We read with interest the study by Roberts et al. indicating that the association between endoplasmic reticulum aminopeptidase 1 (ERAP1) genotypes and ankylosing spondylitis (AS) is by common genotypes rather than rare haplotype combinations (1). The authors also suggest that the data presented by Reeves et al. (2) indicated that only “rare” allotypes/combinations are important in the association of ERAP1 with AS.
However, our data (2) provide prima facie evidence that common allotypes are functionally relevant and may be important in disease. For example, we have shown that the two common haplotypes discussed in Roberts et al. (1) (I/*001 and IV/*008/*011) are both hypofunctional (3). When expressed singly or codominantly with other hypofunctional allotypes, these do not restore surface expression of HLA-B*27:05 expression on ERAP1 knockout fibroblasts (2).
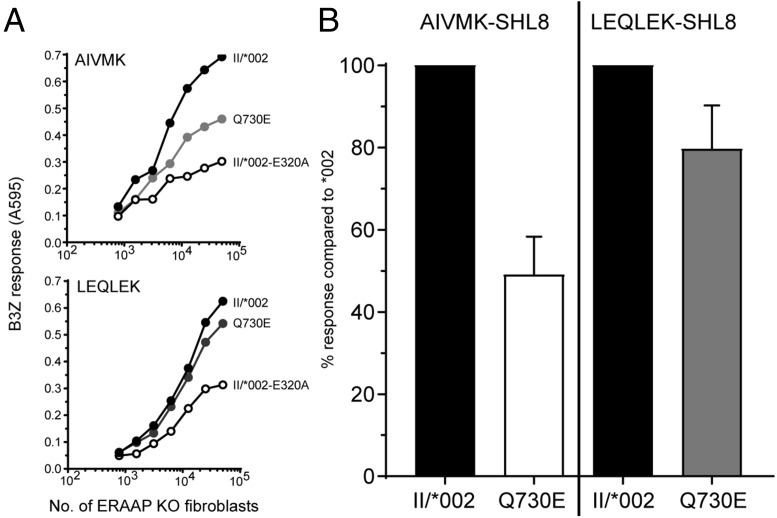
Our work was aimed at understanding the functional consequences of ERAP1 haplotypes containing combinations of SNPs. From this work, it appears that the context in which any polymorphism exists within a haplotype may have profound effects on the whether ERAP1 trims peptides efficiently with all of the consequences that lead from this situation. We agree with Roberts et al. that future functional studies focusing on the K528R variant are likely to be informative because several laboratories, including ours, have shown that this variation leads to a reduction in peptide trimming. However, the extent of this reduction depends upon the haplotype/allotype on which it occurs (3, 4). In addition, other variants have an independent effect on function: for example Q730E, being the only difference between II/*002 and V, alters the efficacy and specificity of the trimming function (ref. 5; Fig. 1). In addition, haplotype I/*001, which has poor trimming function, contains the SNPs N575 and Q725: variants that in other contexts rescue trimming function (in R528/N575) (4) and induce hyperactivity (in Q725/E730) (3), respectively. Furthermore, although haplotype I/*001 has a reduced trimming activity averaged over all substrates, its trimming of some amino acids is unaffected and some are overtrimmed (3). This level of characterization of haplotype trimming activity will be required to identify those individuals that might benefit from the therapeutic use of ERAP1 inhibitors.
Fig. 1.
Q730E affects ERAP1 trimming activity and specificity. (A) ERAAP knockout fibroblasts were transfected with II/*002, Q730E, or II/*002-E320A ERAP1 together with either AIVMK-SHL8 (Top) or LEQLEK-SHL8 (Bottom) and assessed for their ability to stimulate SHL8-specific T-cell hybridoma B3Z. A representative experiment is shown. (B) The relative trimming activity of Q730E was compared to II/*002 for both AIVMK- and LEQLEK-SHL8 minigenes. Data was taken from at least six experiments.
Roberts et al. (1) point out that the frequencies of particular combinations are different between both studies. Because of the smaller number of samples in our study, the relative frequencies of particular allotype combinations may be misrepresentative of the general population/patient population. Importantly, although large genetic studies that more accurately describe the population frequencies of ERAP1 haplotypes/allotypes might help diagnosis, assessment of the functional consequence of the combinations identified seems most likely to produce a greater understanding of AS pathogenesis and direct future diagnostic tests and treatments.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roberts AR, et al. ERAP1 association with ankylosing spondylitis is attributable to common genotypes rather than rare haplotype combinations. Proc Natl Acad Sci USA. 2017;114(3):558–561. doi: 10.1073/pnas.1618856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves E, Colebatch-Bourn A, Elliott T, Edwards CJ, James E. Functionally distinct ERAP1 allotype combinations distinguish individuals with ankylosing spondylitis. Proc Natl Acad Sci USA. 2014;111(49):17594–17599. doi: 10.1073/pnas.1408882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves E, Edwards CJ, Elliott T, James E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191(1):35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martín-Esteban A, Gómez-Molina P, Sanz-Bravo A, López de Castro JA. Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands. J Biol Chem. 2014;289(7):3978–3990. doi: 10.1074/jbc.M113.529610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evnouchidou I, et al. Cutting edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186(4):1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]