Significance
In mammals, when and where a gene is transcribed are primarily regulated by the activity of regulatory DNA elements, or enhancers. Genetic mutation disrupting enhancer function is emerging as one of the major causes of human diseases. However, our knowledge remains limited about the location and activity of enhancers in the numerous and distinct cell types and tissues. Here, we develop a computational approach, regulatory element prediction based on tissue-specific local epigenetic marks (REPTILE), to precisely locate enhancers based on genome-wide DNA methylation and histone modification profiling. We systematically tested REPTILE on a variety of human and mouse cell types and tissues. Compared with existing methods, we found that enhancer predictions from REPTILE are more likely to be active in vivo and the predicted locations are more accurate.
Keywords: enhancer prediction, DNA methylation, bioinformatics, gene regulation, epigenetics
Abstract
Accurate enhancer identification is critical for understanding the spatiotemporal transcriptional regulation during development as well as the functional impact of disease-related noncoding genetic variants. Computational methods have been developed to predict the genomic locations of active enhancers based on histone modifications, but the accuracy and resolution of these methods remain limited. Here, we present an algorithm, regulatory element prediction based on tissue-specific local epigenetic marks (REPTILE), which integrates histone modification and whole-genome cytosine DNA methylation profiles to identify the precise location of enhancers. We tested the ability of REPTILE to identify enhancers previously validated in reporter assays. Compared with existing methods, REPTILE shows consistently superior performance across diverse cell and tissue types, and the enhancer locations are significantly more refined. We show that, by incorporating base-resolution methylation data, REPTILE greatly improves upon current methods for annotation of enhancers across a variety of cell and tissue types. REPTILE is available at https://github.com/yupenghe/REPTILE/.
In mammals, genes are transcribed in a temporally and spatially specific manner during development. The precise regulation of gene expression is primarily driven by the activity of distal regulatory sequences, known as enhancers. Disruption of enhancers can cause developmental abnormalities and diseases (1–6). Moreover, the vast majority of genetic variants associated with human diseases by genome-wide association studies (GWASs) lie in noncoding regions, which potentially affect gene transcription and contribute to diseases through disrupting enhancer activity (7, 8). To identify causal noncoding variants and understand their functional consequences, methods for accurate enhancer annotation are essential.
Enhancers are bound by transcription factors (TFs), which in turn recruit cofactors such as the histone acetyltransferase EP300 to achieve transcription activation of target genes from a distance (9). Active enhancers are generally located in accessible chromatin and marked by enrichment of histone H3 lysine 4 monomethylation (H3K4me1) and H3 lysine 27 acetylation (H3K27ac) (10–12). Enrichment of histone modifications in the genome can be determined by chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq).
Computational approaches have been developed to predict active enhancers from the combinations of these genome-wide profiles [see review (13) for a list of representative methods]. They generally use machine-learning algorithms to learn the histone modification profiles of putative enhancers active in a given cell/tissue type and then predict enhancers in additional cell/tissue types. Although they have proven to be useful, these methods have several important limitations. First, the centers and boundaries of enhancer predictions are not well defined because of the broad enrichment of histone modifications in regions around enhancers. Second, existing methods often perform worse when tested on cells and tissues other than the cell/tissue types used for training of the algorithm. Third, existing methods consider only one cell/tissue type at a time, and thus neglect potentially useful information about the variation between cell/tissue types.
To address these limitations, we developed regulatory element prediction based on tissue-specific local epigenetic marks (REPTILE), an algorithm to predict enhancers by integrating whole-genome, base-resolution cell/tissue-specific DNA methylation data along with histone modification data. Cytosine DNA methylation (mC) is a type of chemical modification that plays critical roles in gene regulation, transposon repression, and the determination of cell identity (14–17). In mammalian genomes, it occurs in both CG and non-CG contexts (18–22) and can be quantified at nucleotide resolution using whole-genome bisulfite sequencing (WGBS) (18). In this study, we consider only the most prevalent form of cytosine methylation (mCG). Transcription factor binding sites (TFBSs) are generally depleted of mCG (18, 23). Whether mCG affects binding affinity is unclear for the majority of TFs, although recent studies suggest that there can be significant alteration of binding affinity (24–26). The anticorrelation of mCG and TF binding is predictive in inferring TFBS (27) and enhancers (23, 28). These observations led us to take advantage of mCG depletion as a high-resolution (∼1 bp depending on density of CG sites) enhancer signature that is complementary to the lower-resolution histone modification data derived from ChIP-seq experiments (with fragment size ranging from 200 to 600 bp after sonication) (29). Our results indicate that, by incorporating mCG data, REPTILE achieves higher prediction accuracy and produces higher-resolution enhancer predictions than existing methods that rely solely on histone modification profiles.
Results
The REPTILE Algorithm.
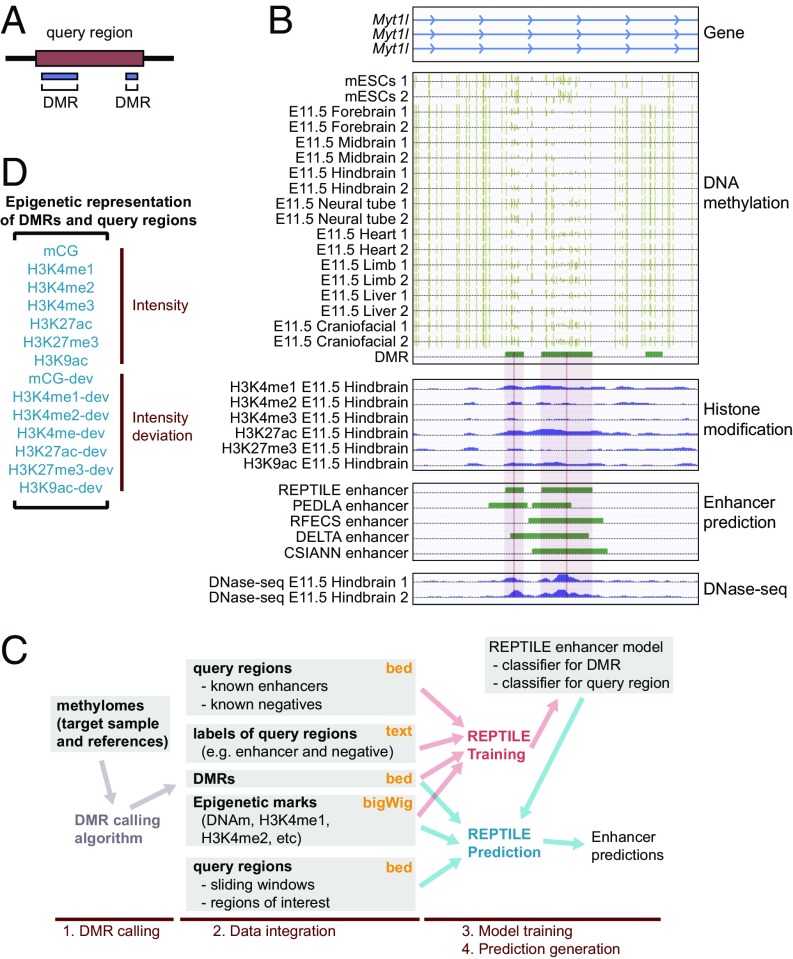
We designed REPTILE based on three observations: (i) active enhancers, which are bound by TFs in certain cells and tissues, show cell/tissue-specific hypomethylated, and such anticorrelation is an informative feature in predicting enhancers. It has been shown that regions that are differentially methylated across diverse cell and tissue types [also known as differentially methylated regions (DMRs)] strongly overlap with enhancers (19, 20, 30). (ii) With base-resolution mCG data, the centers and boundaries of DMRs can be accurately defined, which may be informative in identifying the precise location of enhancers. (iii) The known enhancers (31, 32) (∼2 kb) are generally much larger than TFBSs (∼10–20 bp) and likely include sequences that contribute little to enhancer activity. We used the term “query region” to describe such large regions where a small fraction of the sequences may have a regulatory role. Query regions also refer to negative regions (that showed no observable enhancer activity) and the genomic windows used by enhancer prediction methods. Because a large portion of an active query region may have little contribution to its enhancer activity, the epigenomic signature of the whole active query region may not be an ideal approximation to the epigenomic state of the bona fide regulatory sequences within it. To address this issue, we used DMRs (∼500 bp) to pinpoint the possible regulatory subregions within the query regions and to capture informative local epigenomic signatures in both enhancer model training and prediction generation processes (Fig. 1 A and B).
Fig. 1.
REPTILE improves enhancer identification by incorporating tissue-specific DNA methylation data. (A) Differentially methylated regions (DMRs), typically smaller than query regions, serve as high-resolution enhancer candidates in overlapped query regions. (B) Example of a region (chr12: 29,660,800–29,668,600) where REPTILE uses base-resolution DNA methylation data to improve the resolution of enhancer prediction. Diagram of the gene model (GENCODE M2) in this region is shown at the top (“Gene”). “DNA methylation” displays mCG data of mESCs and eight E11.5 mouse tissues, where ticks represent methylated CG sites and their heights indicate the methylation level. Ticks on the forward strand are projected upward, and ticks on the reverse strand are projected downward. Last track shows DMRs across these samples. “Histone modification” shows the logtwofold change of histone modification ChIP-seq data relative to input. Predictions from four computational methods are visualized in “Enhancer prediction.” Predictions from REPTILE best recapitulate the open chromatin data shown in “DNase-seq.” Light red rectangles mark the REPTILE putative enhancers, whereas the genomic locations of the midpoints (i.e., centers) are highlighted in red. (C) Workflow of REPTILE, including four major steps. (1) DMRs are identified by comparing the CG methylation profiles of target sample and the reference samples. (2) REPTILE integrates data in input files and represents query regions and DMRs as feature vectors (D). Yellow text on the top right corner shows the format for each input data type. (3) REPTILE trains an enhancer model based on the epigenomic signatures of known enhancers and negative sequences as well as the DMRs within them (red arrows). (4) Predictions are generated based on the enhancer model, DMR, query regions, and epigenomic data (blue arrows). (D) Representation of one DMR or query region as a feature vector of intensity or intensity deviation of epigenetic marks. The 14 features used by REPTILE for the benchmark in this paper are shown. The “-dev” features in the vector are the intensity deviation features.
Specifically, the REPTILE algorithm involves four major steps (Fig. 1C). First, DMRs are identified by comparing the mCG profiles of the target sample (in which enhancers will be predicted) and several different cell/tissue types (which serve as reference) (Methods). Next, REPTILE integrates epigenomic data and represents each DMR or query region as a feature vector, where each element is the value of either the intensity or the intensity deviation of an epigenetic mark (Fig. 1D). The intensity deviation feature captures the epigenomic variation between cell/tissue types and is a unique aspect of REPTILE, whereas existing methods rely on data of a single cell/tissue type (Fig. S1A and Methods). In the third step, REPTILE learns a model of enhancer epigenomic signatures from the feature values of (putative) known enhancers and negative regions as well as the DMRs within them. This model contains two random forest (33) classifiers, which predict enhancer activities of query regions and DMRs based on their own epigenomic signature (Methods). In the last step, REPTILE uses the two random forest classifiers to calculate enhancer confidence scores for DMRs and query regions, based on which the final predictions are generated (Methods).
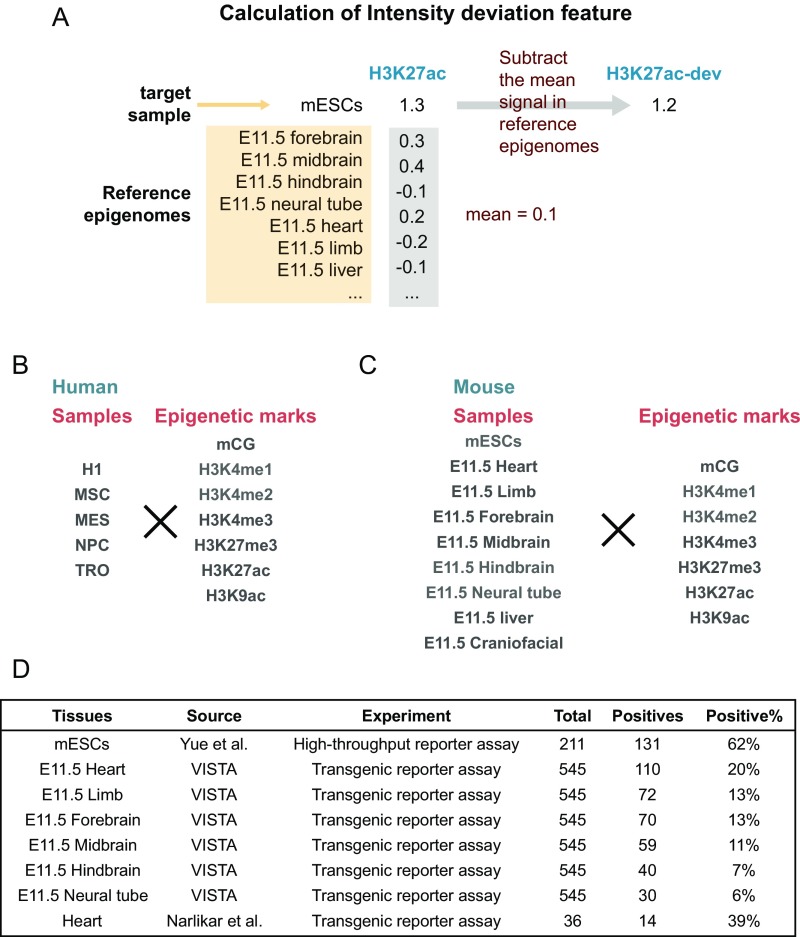
Fig. S1.
Intensity deviation calculation and the enhancer validation and epigenomic data of various samples. (A) An example of calculating intensity deviation. Given one epigenetic mark, the intensity in target sample (where predictions will be generated) is subtracted by the average intensity across reference samples. The result is the intensity deviation, which quantifies how the intensity in target sample is deviated from the default intensity (i.e., average value across reference samples). This feature captures the tissue/cell specificity of epigenetic mark on a given region. (B and C) Human (B) and mouse (C) epigenomic data used in this paper, which includes all epigenetic marks (Left) in all of the samples (Right). (D) Information about the eight enhancer validation datasets. The datasets were collected from Yue et al. (32), VISTA enhancer browser (31), and Narlikar et al. (49). It also shows the basic statistics related to each dataset, including the total number of tested elements (“Total”), number of elements that showed evidence of enhancer activity (“Positives”), and its percentage out of all elements in the dataset (“Positive%”).
Training Computational Models for Human and Mouse Enhancers.
To evaluate the prediction accuracy of REPTILE, we systematically compared REPTILE with four widely used enhancer prediction methods, PEDLA (34), RFECS (35), DELTA (36), and CSIANN (37), using data from a wide variety of human and mouse cells and tissues (Fig. S1 B–D and Methods). These methods all use machine-learning techniques to predict active enhancers based on histone modification profiles, whereas PEDLA also considers evolutionary conservation (SI Methods). Unless specifically stated, six histone modifications were used in these analyses, including H3K4me1, H3K4me2, H3K4me3, H3K27me3, H3K27ac, and H3K9ac (Methods). Notably, REPTILE uses mCG information in addition to histone marks.
For each method, we trained a model (a set of parameters) for human enhancers using epigenomic data from H1 human embryonic stem cells and a model for mouse enhancers using data from mouse embryonic stem cells (mESCs). During the training process, EP300 binding sites were used as putative active enhancers (positive instances), whereas promoters and randomly chosen genomic regions were used as negative instances (SI Methods). When the REPTILE human enhancer model was trained, data of four H1-derived cell types were also included as the reference and DMRs were called for the methylomes of H1 and these cell types. During training of the REPTILE mouse enhancer model, data for eight mouse tissues from embryonic day 11.5 (E11.5) embryo was used as the reference and DMRs were called across the methylomes of mESCs and all of these tissues. In the prediction step, all samples except the target sample were used as the reference. For example, when we applied REPTILE to generate enhancer predictions for E11.5 forebrain, mESCs and the remaining E11.5 tissues were used as the reference.
Unless explicitly stated, all putative enhancers in human cell types and tissues were generated for each method using the human enhancer model, trained using H1 data as described above. Similarly, all enhancer predictions in mouse cell types and tissues were based on the mouse enhancer model, trained using data from mESCs.
REPTILE Shows Superior Prediction Accuracy Compared with Existing Methods.
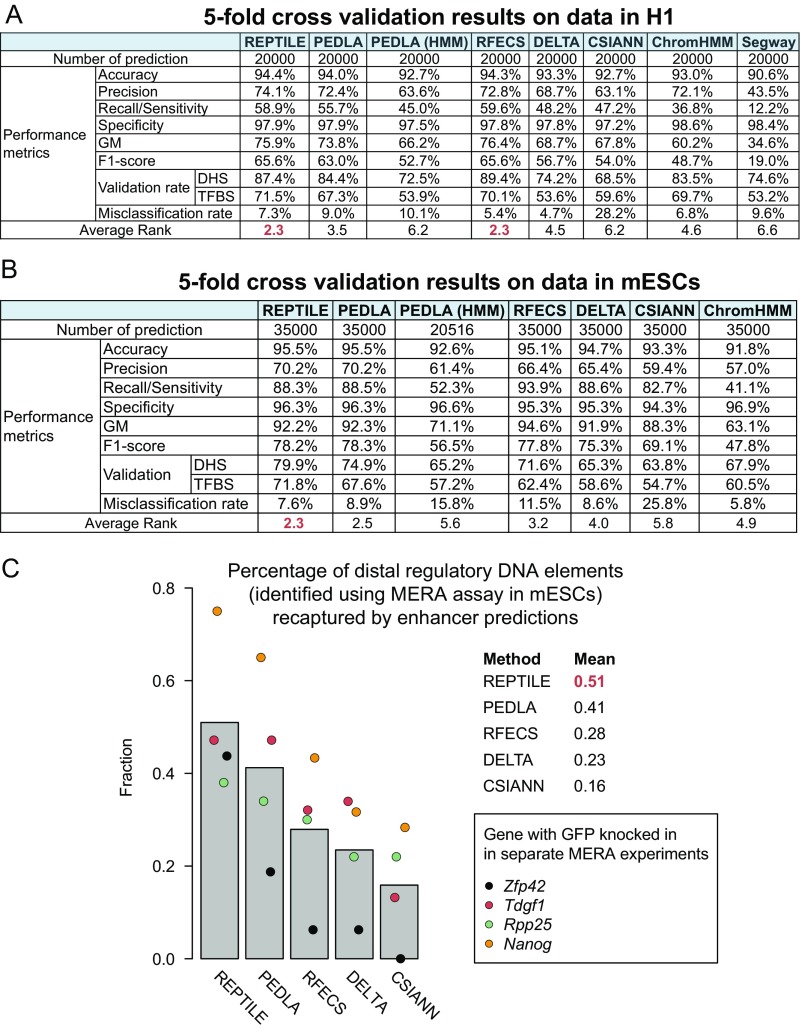
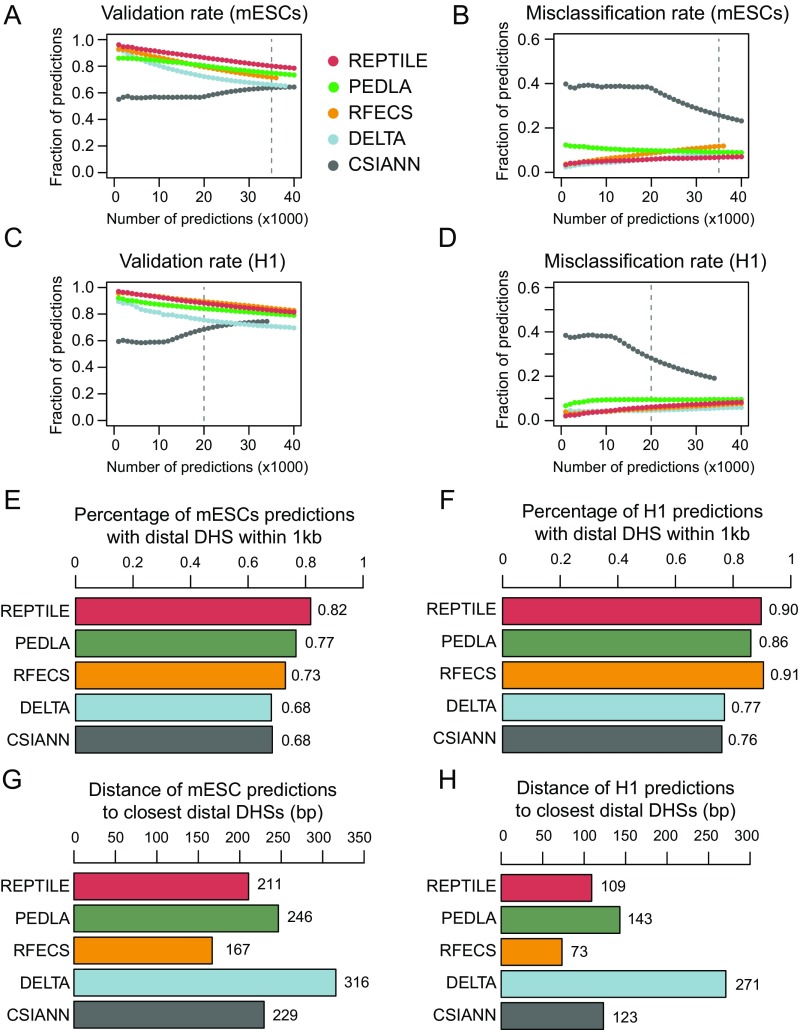
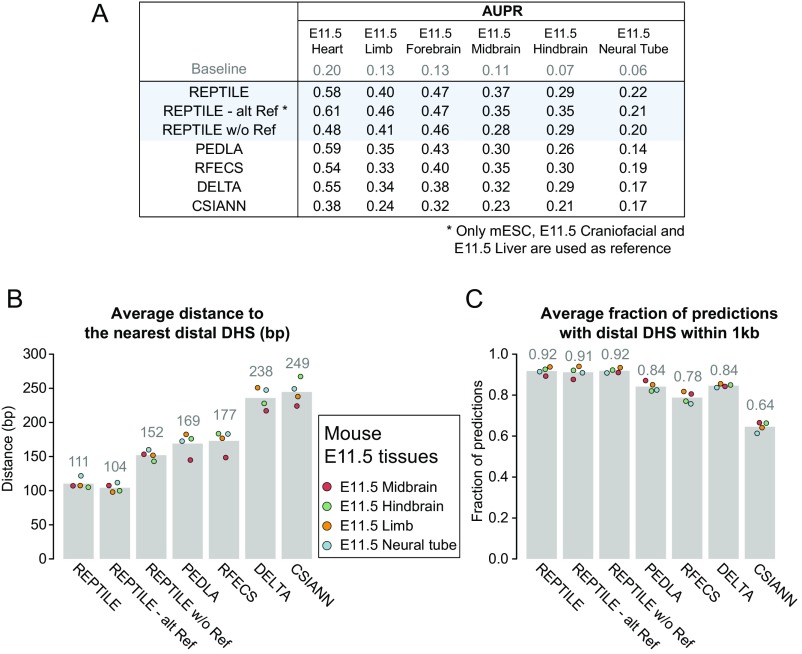
We first used cross-validation to evaluate the learned human enhancer models and mouse enhancer models in H1 and mESCs, where the models were trained. In both cell types, REPTILE showed the best performance among all of the tested methods (Fig. S2 A and B). In addition, we found that, in H1 cells, putative enhancers from REPTILE and RFECS had the greatest overlaps with distal TFBSs and/or distal open chromatin regions [DNase hypersensitivity sites (DHSs)], whereas REPTILE outperformed all other methods in mESCs (Fig. 2 A and B, and SI Methods). Also, REPTILE showed one of the highest validation rates (fraction of predictions that are within 1 kb to distal DHSs but not in promoters) and one of the lowest misclassification rates (fraction of predictions that are within promoters; Fig. S3 A–D). We then tested REPTILE on the 211 experimentally validated regions in mESCs from Yue et al. (32), and it showed superior performance compared with all other methods (Fig. 2C and SI Methods). Furthermore, we found that REPTILE predictions recaptured the most distal regulatory DNA elements that were identified by multiplexed editing regulatory assay (MERA), a high-throughput genome mutation screening approach (38) (Fig. S2C and SI Methods).
Fig. S2.
Cross-validation results and the evaluation of enhancer predictions by MERA data. (A and B) Fivefold cross-validation results on data of H1 (A) and mESCs (B). In each round of cross-validation, we calculated a number of metrics to evaluate the performance of each method. These metrics include accuracy, precision, recall (i.e., sensitivity), GM (geometric mean), F1 score, validation rate(s), and misclassification rate. For each method, the average rank is the mean of ranks in all metrics. Best rank is highlighted in red. (C) Percentage of MERA-identified distal regulatory DNA elements that were recaptured by computational predictions. Bar chart shows the average percentage across all four MERA experiments for each method, whereas each circle shows the percentage in each MERA experiment. See also SI Methods for details.
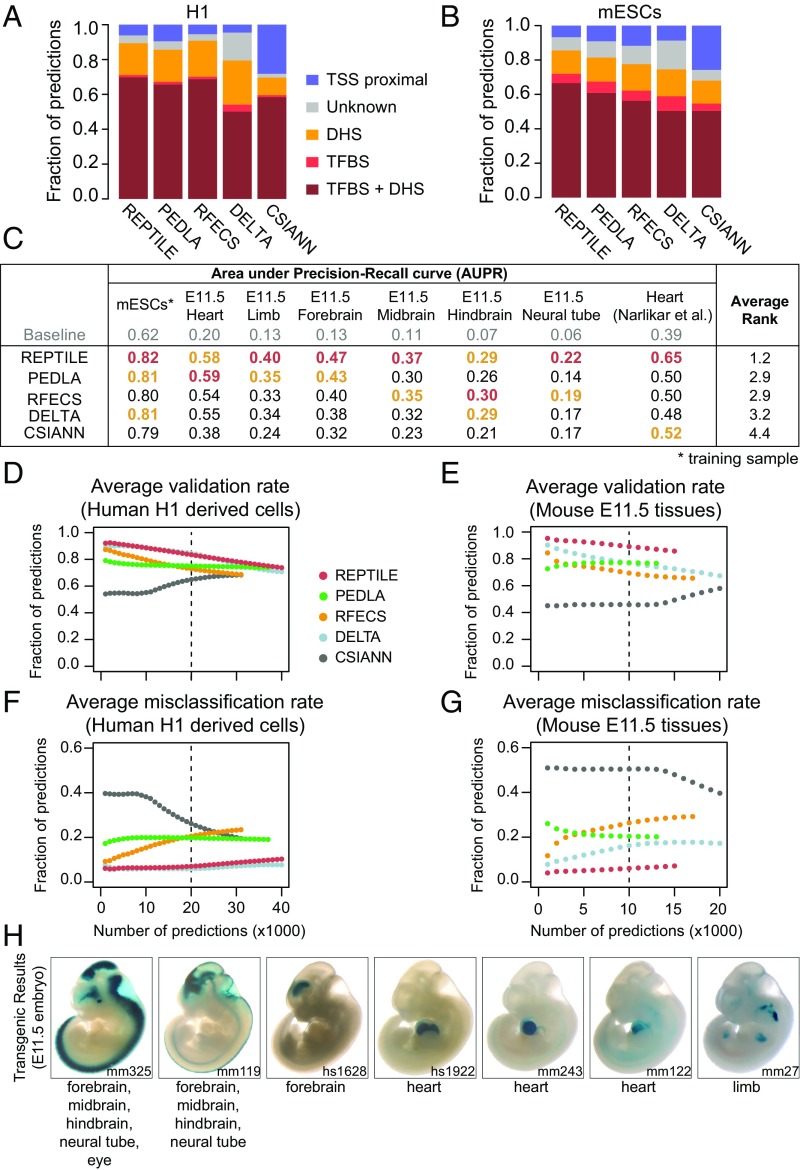
Fig. 2.
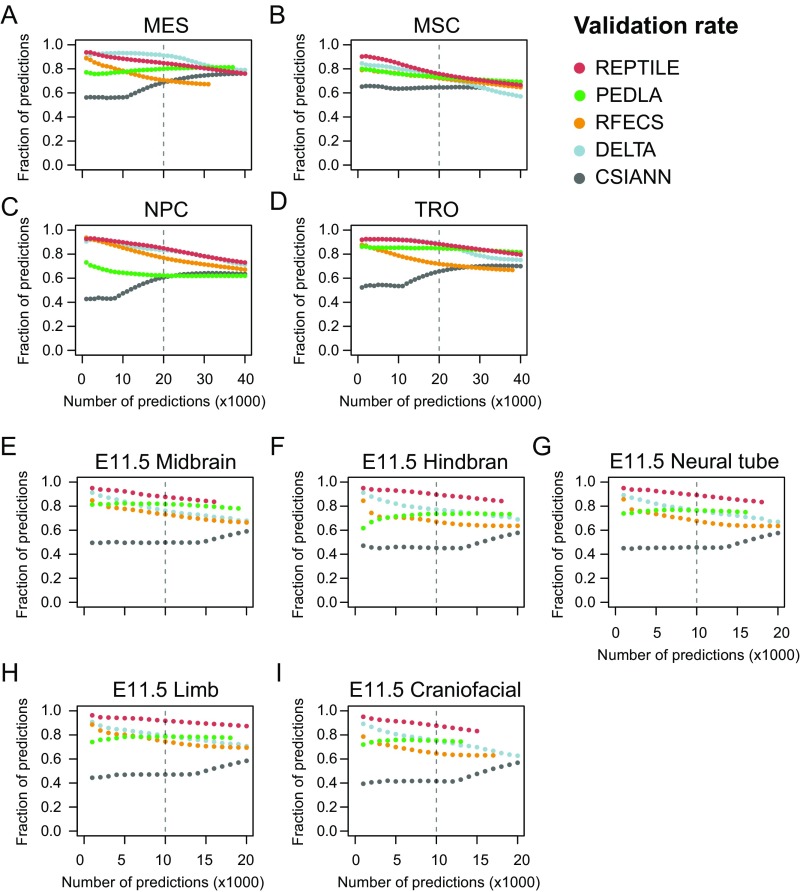
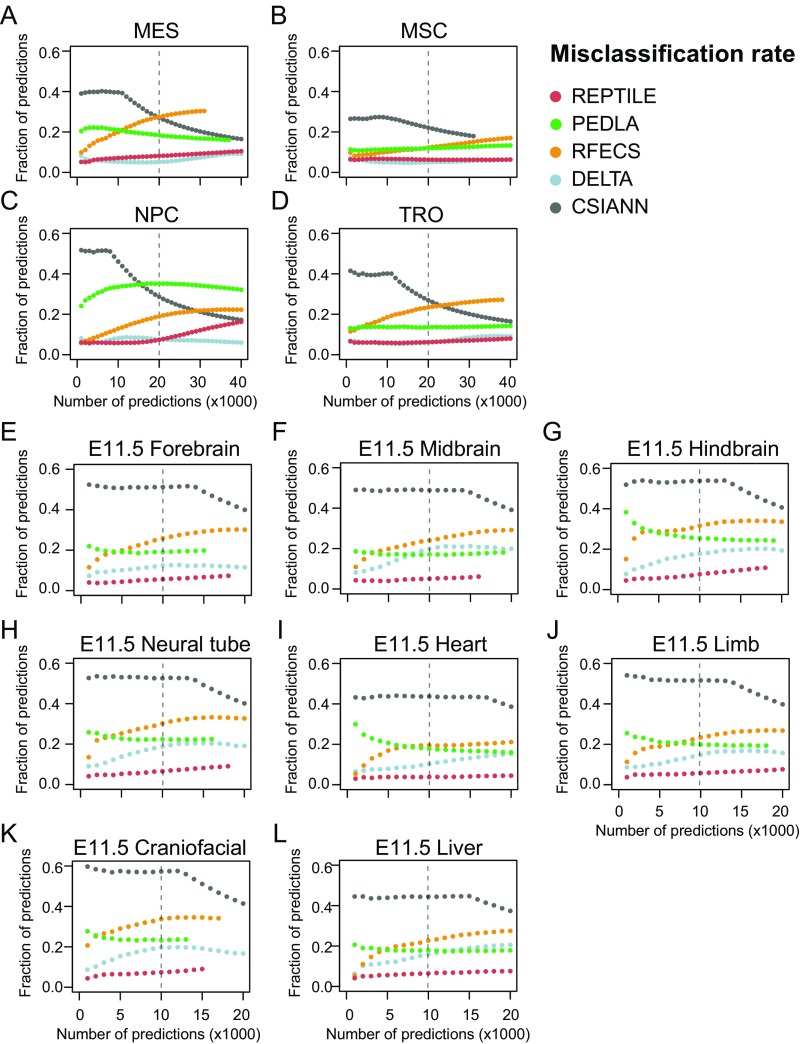
REPTILE shows better enhancer prediction accuracy than existing methods. (A and B) In H1 (A) and mESCs (B), the fractions of enhancers with their centers within 1 kb to TFBS+DHS (dark red, both distal TFBSs and distal DHSs), TFBS (red, only distal TFBSs), DHS (orange, only distal DHSs), TSS proximal (overriding all other categories), or none of the above (gray, labeled as Unknown). Distal TFBS (DHSs) are defined as TFBSs (DHSs) that are at least 1 kb away from any TSSs. “TFBS,” “DHS,” and “TFBS+DHS” are considered as true positives, whereas “TSS proximal” is considered as false positive and misclassification. (C) Performances of all methods in eight test datasets that contain experimentally validated enhancers. Performances are measured by the area under precision-recall curve (AUPR). Best results in each test dataset are highlighted in red, and second best results are marked in orange. The enhancer models used to make predictions in all samples were trained on data of mESCs. The baselines (AUPRs achieved using random guessing) for these datasets are shown in gray. Note that the AUPRs in different datasets cannot be compared because the fractions of validated enhancers are different. Fig. S1D for basic statistics of each dataset. (D and E) The validation rate of each method in human cell lines derived from H1 (D) and mouse tissues from E11.5 embryo (E), at different numbers of predictions. Validation rate is defined as the fraction of predictions whose centers are within 1 kb from distal DHSs and are at least 1 kb away from TSSs. (F and G) The misclassification rate of each method in human cell lines derived from H1 (F) and mouse tissues from E11.5 embryo (G). Misclassification rate is the fraction of predictions whose centers are within 1 kb to TSSs. Vertical dashed lines show the cutoffs used to get the final putative enhancer sets. (H) Examples of newly validated enhancers recapitulated by REPTILE enhancer predictions. Candidate enhancers were tested in transgenic mouse assays at E11.5. The enhancer name (mm or hs number), a representative transgenic embryo, and the tissues showing reproducible reporter gene expression (blue staining) are shown for each enhancer. DHS, DNase hypersensitivity sites; mESCs, mouse embryonic stem cells; TFBS, transcription factor binding site; TSS, transcription start site. See also SI Methods for details.
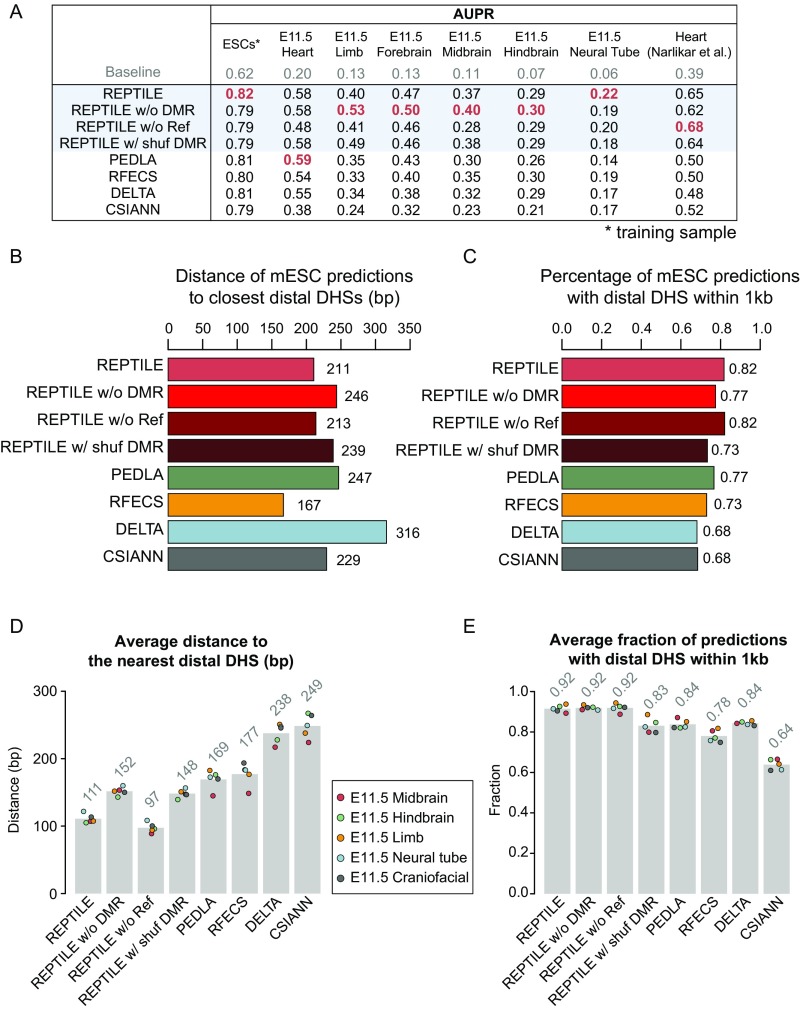
Fig. S3.
Prediction accuracy and resolution in cells where the models were trained. (A) The validation rate of each method in mESCs at different numbers of predictions. Validation rate is defined as the fraction of predictions whose centers are within 1 kb from distal DHSs and are at least 1 kb away from TSSs. (B) The misclassification rate of each method in mESCs. Misclassification rate is the fraction of predictions whose centers are within 1 kb to TSSs. (C) The validation rate of each method in H1. (D) The misclassification rate of each method in H1. (E) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS, in mESCs. (F) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS, in H1. (G) Average distance between the centers of predictions and the closest distal DHSs in mESCs. Predictions whose centers are beyond 1 kb away from the nearest distal DHS were considered as lack of support from open chromatin data and were not included in the calculation. Distal DHSs are at least 1 kb away from any TSSs. (H) Average distance between the centers of predictions and the closest distal DHSs in H1. The metric value in each individual cell/tissue is shown as a point in the bar chart. DHS, DNase hypersensitivity sites; mESC, mouse embryonic stem cell; TSS, transcription start site. See also SI Methods for details.
Because training datasets (e.g., EP300 data) are often not available for the cells or tissues of interest (target samples), it is extremely desirable that the enhancer model learned on one cell/tissue also performs well on other cell/tissue types. To assess this, we applied the models trained on human embryonic stem cell (H1) data to four H1-derived human cell lines and the models trained on mESCs to eight tissues from E11.5 mouse embryo. In human cell types, REPTILE and DELTA show the highest validation rate and the lowest misclassification rate compared with other methods, whereas REPTILE performed the best for mouse enhancer prediction (Fig. 2 D–G and Figs. S4 and S5). REPTILE predictions in E11.5 mouse tissues recapitulated several newly in vivo validated enhancers in E11.5 mouse embryo (Fig. 2H, Table S1, and SI Methods). We then tested REPTILE on in vivo experimentally validated regions and found it achieved the best performance for all test datasets, except in E11.5 midbrain and heart where it ranked second (Fig. 2C). Taken together, these results demonstrate REPTILE’s superior prediction accuracy in both human and mouse cell/tissue types over existing methods, when training and prediction were performed on different samples.
Fig. S4.
Validation rate of enhancer predictions in human and mouse samples. The validation rate of each method in different (A–D) human and (E–I) mouse cells and tissues at different numbers of predictions. Validation rate is defined as the fraction of predictions whose centers are within 1 kb from distal DHSs and are at least 1 kb away from TSSs. DHS, DNase hypersensitivity sites; TSS, transcription start site. See also SI Methods for details.
Fig. S5.
Misclassification rate of enhancer predictions in human and mouse samples. The misclassification rate of each method in different (A–D) human and (E–L) mouse cells and tissues at different numbers of predictions. Misclassification rate is the fraction of predictions whose centers are within 1 kb to transcription start sites (TSSs). See also SI Methods for details.
Table S1.
Elements tested in transgenic mouse assays
| Enhancer | Species tested | Genome coordinates (genome) | Genome coordinates (mm10) | Forward primer | Reverse primer | Tissues showing activity in transgenic assay (reproducibility) |
| hs1628 | Human | chr7:90,765,382–90,768,454 (hg19) | chr5:4,867,060–4,869,936 | TGCCCCATTTCCTTATAGCA | TCTTGAGGGAGCACTGAT | Forebrain (8/8) |
| hs1922 | Human | chr6:150,496,683–150,499,601 (hg19) | chr10:3,393,515–3,396,433 | CAACAATAAAAGTGAACTCTTGAGC | ACACAACCTCACCTGCTGTG | Heart (11/13) |
| mm122 | Mouse | chr12:70,858,396–70,860,249 (mm9) | chr12:69,757,409–69,759,262 | TGCCATTGACCTGTTGGATA | GGGTTAAATCCCCTCTGAGC | Heart (7/9) |
| mm27 | Mouse | chr4:57,555,251–57,559,225 (mm9) | chr4:57,542,379–57,546,353 | TCATCTCTGCCTCTTGCTGA | GTGGCCTCTTGATGGACAGT | Limb (7/10) |
| mm119 | Mouse | chr1:191,011,153–191,012,480 (mm9) | chr1:189,187,274–189,188,601 | CCAGACACTTCCTGGATA | TGAACTATGGACCCTTCTGAAAA | Forebrain (7/9), midbrain (7/9), hindbrain (7/9), neural tube (7/9) |
| mm243 | Mouse | chr17:48,575,655–48,577,860 (mm9) | chr17:48,436,330–48,438,535 | CACCACAGGGTGGTACTGATGA | CCTCACTTTGAAAGCACTCCA | Heart (5/5) |
| mm325 | Mouse | chr15:85,336,360–85,340,036 (mm9) | chr15:85,505,930–85,509,606 | CACCCGTTCTGACCAAGGATAG | AAACAATTAAAACCTCTCGTAGGC | Forebrain (7/10), midbrain (8/10), hindbrain (8/10), neural tube (8/10), eye (5/10) |
The Resolution of REPTILE Predictions Is Better than Existing Methods.
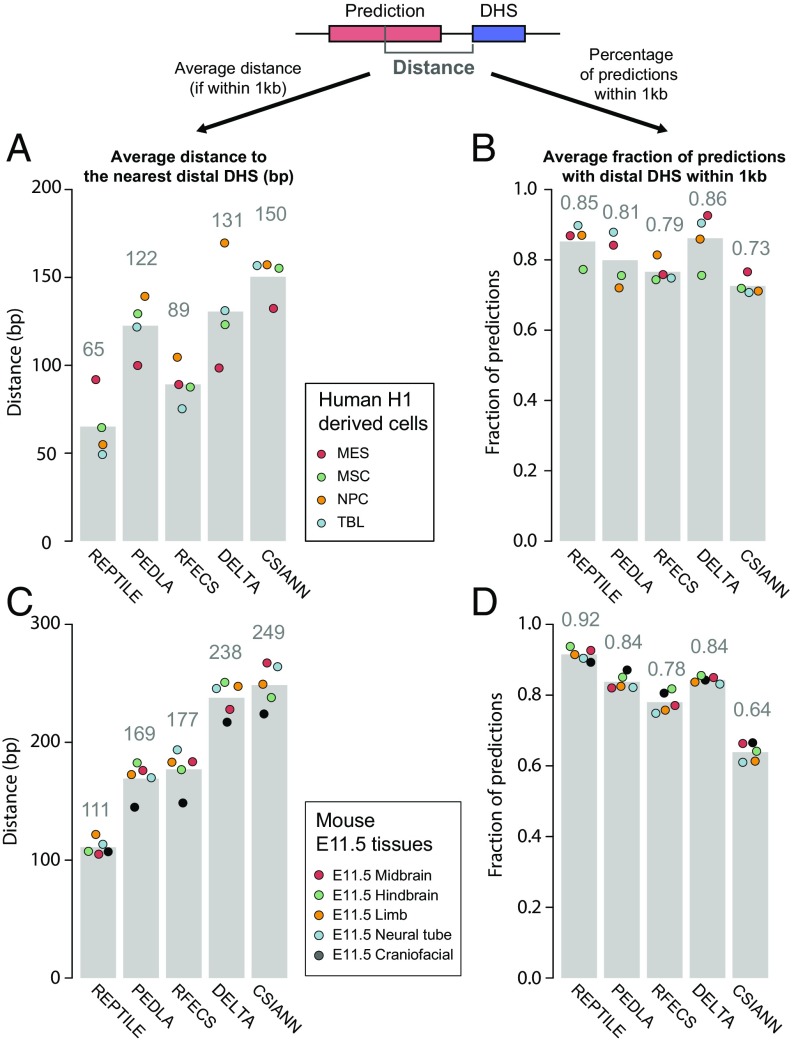
Next, to measure the resolution of enhancer prediction methods, we calculated the average distance between the center of each prediction and the nearest distal DHS (Methods). We found a higher percentage (82%) of REPTILE mESCs predictions had distal DHS nearby (within 1 kb) compared with all other methods (77%; Fig. S3E). For H1 cells, its overlap (90%) ranked second, which is only slightly lower than RFECS predictions (91%) (Fig. S3F). Among these predictions, the centers of RFECS predictions are, on average, 36 bp (H1) and 44 bp (mESCs) closer to the nearest distal DHSs than REPTILE predictions, which ranked second (Fig. S3 G and H). The results highlight RFECS’s superior prediction resolution in the training cell lines (H1 and mESCs), whereas REPTILE’s performance is comparable; both outperformed all other methods.
However, we found that REPTILE achieved much better prediction resolution than all other methods when applied to cell/tissue types different from the training data. In H1-derived human cells, the enhancer predictions made by REPTILE are, on average, over 24 bp closer to the nearest distal DHSs compared with other methods, including RFECS (Fig. 3A). On average, 85% of REPTILE predictions are supported by nearby distal DHSs, which ranked second, only slightly lower than DELTA (86%; Fig. 3B). In tissues from E11.5 mouse embryo, REPTILE predictions are, on average, over 58 bp closer to the nearest distal DHSs than the other methods, and 92% of the REPTILE predictions are close to distal open chromatin regions, outperforming all other methods (84%; Fig. 3 C and D).
Fig. 3.
The resolution of REPTILE predictions exceeds existing methods. (A) Average distance between the centers of predictions and the closest distal DHSs in four human cell types derived from H1. Predictions whose centers are beyond 1 kb away from the nearest distal DHS were considered as lack of support from open chromatin data and were not included in the calculation. Distal DHSs are at least 1 kb away from any TSSs. (B) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS, in human cells derived from H1. (C) Average distance between the centers of predictions and the closest distal DHSs in mouse tissues from E11.5 embryo. (D) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS, in mouse tissues from E11.5 embryo. The metric value in each individual cell/tissue is shown as a point in the bar chart. DHS, DNase hypersensitivity sites; Mes, mesendoderm; MSC, mesenchymal stem cells; NPC, neural progenitor cells; TRO, trophoblast-like cells; TSS, transcription start site. See also SI Methods for details.
Identifying the Transcription Factors Functionally Related to Each Cell Type Using REPTILE Enhancers.
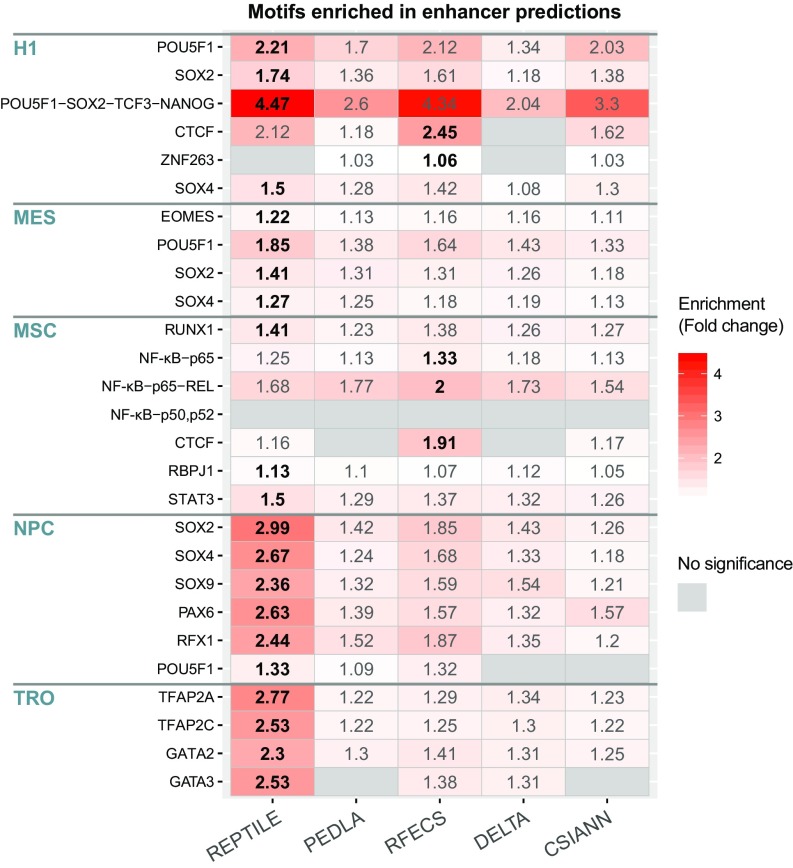
Enhancers are frequently bound by TFs that are critical to the function of cells and tissues. In H1 and H1-derived cell lineages, we found that the predicted enhancers from REPTILE and other methods are enriched for the DNA motifs that are bound by the TFs (or complex) known to function in these cell lines (Fig. 4, Table S2, and SI Methods). Motif analysis of REPTILE enhancers recapitulated the enrichment of TF binding motifs in 25 out of the 27 cases (92.6%). Furthermore, in most cases (21 of 27, 77.8%), the TF binding motif showed stronger enrichment in REPTILE enhancers than in the putative enhancers from other methods. Notably, in the trophoblast-like cell lineage (TRO), the average fold enrichment of the TF motifs nearly doubled in enhancers from REPTILE compared with other methods (2.5-fold versus 1.3-fold; Fig. 4). These results indicate that REPTILE enhancer predictions facilitate the discovery of functionally related TFs in a given cell type by accurately pinpointing the location of their binding motifs.
Fig. 4.
REPTILE enhancers improve the detection of known transcriptional regulators for each cell type. Enrichment of transcription factor–binding-site motifs in the putative enhancers in H1 and H1 derived cells, respectively. Motif enrichments in each cell type were calculated on the predicted enhancers in matched cell type. Enrichment fold change is the fraction of predicted enhancers (target sequences) that contain a certain motif divided by the fraction of background sequences that contain the same motif. Highest enrichment of each motif in each cell type is marked in bold. Not significant enrichment (q value > 0.05) is shown in gray. The transcription factors (complex) listed under each cell type are known to function in that cell type, which were based on the list from Xie et al. (47). SI Methods for details. H1, H1 human embryonic stem cells; Mes, mesendoderm; MSC, mesenchymal stem cells; NPC, neural progenitor cells; TRO, trophoblast-like cells.
Table S2.
Mapping between TF names and motif names
| TF name | Motif name (from Homer) |
| POU5F1 | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer |
| SOX2 | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer |
| POU5F1−SOX2−TCF3−NANOG | OCT4-SOX2-TCF-NANOG(POU,Homeobox,HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer |
| CTCF | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer |
| ZNF263 | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer |
| SOX4 | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer |
| EOMES | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer |
| RUNX1 | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer |
| NF-κB−p65 | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer |
| NF-κB−p65−REL | NFkB-p65-Rel(RHD)/ThioMac-LPS-Expression(GSE23622)/Homer |
| NF-κB−p50,p52 | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber_et_al.)/Homer |
| RBPJ1 | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer |
| STAT3 | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer |
| SOX9 | Sox9(HMG)/Limb-SOX9-ChIP-Seq(GSE73225)/Homer |
| PAX6 | PAX6(Paired,Homeobox)/Forebrain-Pax6-ChIP-Seq(GSE66961)/Homer |
| RFX1 | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer |
| TFAP2A | AP-2alpha(AP2)/HeLa-AP2alpha-ChIP-Seq(GSE31477)/Homer |
| TFAP2C | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer |
| GATA2 | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer |
| GATA3 | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer |
Transcription factor (TF) names and the names of their binding motifs in the motif database of Homer.
REPTILE Enhancers Are Enriched for Noncoding GWAS SNPs and Associated with Increased Expression of Target Genes.
Noncoding disease-associated genetic variants are enriched in the regulatory elements of related cell types and tissues (7). Stronger tissue-specific enrichment of such variants in putative enhancers of related tissues or cell types is likely indicative of better prediction accuracy and resolution. Therefore, we used enrichment as a metric for the evaluation of enhancer prediction methods.
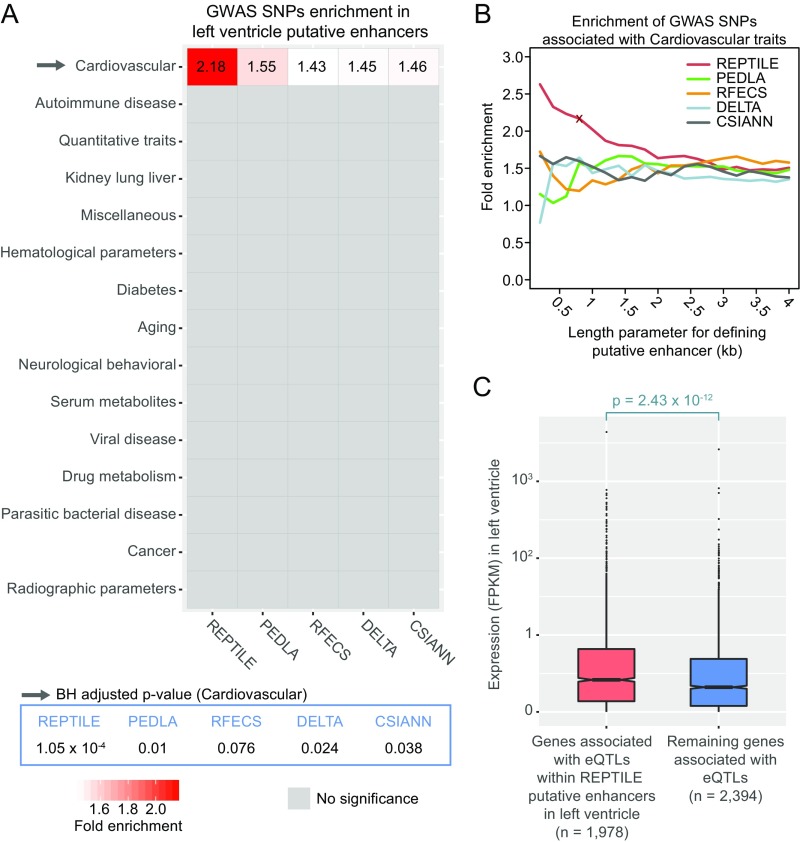
First, we applied all methods to identify enhancers in human heart left ventricle. Because data are available for only some of the epigenetic marks in this tissue, we retrained all methods to generate the enhancer predictions (SI Methods for more details). Then, we tested the enrichment of noncoding GWAS SNPs in these putative enhancers. Consistent with previous findings, only SNPs associated with traits in “Cardiovascular” category showed significant enrichment, indicating that the predicted enhancers are of reasonable quality (Fig. S6A). However, we found that these SNPs were most enriched in REPTILE predicted enhancers, suggesting its better resolution and accuracy compared with other methods (Fig. S6 A and B).
Fig. S6.
REPTILE enhancers are enriched for noncoding GWAS SNPs and associated with increased expression of target genes. (A) Enrichment of noncoding GWAS SNPs in putative enhancers of human heart left ventricle. Noncoding GWAS SNPs were categorized based on the associated traits. One-tail hypergeometric test was used to test for significance. Benjamini–Hochberg approach was then used for multiple testing corrections. Noncoding GWAS SNPs and trait categories are from Maurano et al. (7). See SI Methods for more details. (B) Fold enrichment of the noncoding GWAS SNPs associated with “Cardiovascular” category in the left ventricle putative enhancers given different length parameters. The fold enrichment was calculated using putative enhancers that are defined as genomic regions of given length and centered at the predicted enhancer centers from each method. Red cross indicates the data point of REPTILE enhancers whose enhancer boundary is determined using DMR information. (C) In human heart left ventricle, genes associated with REPTILE enhancers show significantly higher expression than genes associated with other genomic loci. Expression quantitative trait loci (eQTLs) data were used to link REPTILE enhancers to the target genes. eQTLs that are within 2 kb to any TSSs were excluded for this analysis. Gene expression level is represented in fragments per kilobase of transcript per million mapped reads (FPKM).
Enhancers are expected to increase the transcription of target genes. To test this, we linked REPTILE putative enhancers to their target genes using expression quantitative trait loci (eQTLs) data of left ventricle tissue from Genotype–Tissue Expression (GTEx) Project (SI Methods). We found that indeed genes linked to REPTILE enhancers showed significantly higher expression than genes linked to other genomic loci (Fig. S6C).
REPTILE Score Correlates Better with in Vivo Enhancer Activity than Open Chromatin.
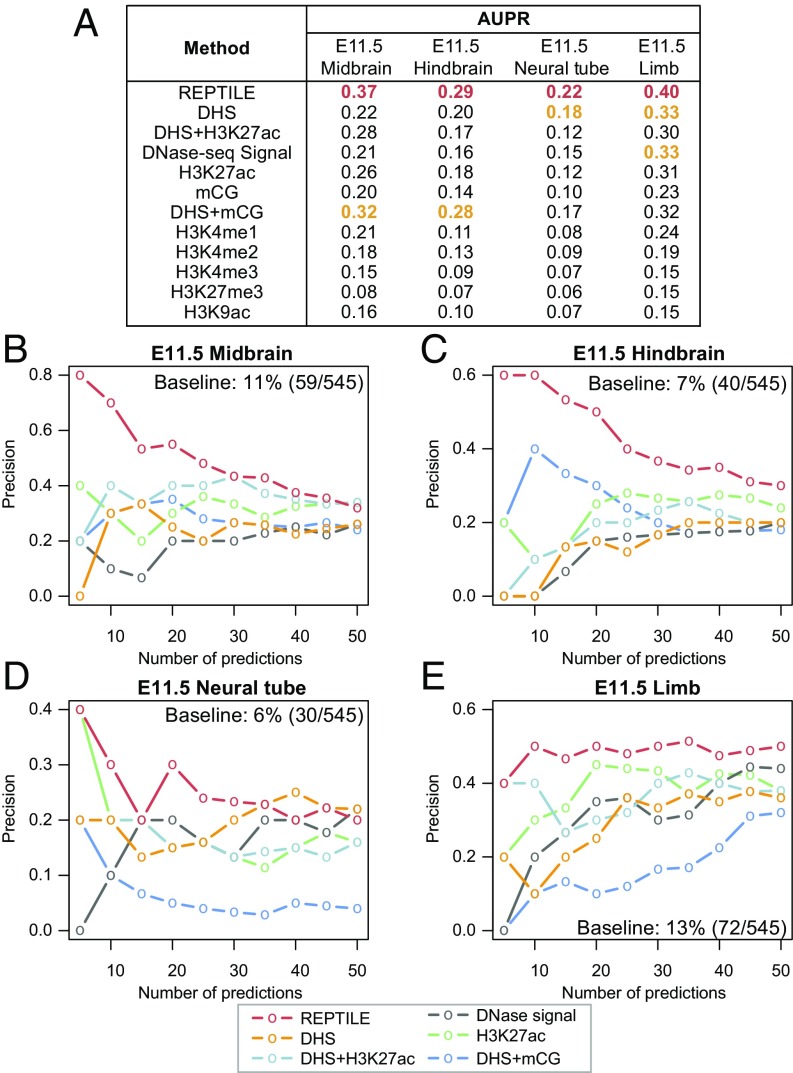
Although open chromatin signatures using DNase-sEq (39)/ATAC-sEq (40) were used for validation in this study, we found that REPTILE score is more predictive of the in vivo activity of DNA elements from VISTA database than open chromatin data (Fig. 5A and SI Methods). Two recent studies showed that low CG methylation in candidates of regulatory regions is an indicator of enhancers (41, 42). To test this idea, we implemented an approach to predict enhancers based on the CG methylation level in DHSs (DHS+mCG; SI Methods). Although useful, this approach does not provide better performance than REPTILE predictions (Fig. 5A). We further tested other single histone marks as well as the H3K27ac signal in DHSs and found that none of these is as predictive as the REPTILE score (Fig. 5A). Consistently, the enhancer predictions based on REPTILE score consistently achieved the best precision given different score cutoffs (Fig. 5 B–E and SI Methods). These results highlight the value of a method that uses integrative data. At the same time, it suggests that open chromatin regions may not be the ideal data type to validate predicted enhancers.
Fig. 5.
REPTILE enhancer confidence score is more predictive of enhancer activity than open chromatin or any single epigenetic mark. (A) Performance of REPTILE and several enhancer prediction methods that are based on open chromatin, single epigenetic mark, or the H3K27ac signal or mCG in open chromatin regions. The benchmark was done in four test datasets, where DNase-seq data are available in the corresponding samples. Performance is measured by the area under precision-recall curve (AUPR). For each test dataset, the best performance(s) are highlighted in red, and the second best are marked in orange. “REPTILE” generated scores on elements based on the enhancer model trained on data of mouse embryonic stem cells. “DHS” method assigned score to each element as the maximum normalized DNase-seq read count across all overlapping DHSs. The score is 0 if the region contains no overlapping DHS. “DHS+H3K27ac” and “DHS+mCG” are similar to “DHS,” but instead of DHS signal, it uses H3K27ac fold enrichment or CG methylation level as signal. The rest of the methods except mCG, DHS+mCG, and H3K27me3 methods use the fold enrichment in whole elements as score. In contrast, mCG, DHS+mCG, and H3K27me3 methods uses the signal values with reversed sign (i.e., depletion) because mCG and H3K27me3 are known to be repressive. (B–E) Precision of predicted enhancers that is based on the scores from REPTILE (red), DHS (orange), DHS+H3K27ac (light blue), DNase signal (gray), H3K27ac (green), and DHS+mCG (blue) in E11.5 midbrain (B), hindbrain (C), neural tube (D), and limb (E). Precision is defined as the percentage of enhancer predictions that showed enhancer activity in vivo. DHS, DNase hypersensitivity sites. See also SI Methods for details.
SI Methods
Whole-Genome Bisulfite Sequencing Data.
The raw reads of MethylC-seq or whole-genome bisulfite sequencing (WGBS) data of eight mouse tissues from E11.5 embryo were downloaded from the ENCODE website (https://www.encodeproject.org/). Mouse embryonic stem cells (mESCs) WGBS data were obtained from Gene Expression Omnibus (GEO). The accession numbers of the two mESCs replicates are GSM1162043 and GSM1162044. The paired-end data (GSM1162045) of the second replicate was not included to avoid potential bias due to different data type (paired end versus single end). WGBS raw reads of human cell lines, H1 human embryonic stem cells (H1), mesendoderm (Mes), mesenchymal stem cells (MSC), neural progenitor cells (NPC), and trophoblast-like cells (TRO), were obtained from SRA (accession number SRP000941). For MSC, whose methylome had been sequenced in paired end, we mapped the first read in each pair to avoid problems in processing overlapping reads similar to Schultz et al. (19). WGBS data of human heart left ventricle was downloaded from GEO (GSM983650). The sources of all of the WGBS data can be found in Table S3.
Table S3.
Information of external WGBS data
| Organism | Sample | Assay | Accession |
| Mouse | mESCs | Whole-genome bisulfite sequencing | GSM1162043, GSM1162044 |
| Mouse | E11.5 forebrain | Whole-genome bisulfite sequencing | ENCSR271HQP |
| Mouse | E11.5 midbrain | Whole-genome bisulfite sequencing | ENCSR091VFX |
| Mouse | E11.5 hindbrain | Whole-genome bisulfite sequencing | ENCSR398UCM |
| Mouse | E11.5 heart | Whole-genome bisulfite sequencing | ENCSR633CON |
| Mouse | E11.5 limb | Whole-genome bisulfite sequencing | ENCSR916GKL |
| Mouse | E11.5 liver | Whole-genome bisulfite sequencing | ENCSR033PGF |
| Mouse | E11.5 craniofacial | Whole-genome bisulfite sequencing | ENCSR950OMB |
| Mouse | E11.5 neural tube | Whole-genome bisulfite sequencing | ENCSR613BMI |
| Human | H1 | Whole-genome bisulfite sequencing | SRP000941 |
| Human | Mes (mesendoderm) | Whole-genome bisulfite sequencing | SRP000941 |
| Human | MSC (mesenchymal stem cells) | Whole-genome bisulfite sequencing | SRP000941 |
| Human | NPC (neural progenitor cells) | Whole-genome bisulfite sequencing | SRP000941 |
| Human | TRO (trophoblast-like cells) | Whole-genome bisulfite sequencing | SRP000941 |
| Human | Heart left ventricle | Whole-genome bisulfite sequencing | GSM983650 |
Accessions started with “GSM” are GEO identifiers (www.ncbi.nlm.nih.gov/geo/). Accessions started with “SRP” are for SRA database (www.ncbi.nlm.nih.gov/sra/). Accessions started with “ENC” are identifiers of ENCODE Project (https://www.encodeproject.org/).
WGBS data were processed as previously described (52), using mm10 reference for mouse data and hg19 reference for human data. Only autosomes, sex chromosomes, mitochondrial chromosomes, and the genome sequence of lambda phage (as control) are included in the reference genome. The sequences were downloaded from University of California, Santa Cruz (UCSC), genome browser (53). For each sample, if biological replicates were available, the data of replicates were combined. To quantify the methylation landscape, we divided the genome into 100-bp bins and calculated the (weighted) methylation level (54) for each bin. Weighted methylation level is also called as the CG methylation (mCG) intensity, and it is defined as the ratio of the sum of methylated basecall counts over the sum of both methylated and unmethylated basecall counts across all CG sites in a given region (54). For each sample, these values were used to generate a file (in bigWig format), which stores the methylation levels of all bins (see also https://genome.ucsc.edu/goldenpath/help/bigWig.html for more about bigWig format). In all of the analyses in this paper, the methylation levels of any region was obtained from these bigwig files using the “bigWigAverageOverBed” executable from UCSC genome browser (53).
Identification of DMRs.
DMR calling was done using very similar procedure as Schultz et al. (19). We included (and rephrased) the entire description here and highlighted the modifications we made. In the procedure, we considered bisulfite sequencing as a binomial process and defined a stochastic model in which, at each position, the observed number of reads supporting methylated cytosine in each sample is drawn from a binomial distribution. The true fraction of methylated alleles in the population in given sample at given cytosine in CG context, , is the parameter of the binomial distribution, where i denotes the position of cytosine and n denotes the sample. The null hypothesis is that the methylation level () at this position is equal across all samples: for all n.
Our procedure is designed to test whether the observed data are consistent with the null hypothesis, or alternatively if there is a significant deviation from equal methylation levels. To do this, we compute a goodness-of-fit statistic, s, introduced by Perkins et al. (55). We arrange the observed data in an N × 2 table, with each row for each of the N samples and the two columns for the number of reads supporting methylated and unmethylated cytosines, respectively. The number of observed reads in sample n at position i is , where j = 1 for methylated reads and j = 2 for unmethylated reads. The expected number of reads in sample n with methylation state j under the null hypothesis is :
where is the total number of reads in all samples. The statistic for the goodness of fit is as follows:
Next, we simulated read count data under our stochastic model assuming the null hypothesis in the following way: (i) Set all cell counts in the table to zero. (ii) Randomly select a cell in the table with probability equal to the expected counts divided by the total number of counts in the table (). Increment the value in this cell by 1. (iii) Repeat this procedure times. (iv) Finally, calculate the value of the statistic, , for the randomly generated table.
This randomization procedure was repeated until we observed 100 iterations with a value of that was at least as extreme as that of the observed data, s, up to a maximum of 3,000 iterations. The P value at position i was then computed as follows:
where Ri is the number of randomization where a statistic greater than or equal to the original table’s statistic was observed. Ti is the total number of randomizations that were conducted. Our adaptive permutation procedure ensures that any sites that we may potentially identify as significantly differentially methylated with will be sampled 3,000 times. At other sites, we have observed an appreciable number (100) of permutations more extreme than our original test statistic (s ≥ sshuff) and the P value for these sites will be P ≥ (100 + 1)/3,000 = 0.034; these sites will therefore not be called as differentially methylated.
To control the false-discovery rate (FDR) at our desired rate of 1%, we used a procedure designed for permutation-derived P values (56). First, we generated a histogram of the P values across all cytosines in CG context as described before. Next, we calculated the expected number of P values to fall in a particular bin under the null hypothesis. This expected count is computed by multiplying the width of the bin by the current estimate for the number of true null hypotheses (m0), which is initialized to the number of tests performed. We then identified the first bin (starting from the most significant bin) where the expected number of P values is greater than or equal to the observed value. The differences between the expected and observed counts in all of the bins up to this point are summed, and a new estimate of m0 is generated by subtracting this sum from the current total number of tests. This procedure was iterated until convergence, which we defined as a change in the m0 estimate less than or equal to 0.01. With this m0 estimate, we were able to estimate the FDR corresponding to a given P value cutoff by multiplying the P value by the m0 estimate (the expected number of positives at that cutoff under the null hypothesis) and dividing that product by the total number of significant tests we detected at that P value cutoff. We chose the largest P value cutoff that still satisfied our FDR requirement.
Next, we combined significant sites [differentially methylated sites (DMSs)] into blocks if they were within 250 bp and showed methylation changes in the same direction (e.g., sample A was hypermethylated and sample B was hypomethylated at both sites). A sample was considered hypomethylated or hypermethylated if the deviation of observed counts from the expected counts was in the top or bottom 1% of deviations. These residuals were calculated for a position i using the following formula for a given cell in row n and column j of the table:
The distinction between hypermethylation and hypomethylation was made based on the sign of the residuals. For example, if the residual for the methylated read count of sample A was positive, it was counted as hypermethylation. Furthermore, blocks that contained fewer than two DMSs were discarded. Instead of the 10-DMS cutoff used in original procedure, we used a more lenient 2-DMS cutoff to get a more comprehensive list of DMRs (enhancer candidates) to feed REPTILE. As an additional step to the original procedure, we next extended the remaining blocks by 150 bp from both sides and defined them as DMRs. The purpose of this extra step is to include regions where the histone modifications generally occur—the upstream and downstream nucleosomes flanking putative enhancers.
Applying the DMR Calling Algorithm on Human and Mouse Cells and Tissues.
To obtain DMRs for mouse samples, we applied the above calling algorithm on the mCG profiles of mESCs and eight E11.5 mouse tissues. In total, 542,139 DMRs were identified, with average length of 484 bp and covering over 262 Mb or ∼10% of the genome. We found that 97% of the experimentally validated enhancers (246 out of 253) in VISTA enhancer browser (31) overlap with DMRs. By contrast, out of the 45 elements in VISTA enhancer browser that did not overlap with any DMRs, 38 (86%) did not show any enhancer activity, implying that differential methylation is a significant enhancer signature.
We applied the same procedure to call DMRs across the mCG profiles of all human cell lines. We identified 159,474 DMRs and their average length is 439 bp. These DMRs covered ∼2% of the genome.
Chromatin and Transcription Factor ChIP-Seq Data.
For the eight E11.5 mouse tissues, we downloaded the ChIP-seq data of six previously identified enhancer-related histone marks (H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K27me3, and H3K9ac) and the corresponding control from the ENCODE project website (https://www.encodeproject.org/). For mESCs, ChIP-seq data of the same histone modifications and the corresponding controls were downloaded from GEO (Table S4). In addition, ChIP-seq data of EP300 and its corresponding control data were downloaded from GEO (Table S4). We also downloaded ChIP-seq data of 12 transcription factors in mESCs from GEO (Table S4).
Table S4.
Information of external ChIP-seq data
| Organism | Sample | ChIP-seq target | ID | Accession | Note |
| Mouse | mESCs | H3K27ac | mESC_H3K27ac | GSM1000099 | |
| Mouse | mESCs | H3K27me3 | mESC_H3K27me3 | GSM1000089 | |
| Mouse | mESCs | H3K4me1 | mESC_H3K4me1 | GSM769009 | |
| Mouse | mESCs | H3K4me3 | mESC_H3K4me3 | GSM769008 | |
| Mouse | mESCs | H3K9ac | mESC_H3K9ac | GSM1000127 | |
| Mouse | mESCs | Control | mESC_control_1 | GSM918754 | Control for marks above including mESC_H3K27ac, mESC_H3K27me3, mESC_H3K4me1, mESC_H3K4me3 and mESC_H3K9ac |
| Mouse | mESCs | EP300 | mESC_p300 | GSM723018 | |
| Mouse | mESCs | Control | mESC_control_2 | GSM723020 | Control for mESC_p300 |
| Mouse | mESCs | H3K4me2 | mESC_H3K4me2 | GSM747543,GSM747544 | |
| Mouse | mESCs | Control | mESC_control_3 | GSM747545,GSM747546 | Control for mESC_H3K4me2 |
| Mouse | mESCs | Nanog | mESC_Nanog | GSM288345 | TF ChIP-seq |
| Mouse | mESCs | Oct4 | mESC_Oct4 | GSM288346 | TF ChIP-seq |
| Mouse | mESCs | Sox2 | mESC_Sox2 | GSM288347 | TF ChIP-seq |
| Mouse | mESCs | Smad1 | mESC_Smad1 | GSM288348 | TF ChIP-seq |
| Mouse | mESCs | E2f1 | mESC_E2f1 | GSM288349 | TF ChIP-seq |
| Mouse | mESCs | Tcfcp2I1 | mESC_Tcfcp2I1 | GSM288350 | TF ChIP-seq |
| Mouse | mESCs | Zfx | mESC_Zfx | GSM288352 | TF ChIP-seq |
| Mouse | mESCs | STAT3 | mESC_STAT3 | GSM288353 | TF ChIP-seq |
| Mouse | mESCs | Klf4 | mESC_Klf4 | GSM288354 | TF ChIP-seq |
| Mouse | mESCs | Esrrb | mESC_Esrrb | GSM288355 | TF ChIP-seq |
| Mouse | mESCs | cMyc | mESC_cMyc | GSM288356 | TF ChIP-seq |
| Mouse | mESCs | nMyc | mESC_nMyc | GSM288357 | TF ChIP-seq |
| Mouse | mESCs | Control | mESC_control_4 | GSM288358 | Control for TF ChIP-seq |
| Mouse | E11.5 forebrain | All chromatin marks | ENCSR415TUB | ||
| Mouse | E11.5 midbrain | All chromatin marks | ENCSR843IAS | ||
| Mouse | E11.5 hindbrain | All chromatin marks | ENCSR501OPC | ||
| Mouse | E11.5 heart | All chromatin marks | ENCSR016LTR | ||
| Mouse | E11.5 limb | All chromatin marks | ENCSR283NCE | ||
| Mouse | E11.5 liver | All chromatin marks | ENCSR231EPI | ||
| Mouse | E11.5 craniofacial | All chromatin marks | ENCSR800JXR | ||
| Mouse | E11.5 neural tube | All chromatin marks | ENCSR215ZYV | ||
| Human | H1 | All chromatin marks | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) | |
| Human | Mes (mesendoderm) | All chromatin marks | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) | |
| Human | MSC (mesenchymal stem cells) | All chromatin marks | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) | |
| Human | NPC (neural progenitor cells) | All chromatin marks | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) | |
| Human | TRO (trophoblast-like cells) | All chromatin marks | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) | |
| Human | Heart left ventricle | H3K4me1, H3K4me3, H3K27ac, H3K27me3 and input | egg2.wustl.edu/roadmap/web_portal/ | Unconsolidated epigenomes (uniform mappability) |
Accessions started with “GSM” are GEO identifiers (www.ncbi.nlm.nih.gov/geo/). Accessions started with “ENC” are identifiers of ENCODE Project (https://www.encodeproject.org/).
All mouse ChIP-seq data were processed using the ENCODE uniform processing pipeline for ChIP-seq: First, reads were mapped to the mm10 reference using bwa (57) (version 0.7.10) with parameters “-q 5 -l 32 -k 2.” The mm10 reference only contains autosomes, sex chromosomes, and mitochondrial sequences. It is also called as “mm10-minimal” in the ENCODE website. Then, Picard tool (broadinstitute.github.io/picard/, version 1.92) was used to remove PCR duplicates using parameter “REMOVE_DUPLICATES=true.”
For chromatin ChIP-seq data of human cell lines and heart left ventricle tissue, we directly downloaded the alignment files [labeled as “Unconsolidated Epigenomes (Uniform mappability)”] from the data portal of the NIH Roadmap Epigenomics Mapping Consortium (egg2.wustl.edu/roadmap/web_portal/index.html). We obtained ChIP-seq data of H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K27me3, H3K9ac, and corresponding control for all five human cell lines. For heart left ventricle, we downloaded H3K4me1, H3K4me3, H3K27ac, H3K27me3, and control because other histone marks were not available.
For each histone modification mark in human and mouse samples, we represented it as continuous enrichment values of 100-bp bins across the genome. Specifically, we first extended reads to 300 bp (expected fragment length) using the -r option (along with -s and -l 0 options) in slopBed from bedtools (58). We then divided the mouse genome into 100-bp bins, and for each bin, we calculated log2 reads per million mapped reads (RPM) fold enrichment relative to control. RPM for control experiment in each bin is smoothed by averaging it over the RPMs of two bins upstream and two bins downstream. RPM for a given bin is defined as the number of mapped reads that overlap (1 bp) with the bin divided by the total number (in million reads) of the uniquely mapped reads in the genome.
For the ChIP-seq data of TFs and EP300 in mESCs, we used MACS (59) (1.4.2) to call peaks with default parameters. The reported TF peaks were filtered out if they are within 1 kb to any transcription start sites (TSSs) of genes in mouse GENCODE (60) annotation (M2).
EP300 and Transcription Factor Binding Sites in H1.
We downloaded the binding sites of DNA-binding proteins in H1 from ENCODE data portal in the UCSC genome browser (hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wgEncodeRegTfbsClusteredWithCellsV3.bed.gz). The binding sites of EP300 were used as positive instances (i.e., putative active enhancers) in the training of enhancer prediction methods. The distal binding sites of remaining DNA-binding proteins, excluding CTCF, were used to validate the prediction in H1 in Fig. 2A (see later section for details). The reason to exclude CTCF was that CTCF played a major role in shaping the chromatin architecture and its binding sites included insulators (61). Distal binding sites are at least 1 kb away from any TSSs in the human GENCODE annotation (release 19).
Enhancer Validation Data.
To evaluate the enhancer prediction accuracy, we collected publicly available data of experimentally validated enhancers and negative sequences (sequences that showed no detectable enhancer activity) from three sources (Fig. S1D). The in vivo and in vitro data were used to construct the eight test datasets used in benchmark (Fig. S1D).
-
i)
From Yue et al. (32), we downloaded 212 regions that were tested for in vitro enhancer activity by luciferase reporter assay in mESCs. The original coordinates of these regions were in mm9 reference and they were liftover to mm10 using liftOver utility from UCSC genome browser (53). One region was filtered out in this process. Out of the remaining 211 tested regions, 131 showed enhancer activity in mESCs and were labeled as positive, whereas the rest were labeled as negative.
-
ii)
In addition, we obtained in vivo enhancer validation data from VISTA enhancer browser (31) (October 24, 2015). In total 546 mouse sequences were tested for in vivo enhancer activity in E11.5 mouse embryo using transgenic reporter assay. Their mm9 coordinates were liftover to mm10 and one region was removed. In the eight E11.5 mouse tissues where epigenomic data are available, six of them had a reasonable number (≥30) of validated enhancers. We used the data of these tissues (forebrain, midbrain, hindbrain, heart, limb, and neural tube) to build six test datasets (Fig. S1D). In this study, we only included the mouse sequences in VISTA database and excluded all human sequences. The rationale is that the in vivo enhancer activity of human sequences may be different from the activity of their mouse counterparts (orthologs), preventing them from being good validations.
-
iii)
We also included 36 in vivo validated sequences that were tested in vivo in the heart of zebrafish embryo from Narlikar et al. (49). The enhancer activity in the embryonic heart of zebrafish was shown to be conserved in mouse embryo (49). Based on this, we used these regions as approximation of enhancers in E11.5 mouse heart. The original dataset included 46 regions with coordinates in hg18 human reference genome. The hg18 coordinates were first liftover to hg19, which were then converted to mm10. In this process, 10 regions were eliminated and the remaining 36 were included in later analysis.
DNase-Seq Data.
The DNase hypersensitivity sites (DHSs) identified based on DNase-seq data were used to validate enhancer predictions. DHS calls of all five human cell lines were obtained from the NIH Roadmap Epigenomics Mapping Consortium (egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/narrowPeak/). We downloaded the narrow DHS peaks from MACS2 (59) (files whose names ended with “-DNase.macs2.narrowPeak.gz”).
DHS calls of mESCs were downloaded from UCSC genome browser (hgdownload.cse.ucsc.edu/goldenPath/mm9/encodeDCC/wgEncodeUwDgf/wgEncodeUwDgfEscj7129s1ME0PkRep1.narrowPeak.gz). The coordinates of these elements (mm9) were liftover onto mm10. DNase-seq data and DHSs in E11.5 mouse tissues were downloaded from the ENCODE project website (https://www.encodeproject.org/). We found that the DNase-seq data were available for five E11.5 tissues. The tissues and the corresponding accessions in the ENCODE project website are E11.5 craniofacial (ENCSR196VDE), E11.5 neural tube (ENCSR312QVY), E11.5 midbrain (ENCSR292QBA), E11.5 hindbrain (ENCSR358ESL), and E11.5 limb (ENCSR661HDP). Narrow peak files were downloaded and each peak call was defined as one DHS.
DNase-seq data were available for two biological replicates of each mouse E11.5 tissue. The DHSs of two biological replicates were combined using bedops (bedops.readthedocs.io/en/latest/content/usage-examples/master-list.html) (62). Below is the procedure description adapted from the text in the bedops webpage. The procedure starts with the union of DHSs called in both replicates (i.e., original elements) and an empty master list, which stores the final result.
-
i)
Original elements not yet in the master list are merged into nonoverlapping intervals (using bedops -m).
-
ii)
For each merged interval, the original element of highest score within the interval is selected to go into the master list.
-
iii)
Any original elements that overlap the selected element are thrown out.
-
iv)
Repeat steps 1, 2, and 3 until no original element is left. Then the master list is reported as the final DHS list.
Existing Enhancer Prediction Approaches.
To evaluate the performance of REPTILE, it was compared with four publicly available methods, PEDLA (34), RFECS (35), DELTA (36), and CSIANN (37). All of these methods are supervised learning approaches, meaning that they learned the profiles of enhancers from data with labels and then classify unseen regions. Specifically, they first represent genomic regions using histone modification data (and possibly other data types). Then, machine-learning technique is used to learn the histone modification signatures of (putative) enhancers and background regions. Last, the trained computational model is used to classify unknown regions into enhancers or negative regions.
Their differences mainly lie in the distinct strategy used to represent genomic regions and their different underlying machine learning framework.
-
i)
PEDLA used the histone modification signals and evolutionary conservation score as low-level features and it is capable of incorporating additional data types. Then, PEDLA applies deep neural network (DNN), in an unsupervised fashion, to extract high-level features from these low-level features in all 200-bp nonoverlapping bins across the genome. Last, the DNN is used to learn the feature signatures of enhancers and background sequencers (supervisedly) and then makes predictions.
-
ii)
RFECS represents the shape and intensity of each histone modification (ChIP-seq) signal in each 2-kb genomic window using a feature vector of length 20. Specifically, RFECS divides the 2-kb window equally into 20 100-bp nonoverlapping bins and the values in the feature vector correspond to the signal values in the 20 100-bp bins. Next, a random forest classifier (33) with linear separator is trained on this type of data on putative enhancers and background sequences. This model is then used to delineate enhancer-like chromatin signatures from genomic background.
-
iii)
DELTA defines four shape features to describe the histone modification (ChIP-seq) signature and then uses AdaBoost algorithm (63) to distinguish enhancers from negative regions based on this representation schema.
-
iv)
CSIANN was built on neural network framework, and it makes predictions based on the histone modification signals of 2-kb nonoverlapping genome windows.
Running REPTILE and Existing Enhancer Prediction Methods.
REPTILE and the four existing methods were trained in mESCs (for mouse enhancer prediction) or in H1 (for human enhancer prediction) by learning the epigenomic signatures of known/putative enhancers (EP300 binding sites) and negative regions (promoters and genomic background). The promoters are defined as 2-kb regions centered at TSSs and the TSSs were based on GENCODE annotation (mouse, M2; human, release 19).
-
i)
REPTILE: The training dataset for REPTILE was constructed using a similar strategy used for training RFECS previously (35). The training dataset for mouse enhancer prediction is composed of 5,000 positive instances (enhancers) and 35,000 negatives (negative regions). Positives were the ±1-kb regions around the summits of top 5,000 EP300 peaks in mESCs. Negatives included 5,000 randomly selected promoter regions and 30,000 (six times than number of positives) randomly chosen 2-kb bins. The 2-kb bins have no overlap with promoters, top 5,000 EP300 binding regions, or any regions in the mESCs test dataset. The training dataset for human enhancer prediction was constructed similarly. It includes 5,476 distal EP300 binding sites in H1 as positives and equal number of randomly chosen promoters and 32,856 (six times than number of positives) 2-kb bins. Score cutoff of 0.5 was used to generate genome-wide enhancer predictions for both human and mouse samples.
-
ii)
PEDLA: The training dataset for PEDLA was constructed similarly as REPTILE. The only difference is that the number of 2-kb bins is nine times of the number of positives to be consistent with how PEDLA was trained (34). We benchmarked various parameters of PEDLA and found that single layer with 500 neurons performed well in both human and mouse data (data not shown). This setting was used for running PEDLA. In the current implementation of PEDLA, hidden Markov model (HMM) is used to generate the final enhancer prediction based on the scores from the artificial neural network model. Score is defined as the observatory probability conditioned on enhancer state divided by prior probability of enhancer state (i.e., base rate). However, its performance was not as good as other methods [labeled as “PEDLA (HMM)” in Fig. S2 A and B]. Therefore, we implemented an alternative enhancer calling approach by applying the peak-calling algorithm used by REPTILE on the scores directly. We called this approach “PEDLA” in this study. It showed better performance than the current PEDLA implementation (Fig. S2 A and B). Score cutoff of 5 was used to generate enhancer calls.
-
iii)
RFECS: RFECS were trained on the same dataset as REPTILE. The default cutoff of 0.5 was used to generate genome-wide enhancer predictions in mESCs and all human cell types. In E11.5 tissues, we used cutoff of 0.2 to ensure that the number of putative enhancers was practically useful and enough (>10,000) for validation.
-
iv)
DELTA: For mouse enhancer prediction, the training dataset for DELTA were composed of the top 5,000 EP300 binding sites in mESCs and all promoters in mouse genome. For human enhancer prediction, the training dataset includes the 5,476 EP300 binding sites in H1 and all promoters in human genome. In the step of generating genome-wide predictions, we switched to the peak-calling algorithm used by REPTILE. It is because the default peak-calling algorithm in DELTA does not consider the spacing between peaks and thus generates a large number of predictions that are within 100 bp to each other, which is not desirable in practice. Score cutoffs of 0.1 in mESCs and human samples, whereas 0.05 in E11.5 tissues, were used to generate enough genome-wide predictions for validation.
-
v)
CSIANN: For mouse enhancer prediction, top 500 EP300 binding sites in mESCs and gene annotation from GENCODE (M2) were used as input for CSIANN training. Similarly, for human enhancer prediction, top 500 EP300 binding sites in H1 and gene annotation (human GENCODE release 19) were used for training. The small number of positives in the input was due to the fact that current CSIANN implementation imposed a size limit on the training data. Default settings were used for both model training and prediction generation.
Evaluating the Performance of Methods Using Cross-Validation.
In mESCs and H1, we used cross-validation to evaluate the performance of each method similar to Liu et al. (34). Results are shown in Fig. S2 A and B. The training data for PEDLA were used because it contains the most regions. We used fivefold stratified cross-validation, in which the ratio of positives to negatives was maintained in each round. Note that the current implementation of RFECS, DELTA, and CSIANN did not allow users to specify the negative regions for training. Therefore, we just changed the positives for training RFECS and DELTA in cross-validation, whereas we always used the top 500 positives to training CSIANN due to its limit in current implementation. In addition to these methods, we also included the chromatin states of mESCs and H1 (if available). The chromatin state of H1 was downloaded from the ENCODE portal at UCSC genome browser. The chromatin state of mESCs was downloaded from github (https://github.com/gireeshkbogu/chromatin_states_chromHMM_mm9/raw/master/mESC_cStates_HMM.zip). Strong enhancers in the chromatin state map were regarded as enhancer predictions.
To ensure a fair comparison, we selected equal number of predictions from each method (if possible) to evaluate and then resized them to 2-kb regions while maintaining their center. Predictions from REPTILE, PEDLA, RFECS, DELTA, and CSIANN were ranked, and the top ones were selected. Because the enhancer predictions from ChromHMM (64) and Segway (65) cannot be ranked, we randomly chose the same number of putative strong enhancers from their annotations.
To evaluate the prediction results, we first defined the following: True positives (TP) are positives that are overlapped with predicted enhancers. False positives (FP) are negatives that are overlapped with predicted enhancers. True negatives (TN) are negatives that do not overlap any enhancer predictions. The remaining are false negatives (FN), which are positives that are not predicted as enhancers. Next, we calculated the below metrics for the predictions from each methods:
DHS is the fraction of enhancer predictions that are overlapped with DHS but not any TSSs.
TFBS is the fraction of enhancer predictions that are overlapped with distal TFBSs but not any TSSs.
Misclassification is the fraction of enhancer predictions that are overlapped with any TSSs.
Evaluating the Prediction Accuracy on Data of Validated Enhancers.
We also validated the predictions using experimentally validated regions; we applied all of the methods to predict the enhancer activity of tested regions in the eight test datasets, which contain validated enhancers, and negative regions (Fig. S1D): First, we ran all methods to generate scores for 2-kb sliding windows in the genome. Then, the score of each tested region is assigned as the score of the sliding window whose center is the closest to the center of the tested region. If the centers of two sliding windows are equally close to the center of one tested region, the maximum score is used.
The reason behind this procedure is that RFECS, DELTA, and CSIANN were designed to predict the enhancer activity of 2-kb sliding windows in the genome, and their current implementations were unable to calculate scores for predefined regions. The strategy of test PEDLA was different because it made predictions based on the chromatin profiles of 200-bp bins, which is much smaller than 2 kb. To address this issue, for each 2-kb sliding window, we used the maximum PEDLA score among scores of overlapping 200-bp bins as the score of the 2-kb window. Also, to ensure the prediction results from all methods are comparable, we chose to run REPTILE to predict enhancer activity of 2-kb sliding windows in the genome as well: REPTILE will first generate multiple enhancer confidence scores for each 2-kb sliding window based on the epigenomic signature of the whole region as well as that of the DMRs within the region, and then the highest is assigned as the final score for the window.
Then, the area under the precision-recall curve (AUPR) was used to measure the performance of each method in the test datasets. Precision is defined as the fraction of predictions that are real enhancers, that is, (True positives)/(True positives + False positives). Recall is defined as the percentage of real enhancers that are predicted as positive, that is, (True positives)/(True positives + False negatives). Precision-recall curve can be drawn by changing the score cutoff. AUPR is defined as the (area) integral between the curve and two axes. R package “flux” (0.3.0) was used to implement the calculation of AUPR.
Validating Enhancer Prediction with Distal TFBSs and Distal DHSs.
We overlapped the mESCs and H1 predictions with the distal DHSs and the distal transcription factors binding sites (TFBSs). We calculated the distance between the center of each prediction and the closest distal DHS (or the closest distal TFBSs). If the distance is no greater than 1 kb, we see it as an overlap. Similar analysis was done to measure the overlaps with TSSs. If the center of certain prediction is within 1 kb to any TSSs, it was counted as overlapping. Based on the overlap patterns, we divided the mESCs predictions into five categories: “TSS proximal” (overlap with TSSs), “DHS” (overlap with distal DHS only), “TFBS” (overlap with distal TFBS only), “TFBS+DHS” (overlap with both distal DHS and distal TFBS), and “Unknown” (none of the above). If a prediction is within 1 kb to any TSSs, it will be consider as “TSS proximal” regardless of its distance to DHSs or TFBSs. “TFBS,” “DHS,” and “TFBS+DHS” are considered as true positives, whereas “TSS proximal” is considered as false positive.
Validating Enhancer Prediction Using MERA Identified Regulatory Elements.
To validate the enhancer predictions using diverse source of evidence, we acquired the data of regulatory elements identified by a genome mutation screening approach, multiplexed editing regulatory assay (MERA) (38). Briefly, GFP was knocked in to a selected gene, and then CRISPR-Cas9 system was used to disrupt regions that are likely to have regulatory function on the selected gene. Next, the targeted regions of the guide RNA (gRNA) that significantly reduced the GFP signal were identified as regulatory elements.
We downloaded the data from previous publication (38), where MERA assay was conducted in mESCs on four genes, Tdgf1, Zfp4, Nanog, and Rpp25, separately. We used the same procedure as in the publication (38) to select gRNAs that were statistically significantly overexpressed in GFP-negative cells. Only the gRNAs that showed significance in all replicates were considered. Next, we merged the targeted regions of these gRNAs if they were within 100 bp, and we then filtered out the merged elements that were within 1 kb to any TSSs. Then, we overlapped the (top 35,000) mESCs enhancer predictions from each method with these distal merged elements. Last, we calculated the percentage of the distal merged elements that were within 500 bp to the center of any enhance predictions (Fig. S2C).
Evaluation of Genome-Wide Enhancer Predictions.
We evaluated the quality of genome-wide enhancer predictions by measuring the fraction of predictions that show evidence of distal open chromatin, how close the predictions are to nearest distal open chromatin regions (DHSs), and the percentage of predictions that are more likely to be (misclassified as) promoters. Before calculating these metrics, we selected the same number of predictions from each method to ensure a fair comparison. In human cell lines, the top 20,000 predictions were considered, which is similar to the strategy used in a recent study (34). In mESCs, the top 35,000 putative enhancers were selected. In E11.5 tissues, the top 10,000 were selected because generally fewer predictions were generated in these samples than in mESCs. In total, three metrics were calculated.
-
i)
First, we measured the fraction of predictions whose centers were within 1 kb to distal DHSs (1 kb from any TSSs) and were at least 1 kb away from any TSS. We called this metric as “validation rate.”
-
ii)
In addition to the “validation rate,” we calculate a metric “misclassification rate” as the fraction of predictions that are within 1 kb to TSS. These predictions are likely to be promoters and thus are misclassified.
-
iii)
Furthermore, we measured the average distance between the centers of predictions and distal DHSs if the distance is no greater than 1 kb. We intend to use this metric to measure the resolution of predictions and the ability of the method to accurately locate enhancer regions with little influence by false positives. Therefore, the predictions whose centers are 1 kb away from DHS were not included in the calculation as they are considered as false positives.
Transgenic Mouse Experiments.
Enhancer names (mm and hs numbers) are the unique names used in the VISTA Enhancer Browser (https://enhancer.lbl.gov/) (31). Enhancer sequences were amplified from human (hs numbers) or mouse (mm numbers) genomic DNA and cloned into an hsp68-lacZ expression vector (66). Genome coordinates and primer sequences for all elements are listed in Table S1. Transgenic mouse assays were performed as previously described (66, 67) in Mus musculus FVB strain mice. All animal work was reviewed and approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
We then overlapped these newly validated VISTA enhancers with REPTILE predictions in E11.5 tissues. The murine VISTA elements (mm9) were lifted to mm10 using “minMatch=0.95” using liftOver, whereas the human ones (hg19) were lifted to mm10 using “minMatch=0.10.” The resulting mm10 coordinates were intersected with the REPTILE predictions in E11.5 tissues and elements that were overlapped by at least 1 bp were reported.
TF–Binding-Site Motif Enrichment Analysis on Predicted Enhancers of H1 and H1 Derived Cell Lines.
To test whether the higher resolution of REPTILE enhancers improves TF–binding-site motif discovery, we conducted motif analysis on the REPTILE enhancer predictions in each human cell lineage. Homer (version 4.8.3) (68) was used to identify the TF–binding-site motifs that were enriched in predicted enhancers in each human cell lineage. For each cell line, predicted enhancers were used as foreground (target) sequencers and Homer automatically selected the background sequencers (i.e., the default option). “mm10” was used as the reference genome, and we included the “-nomotif” option such that Homer only considered known motifs. In the next step, we selected motifs with q value less than or equal to 0.05 as significantly enriched motifs. For each motif and predicted enhancers in each cell type, we calculated the degree of enrichment, which was defined as follows:
where “Target Sequences” refer to predicted enhancers and “Background Sequencers” are background regions automatically selected by Homer.
We asked whether this analysis could recapture motifs of the TFs known to function in that cell type. We obtained the list of known transcription regulators for each human cell line from Xie et al. (47). The mapping between TF names in the list and the motif names in Homer is available in Table S2. We also conducted this analysis on the top 20,000 enhancer predictions from REPTILE and other existing methods in each human cell lineage. The results are shown in Fig. 4.
Note that the lengths of enhancer predictions are different. As described in previous section, REPTILE enhancers have various lengths—they have either the size of a DMR or 2 kb (the length of sliding windows), depending how each of them was called as enhancer. Enhancer predictions from RFECS, DELTA, and CSIANN are 2-kb regions centered on the predicted enhancer centers. PEDLA made prediction on 200-bp bins such that the size of PEDLA enhancers is 200 bp.
Enhancer Prediction By Single Data Type.
To understand how informative single data type is, we used single epigenetic mark or only the open chromatin signature to predict the enhancer activity of regions in the test datasets (Fig. 5A). We first calculated the enrichment score of an active mark (including open chromatin) or the depletion score of a repressive mark (mCG or H3K27me3) in tested regions. Then, we rank regions by their score and use AUPR to measure the how well the ranking distinguish active enhancers from negative regions. One common combination, DHS and H3K27ac, was also tested. The details of each approach are as follows:
-
i)
DHS: The score of a tested region is the highest score of DHSs that overlap with it. If no overlapping DHS is found, its score is set to be negative infinity. The score of DHS corresponds to the signal value in the narrow peak format (https://genome.ucsc.edu/FAQ/FAQformat.html#format12).
-
ii)
DHS+H3K27ac: The score of a tested region is the highest H3K27ac enrichment score in the DHSs that overlap with the tested region. If no DHS overlap is found, its score is set to be negative infinity. H3K27ac signal is the log2 RPM fold enrichment relative to control.
-
iii)
DNase-seq signal: RPM of DNase-seq data for the tested region. The mean of values from replicates was used.
-
iv)
H3K27ac: Average H3K27ac log2 RPM fold enrichment relative to control.
-
v)
mCG: (–1) × methylation level of tested region.
-
vi)
DHS+mCG: The score of a tested region is the largest negative CG methylation level in the DHSs that overlap with the tested region. If no DHS overlap is found, its score is set to be minus infinity.
-
vii)
H3K4me1: H3K4me1 log2 RPM fold enrichment relative to control.
-
viii)
H3K4me2: H3K4me2 log2 RPM fold enrichment relative to control.
-
ix)
H3K4me3: H3K4me3 log2 RPM fold enrichment relative to control.
-
x)
H3K27me3: (-1) × H3K27me3 log2 RPM fold enrichment relative to control.
-
xi)
H3K9ac: H3K9ac log2 RPM fold enrichment relative to control.
Calling Enhancers in Human Heart Left Ventricle.
We specifically trained a human enhancer model for each method to generate enhancer prediction for human heart left ventricle because not all six previously used histone modifications are available in this tissue. For PEDLA, DELTA, RFECS, and CSIANN, this (re)training is almost identical to the previous training procedure, but we limited the histone modifications to H3K4me1, H3K4me3, H3K27ac, and H3K27me3, the histone marks that are available in both H1 and left ventricle. PEDLA also incorporated evolutionary conservation. The new enhancer models were retrained on the data of H1. REPTILE was trained on mCG data and histone modification data of H1, whereas left ventricle and H1-derived cells were used as reference. The DMR input for REPTILE was obtained by comparing the methylomes of left ventricle, H1, and H1-derived cells. In the prediction step, REPTILE used H1 and H1-derived cells as reference. Last, we applied these methods to generate enhancer predictions for left ventricle, and the top 50,000 putative enhancers from each method were selected for later analyses.
Enrichment of Disease-Associated Genetic Variants in Putative Enhancers.
To test for the enrichment of disease-associated SNPs in putative enhancers, we first downloaded the data of 5,654 noncoding GWAS SNPs from Maurano et al. (7). The 5,654 SNPs were originally grouped into 15 categories based on the associated traits/diseases. We applied one-tail hypergeometric test to test the enrichment of SNPs from each category in the putative enhancers of left ventricle.
Specifically, for category , the total number of SNPs in is denoted as and the total number of SNPs is . Given a list of putative enhancers, the observed number of overlapped SNPs in category is and total observed number of overlapped SNPs is . The P value for SNPs in is calculated as follows:
where is a random variable representing the number of SNPs that are in category and overlapped with putative enhancers. The fold enrichment of SNPs from category in putative enhancers is calculated as follows: .
Using the statistical test described above, for SNPs in each category, we tested for their enrichment in putative enhancers. We then used Benjamini–Hochberg approach to adjust P values for multiple testing. P value cutoff given 1% false-discovery rate (FDR) was used to call significant enrichment. This procedure was conducted separately for the putative enhancers from each method.
Linking REPTILE Enhancers to Target Genes in Left Ventricle.
To identify the target genes of REPTILE enhancers in left ventricle, we downloaded expression quantitative trait loci (eQTLs) data of left ventricle from the Genotype–Tissue Expression (GTEx) Project (version: v6p; file: Heart_Left_Ventricle_Analysis.v6p.signif_snpgene_pairs.txt.gz from GTEx_Analysis_v6p_eQTL.tar). The eQTLs that are within 2 kb to any TSSs were filtered out. Then, we overlapped the REPTILE enhancers in left ventricle with the remaining eQTLs and assigned each putative enhancer to the gene linked to overlapping eQTL (if any). If multiple eQTLs are within one putative enhancer, the putative enhancer is assigned to all of the genes linked to these eQTLs.
Next, based on the enhancer-gene assignment, we separated the genes that are linked to eQTLs into two groups. The first group consists of genes that are linked to at least one REPTILE enhancers, whereas the second group contains genes that are only linked to eQTLs outside of REPTILE enhancers. We then compared the expression levels of genes from these two groups. The gene expression data of left ventricle (from donor STL003) were obtained from Schultz et al. (19), and the expression level is represented in fragments per kilobase of transcript per million mapped reads (FPKM). Two-tailed Mann–Whitney test was conducted to test for the significance of difference in the median expression levels of two groups of genes.
Testing the Robustness of REPTILE Given Various Input Data.
To test how sensitive REPTILE’s performance is to various inputs, we ran REPTILE without DMRs (REPTILE w/o DMR), without reference epigenome (REPTILE w/o Ref), and with shuffled DMRs (REPTILE w/ shuf DMR), respectively. Because enhancer validation data are available for mouse samples, this test was done using mouse data. REPTILE w/o DMR performed prediction solely based on the epigenomic signature of query regions (e.g., 2-kb sliding windows across the genome). REPTILE w/o Ref only uses the data of target sample (where prediction is generated) and does not calculate intensity deviation to describe tissue specificity of epigenetic marks. Its enhancer model only uses the intensity of seven marks as features. REPTILE w/ shuf DMR takes shuffled DMRs as input, but its enhancer model is learned using unshuffled DMRs. We obtained the shuffled DMRs by shuffling the coordinates of DMRs within the genome while maintaining their lengths, which was done by using shuffleBed in bedtools (58).
REPTILE includes data of reference samples to capture the information in cell/tissue-specific epigenomic variation. However, it is unclear how the choice of reference would affect the prediction performance. To address this question, we implemented and benchmarked a different strategy of choosing reference. The new setup is called REPTILE alt Ref. In the new strategy, REPTILE always used mESCs, E11.5 craniofacial, and E11.5 liver as reference samples for generating prediction. We applied this new setup to predict enhancers in E11.5 forebrain, midbrain, hindbrain, heart, limb, and neural tube. For each target sample, the enhancer model was trained on data of mESCs using target sample, E11.5 craniofacial, and E11.5 liver as reference, which corresponds to a scenario in which only the data of the target sample and reference samples are available. The analysis of the prediction results is identical to the evaluation of the results of the original setup.
Discussion
In this study, we describe an algorithm, REPTILE, which is able to predict active enhancers by integrating tissue-specific histone modification data and base-resolution mCG data. We found that the overall accuracy and resolution of REPTILE predictions exceeds other methods, especially when applied to cell/tissue types different from the training data. Further benchmarking revealed that REPTILE’s performance is robust to different DMR inputs and reference choices (Figs. S7 and S8; SI Notes, Performance of REPTILE Is Robust to Choice of Reference and Suboptimal Differentially Methylated Region Calling). In summary, by incorporating DNA methylation data produced by whole-genome bisulfite sequencing and using information of cell/tissue type-specific variation of epigenetic marks, REPTILE greatly improves upon current methods for annotation of enhancers across a variety of cell and tissue types (see also Figs. S7 and S9; SI Notes, Epigenomic Variation Information Improves Enhancer Prediction Resolution and Accuracy).
Fig. S7.
REPTILE’s performance is robust to various subsets of input data. (A) Performance of REPTILE models with various inputs and the four published methods in all test datasets. REPTILE w/o DMR has no DMR input. REPTILE w/o Ref makes predictions only based on the epigenomic data of target sample and thus does not include the intensity deviation features. REPTILE w/ shuf DMRs takes shuffled DMRs as input and makes prediction based on an enhancer model pretrained in mESCs with DMRs before shuffling. For all methods, prediction results were generated by enhancer model trained in mouse embryonic stem cells. Model performance is measured by the area under precision-recall curve (AUPR). For each test dataset, the best performance(s) are highlighted in red. The baselines are the AUPRs from random guessing for these datasets and are shown in gray. Note that the AUPRs in different datasets cannot be compared because the fractions of validated enhancers are different. See Fig. S1D for the fraction in each dataset. (B and C) Bar charts showing the average distance from the center of mESCs enhancer predictions to nearest distal DHSs (B) and the fraction of predictions whose centers are within 1 kb to distal DHSs (C). (D) Average distance between the centers of predictions and the nearest distal DHS. Predictions whose centers are beyond 1 kb away from the nearest distal DHS were considered as lacking support from open chromatin data and were not included in the calculation. (E) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS. The results shown (D and E) are based on the enhancer predictions in E11.5 midbrain (red), hindbrain (green), limb (orange), neural tube (blue), and craniofacial (dark gray), where DNase-seq data were available. DHS, DNase hypersensitivity sites. See SI Methods for details.
Fig. S8.
REPTILE’s performance is robust to the choice of reference. (A) Performance of REPTILE models with different references and the four published methods in all test datasets. REPTILE alt Ref uses mESCs, E11.5 craniofacial, and E11.5 liver as the reference in generating enhancer predictions. REPTILE w/o Ref does not use any reference. Model performance is measured by the area under precision-recall curve (AUPR). The baselines are the AUPRs from random guessing for these datasets and are shown in gray. Note that the AUPRs in different datasets cannot be compared because the fractions of validated enhancers are different. (B) Average distance between the centers of predictions and the nearest distal DHS. Predictions whose centers are beyond 1 kb away from the nearest distal DHS were considered as lacking support from open chromatin data and were not included in the calculation. (C) Average percentage of predictions whose centers are within 1 kb to the closest distal DHS. The results shown (B and C) are based on the enhancer predictions in E11.5 midbrain (red), hindbrain (green), limb (orange), and neural tube (blue). DHS, DNase hypersensitivity sites. See SI Methods for details.
Fig. S9.
Importance of features in the REPTILE enhancer model trained on mESCs and H1. (A and B) In the mouse REPTILE enhancer model, importance of features in the random forest classifiers for differentially methylated region (DMR) (A) and query regions (B). (C and D) In the human REPTILE enhancer model, importance of features in the random forest classifiers for (DMR) (C) and query regions (D). The importance is measured by the average decrease of Gini impurity index (meanDecreaseGini) from the “randomForest” R package (69).
Although some methods showed better performance in a few tests, REPTILE’s performance was superior in most tests. Although we tried to evaluate the prediction accuracy of all methods in an unbiased manner, we should point out that these benchmarks might be further improved in several ways. First, the validated regions in mESCs were originally selected based on RFECS predictions, which introduces a potential bias. However, if this bias alters the performance of prediction algorithms, it is likely to inflate the performance of RFECS more than REPTILE. Second, the number of validated enhancer elements is currently limited, although this issue may be resolved in the near future, as more elements will be tested for in vivo function. Third, the negative datasets obtained from the VISTA enhancer database were mostly “putative” enhancer elements from previous studies and therefore may be very similar to true enhancers in many aspects, such as the degree of evolutionary conservation (43). As a result, the prediction accuracy on VISTA enhancer dataset is likely to be lower than the accuracy of whole-genome prediction because many of the “negatives” in the VISTA database actually have some enhancer-like characteristics, which likely makes them harder to differentiate from true positives. Although improvements are possible (such as benchmarking of methods on genomic regions tested in high-throughput enhance assay and incorporating more sophisticated features in the REPTILE model), our results show that REPTILE outperforms existing enhancer prediction methods, especially for samples where training data are unavailable.
As epigenomic information of a larger number of cell/tissue types continues to be comprehensively profiled by the effort of Encyclopedia of DNA Elements (ENCODE) (32, 44, 45), Roadmap Epigenomics Mapping Consortium (REMC) (46), International Human Epigenome Consortium (IHEC), and other consortia, we envision that REPTILE will be a valuable tool to generate accurate enhancer annotations for these datasets, facilitating better regulatory DNA predictions and fueling biological insights.
SI Notes
Performance of REPTILE Is Robust to Choice of Reference and Suboptimal Differentially Methylated Region Calling.
Compared with other methods, REPTILE uses differentially methylated regions (DMRs) to improve prediction resolution. However, there is no consensus DMR definition, and different algorithms may identify different regions as DMRs (51). We test the robustness of REPTILE to DMR input using the mouse data because experimentally validated enhancers are available in mouse samples (Fig. S1D). First, we shuffled the genomic location of DMRs and used these shuffled DMRs in the prediction step, whereas the enhancer model was learned using unshuffled DMRs (REPTILE w/ shuf DMR) (SI Methods). We found that the prediction accuracy remained superior to existing methods in four out of the eight test datasets (Fig. S7A). As expected, without meaningful DMRs, this method has worse prediction resolution than REPTILE with complete input set (Fig. S7 B and D). We also found that fewer predictions were near distal DHS compared with REPTILE with full inputs (Fig. S7 C and E). However, the REPTILE w/ shuf DMR predictions remain comparable with existing methods, indicating that REPTILE’s performance is robust to suboptimal DMR input given a correctly pretrained enhancer model. Inspired by these results, we provide pretrained enhancer models along with the software to facilitate the use of REPTILE. This robust performance is likely because REPTILE generates good enhancer prediction solely based on epigenomic signatures of the query regions (REPTILE w/o DMR) and the DMR input simply improves upon an already relatively accurate prediction.
Next, we asked whether the performance of REPTILE is robust to the choice of reference samples. To test this, we ran REPTILE with a different strategy of choosing the reference (Fig. S8). Instead of using all nontarget samples as the reference, we only used mESCs, E11.5 craniofacial, and E11.5 liver as reference in the prediction step and we called this setup “REPTILE alt Ref.” We then evaluated this strategy using mouse data. Specifically, we ran REPTILE to predict in six E11.5 tissues the enhancer activity of elements from VISTA enhancer browser (Fig. S8A). For each target sample, we only used data of the target sample, mESCs, E11.5 craniofacial, and E11.5 liver. We trained an enhancer model for each target sample on data of mESCs with the target sample, E11.5 craniofacial, and E11.5 liver as reference (SI Methods). In the prediction step, mESCs, E11.5 craniofacial, and E11.5 liver were used as the reference (SI Methods). For each target sample, DMRs were called across methylomes of the four samples. We found that even if we changed the reference, REPTILE (REPTILE alt Ref) showed performance as good as the previous setup (REPTILE; Fig. S8). We further used DHS data to validate the enhancer predictions from these two setups and they showed similar prediction accuracy and resolution. Collectively, these results demonstrate that REPTILE’s performance is robust to different reference choices.
Epigenomic Variation Information Improves Enhancer Prediction Resolution and Accuracy.
Use of the random forest algorithm allowed us to identify key epigenetic features in the enhancer prediction model (Fig. S9 and SI Methods). In the mouse enhancer model, we found that mCG was the most informative feature for predicting enhancer activities of DMRs, whereas H3K27ac was the most predictive mark for query regions. This is likely due to the fact that hypomethylation tends to be restricted within DMRs, and thus becomes less predictive in larger query regions where hypomethylation pattern is washed out. In the human enhancer model trained on H1 cells, H3K4me2 is the most informative feature in both classifiers. We also found several other high-ranking features including intensity deviation features, such as H3K4me2-dev and H3K27me3-dev, indicating the necessity to capture the tissue specificity of epigenetic marks for enhancer prediction (Fig. S9). When the intensity deviation features were removed (REPTILE w/o Ref), REPTILE prediction accuracy decreased, even though the results remained comparable or superior to other methods (Fig. S7).
Next, to understand the contribution of DMRs, we tested REPTILE without DMR input (REPTILE w/o DMR). We found that the midpoint genomic locations of these predictions were ∼30–40 bp further from the closest distal DHS compared with the predictions made by REPTILE with DMR input (Fig. S7 B and D). Also, the percentage of DHS-supported predictions slightly decreases (Fig. S7 C and E). However, in the enhancer validation datasets, the prediction accuracy without DMR input remains as good as the REPTILE method with all inputs (Fig. S7A). These results indicate that the inclusion of tissue specificity information improves prediction accuracy, whereas DMRs are necessary for more refined prediction of enhancer locations.
Methods
Overview of Data Acquisition.
To systematically benchmark REPTILE, we collected epigenomic data of various human and mouse cells and tissues. These epigenetic marks included base-resolution DNA methylation data (WGBS) and six histone modifications: H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K27me3, and H3K9ac (Fig. S1 B and C, and Tables S3 and S4). We downloaded data of five human cell lines from Xie et al. (47): H1 human embryonic stem cells (H1), mesendoderm (Mes), mesenchymal stem cells (MSCs), neural progenitor cells (NPCs), and trophoblast-like cells (TRO). Human data also contain WGBS of heart left ventricle from Schultz et al. (19) and histone modification data of the same tissue from Leung et al. (48). In addition, we included data of nine mouse samples: mESCs and eight mouse tissues from E11.5 embryo (SI Methods).
Next, to train the computational enhancer prediction methods, we obtained EP300 binding data from mouse and human ESCs (SI Methods). It has been shown that EP300 binding is a key feature of a fraction of active enhancers but computational approaches are able to learn the chromatin signatures of these enhancers and predicts other active enhancers without EP300 binding (10, 11). In this regard, we used EP300 binding sites as putative active enhancers in the training datasets.
To validate the enhancer predictions from these methods, we downloaded in vivo enhancer validation data in E11.5 embryonic mouse tissues from the VISTA enhancer browser (31) as well as high-throughput report assay data in mESCs from Yue et al. (32). We also included in vivo validated embryonic heart enhancers from Narlikar et al. (49). In total, eight test datasets were used (Fig. S1D). In addition, in all five human cell lines, mESCs, and five E11.5 mouse tissues, we downloaded publicly available DNase-seq data to validate enhancer predictions, assuming the actual location of enhancers to coincide with distal DHSs in the corresponding cell/tissue types. See also SI Methods for more details.
REPTILE.
REPTILE is an algorithm that generates high-resolution prediction of active enhancers genome-wide by integrating mCG and histone modification data. REPTILE uses the DMRs that are identified across all samples as high-resolution enhancer candidates, and it is able to capture local epigenomic signatures that may otherwise be washed out in the signal of larger region. In addition, it takes into account the tissue specificity of enhancers as features to further improve its performance; REPTILE predicts enhancers based on epigenomic data of not only the target sample (where enhancer predictions are generated) but also additional reference samples to exploit the useful information in variation between cells and tissues.
The overview of REPTILE workflow is shown in Fig. 1C, which includes four major steps:
-
i)
DMR calling: DMRs are identified by comparing the DNA methylomes of input samples. We first called differentially methylated sites (DMSs). Next, we merged DMSs into blocks if they both show similar sample-specific methylation patterns and are within 250 bp. These two steps were performed as previously described (19) (see also SI Methods for details). We then filtered out the blocks that contain only one DMS. The remaining blocks were then extended 150 bp from each side to include the two regions covered by first upstream and first downstream nucleosomes, respectively. These extended blocks are defined as DMRs, which were used in later steps.
-
ii)
Data integration: Then, REPTILE integrates various types of input data to obtain the epigenomic signatures of DMRs and query regions, in preparing for the next two steps: enhancer model training and prediction generation. Specifically, each DMR or query region is represented as a feature vector and each variable in the vector corresponds to the intensity or intensity deviation of one epigenetic mark (Fig. 1D). In this study, the intensity of each histone modification is defined as the logtwofold change in reads per million mapped reads (RPM) relative to control and the intensity of mCG is the CG methylation level. Note that different definitions of intensity can also be used, such as the RPM with subtraction of control or simply RPM of ChIP-seq itself. It makes REPTILE more flexible and allows various way of normalization to be imposed on the input data. Intensity deviation of an epigenetic mark is defined as the intensity in target sample subtracted by its mean intensity in reference samples (i.e., reference epigenome), and this type of feature quantifies the tissue specificity of the epigenetic mark (Fig. S1A). Because the data of reference samples is only used to calculate the mean signal value, REPTILE does not require that all epigenetic marks are available in all reference samples, that is, missing data are allowed. However, the target sample, where enhancer predictions are generated, must contain the data of all of the epigenetic marks. In this study, we used seven epigenetic marks (DNA methylation and six histone modifications), and thus the complete REPTILE model contains in total 14 features (two features, intensity and intensity deviation, for each mark; Fig. S9).
The input data vary according to the next step. (i) The training step requires data of known/putative enhancers (such as EP300 binding sites) and known negative regions as well as the DMR list and the epigenomic data of target sample and reference samples. (ii) Prediction generation takes the enhancer model obtained from the training step, together with the DMRs, the epigenomic data, as input. It also requires query regions. The query regions can be 2-kb sliding windows with step size 100 bp across the genome for generating genome-wide enhancer predictions (see below). They can also be predefined regions, such as conserved elements in the genome, where their enhancer activity is of interest. More details about REPTILE input preparation are available at https://github.com/yupenghe/REPTILE/.
-
iii)
Model training: In the next step, REPTILE enhancer model are trained by learning the epigenomic signatures of query regions, including known enhancers and negatives, as well as the DMRs within them. Specifically, one random forest classifier is trained to learn the epigenomic profiles of the labeled query regions, whereas another random forest classifier is trained to learn epigenomic features in the DMRs that overlap with the query regions. Both classifiers use same 14 features, but the values of these features are calculated differently. The classifier for query regions computes feature values based on the epigenomic data of whole query regions, whereas the classifier for DMRs is trained and applied on the data of DMRs.
The random forest classifier for query regions can be trained on data of known active enhancers and negative regions. However, the classifier for DMRs cannot be trained in such a straightforward way due to the lack of labels for DMRs. To circumvent this, we label all DMRs that are within known enhancers as active, and we label the ones that are within negative regions as inactive. Then, we use these labels to train the random forest classifier for DMRs in a similar fashion as in the training of classifier for query regions. The rationale behind this is that (we assume that) DMRs within negative regions are inactive and part of the DMRs within active enhancers can be inactive. In the training dataset where negative regions greatly outnumber active enhancers, we expect that there are many more DMRs labeled as inactive than active. Therefore, although the inactive DMRs within active enhancers might be incorrectly labeled as active, they only compose a small portion of DMRs. In this paper, the ratio of negatives to positives in the training datasets is at least 7:1 (SI Methods). The random forest model can be successfully trained on such data with a small fraction of instances incorrect labeled, which has been demonstrated by the better performance of REPTILE than existing methods. The implementation of random forest model is built on the R (version 3.2.1) package “randomForest” (version 4.6.12) with parameter “ntree=2000, nodesize=1.”
-
iv)
Prediction generation: Last, we apply the enhancer model learned in the training step to generate enhancer predictions. Specifically, for every query region or DMR, the corresponding random forest classifier will generate an enhancer confidence score, which is defined as the fraction of decision trees in the random forest model that vote in favor of the active enhancer class.
Given a set of regions of interest, REPTILE is able to predict their enhancer activity. First, REPTILE generates one enhancer confidence score based on the epigenomic signature of certain query region and also multiple scores based on the data of DMRs within it. Then, the maximum is assigned as the final score for this region. In this design, data of DMRs are used to complement the prediction based on query regions. We found that, with correct enhancer model, even if the DMRs were not correctly identified, the prediction performance did not decrease much (see REPTILE w/ shuf DMR in Fig. S7). It is because the incorrect DMRs are not likely to show enhancer-like epigenomic signatures and low enhancer confidence scores will be assigned to them. In this case, the prediction will be dominated by the enhancer confidence score calculated based on the data of whole query regions (see REPTILE w/o DMR in Fig. S7).
REPTILE can also generate enhancer predictions across the genome. In this study, we used REPTILE to first calculate enhancer scores for all DMRs in the genome as well as all 2-kb sliding windows with 100-bp step size across the whole genome. The empirical choices of window size of 2 kb and step size of 100 bp are based on the benchmark results from previous study (35, 50). Then, DMRs with score higher than a given cutoff (0.5 is used in this study) are predicted to be enhancers (termed “enhancer-like DMRs”). To generate nonoverlapping enhancer predictions, overlapping enhancer-like DMRs are merged into single prediction and its score is the highest score of all enhancer-like DMRs that are merged to form this prediction. Next, to capture the enhancers with no detectable mCG variation, REPTILE calls peaks of the enhancer scores across the sliding windows that pass the given score cutoff using the following procedure: (i) All sliding windows that pass the cutoff are labeled as enhancer candidates. Candidates that are within 1 kb to each other are grouped into clusters. (ii) For each cluster, the candidate with maximum score is set as a peak. If multiple candidates share the highest score, we randomly select one of them as the peak. (iii) For each cluster, the peak and all candidates that are within 1 kb of the peak are excluded from the candidate list. (iv) Steps 2 and 3 are repeated until the candidate list in each cluster is empty.
After this process, all sliding windows that have score greater than threshold are either peaks or within 1 kb to peaks. The rationale behind this is that the sliding windows adjacent to a peak are part of the peak. Last, the final predictions are the union of the enhancer-like DMRs and the sliding windows that are called as peaks but have no overlap with any enhancer-like DMRs. Similar to the prediction on given regions, this procedure is robust to incorrect DMRs because the enhancers that can be identified using the epigenomic mark of sliding windows will still be called.
Software Availability.
The REPTILE software is published under the BSD 2-Clause License. It was written in R and Python. The R code was submitted as an independent R package, called “REPTILE,” in the Comprehensive R Archive Network (CRAN). The source code, pretrained enhancer models, use, and further details of the complete pipeline are available in https://github.com/yupenghe/REPTILE.
Acknowledgments
We thank Dr. John A. Stamatoyannopoulos for generously sharing the DNase-seq data of five E11.5 mouse tissues. We specifically thank Dr. Nisha Rajagopal for kindly helping with the data from MERA. We thank Drs. Shao-shan Carol Huang, Chongyuan Luo, and Manoj Hariharan for their critical comments. Y.H. is supported by the H. A. and Mary K. Chapman Charitable Trust. D.U.G. is supported by the A. P. Giannini Foundation and NIH Institutional Research and Academic Career Development Award K12 GM068524. Transgenic mouse work was conducted at the E. O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. J.R.E. is an Investigator of the Howard Hughes Medical Institute and is supported by grants from the Gordon and Betty Moore Foundation (GBMF3034), the NIH (R01 MH094670 and U01 MH105985), and the California Institute for Regenerative Medicine (GC1R-06673-B). This work was funded by NIH Grant U54 HG006997.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618353114/-/DCSupplemental.
References
- 1.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 2.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132(4):797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470(7333):264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleinjan DA, van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13(1):238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tak YG, Farnham PJ. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN β enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1(2):277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 10.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 12.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleftogiannis D, Kalnis P, Bajic VB. Progress and challenges in bioinformatics approaches for enhancer identification. Brief Bioinform. 2015;17(6):967–979. doi: 10.1093/bib/bbv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 17.Smith ZD, Meissner A. DNA methylation: Roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 18.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varley KE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Ecker JR. Non-CG methylation in the human genome. Annu Rev Genomics Hum Genet. 2015;16:55–77. doi: 10.1146/annurev-genom-090413-025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2012;484:550. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 24.Sayeed SK, Zhao J, Sathyanarayana BK, Golla JP, Vinson C. C/EBPβ (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim Biophys Acta. 2015;1849(6):583–589. doi: 10.1016/j.bbagrm.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165(5):1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens DC, Poon GMK. Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res. 2016;44(18):8671–8681. doi: 10.1093/nar/gkw528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu T, et al. Base-resolution methylation patterns accurately predict transcription factor bindings in vivo. Nucleic Acids Res. 2015;43(5):2757–2766. doi: 10.1093/nar/gkv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang W, Oliver VF, Merbs SL, Zhu H, Qian J. Prediction of promoters and enhancers using multiple DNA methylation-associated features. BMC Genomics. 2015;16(Suppl 7):S11. doi: 10.1186/1471-2164-16-S7-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45(10):1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue F, et al. Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 34.Liu F, Li H, Ren C, Bo X, Shu W. PEDLA: Predicting enhancers with a deep learning-based algorithmic framework. Sci Rep. 2016;6:28517. doi: 10.1038/srep28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajagopal N, et al. RFECS: A random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9(3):e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Qu W, Shan G, Zhang C. DELTA: A distal enhancer locating tool based on AdaBoost algorithm and shape features of chromatin modifications. PLoS One. 2015;10(6):e0130622. doi: 10.1371/journal.pone.0130622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010;26(13):1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34(2):167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao L, Shen H, Laird PW, Farnham PJ, Berman BP. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:105. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhie SK, et al. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin. 2016;9:50. doi: 10.1186/s13072-016-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erwin GD, et al. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol. 2014;10(6):e1003677. doi: 10.1371/journal.pcbi.1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birney E, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kundaje A, et al. Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung D, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518(7539):350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narlikar L, et al. Genome-wide discovery of human heart enhancers. Genome Res. 2010;20(3):381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Won K-J, Chepelev I, Ren B, Wang W. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, et al. Statistical methods for detecting differentially methylated loci and regions. Front Genet. 2014;5:324. doi: 10.3389/fgene.2014.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma H, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511(7508):177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz MD, Schmitz RJ, Ecker JR. “Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28(12):583–585. doi: 10.1016/j.tig.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins W, Tygert M, Ward R. Computing the confidence levels for a root-mean-square test of goodness-of-fit. Appl Math Comput. 2011;217:9072–9084. [Google Scholar]
- 56.Bancroft T, Du C, Nettleton D. Estimation of false discovery rate using sequential permutation p-values. Biometrics. 2013;69(1):1–7. doi: 10.1111/j.1541-0420.2012.01825.x. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ong C-T, Corces VG. CTCF: An architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neph S, et al. BEDOPS: High-performance genomic feature operations. Bioinformatics. 2012;28(14):1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freund Y, Schapire RE. European Conference on Computational Learning Theory. Springer; Berlin: 1995. A decision-theoretic generalization of on-line learning and an application to boosting; pp. 23–37. [Google Scholar]
- 64.Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9(5):473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 67.Kothary R, et al. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105(4):707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]