amin GA, Youssef NA, Bazaid S, Saleh WD. Assessment of insecticidal activity of red pigment produced by the fungus Beauveria bassiana. World J Microbiol Biotechnol. 2010;26(12):2263–2268. [Google Scholar]
- 18.Alurappa R, Bojegowda MRM, Kumar V, Mallesh NK, Chowdappa S. Characterisation and bioactivity of oosporein produced by endophytic fungus Cochliobolus kusanoi isolated from Nerium oleander L. Nat Prod Res. 2014;28(23):2217–2220. doi: 10.1080/14786419.2014.924933. [DOI] [PubMed] [Google Scholar]
- 19.Pegram RA, Wyatt RD. Avian gout caused by oosporein, a mycotoxin produced by Caetomium trilaterale. Poult Sci. 1981;60(11):2429–2440. doi: 10.3382/ps.0602429. [DOI] [PubMed] [Google Scholar]
- 20.Michelitsch A, et al. Accurate determination of oosporein in fungal culture broth by differential pulse polarography. J Agric Food Chem. 2004;52(6):1423–1426. doi: 10.1021/jf0307494. [DOI] [PubMed] [Google Scholar]
- 21.Xiao G, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson DM, Donzelli BGG, Krasnoff SB, Keyhani NO. Discovering the secondary metabolite potential encoded within entomopathogenic fungi. Nat Prod Rep. 2014;31(10):1287–1305. doi: 10.1039/c4np00054d. [DOI] [PubMed] [Google Scholar]
- 23.Feng P, Shang Y, Cen K, Wang C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc Natl Acad Sci USA. 2015;112(36):11365–11370. doi: 10.1073/pnas.1503200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Z, et al. Bbmsn2 acts as a pH-dependent negative regulator of secondary metabolite production in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol. 2015;17(4):1189–1202. doi: 10.1111/1462-2920.12542. [DOI] [PubMed] [Google Scholar]
- 25.Fang W, et al. Agrobacterium tumefaciens-mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J Invertebr Pathol. 2004;85(1):18–24. doi: 10.1016/j.jip.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Liao XG, et al. Characterization of a highly active promoter, PBbgpd, in Beauveria bassiana. Curr Microbiol. 2008;57(2):121–126. doi: 10.1007/s00284-008-9163-3. [DOI] [PubMed] [Google Scholar]
- 27.Brakhage AA, Schroeckh V. Fungal secondary metabolites: Strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48(1):15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Deepika VB, Murali TS, Satyamoorthy K. Modulation of genetic clusters for synthesis of bioactive molecules in fungal endophytes: A review. Microbiol Res. 2016;182:125–140. doi: 10.1016/j.micres.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Fox EM, Howlett BJ. Secondary metabolism: Regulation and role in fungal biology. Curr Opin Microbiol. 2008;11(6):481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Jain S, Keller N. Insights to fungal biology through LaeA sleuthing. Fungal Biol Rev. 2013;27(2):51–59. [Google Scholar]
- 31.Bills GF, Gloer JB, An Z. Coprophilous fungi: Antibiotic discovery and functions in an underexplored arena of microbial defensive mutualism. Curr Opin Microbiol. 2013;16(5):549–565. doi: 10.1016/j.mib.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Karwehl S, Stadler M. 2016. Exploitation of Fungal Biodiversity for Discovery of Novel Antibiotics. Current Topics in Microbiology and Immunology (Springer, Berlin), Vol 398, pp 1–36.
- 33.Oide S, et al. NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell. 2006;18(10):2836–2853. doi: 10.1105/tpc.106.045633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brosch G, Ransom R, Lechner T, Walton JD, Loidl P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell. 1995;7(11):1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charnley AK, Collins SA. 2007. Entomopathogenic fungi and their role in pest control. Environmental and Microbial Relationships, The Mycota, eds Kubicek PC, Druzhinina SI (Springer, Berlin), Vol IV, Ed 2, pp 159–187.
- 36.Xu Y, et al. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem Biol. 2008;15(9):898–907. doi: 10.1016/j.chembiol.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, et al. Cytotoxic and antihaptotactic beauvericin analogues from precursor-directed biosynthesis with the insect pathogen Beauveria bassiana, ATCC 7159. J Nat Prod. 2007;70(9):1467–1471. doi: 10.1021/np070262f. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Wijeratne EMK, Espinosa-Artiles P, Gunatilaka AAL, Molnár I. Combinatorial mutasynthesis of scrambled beauvericins, cyclooligomer depsipeptide cell migration inhibitors from Beauveria bassiana. ChemBioChem. 2009;10(2):345–354. doi: 10.1002/cbic.200800570. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, et al. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet Biol. 2009;46(5):353–364. doi: 10.1016/j.fgb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Eley KL, et al. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana. ChemBioChem. 2007;8(3):289–297. doi: 10.1002/cbic.200600398. [DOI] [PubMed] [Google Scholar]
- 41.Halo LM, et al. Late-stage oxidations during the biosynthesis of the 2-pyridone tenellin in the entomopathogenic fungus Beauveria bassiana. J Am Chem Soc. 2008;130(52):17988–17996. doi: 10.1021/ja807052c. [DOI] [PubMed] [Google Scholar]
- 42.Halo LM, et al. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. ChemBioChem. 2008;9(4):585–594. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- 43.Yakasai AA, et al. Nongenetic reprogramming of a fungal highly reducing polyketide synthase. J Am Chem Soc. 2011;133(28):10990–10998. doi: 10.1021/ja204200x. [DOI] [PubMed] [Google Scholar]
- 44.Cho EM, Boucias D, Keyhani NO. EST analysis of cDNA libraries from the entomopathogenic fungus Beauveria (Cordyceps) bassiana, II: Fungal cells sporulating on chitin and producing oosporein. Microbiology. 2006;152(Pt 9):2855–2864. doi: 10.1099/mic.0.28845-0. [DOI] [PubMed] [Google Scholar]
- 45.Lewis MW, Robalino IV, Keyhani NO. Uptake of the fluorescent probe FM4-64 by hyphae and haemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology. 2009;155(Pt 9):3110–3120. doi: 10.1099/mic.0.029165-0. [DOI] [PubMed] [Google Scholar]
- 46.Wanchoo A, Lewis MW, Keyhani NO. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology. 2009;155(Pt 9):3121–3133. doi: 10.1099/mic.0.029157-0. [DOI] [PubMed] [Google Scholar]
- 47.Tan G, et al. SiteFinding-PCR: A simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 2005;33(13):e122. doi: 10.1093/nar/gni124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl Environ Microbiol. 2009;75(11):3787–3795. doi: 10.1128/AEM.01913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan Y, Ortiz-Urquiza A, Garrett T, Pei Y, Keyhani NO. Involvement of a caleosin in lipid storage, spore dispersal, and virulence in the entomopathogenic filamentous fungus, Beauveria bassiana. Environ Microbiol. 2015;17(11):4600–4614. doi: 10.1111/1462-2920.12990. [DOI] [PubMed] [Google Scholar]
- 50.Fan Y, Zhang S, Kruer N, Keyhani NO. High-throughput insertion mutagenesis and functional screening in the entomopathogenic fungus Beauveria bassiana. J Invertebr Pathol. 2011;106(2):274–279. doi: 10.1016/j.jip.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Zhou YH, et al. Selection of optimal reference genes for expression analysis in the entomopathogenic fungus Beauveria bassiana during development, under changing nutrient conditions, and after exposure to abiotic stresses. Appl Microbiol Biotechnol. 2012;93(2):679–685. doi: 10.1007/s00253-011-3561-3. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, et al. Expression of a Toll signaling regulator serpin in a mycoinsecticide for increased virulence. Appl Environ Microbiol. 2014;80(15):4531–4539. doi: 10.1128/AEM.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.