Significance
Nitrate is an essential nutrient and a critical signal for plant growth, development, and stress responses. Nitrate signaling underlies a myriad of physiological, morphological, and developmental processes. Here we report that interacting teosinte branched1/cycloidea/proliferating cell factor and NIN-like protein transcription factors constitute a molecular link between nitrate signaling and the control of the cell-cycle progression gene CYCB1;1 and root meristem growth. Our findings shed light on the regulatory mechanisms underlying an important plant adaptive process for coping with and surviving environmental challenges.
Keywords: TCP, NIN-like protein, nitrate signaling, cell cycle, root growth
Abstract
Plants have evolved adaptive strategies that involve transcriptional networks to cope with and survive environmental challenges. Key transcriptional regulators that mediate responses to environmental fluctuations in nitrate have been identified; however, little is known about how these regulators interact to orchestrate nitrogen (N) responses and cell-cycle regulation. Here we report that teosinte branched1/cycloidea/proliferating cell factor1-20 (TCP20) and NIN-like protein (NLP) transcription factors NLP6 and NLP7, which act as activators of nitrate assimilatory genes, bind to adjacent sites in the upstream promoter region of the nitrate reductase gene, NIA1, and physically interact under continuous nitrate and N-starvation conditions. Regions of these proteins necessary for these interactions were found to include the type I/II Phox and Bem1p (PB1) domains of NLP6&7, a protein-interaction module conserved in animals for nutrient signaling, and the histidine- and glutamine-rich domain of TCP20, which is conserved across plant species. Under N starvation, TCP20-NLP6&7 heterodimers accumulate in the nucleus, and this coincides with TCP20 and NLP6&7-dependent up-regulation of nitrate assimilation and signaling genes and down-regulation of the G2/M cell-cycle marker gene, CYCB1;1. TCP20 and NLP6&7 also support root meristem growth under N starvation. These findings provide insights into how plants coordinate responses to nitrate availability, linking nitrate assimilation and signaling with cell-cycle progression.
Nitrate is the main form of inorganic nitrogen (N) in aerobic soils and is usually the most growth-limiting plant nutrient. Nitrate is also a potent signal that regulates plant metabolism, growth, and development. As sessile organisms, plants have evolved an elaborate regulatory network in response to nitrate, which can fluctuate spatiotemporally in soil solution by up to four orders of magnitude (1–3). In Arabidopsis, nitrate induces the primary nitrate response (PNR) in roots and shoots, where rapid, broad-ranged modulation of gene expression affects over 1,000 genes (4, 5). Using a nitrate reductase-null mutant, our previous work further showed that 595 genes responded to nitrate alone, independent of nitrate reduction. Those genes are most overrepresented in the categories of energy and metabolism, including glycolysis and gluconeogenesis, amino acid metabolism, nitrogen and sulfur utilization, and transport facilitation (6). Furthermore, the PNR is accompanied by adaptive processes in nitrate transport activity and remobilization and modulations of root growth (2).
Underlying N-regulated, adaptive responses in metabolism and development are transcription factors (TFs), which play crucial roles in regulating nitrate-responsive genes, particularly the sentinel genes such as NPF6.3 (CHL1/NRT1.1), NRT2.1, NIA1, NIA2, or NiR (2, 7). Many TFs, including ANR1, NLP6/7, LBD37/38/39, SPL9, NAC4, bZIP1, TGA1/4, TCP20, and NRG2, have been thus far identified in mediating nitrate responses (8–17). However, the transcriptional mechanisms that coordinate the regulation of nitrate assimilation and signaling and of plant growth remain enigmatic (7, 18). Among the TFs, direct interactions of NLP6/7, TGA1, bZIP1, and TCP20 with target gene promoters have been verified. No protein–protein interactions involved in nitrate signaling have yet been reported.
TCP20 and NLP6&7 belong to two ancient gene families, the protein sequences of which contain multiple, deeply conserved motifs in plants (19, 20). NLP6&7 proteins are expressed in almost all organs (21–23). NLP7 is an important regulator of PNR (10, 23). Nitrate does not induce NLP7 mRNA but instead induces a rapid accumulation of NLP7 in the nucleus by nuclear retention, leading to induction of nitrate-regulated genes (10). Transcriptomic analysis revealed that NLP7 binds to and regulates 91 nitrate-regulated genes (10). In another study, the synthetic transcription activator, NLP6-VP16, was found to promote the expressions of nitrate-inducible genes; and the synthetic transcription repressor, NLP6-SUPRD, was found to suppress the expressions of nitrate-inducible genes (9). However, no phenotypes have been described for NLP6 single mutants so its regulatory roles and functional relationship with NLP7 have not been fully elucidated, and no nlp6nlp7 double mutants have been reported. Interestingly, NLPs carry PB1 domains at their C termini, which are protein–protein interaction domains conserved in animals, fungi, amoebas, and plants and which are involved in responses to nutrients, growth factors, and stress (20, 24, 25). TCP20 is a member of the class I TCP gene family (19). Throughout developmental stages, TCP20 protein is expressed in embryos, seedlings, leaves, flower buds, and roots (22, 26, 27) and was found to regulate pavement cell size during early leaf development and onset of leaf senescence by promoting jasmonic acid synthesis. Although TCP20 has no obvious role in PNR (16), it was found to bind to the DNA of over 100 nitrate-regulated genes (6, 26, 27), and it is the only gene identified in both local and systemic regulations of N root foraging (7, 16). In plant defense, TCP20 was recently identified among the TCP factors that are targets of pathogenic effectors (28). TCP20’s DNA-binding properties also suggest its regulatory role in cell division, expansion, and differentiation (29, 30). In particular, TCP20 was reported to regulate mitotic cyclin gene CYCB1;1 and ribosomal protein genes by binding to the GCCCR motif in their promoters in vitro and in vivo (30). CYCB1;1 is an effector of growth control at the G2-to-M phase of the cell cycle. It is a marker of mitotic activity, particularly being a division marker of apical meristems, and also a DNA stress marker as an index of G2/M checkpoint arrest (31, 32).
In this paper, we demonstrate that TCP20 and NLP6&7 physically interact under continuous nitrate and N-starvation conditions, forming heterodimers in different compartments of the cell. These interacting regulators were found to play an important role in controlling the expression of key nitrate-responsive genes and the G2/M cell-cycle marker gene, CYCB1;1. NLP6, like NLP7, is retained in the nucleus in the presence of nitrate, and, based on single- and double-mutant analyses, serves as a partially redundant activator along with NLP7. Under N starvation, TCP20-NLP6&7 complexes accumulate in the nucleus, which coincides with TCP20, NLP6&7-dependent regulation of nitrate assimilation and signalinggenes, of the cell-cycle progression gene CYCB1;1, and of root meristem growth.
Results
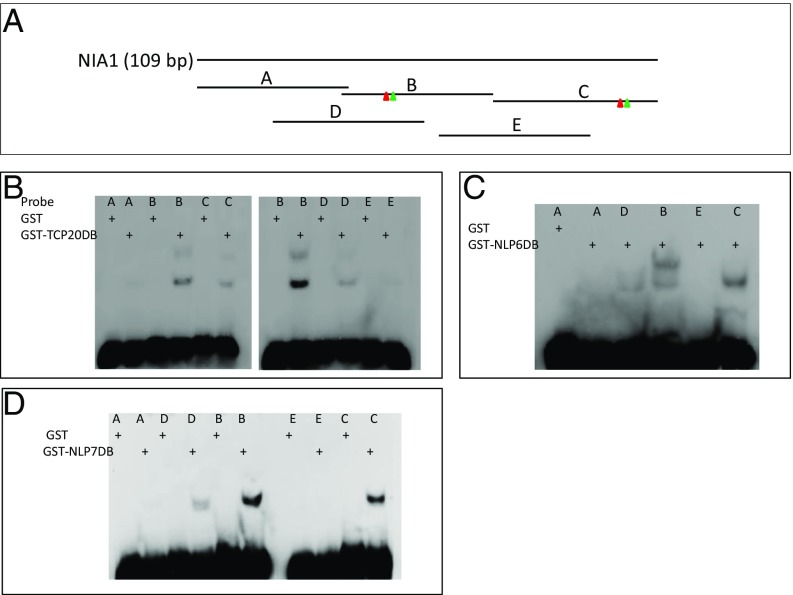
The Binding Sites of TCP20, NLP6, and NLP7 on a 109-bp NIA1 Enhancer Fragment Are in Close Proximity to Each Other.
In our search for direct nitrate regulators, a 109-bp cis regulatory module was identified as nitrate enhancer in the NIA1 promoter (33). The 109-bp NIA1 enhancer fragment was incorporated into a nitrate-inducible reporter construct–YFP and used in a forward genetic screen, which yielded mutations in NRT1.1, a nitrate transceptor (34, 35), and NLP7 (34). The enhancer fragment was also used in yeast one-hybrid screens, which identified TCP20 (16) and NLP6. In this study, we first examined TCP20, NLP6, and NLP7 binding to the 109-bp NIA1 enhancer fragment using electrophoretic mobility shift assays (EMSA). The DNA-binding domains of all three proteins (TCP20-DB, NLP6-DB, and NLP7-DB) bound to the same subfragments of the 109-bp NIA1 enhancer fragment (Fig. 1). Furthermore, a set of mutant probes was used to identify DNA-specific cis-elements that interact with these three proteins (SI Appendix, Fig. S1 and Table S1). We found that the TCP20 and NLP6&7 sites either overlap or are in close proximity within two subfragments (probes B and C) (Fig. 1 and SI Appendix, Fig. S1). Interestingly, in our previous study of nitrate regulatory elements in the 109-bp NIA1 enhancer fragment, the CGCCACT sequence, which overlaps TCP20-NLP6&7 binding sites in probe C, was found to be critical for nitrate induction (19), whereas none of these nitrate-responsive elements was identified in probe B (19). In addition, the close proximity of the TCP20 and NLP6&7 binding sites suggests that each NLP might interact with TCP20 when binding to the 109-bp fragment.
Fig. 1.
Binding of TCP20 DNA-binding domain (DB), NLP6-DB, and NLP7-DB to the fragments of NIA1 (109 bp) DNA was determined by EMSA. (A) Diagram of probes. Red and green triangles represent TCP20 (red) and NLP6/7(green) binding sites, respectively. (B–D) Binding of TCP20DB, NLP6DB, and NLP7DB to A, B, C, D, and E probes. The probes are listed in SI Appendix, Table S1.
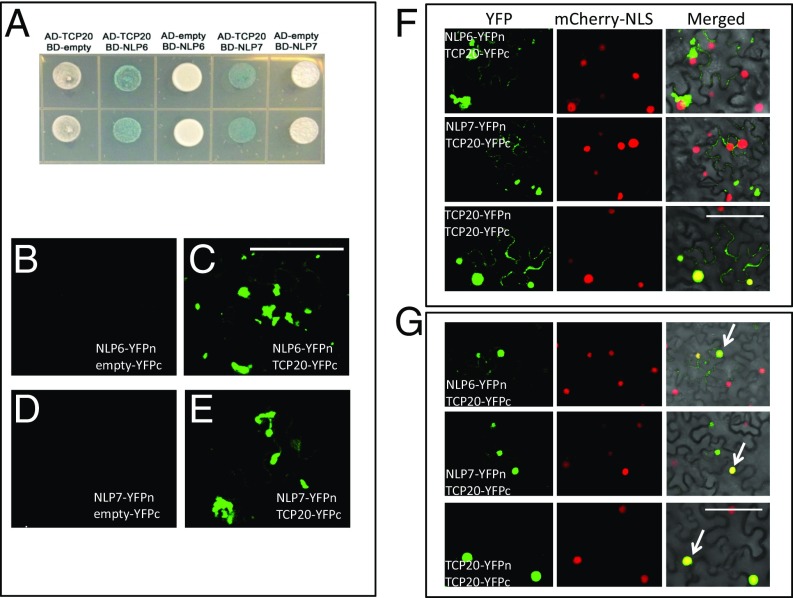
TCP20, NLP6, and NLP7 Physically Interact with Each Other and Subcellular Localizations of Their Interactions Depend on Nitrate Availability.
We next tested for protein interactions using both yeast two-hybrid (Fig. 2A) and bimolecular fluorescence complementation (BiFC) assays (Fig. 2B). In the yeast two-hybrid assays, both NLP6 and NLP7 strongly interact with TCP20 as seen by the blue yeast colonies (Fig. 2A). These interactions were further confirmed by BiFC assays in Nicotiana benthamiana by agroinfiltration, which showed green fluorescence indicative of heterodimer formation (Fig. 2 B–E; the mCherry-NLS was used as a marker for nuclei). The BiFC constructs of NLP6&7-nYFPs and TCP20-cYFPs were transformed and tested in transgenic Arabidopsis plants. The results in transgenic Arabidopsis plants validated the presence of the TCP20-NLPs interactions and showed that the interactions occurred in roots (SI Appendix, Fig. S2). We also examined self-interactions for TCP20 (SI Appendix, Fig. S3), NLP6, and NLP7 (SI Appendix, Fig. S4 A–C). TCP20 has been reported to be a dimeric protein located mainly in the nucleus (27). Our data confirm this finding in that TCP20-YFPn and TCP20-YFPc indeed interact, primarily in the nucleus and but in other locations as well. Additionally, NLP6 and NLP7 each interact with themselves and with each other (SI Appendix, Fig. S4).
Fig. 2.
Protein–protein interactions among TCP20 and NLP6&7. (A) Blue and white colonies in yeast two-hybrid assay for testing interaction between TCP20 and NLP6 and TCP20 and NLP7. (B–E) BiFC in N. benthamiana. (F and G) The subcellular locations of TCP20–NLP6/NLP7 interactions are affected by N status. Red mCherry was used as a marker for nuclei. BiFC was conducted in N. benthamiana grown on 5 mM KNO3 as the sole N source (F) or the plants grown on 5 mM KNO3 and then grown on N-free medium for 4 days (G, 0 mM KNO3). White arrow indicates nuclear localization. Because the nuclear marker (mCherry-NLS) and the NLP marker (NLP-YFP) are carried on separate plasmids, overlapping signals (yellow) occur only in cells that have been transformed with both plasmids. (Scale bars: 100 µm.)
We next tested the effects of N growth conditions on the subcellular localizations of the TCP20 and NLP6&7 proteins and of the TCP20-NLP heterodimers. First, cellular localizations were examined under two treatment conditions: continuous nitrate (no ammonium) (SI Appendix, Fig. S5 A–C) and N starvation (after growth on KNO3) (SI Appendix, Fig. S5 D–F). Each protein was fused to YFP and then transiently expressed in N. benthamiana by using Agrobacterium with DNA constructs driven by the CaMV 35S promoter. TCP20 was found primarily in the nucleus under both conditions (SI Appendix, Fig. S5 C and F), consistent with a previous report for transgenic Arabidopsis plants (27). Nuclear retention was also found for NLP6 and NLP7 in nitrate-grown plants with some additional signal outside the nucleus (SI Appendix, Fig. S5 A and B). Under N starvation, signals for NLP6 and NLP7 were very low. A few NLP6 or 7-YFP fusion proteins were found in the nucleus, whereas a majority of the signals were found outside the nucleus (SI Appendix, Fig. S5 D and E). We validated the findings in the roots of NLP6/7-YFP transgenic plants. (SI Appendix, Fig. S5 G–L). Our findings are consistent with those of Marchive et al. (10) who showed nuclear retention of NLP7 in nitrate-treated plants. In addition, our data indicate that NLP6, like NLP7, is retained in the nucleus in the presence of nitrate.
Next, the effect of N growth conditions on TCP20–NLP interactions was tested. Under both conditions (continuous nitrate versus N starvation), NLP6 and NLP7 interacted with TCP20 in the BiFC experiments (Fig. 2 F and G and SI Appendix, Fig. S6); however, the primary locations of NLP6&7-TCP20 heterodimers varied dramatically. Under nitrate-grown conditions, NLP6&7-TCP20 heterodimers reside primarily outside the nucleus (Fig. 2F and SI Appendix, Fig. S6), whereas under N starvation, the heterodimers are primarily located in the nucleus (Fig. 2G). We validated the findings in transgenic Arabidopsis roots (SI Appendix, Fig. S7). This is the opposite of what was found for the NLP single proteins (i.e., NLP6&7-YFP fusions) (SI Appendix, Fig. S5) (10). The TCP20 homodimers, however, were found primarily in the nucleus under both conditions with additional signal outside the nucleus in nitrate-grown plants (Fig. 2 F and G). NLP6 and NLP7 homodimers and heterodimers were found outside the nucleus in the presence of nitrate (SI Appendix, Fig. S4), whereas no signal was detected in N-starved plants (SI Appendix, Fig. S8). These results suggest that NLP 6&7 homodimers and heterodimers are not functioning as the active complexes in nuclei to induce gene expression in the presence of nitrate.
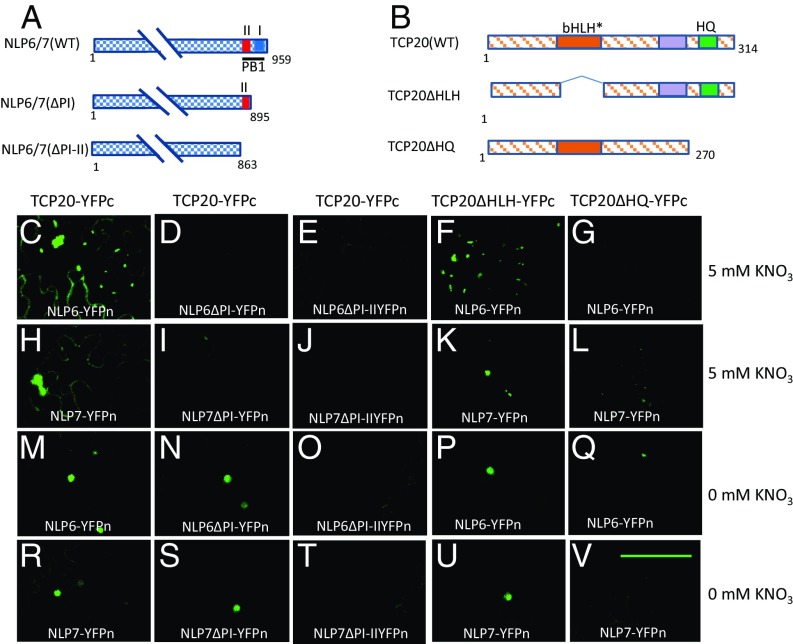
The Type I/II PB1 Domains of NLP6&7 and the Glutamine-Rich Domain of TCP20 Are Required for Protein–Protein Interactions.
We also investigated potential interaction domains in NLP6&7 and TCP20. The NLP6&7 PB1 domains display both type I and type II structures containing key signature residues: a conserved Lys residue in the type II structure and an OPCA [the octicosapeptide repeat (OPR), p40phox and budding yeast Cdc24p (PC), and atypical protein kinase C-interaction domain (AID)] motif in the type I structure (SI Appendix, Fig. S9A). As an example, the type I/II PB1 domain is present in the mammalian scaffold protein p62, a crucial regulator linking amino acids to the activation of the mammalian target of rapamycin complex 1 (mTORC1) to control cell size and proliferation (24, 36, 37). TCP20 contains a basic helix-loop-helix–like motif (bHLH*) that is involved in dimerization (38). We also identified two other candidate domains in the TCP20 C terminus. One is a histidine- and glutamine-rich (HQ-rich); the other is next to the HQ-rich domain and within a larger glycine-rich domain (SI Appendix, Fig. S9B). Both of these domains contain residues that are deeply conserved across plant species. We hypothesized that these domains could be involved in the TCP20-NLP6&7 protein interactions.
To determine the function of these domains, various deleted derivatives of TCP20 and NLP6&7 were constructed (Fig. 3 A and B) and then examined in BiFC assays. Deletion constructs of NLP6&7 were tested first. Deleting the OPCA motif (ΔPI) of NLP6 and NLP7 (leaving the first lysine-region intact) or both regions of PB1 (ΔPI-II) abolished the TCP20-NLP6&7 interactions under 5 mM KNO3 growth conditions (Fig. 3 D, E, I, and J). Interestingly, under N-starvation conditions, TCP20-NLP6&7 interactions were abolished only when both PB1 motifs (ΔPI-II) were deleted (Fig. 3 O and T) because interaction was still evident in the single OPCA motif deletion construct (ΔPI) (Fig. 3 N and S). For NLP6&7 homodimer interactions, deletion of the OPCA region alone (ΔPI) or both regions of PB1 (ΔPI-II) abolished the interactions (SI Appendix, Fig. S10). These results are consistent with previous findings showing that type I/II PB1 domains are critical for the formation of homo- and heterodimers and in interactions with proteins lacking PB1 domains (24, 25). Next, deletion constructs of TCP20 were analyzed. Deletion of the N terminus of TCP20 (TCP20Δ1–78) led to a small decrease in the TCP20–NLP6&7 interactions under nitrate growth or N starvation (SI Appendix, Fig. S11). Deletion of bHLH-like motif in TCP20 (TCP20ΔHLH; Fig. 3 F, K, P, and U) had no impact on the interactions under both conditions. However, deleting the amino acids from 270 to 314 (TCP20ΔHQ) in the C-terminal domain of TCP20 significantly decreased or abolished the TCP20–NLP6&7 interactions under both nitrate-grown (Fig. 3 G and L) and N starvation (Fig. 3 Q and V) conditions. Further deletion of the other conserved region (TCP20Δ230–314) showed the same results (SI Appendix, Fig. S11). Overall, these results indicate that both the PB1 domains of NLP6&7 and the C-terminal HQ-rich domain of TCP20 are necessary for the TCP20–NLP6&7 interactions. The N terminus and bHLH-like domain of TCP20 are dispensable.
Fig. 3.
PB1 domain of NLP6/7 and a C-terminal HQ region of TCP20 are required for TCP20–NLP6/7 interaction. (A and B) Diagrams of NLP6/7 and TCP20 derivatives. The numbers shown for NLP6/7 are for NLP7. Corresponding number for NLP6 are the following: full length, 841 amino acids; deletion derivatives, 741 or 774 amino acids. For NLPs, red and dark-blue boxes indicate two modules (II and I) in PB1 domain. For TCP20, orange and green boxes present bHLH* and HQ, respectively. Purple box presents a conserved region next to HQ. (C–V) BiFC was conducted in leaves of N. benthamiana under 5 mM KNO3 growth condition (C–L) or the plants grown on 5 mM KNO3 and then grown on N-free medium for 4 days (M–V). (Scale bar: 100 µm.)
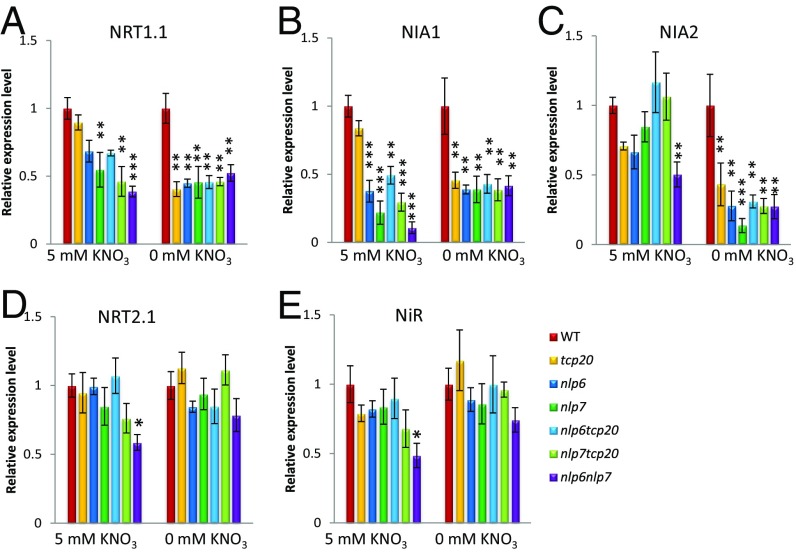
Analysis of tcp20, nlp6, and nlp7 Single and Double Mutants Reveals Distinctive Regulatory Roles of the TFs in Nitrate Assimilation and Signaling Under Continuous Nitrate and N-Starvation Conditions.
To dissect individual and combinatorial molecular functions of NLP6, NLP7, and TCP20 on expression of genes involved in nitrate transport, assimilation, and signaling, we conducted quantitative RT-PCR on whole roots of wild-type (WT) and single- and double-mutant lines under continuous nitrate-grown and N-starvation conditions (Fig. 4). First, nlp6 and nlp7 single mutants showed lower levels of NRT1.1, NIA1, and NIA2 mRNAs under nitrate-grown conditions (Fig. 4 A–C); levels of NRT2.1 and NiR mRNAs, however, were similar to WT under these conditions (Fig. 4 D and E). nlp6nlp7 double mutants showed significant reductions in mRNA levels compared with WT across five target genes as follows: NIA1 (∼90%), NIA2 (∼50%), NRT1.1 (∼61%), NiR (∼50%), and NRT2.1 (∼40%) (Fig. 4). Compared with nlp6 and nlp7 single mutants, the further significant reductions of mRNA levels in nlp6nlp7 double mutants were observed across all of the target genes compared with at least one of the single mutants (Fig. 4 A–E and SI Appendix, Fig. S12). These are phenotypes of nlp6 single and nlp6nlp7 double mutants. The results suggest that NLP6 and NLP7 are partially redundant activators of key nitrate transport, assimilation, and signaling genes in the presence of nitrate and function additively. Consistent with this proposal, nlp6nlp7 double mutants showed severe growth defects with nitrate as the sole N source but not with ammonium (SI Appendix, Fig. S13), which is similar to the propagation and growth properties observed in NR-null (nia1nia2) mutants (6). Next, tcp20 single and tcp20nlp6 and tcp20nlp7 double mutants were examined under nitrate-grown conditions. The tcp20 single mutation had little effect on mRNA levels across the target genes (Fig. 4), which is consistent with what was reported in a previous study (30). The tcp20nlp6 and tcp20nlp7 double mutants displayed no obvious additive effects (Fig. 4).
Fig. 4.
The interacting transcriptional factors, TCP20 and NLP6&7, regulate key genes of nitrate assimilation and signaling in response to nitrate status. (A–E) RT-PCR quantification of NRT1.1, NIA1, NIA2, NRT2.1, and NiR under nitrate-grown condition and N starvation. WT mRNA level was set to 1 for each condition. Error bars show SEM (n = 3 biological replicates). Arabidopsis seedlings were grown on 5-mM KNO3 plates for 6–7 days and then were transferred to new 5 mM KNO3 plates or N-free plates for 3 d. Total roots were harvested for analysis. For each experiment, within each nitrate condition, one-way ANOVA was performed and followed by a t test (two-side, using WT as control). Black asterisks represent statistical difference between WT and mutants under same nitrate condition. *P < 0.05, **P < 0.01, ***P < 0.001. (Scale bar: 50 µm.)
Under N starvation, however, the mRNA levels of NRT1.1, NIA1, and NIA2 were significantly and uniformly lower across all of the mutant lines (∼50–60% reduction) (Fig. 4 A–C). However, no evident reduction in mRNA levels of NRT2.1 and NiR was observed (Fig. 4 D and E). Especially interesting was the effect of tcp20 mutations, which showed strong reductions in mRNA levels of NRT1.1, NIA1, and NIA2, similar to what was observed for nlp6nlp7 double mutants (Fig. 4 A–C). This contrasts to the lack of effect of TCP20 in the presence of nitrate. These results are consistent with the nuclear TCP20–NLP6&7 interactions under N starvation (Fig. 2G), suggesting that these proteins may function as a complex in regulating expression of NRT1.1, NIA1, and NIA2 under N starvation. In addition to strongly regulating NR genes, the interacting transcriptional regulators also control the mRNA levels of NRT1.1 (CHL1/NPF6.3), which plays a versatile and central role in adaptive responses to nitrate availability (35, 39, 40).
A two-way ANOVA analysis revealed that both main effects of N conditions and genotypes are statistically significant in regulating expression of NRT1.1 (***P < 0.001), NIA1 (***P < 0.001), and NIA2 (***P < 0.001), and an interaction effect between N treatment and genotypes is also statistically significant in regulating expression of NRT1.1 (*P < 0.05), NIA1 (*P < 0.05), and NIA2 (***P < 0.001) (SI Appendix, Table S4). These results demonstrate that these interacting regulators control the expression of the key genes NRT1.1, NIA1, and NIA2 and that the regulatory controls are distinct between nitrate-grown conditions and N starvation. The expression of NRT1.1, NIA1, and NIA2 genes under N starvation corresponds to nuclear localization of the interacting transcriptional regulators and supports the idea that these regulators are working in concert to promote the expression of these genes under N starvation.
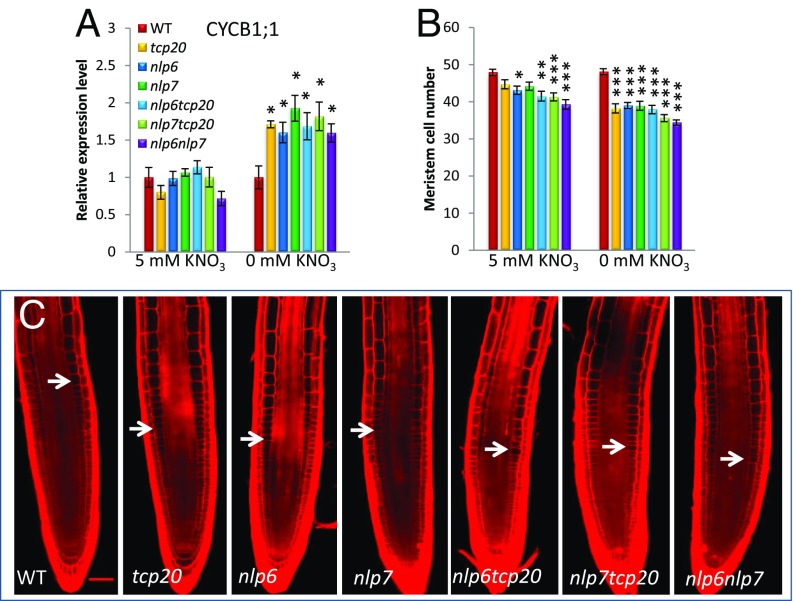
Interacting TCP20 and NLP6&7 Are Required for Proper Expression of the Cell-Cycle Progression Gene CYCB1;1 and Root Meristem Growth Under N Starvation.
We next addressed the question, What additional regulatory roles do TCP20 and NLP6&7 play in response to nitrate availability beyond regulating the nitrate assimilation and signaling genes? TCP20 is known to bind to the GCCCR motif in the promoter of mitotic cyclin gene CYCB1;1 in vitro and in vivo (30). We further examined the CYCB1;1 promoter region containing the GCCCR motif, which, interestingly, led to the identification of two potential NLP6&7-binding sites (GCCCACTT and CGGGCCTT) in the same region (SI Appendix, Fig. S14). The two consensus sequences only have a two-nucleotide mismatch compared with the sequences of the 109-bp NIA1 promoter fragment (SI Appendix, Fig. S1), and each contains a GCCCR motif recognized by TCP20 (SI Appendix, Fig. S14). In addition, in close proximity to the putative NLP6&7-binding sites were another two GCCCR motifs and an auxin response factor (ARF)-binding site (SI Appendix, Fig. S14) (30). As shown in Fig. 5A, under nitrate-grown conditions there was some reduction of CYCB1;1 mRNA levels measured in whole roots of the nlp6nlp7 double- and tcp20 single-mutant lines, but neither was statistically significant. No reduction in other mutant lines was observed. In contrast, under N starvation, the mRNA levels of CYCB1;1 measured in whole roots were significantly and uniformly higher across all of the mutant lines (Fig. 5A). A two-way ANOVA analysis revealed that, in regulating expression of CYCB1;1, both main effects of N conditions and genotypes are statistically significant (***P < 0.001; *P < 0.05), and an interaction effect between N treatment and genotypes is also statistically significant (*P < 0.05) (SI Appendix, Table S4). These results also indicate that the regulation of this key cyclin gene by the interacting nitrate regulators is distinct between nitrate-grown conditions and N starvation. The close proximity of the confirmed binding sites of TCP20 and NLP6&7 in the NIA1 promoter fragment (Fig. 1 and SI Appendix, Fig. S1) and their putative binding sites found in the CYCB1;1 promoter region (SI Appendix, Fig. S14) suggest that the interaction of these regulators might play a role in responses to nitrate availability for both nitrate assimilation and signaling and cyclin genes.
Fig. 5.
The interacting transcriptional factors TCP20 and NLP6&7 regulate the cell-cycle gene CYCB1;1 and control cell division in the root meristem in response to nitrate status. (A) CYCB1;1 gene expression level from total roots in WT and mutant lines (conditions here are the same as those of A–C in Fig. 4). Error bars show SEM (n = 3 biological replicates). (B) Meristem cell number in WT and mutant lines under 5 mM KNO3 growth condition and N starvation for 3 d. Root meristem number was measured by counting the cortical cells from the quiescent center to the first cell that clearly doubles its volume. Error bars represent SEM (n = 18–20). (C) Confocal images of WT, tcp20, nlp6, nlp7, nlp6tcp20, nlp7tcp20, and nlp6nlp7 root tips stained with propidium iodide. White arrows indicate the boundary of the apical meristem region. Seedlings were grown on 5 mM KNO3 medium for 3 d and then were transferred to N-free medium for 3 d. The same statistical test used in Fig. 4 has been used here. *P < 0.05, **P < 0.01, ***P < 0.001. (Scale bar: 50 µm.)
To investigate further the linkage between nitrate regulation and growth via cell-cycle control, primary root growth phenotypes in the single and double mutants of TCP20 and NLP6&7 were examined (SI Appendix, Fig. S15). Overall, all of the mutants had shorter roots than the WT. The mutants under N starvation displayed significantly greater reductions in primary root length than the mutants under nitrate-grown conditions (SI Appendix, Fig. S15). In addition, under nitrate-grown conditions, nlp6nlp7 double mutants showed stronger deleterious effects on primary root length than the single mutants (SI Appendix, Fig. S15 A and C), consistent with our previous finding in the mRNA levels of nitrate sentinel genes. Meristem size and cell numbers, which are controlled by cell division and are a determinant of root growth, were then examined in primary roots under two distinct nitrate conditions. All single and double mutants showed reduced levels of cell numbers and meristem length (Fig. 5 B and C; SI Appendix, Fig. S15D). The phenotypes were more conspicuous under N starvation than those under nitrate-grown conditions (Fig. 5 B and C; SI Appendix, Fig. S15D). A two-way ANOVA analysis revealed that an interaction effect between N treatment and genotypes is statistically significant in regulating these growth phenotypes: primary root meristem cell number and length (*P ≤ 0.05) and primary root length (***P < 0.001); and both main effects of N conditions and genotypes are statistically significant (***P < 0.001) (SI Appendix, Table S4). The mutation effects on meristem size and cell numbers are largely consistent with what we observed in primary root length, supporting the idea that there is a direct correlation between nitrate-regulated CYCB1:1 levels and primary root growth. The higher mRNA levels of CYCB1;1 under N starvation in the mutants could result in one of two outcomes: increased cell production rate in apical meristem or arrest at G2/M checkpoint in cell cycle as has been observed under stress conditions (32, 41). The elevated mRNA levels of CYCB1;1 coincided with the decreased number of cells in the root apical meristem in our experiments, supporting G2/M cell-cycle arrest as the reason for the observed effects.
Discussion
We have demonstrated that TCP20 physically interacts with both NLP6 and NLP7 under nitrate-grown and N-starvation conditions and that both the type I/II PB1 domain of NLP6&7 and the histidine- and glutamine-rich domain of TCP20 are necessary for these interactions. Under N starvation, TCP20-NLP6&7 heterodimers accumulate in the nuclei, which correlates with the regulation of nitrate assimilation and signaling genes and the mitotic cyclin gene CYCB1;1 to counteract N-starvation stress. Mutants show significantly stunted root growth and decreased root meristem size and meristem cell numbers, which we suggest is a result of premature cell-cycle exit due to the misregulation of CYCB1;1. In addition, NLP6 and NLP7 physically interact with each other and themselves via the PB1 domain and are partially redundant activators in the presence of nitrate. These results suggest that these three transcription factors form a regulatory nexus integrating the regulation of PNR genes in the presence and the absence of nitrate and linking nitrate responses to root growth.
The regulatory roles of AtTCP20 in response to nitrate availability compare with those of AtTCP21/CHE (for CCA1 hiking expedition) in the circadian oscillator (42) in that both these class I TCP genes are crucial for plant responses to variations in environmental cues, i.e., nutrients, light, and temperature. The type I/II PB1 domains that we found are required for forming TCP20-NLP6&7 and NLP6&7 self/hetero-complexes are also found to be important for other key interactions. They are needed for the homo- and hetero-oligomerization of auxin response factor (ARF) transcription factors and auxin/indole 3-acetic acid (Aux/IAA) repressor proteins (25, 43) as well as for organizing growth factor and nutrient signaling within the mTORC1 pathway in animals (37). Thus, the type of interactions uncovered for TCP20, NLP6, and NLP7 regulators are part of a more general pattern used for signaling in both plants and animals.
In plants, growing evidence indicates that complex crosstalk of nitrate and hormone signaling, and a close connection between TCP regulation and hormone signaling, underlie the integrated control of plant growth, development, and defense response (7, 44, 45). Nitrate controls hormonal pathways in biosynthesis, transport, and signal transduction, and, conversely, hormonal signaling regulates nitrate/nitrogen transport and assimilation to adjust nutritional status to growth (7, 44). On the other hand, in addition to acting as transcriptional modulators of cell division, TCPs affect hormone synthesis, transport, and signal transduction by controlling gene expression through their context-dependent interaction with tissue-specific cellular components (45). Our study reveals that TCP regulation is part of nitrate signaling, providing a transcriptional control of cell-cycle genes when plants face loss of nitrate availability. Our findings offer insight into the close interplays and convergent regulations between nitrate- and hormone-signaling pathways in plants. It will be interesting to investigate in the future whether PB1 domain-dependent signaling complexes may provide additional links between nitrate- and hormone-signaling pathways and whether other members of the TCP family function in a similar fashion as TCP20 in nitrate signaling.
Materials and Methods
Details of plant material and growth conditions, plasmid constructions, protein expression and purification, EMSA, yeast two-hybrids, BiFC and transient expression, transgenic lines, confocal microscopy, and quantitative PCR are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Martin F. Yanofsky for his support and for sharing published reagents and Mark Estelle and Yi Zhang for use of the confocal microscope. Research reported in this publication was supported by the National Science Foundation [Grant IOS-1021380 (to N.M.C.)], a UCSD General Campus Research grant, and Division of Biological Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615676114/-/DCSupplemental.
References
- 1.Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3(10):389–395. [Google Scholar]
- 2.Krouk G, Crawford NM, Coruzzi GM, Tsay YF. Nitrate signaling: Adaptation to fluctuating environments. Curr Opin Plant Biol. 2010;13(3):266–273. doi: 10.1016/j.pbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Nacry P, Bouguyon E, Gojon A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil. 2013;370(1):1–29. [Google Scholar]
- 4.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132(2):556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medici A, Krouk G. The primary nitrate response: A multifaceted signalling pathway. J Exp Bot. 2014;65(19):5567–5576. doi: 10.1093/jxb/eru245. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136(1):2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien JA, et al. Nitrate transport, sensing, and responses in plants. Mol Plant. 2016;9(6):837–856. doi: 10.1016/j.molp.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279(5349):407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 9.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 10.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 11.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21(11):3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11(12):R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal EA, Álvarez JM, Gutiérrez RA. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal Behav. 2014;9(3):e28501. doi: 10.4161/psb.28501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Para A, et al. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(28):10371–10376. doi: 10.1073/pnas.1404657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez JM, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014;80(1):1–13. doi: 10.1111/tpj.12618. [DOI] [PubMed] [Google Scholar]
- 16.Guan P, et al. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc Natl Acad Sci USA. 2014;111(42):15267–15272. doi: 10.1073/pnas.1411375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu N, et al. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell. 2016;28(2):485–504. doi: 10.1105/tpc.15.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal EA, Álvarez JM, Moyano TC, Gutiérrez RA. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr Opin Plant Biol. 2015;27:125–132. doi: 10.1016/j.pbi.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Martín-Trillo M, Cubas P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010;15(1):31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Schauser L, Wieloch W, Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol. 2005;60(2):229–237. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- 21.Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J Exp Bot. 2014;65(19):5577–5587. doi: 10.1093/jxb/eru261. [DOI] [PubMed] [Google Scholar]
- 22.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castaings L, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57(3):426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 24.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007(401):re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 25.Guilfoyle TJ. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell. 2015;27(1):33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hervé C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009;149(3):1462–1477. doi: 10.1104/pp.108.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danisman S, et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012;159(4):1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SH, et al. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014;78(6):978–989. doi: 10.1111/tpj.12527. [DOI] [PubMed] [Google Scholar]
- 29.Trémousaygue D, et al. Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003;33(6):957–966. doi: 10.1046/j.1365-313x.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102(36):12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20(4):503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 32.De Schutter K, et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell. 2007;19(1):211–225. doi: 10.1105/tpc.106.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, et al. Multiple regulatory elements in the Arabidopsis NIA1 promoter act synergistically to form a nitrate enhancer. Plant Physiol. 2010;154(1):423–432. doi: 10.1104/pp.110.162586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Xing X, Wang Y, Tran A, Crawford NM. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009;151(1):472–478. doi: 10.1104/pp.109.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138(6):1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44(1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linares JF, et al. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade. Cell Reports. 2015;12(8):1339–1352. doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999;18(2):215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 39.Remans T, et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA. 2006;103(50):19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krouk G, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18(6):927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Beemster GT, Baskin TI. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998;116(4):1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krouk G. Hormones and nitrate: A two-way connection. Plant Mol Biol. 2016;91(6):599–606. doi: 10.1007/s11103-016-0463-x. [DOI] [PubMed] [Google Scholar]
- 45.Nicolas M, Cubas P. TCP factors: New kids on the signaling block. Curr Opin Plant Biol. 2016;33:33–41. doi: 10.1016/j.pbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.