Significance
We have tested the hypothesis that the placenta of early pregnancy might be more easily breached by the Zika virus (ZIKV) than the relatively resistant outer cells of the mature placenta. Colonies of placental lineage cells derived from embryonic stem cells, which are probably analogous to the primitive placenta at implantation, were lysed more rapidly by an African strain of ZIKV, considered relatively benign, than by an Asian strain linked to fetal brain abnormalities. We conclude that the human fetus may be most vulnerable to ZIKV very early in pregnancy and that the African strain may threaten a pregnancy more strongly than previously believed.
Keywords: embryonic stem cell, placenta, pregnancy, trophoblast, Zika virus
Abstract
Infection of pregnant women by Asian lineage strains of Zika virus (ZIKV) has been linked to brain abnormalities in their infants, yet it is uncertain when during pregnancy the human conceptus is most vulnerable to the virus. We have examined two models to study susceptibility of human placental trophoblast to ZIKV: cytotrophoblast and syncytiotrophoblast derived from placental villi at term and colonies of trophoblast differentiated from embryonic stem cells (ESC). The latter appear to be analogous to the primitive placenta formed during implantation. The cells from term placentas, which resist infection, do not express genes encoding most attachment factors implicated in ZIKV entry but do express many genes associated with antiviral defense. By contrast, the ESC-derived trophoblasts possess a wide range of attachment factors for ZIKV entry and lack components of a robust antiviral response system. These cells, particularly areas of syncytiotrophoblast within the colonies, quickly become infected, produce infectious virus and undergo lysis within 48 h after exposure to low titers (multiplicity of infection > 0.07) of an African lineage strain (MR766 Uganda: ZIKVU) considered to be benign with regards to effects on fetal development. Unexpectedly, lytic effects required significantly higher titers of the presumed more virulent FSS13025 Cambodia (ZIKVC). Our data suggest that the developing fetus might be most vulnerable to ZIKV early in the first trimester before a protective zone of mature villous trophoblast has been established. Additionally, MR766 is highly trophic toward primitive trophoblast, which may put the early conceptus of an infected mother at high risk for destruction.
Although the placental syncytiotrophoblast (STB) is an effective barrier to many pathogens (1, 2), including viruses (3), the extent to which it protects against vertical transmission of a flavivirus, such as the Zika virus (ZIKV: Flaviviridae; Flavivirus) at different stages of pregnancy is largely unknown (4). In the case of ZIKV, clinical symptoms in adults are generally mild, yet infection of a woman during her pregnancy has been associated with fetal death, placental insufficiency, fetal growth restriction, and central nervous system abnormalities (5). In particular, there is strong link to fetal and pediatric microcephaly (6–8). The likely involvement of ZIKV in neurological disease is consistent with its ability to infect and replicate within human cortical neuronal progenitor cells and slow their growth (9, 10), infect human neurospheres and brain organoids (9, 11–13), and colonize the developing brain of mouse fetuses injected directly with the virus (14). Importantly, an African lineage strain (MR766) of ZIKV from Uganda (ZIKVU) is as at least as capable of infecting human neural progenitor cells in vitro as strains of Asian origin (10, 12, 13, 15), even though there is no indication that the former causes brain developmental abnormalities in infants.
Because ZIKV strains of Asian lineage are linked to microcephaly in humans, the virus is generally believed to cross the placental barrier early in gestation when the brain is beginning to form. However, there have been reports of fetal brain ultrasound anomalies and abnormal brain development in women considered to have become infected by ZIKV late in their pregnancies (5, 16). On the other hand, the overall risk for microcephaly and other brain abnormalities in infants born to a larger cohort of US women exposed to ZIKV (n = 442) was 5.9% (17), and, of these, there were no cases noted among the women known to have been infected during their second or third trimesters. Clearly, there remains some uncertainty regarding the consequences of maternal ZIKV infection in the later stages of pregnancy. One factor that may have affected the data from Rio de Janeiro is that almost 90% of those women were serologically positive for Dengue (DENV), presumably due to past infections with one or more of the known DENV strains (DENV1–4). Sero-cross-reactivity between DENV1–4 and ZIKV (18, 19) may allow the latter to be piggybacked across the protective STB layer of the mature villous placenta. Such transcytosis of maternal IgG appears to be initiated around the beginning of the second trimester and continues to term (20).
There are additional reasons to believe that the fetus may be most vulnerable to ZIKV infection and microcephaly early in a pregnancy, and then quite rarely. Epidemiological studies performed following the French Polynesian outbreak of 2013–2014 (21) and the one in the Bahia region of Brazil in 2015 (22), locations where the virus swept rapidly through the population and infection rates were high, indicated that there was a strong association between the risk for microcephaly and infection of the mother during the first trimester. These authors suggest a negligible risk for the second and third trimester infections. Clearly, more information is needed to refine these data and further narrow the window of susceptibility. Ideally, such analyses will also take into account other adverse outcomes of maternal ZIKV infection, such as fetal growth restriction and fetal loss, and whether these complications are also linked to vulnerability of the placenta at a particular gestational stage or indirectly to the severity of maternal disease.
There is mounting evidence that, by term, the human placenta provides a relatively robust barrier to virus transmission. Cultures of primary term human placental cytotrophoblast (CTB) recovered and allowed to differentiate into STB in vitro are resistant to infection by a range of diverse viruses, including vesicular stomatitis virus (Rhabdoviridae; Vesiculovirus), poliovirus (Picornaviridae; Enterovirus; enterovirus C), vaccinia virus (Poxviridae; Chordopoxvirinae; Orthopoxvirus), human cytomegalovirus (Herpesviridae; Betaherpesvirinae; Cytomegalovirus) (23), and, as shown more recently, to two strains of ZIKV and DENV2 (Flaviviridae; Flavivirus) (24). Whether placental STB from earlier in gestation is comparably resistant to ZIKV and related viruses remains unclear. Mouse models have not been altogether helpful in this respect, in part because the development of the placenta follows a different progression than in humans and also because many mouse strains are resistant to ZIKV infection, including the C57/B6 strain used so widely (9). However, mice engineered or treated in such a manner that they are unable to mount a normal antiviral response are susceptible (25, 26) and can transmit the virus to their fetuses, allowing effects on the brain to be assessed (27, 28). The latter studies also implicated an appropriate IFN response by the placenta as being at least partially responsible for protecting the fetus from ZIKV.
It is now well established that human embryonic stem cells (ESC) and induced pluripotent stem cells can be driven along the trophoblast lineage by exposing them to BMP4 and inhibiting the signaling pathways that maintain the pluripotent phenotype (29–31). Within 5–6 d of initiating differentiation, areas of syncytium expressing typical markers of STB begin to appear within the colonies. Based on RNAseq data, we believe that the STB generated in vitro from ESC likely corresponds to a primitive type of STB observed during the early stages of a human pregnancy before the villous placenta has fully formed (32). The availability of RNAseq data from CTB and STB derived from placentas at term and analogous cells generated from ESC has also provided the opportunity to make inferences about whether or not these cell types might be protected from ZIKV entry and replication. Here we tested the hypothesis that trophoblasts from a mature placenta are more resistant to a virus like ZIKV than the ESC-derived trophoblasts representing the very early conceptus.
Results
Trophoblast Phenotype of Cells.
Our starting point for all of the comparisons made with the RNAseq data study was trophoblast derived from normal-term placentas or generated from H1 ESC (WA01) after an 8-d exposure to BMP4/A83-01/PD173074 (BAP treatment) (29). The placental cells consisted of two cell populations: (i) undifferentiated primary human trophoblasts (PHTu), which are mononucleated CTB that have been purified after proteolytic dispersion of placental tissue (3) and then cultured for 8 h in presence of FBS to allow live cells to attach to the culture plates; and (ii) differentiated STB (PHTd), which are the PHTu cultured for 48 h to allow them to fuse and differentiate into STB. The colonies derived from undifferentiated ESC (ESCu) by BAP treatment also consisted of two populations, a mononucleated CTB population that passed through a 40-μm screen [ESC-derived trophoblast (ESCd) < 40] and a larger STB population that had failed to pass through a wider mesh 70-μm screen (ESCd > 70) (32). SI Appendix, Table S1 lists the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values in these four cell types for over 100 genes, the expression of which has been associated with the trophoblast lineage of mammals, including ones recently recommended to be included in any phenotypic definition of trophoblast (33). Of note, however, four genes encoding transcription factors (CDX2, ELF5, EOMES, ASCL1), which are generally regarded as markers of trophoblast stem cells (34), are barely expressed in either the two groups of ESC-derived cells at day 8 of BAP treatment or the PHTu (CTB) and PHTd (STB) generated from term placentas, suggesting that all these cells had differentiated beyond the trophoblast stem-cell stage. Consistent with this inference is the down-regulation of proliferation-related genes in the ESC- and placenta-derived trophoblast cells relative to the ESCu controls (SI Appendix, Fig. S1). Genes down-regulated in trophoblast cells demonstrated significant enrichment in the category “mitotic cell cycle process” (Benjamini–Hochberg corrected P value < 1e-20) and related gene ontology (GO) categories.
Putative Flavivirus Attachment Factor Expression.
Surface exposure of phosphatidylserine on the phospholipid bilayer of enveloped viruses appears to aid viral entry into cells (35, 36). The expressions of five genes encoding potential flavivirus attachment factors [TYRO3, AXL, MERTK, CD209 (DC-SIGN), HAVCR1 (TIM1; hepatitis C receptor] (37–39) were compared in ESCu, the four different trophoblast cell types (ESCd < 40; ESCd > 70; PHTu; PHTd), and trophoblasts from the Roadmap project (40). We have additionally profiled expression levels of these genes in publicly available RNAseq data from chorionic villi samples from late first and early second trimester pregnancies (24) and human neuronal progenitor cells (NPC) (Fig. 1A). The latter served as ZIKV-susceptible controls (10). Additionally, we have compared the expression of these same genes in placenta-associated tissues (amnion, chorion, and maternal decidual tissues) from the data acquired by Kim et al. (42), plus an additional preparation of placental trophoblast (PHT) cells differentiated after retrieval from term placentas (24) (SI Appendix, Fig. S2). Of the genes examined, three, namely those encoding the so-called TAM receptors (TYRO3, AXL, and MERTK), exhibited low FPKM values in the trophoblast that had originated from term placenta (PHTu, PHTd) but significantly higher values in the ESC-derived trophoblast cells (ESCd < 40, ESCd > 70) (Fig. 1A; SI Appendix, Table S2). HAVRCR1 and CD209 were barely expressed in any of the tissues analyzed including amnion and chorion (SI Appendix, Fig. S2). As expected, NPC expressed TYRO3 and AXL, although MERTK, which has been implicated in ZIKV infection of cortical neuronal progenitor cells of the mouse (9), was only expressed weakly. AXL transcripts were high in chorion, but low in amnion and maternal decidual tissue at term. The low expression of TYRO3, AXL, and MERTK genes in trophoblast generated from term pregnancy samples relative to the robust expression in ESC-generated trophoblast is consistent with protein data obtained by Western blotting experiments (Fig. 1B). Western blotting also confirmed the relatively low expression of MERTK and AXL and the relatively high expression of TYRO3 in ESCu.
Fig. 1.
Expression of candidate attachment factors and bridging proteins implicated in ZIKV infection of human cells. (A) Expression levels (in FPKM) of genes for five candidate receptors in ESCu; in CTB (ESCd < 40) and STB (ESCd > 70) fractions from BAP-treated H1 ESCs; in trophoblasts from BMP4-treated H1 ESC (Roadmap Epigenomics Project H1 BMP4 hTBCs) (40); in human chorionic villi collected from late first and early second trimester [samples N05–N10 from (41) in human term placenta-derived CTB (PHTu) and STB (PHTd)]; and in in vitro-derived human NPC, mock-infected samples) (10). (B) Western blots showing expression of TAM receptors (TYRO3, AXL, MERTK) and IFN response proteins (MX2 and IFIT3) in ESCu, ESCd after 4 d and 8 d of differentiation with BAP, and STB derived from term placenta at 24 h and 48 h of in vitro differentiation (PHTd). “ESCd-8d” correlates with the samples in A, consisting of both ESCd < 40 and ESCd > 70 fractions combined. “ESCd-4d” correlates with samples (see Figs. 5 and 6), as it is the time point of initial ZIKV infections. “PHTd-48h” correlates with the sample in A named PHTd. The loading control is TUBA. All data are from the same blot. (C) Expression levels of PROS1 and GAS6 in the same cell types as in A. For TYRO3, AXL, MERTK, PROS1, and GAS6, the differences between ESCd < 40 and PHTu and between ESCd > 70 and PHTd cells were statistically significant (two-tailed t test, equal variance, P < 0.01). Error bars show SD.
PROS1 and GAS6 are natural ligands of the TAM receptors (43, 44) and participate in the cellular uptake of flaviviruses by forming a bridge between the virus and the receptor. Their genes were also expressed significantly higher (P < 0.01) in ESCu-derived trophoblast (ESCd < 40, ESCd > 70) than in trophoblasts from term placental samples (PHTu, PHTd) (Fig. 1C).
Cellular proteoglycans, especially heparan sulfates, form primary attachment sites for many kinds of viruses (45–48), including DENV (49). A major distinction between the trophoblast derived from ESC (ESCd < 40 and ESCd > 70) and that from term placentas (PHTu and PHTd) is the high expression of genes encoding proteins that possess glycosoaminoglycan side chains in ESCd (SI Appendix, Table S3). The ESC-derived trophoblasts, for example, express high concentrations of the mRNAs for HSPG2 (perlecan; heparan sulfate proteoglycan 2, SI Appendix, Fig. S2), DCN (decorin proteoglycan), AGRN (agrin), and LUM (lumican; keratan sulfate proteoglycan, SI Appendix, Table S3), whereas PHTu and PHTd do not express these genes to any significant extent.
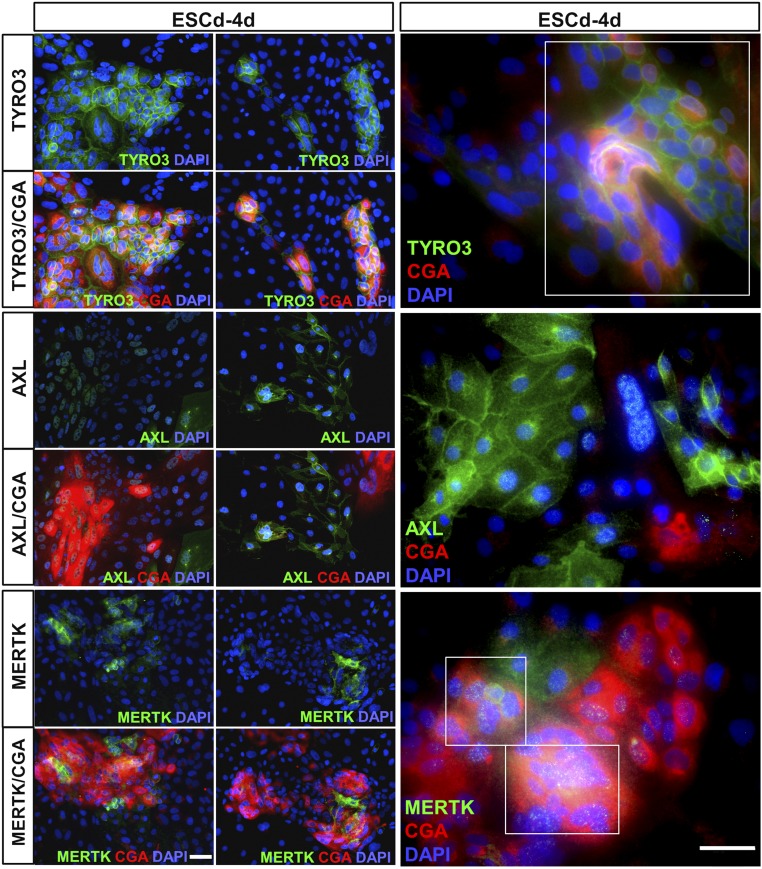
Immunolocalization of the TAM receptors in colonies of ESCu that had undergone BAP differentiation for 4 d (the stage that was subsequently used for ZIKV infection) indicated that MERTK and TYRO3 tended to be coexpressed in areas that were positive for CGA, a marker of presumptive STB. By contrast, AXL expression was most abundantly expressed in the cells not associated with STB (Fig. 2).
Fig. 2.
Putative ZIKV attachment expression in ESCd-4d and its correlation with CGA, a marker for emerging STB. Two representative images have been selected for each receptor at low magnification (Left panels) and one representative image at higher magnification (Right panels). The sample “ESCd-4d” correlates with other samples (see Figs. 5 and 6) as it is the time point of initial ZIKV infections. Immunofluorescent detection of TYRO3 and MERTK correlates with CGA, whereas AXL expression is mainly outside of the CGA+ areas. The far Right panels further highlight the coexpression of TYRO3 and MERTK with CGA, indicated by white boxes. Red, CGA; green, receptors; blue, DAPI. (Scale bars, 50 μm.)
Expression of Genes Indicative of an IFN Response.
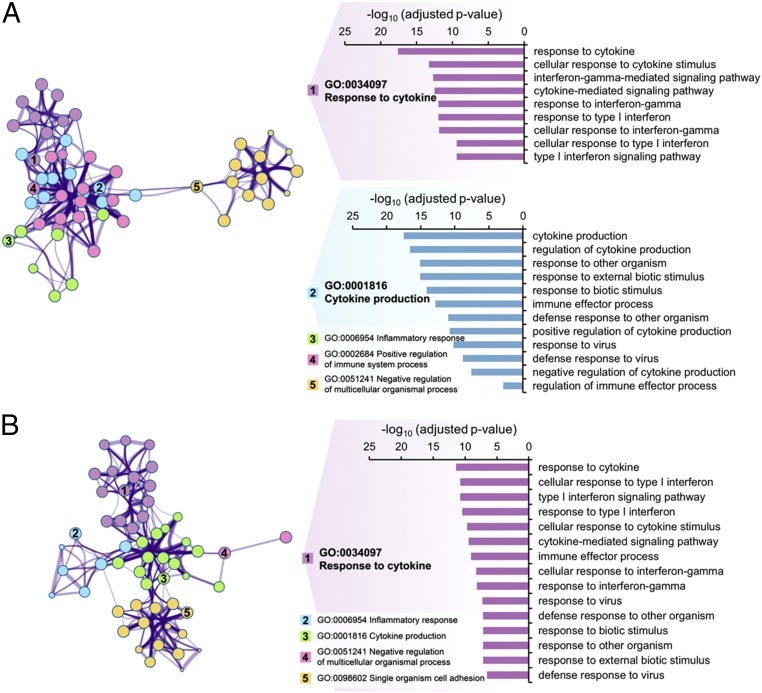
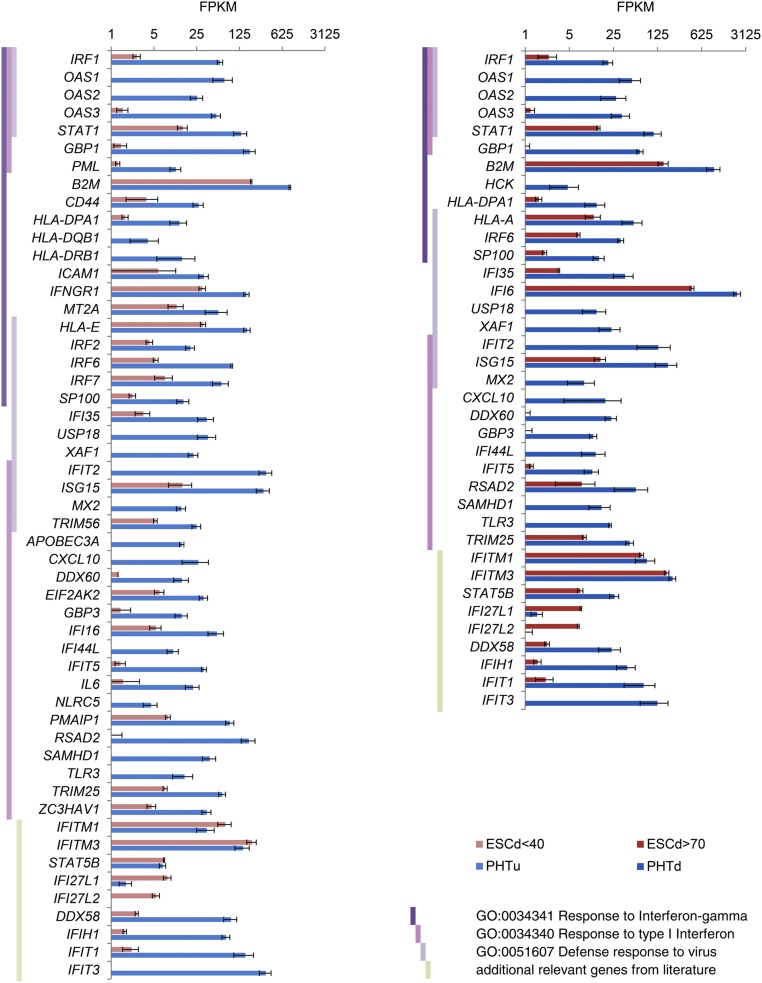
An examination of previously published RNAseq data (32) through use of Biological Process GO terms indicated an enrichment of terms associated with responses to pathogens and type I and type II IFN in PHTu and PHTd cells relative to ESCd < 40 and ESCd > 70 cells (Fig. 3). Fig. 4 compares expression values in FPKM for genes that were differentially expressed in these GO categories, namely Response to IFN gamma, Response to Type I IFN, and Defense response to virus. These genes were highly up-regulated in the PHTu and PHTd cells relative to ESCd < 40 and ESCd > 70 (Dataset S1), respectively. Transcripts for many of the genes analyzed were barely detectable (FPKM values < 1) in the trophoblast cells generated from ESC, whereas transcripts for the majority of others were at least 10-fold higher in placenta-derived CTB and STB. The only exceptions to these generalities were the two IFN-inducible proteins IFI27L1 and IFI27L2, which exhibited higher expression in ESCd < 40 and ESCd > 70 than in the placenta-derived cells (PHTu and PHTd), and the IFN-induced transmembrane proteins IFITM1, IFITM3, and STAT5B, which were not differentially expressed. STAT2 and IRF2, which do not appear in Fig. 4 but encode proteins involved in IFN signaling, were also not differentially expressed.
Fig. 3.
GO terms of genes associated with responses to pathogens and down-regulated in ESC-derived trophoblast cells relative to placenta-derived trophoblast cells. The figures represent clusters of related, enriched Biological Process GO terms of genes that showed strong significant down-regulation (q-value < 0.01, fold change > 4) in ESCd < 40 versus PHTu cells (A) and in ESCd > 70 versus PHTd cells (B). Note that these GO terms are related to responses to pathogens. The network view on thte Left represents GO categories by nodes; shared genes between nodes are linked by lines. The number of genes in a given GO category is represented by the size of the corresponding node, and the number of shared genes between two given GO categories is reflected by the thickness of the line that connects the nodes. Shown are the five most significantly enriched clusters, which are named in accordance with the most significantly enriched GO term within each one of the categories. Genes in GO categories Response to IFN gamma, Response to Type I IFN, and Defense response to virus are discussed in the text (see Fig. 4 for representative gene expressions). These genes are located either in cluster 1 or 2 in A, but all belong to cluster 1 in B. The bar graphs on the Right show enrichment significance of all GO categories that belong to these clusters. Enrichments for all GO terms within the shown clusters are presented in SI Appendix, Table S2.
Fig. 4.
Expression levels (in FPKM) for representative genes from Response to IFN gamma, Response to type I IFN, and Defense response to virus GO categories. These genes showed strong, significant down-regulation (q-value < 0.01, fold change > 4) in the ESCd < 40 and ESCd > 70 cell fractions relative to PHTu and PHTd cells, respectively. Vertical dark violet, pink, and light-violet bars indicate affiliation of a given gene with the GO categories mentioned in the key at the lower right. Light green vertical bars indicate that a given gene was added based on published data regarding its role in immune response. Error bars show SD.
Two known viral defense proteins, MX2 and IFIT3, whose genes were up-regulated in the PHTu and PHTd cells (Fig. 4), could be detected by Western blots of extracts of these cells prepared from term placentas, whereas they were absent in the ESC-derived cells (Fig. 1B). Together, these data suggest that the cells derived from term placentas and differentiated in vitro possess a poised innate immune system, encompassing a large number of genes normally responsive to type I and type II IFN.
Expression of IFN and IFN Receptors.
Both the ESC-derived and term placenta-derived trophoblast cells showed relatively low expression of genes encoding receptor subunits for type I IFN (IFNAR1 and IFNAR2) and the IFNLR1 subunit for type III IFN (IFNλ; IFNL) (SI Appendix, Fig. S3 and Table S2). These data suggest that the ESC-derived cells, in particular, have a poor potential to respond to type I or type III IFN, despite the fact that the gene encoding IL10RB, the second subunit for IFNL, was modestly up-regulated in the cells derived from term placenta (PHTu and PHTd) and placental villi from earlier in pregnancy. On the other hand, PHTu and PHTd, but not early chorionic villi, ESCd < 40 and ESCd > 70, strongly expressed IFNGR1, the gene encoding the main binding subunit for type II IFN (SI Appendix, Fig. S3). IFNG itself was not, however, markedly up-regulated in PHTu and PHTd (SI Appendix, Fig. S4). Expression of genes for two type I IFN (IFNE and IFNB1) and three type III IFN (IFNL1–3) was detectable but quite low in all of the cell fractions (SI Appendix, Fig. S4). Although ESCd < 40 and ESCd > 70 were marked by expression of IFNE, PHTu and PHTd predominantly expressed IFNB, IFNL, and IFNG. Transcripts for IFNA genes were barely detectable in any cell fractions.
Relative Susceptibilities of ESC and ESC-Derived Trophoblast to ZIKV Infection.
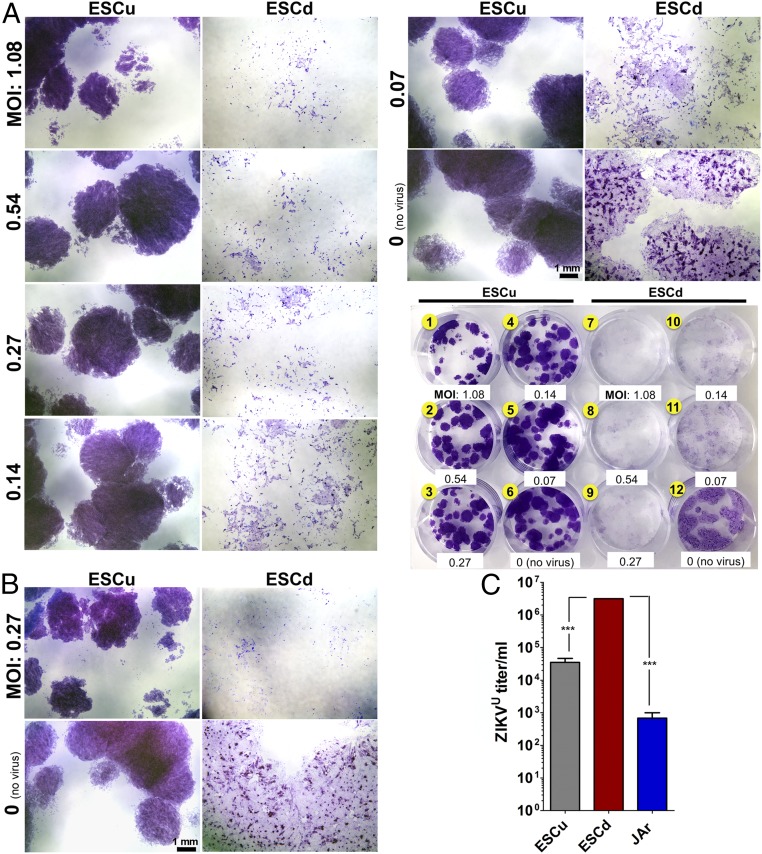
PHTu and PHTd from term placentas are highly resistant to infection by viruses, including the flaviviruses ZIKV and DENV (23, 24), an observation consistent with the high expression of a full range of viral defense genes described above. Here we describe the effects of two strains of ZIKV—the African Lineage strain MR766 (ZIKVU) (50) and the Asian Lineage strain Cambodia FSS13025 (ZIKVC) (51)—on ESCu and on ESC after their differentiation to trophoblast (ESCd). The effects of the two ZIKV strains were examined on ESCu cultured in parallel with the same cells exposed to BAP conditions for 4 d, at which time both types of colony were infected at relatively low multiplicities of infection (MOI), ranging from 0.07 to 1.08 (Fig. 5A). Differentiation, although it slowed proliferation, caused the ESCd to increase in surface area, causing the colonies to spread. As a result, the colonies of ESCd also stained less intensely with crystal violet, except in regions in which STB, notable as relatively intense purple zones, had formed. Within 48 h, cultures of BAP-differentiated colonies exposed to ZIKVU at all of the tested MOI showed a loss of structural integrity (Fig. 5A, wells 7–11), whereas BAP-differentiated colonies exposed to ZIKVC showed minimal cell lysis even at the highest MOI (SI Appendix, Fig. S5A). At 72 h, colonies exposed to 0.27 and 0.027 MOI of ZIKVU had largely disintegrated, with considerable destruction also observed at an MOI of 0.0027 (Fig. 5A and SI Appendix, Fig. S6C, wells 7–9). By contrast, ZIKVC was much less lytic. At 48 h, little or no morphological effects were visible in any of the treatments, and, even by 72 h, only those colonies infected at an MOI of 0.27 demonstrated limited signs of structural damage (SI Appendix, Fig. S6 A, B, and D, well 7).
Fig. 5.
Relative susceptibilities of ESCu and ESCd to ZIKVU infection (A and B) and release of infection-competent virus by ESCu and ESCd following ZIKVU infection (C). (A) After passage of ESCu onto a 12-well plate, colonies in six wells (#1–6) were maintained undifferentiated with MEF-conditioned medium supplemented with FGF2 (ESCu). The colonies in the other six wells (wells 7–12) were differentiated into trophoblasts with BAP (ESCd). After 4 d, the cells were infected with ZIKVU (MOIs are presented in the image) for 1 h. Two wells (wells 6 and 12) were respective controls with no virus. At 48 h (A) and 72 h (B) postinfection, cells were stained with crystal violet. Individual wells were imaged by using the Leica M205 FA Stereo Microscope to demonstrate cytopathic effects, and the whole-plate image is featured as a reference. Colonies of ESCu (wells 1–6) showed deeper staining than ESCd because of higher cell density per colony. (B) The images presented are at the median MOI (0.27) to demonstrate the severity of cell lysis in ESCd compared with ESCu 72 h after ZIKVU infection. (Scale bars, 1 mm.) (C) ESCu, ESCd, and JAr cells were infected with ZIKVU at an MOI of 0.27. New medium was placed on the cells 24 h postinfection and then collected 24 h later (48 h postinfection). ZIKV titer was determined by TCID50 by means of three biological replicates of the collected supernatants. The viral titer was higher in ESCd compared with ESCu and JAr cells (***P < 0.0001) (one-way ANOVA).
The controls (ESCu) appeared little affected 48 h after infection with either strain of virus, demonstrated here for ZIKVU (Fig. 5A, wells 1–6). By 72 h, there were some indications that the ESCu colonies were showing loss of integrity after infection at an MOI of 0.27 (Fig. 5B and SI Appendix, Fig. S6C, well 1). The ESCu appeared unaffected by ZIKVC infection at either 48 h or 72 h (SI Appendix, Figs. S5A and S6 A, B, and D, wells 1–6). Clearly, the ESCu were much less susceptible than ESCd to lytic damage by either virus.
JAr choriocarcinoma cells were then infected (MOI 0.27) to determine whether they were equivalently susceptible to ZIKVU and ZIKVC (SI Appendix, Fig. S7) as ESCu and ESCd were, or, like PHTu and PHTd cells (24), completely resistant to virus. Clearly, the dramatic cell lysis observed in ESCd after infection with ZIKVU (Figs. 5A and 6 A and B and SI Appendix, Fig. S6C, wells 7–11) was not seen in the JAr cells infected with either ZIKVU or ZIKVC (SI Appendix, Fig. S7, Lower panels). On the other hand, the JAr cells had not resisted infection completely by either viral strain, as evident by the presence of small areas staining positively for capsid protein (SI Appendix, Fig. S7, Upper panels). JAr cells were clearly more resistant to ZIKV of either strain than were ESCd.
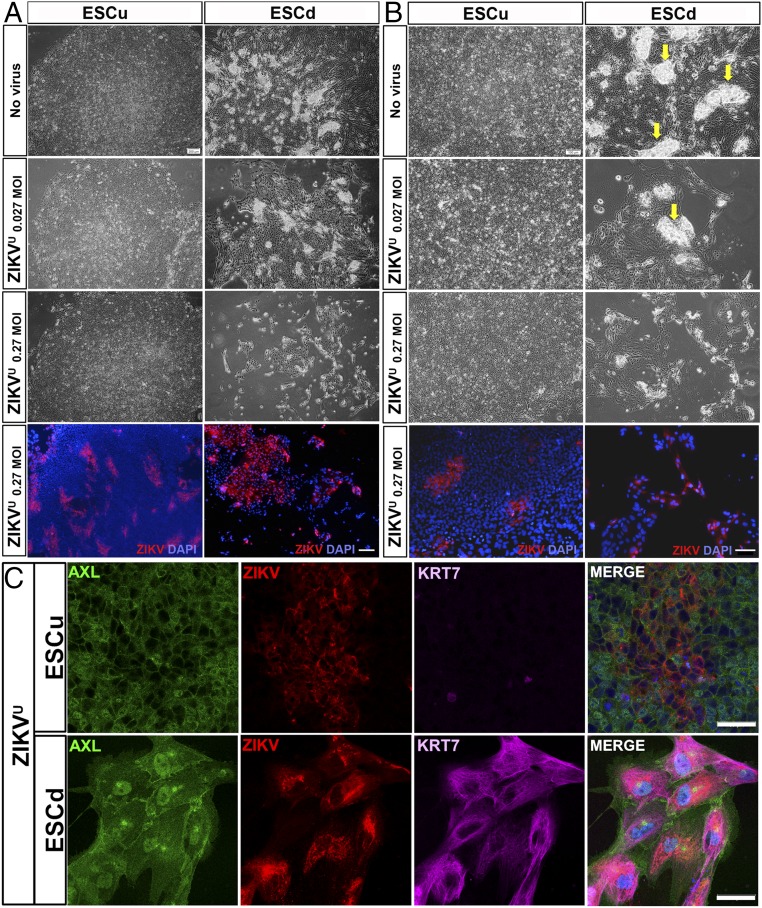
Fig. 6.
Cytopathic effect of ZIKVU on ESCu and ESCd 48 h after infection. Upper three panels of A and B are phase-contrast images of colonies of no virus control (Top) and ZIKVU-infected cells (second and third rows) at low (A) and high (B) magnifications. Although ESCu showed no signs of cell lysis even at the highest virus concentration (0.27 MOI in A and B), a cytopathic effect on ESCd was evident even at the lower virus titer (0.027 MOI in A and B). In ESCd, areas of presumptive STB are selectively lost as virus titer is increased (arrows in B). Bottom panels of A and B are immunofluorescent (IF) images. Cell nuclei were stained with DAPI (blue) and, for ZIKV antigen, with monoclonal antibody 4G2 (red). (C) Confocal images show the presence of AXL (green), ZIKV (red), and KRT7 (magenta) in ESCu and ESCd ZIKVU-infected cells. [Scale bars: 200 μm (A) and 100 μm (B) for both phase-contrast and IF images and 50 μm (C).]
Even when examined under phase microscopy at higher magnification (Fig. 6B), the control cells (ESCu) infected with ZIKVU showed few overt signs of pathology at 48 h, although it was clear from immunolocalization of surface antigen that ZIKV had established itself in restricted areas of the colonies without causing obvious cell destruction (Fig. 6). Similar restricted sites of infection were observed with ZIKVC at a MOI of 0.27, especially at 72 h postinfection (SI Appendix, Fig. S6 A and B). By contrast, ESCd showed clear signs of physical damage after infection with ZIKVU at a MOI of 0.027 (Fig. 6 A and B). At a MOI of 0.27, the colonies had become depleted of the denser STB regions, and only fragments of CTB monolayers remained attached, nearly all of them containing virus (Fig. 6 A and B). ESCd infected with ZIKVC showed minor indications of cell lysis at 72 h postinfection (SI Appendix, Fig. S6 A and B) when virus was detected in the majority of the cells (SI Appendix, Fig. S6 A and B).
When the ESCd were viewed by confocal microscopy after they had been exposed to ZIKVU at a MOI of 0.27 48 h earlier, it was clear that all of the infected cells had remained positive for the trophoblast marker KRT7 and that uninfected AXL-positive cells still mingled with infected cells in the patches of the colonies that remained attached to the substratum (Fig. 6C, Lower panels). Confocal microscopy also provided better definition of the infected areas of ESCu, which also remained positive for AXL (Fig. 6C, Upper panels).
Release of Infection-Competent Virus by ESC and ESC-Derived Trophoblast Following ZIKV Infection.
Here, samples of medium collected in experiments similar to those described in Figs. 5 and 6 and in SI Appendix, Fig. S7, were assessed for the quantity of infectious virus released by TCID50 assays on monolayers of Vero cells. Almost 100-fold more infectious virus was released by the ESCd cells than by the ESCu cells after they had been exposed to 0.27 MOI of ZIKVU 48 h earlier (Fig. 5C). The amount of virus released from JAr cells was lower still, but nevertheless measurable. ESCd also released significantly more infectious viral particles than ESCu, following exposure to ZIKVC (SI Appendix, Fig. S5B), but no infectious virus was detectable in the medium from the JAr cells.
To compare the replication rates between ZIKVU and ZIKVC over time after infecting ESCd colonies, growth curves were established for the period 36–72 h postinfection (SI Appendix, Fig. S5C). ESCd infected with ZIKVU failed to show an increase in viral titer after 36 h, a time likely coinciding with the beginning of colony destruction. ZIKVC titers showed a slight upward trend after 36 h, but were always one to two orders of magnitude below those noted with ZIKVU. These results reinforce the conclusion that ESCd are significantly more susceptible to ZIKVU than to ZIKVC.
Discussion
ZIKV entry into target cells is a complex, multistep process and has implicated several host molecules, including the TAM receptors, HAVCR1 (the hepatitis C receptor, sometimes known as TIM1 or TIMD1), CD209 (DC-SIGN1), attendant linker proteins PROS1 and GAS6 that associate with phosphatidylserine on the capsid surface, and proteoglycans, especially heparan sulfates that appear to form the primary attachment sites for many kinds of viruses (45–48), including DENV2 (49). Villous trophoblast cells obtained from term placentas, which are resistant to a wide range of viruses (23), including ZIKV (24), appear to lack most, if not all, these candidate attachment factors (Fig. 1 A and B and SI Appendix, Fig. S2 and Tables S2 and S3), thus providing one possible explanation for why these cells are not readily infected by ZIKV.
Another explanation for the resistance of the villous trophoblast to ZIKV is that the cells express a poised innate immune system (Figs. 3 and 4). It has long been clear that a preactivated innate immune system, usually induced by IFN after the IFN has been released from infected cells, underpins the ability of an animal to limit the further spread of a virus (52). Among the IFN response genes highly up-regulated in the cells obtained from term placentas are ones induced by ZIKV infection of primary human skin cell cultures (37), which include DDX58 (RIG-1), IFIH1 (MDA5), TLR3, ISG15, and OAS family members (Fig. 4). Many of these same genes are also observed in DENV-infected human keratinocytes (53). It is unusual, however, for tissues to display a constitutive expression of such a network of genes, as noted here with the PHTu and PHTd cells from term placentas (Fig. 3), except under certain pathological conditions (54). However, a comparison of neuronal cell types from the mouse has indicated that aspects of innate immune response networks are differentially expressed in the brain, likely explaining differences in regional permissiveness to neurotropic viruses (55). One possible explanation for the up-regulated immune response pathways in PHTu and PHTd cells (Figs. 3 and 4) is that the virus-resistant state of the cells was acquired by prior IFN exposure. The cells do express transcripts for the type I IFNs, IFNB and IFNE, and for the type III IFN, IFNL (SI Appendix, Fig. S4), but the corresponding levels of receptor mRNA are low (SI Appendix, Fig. S3). On the other hand, the PHTu cells, in particular, express transcripts for both IFNGR1 and IFNGR2 (SI Appendix, Fig. S3), and GO analyses strongly implicate IFNG as a possible upstream regulator of the observed innate response network (Fig. 3). These cells also express members of the chromosome 19 microRNA cluster (56), which have been implicated in conferring viral resistance when introduced into previously susceptible cells (23). Together, these features, namely low expression of genes encoding virus attachment factors and an up-regulated antiviral response system, reinforce the concept that the cells isolated from term placentas are in a strong position to minimize infection and viral replication.
Despite the growing acceptance that ZIKV-associated microcephaly is associated with infection in the first trimester, ambiguity still remains as to whether ZIKV can access the fetus via the placenta throughout pregnancy to cause central nervous system abnormalities and also whether alternative routes not involving STB might exist (57). Additionally, damage to STB, for example, by herpes simplex virus (58), or transcytosis of ZIKV across STB by piggybacking on an Ig directed against another flavivirus, such as one of the DENV serotypes (18, 59), could be other causes of fetal ZIKV infection beyond the first trimester and reasons for geographical and socioeconomic differences in fetal susceptibilities. Whole villi dissected from placentas at different stages of pregnancy and cultured in the presence of ZIKV certainly become infected (57, 58, 60, 61), but STB remains largely unscathed. In the in vitro trophoblast system derived from term placenta, it also seems possible that infection occurs as a result of damage incurred during isolation and culture of the tissue and exposure to infectious virus through the open ends of the explants. In other words, this model might present a false indication of placental susceptibility.
Unlike trophoblast derived from term placentas, ESC differentiated under BAP conditions for 4 d, which we suggest is analogous to the primitive trophoblast of the early first trimester, are highly susceptible to infection by ZIKVU. The colonies are quickly destroyed, with the STB component apparently succumbing more rapidly than the CTB (Figs. 5 and 6). The RNAseq data provide a possible explanation for this vulnerability. First, unlike the PHTu and PHTd cells, ESCd < 40 and ESCd > 70 do not possess an already poised antiviral defense system (Figs. 3 and 4). Rather, with a few exceptions, there is low expression of genes associated with the innate immune response (Fig. 4). It is also uncertain whether these cells could even mount a timely antiviral response because transcript levels of genes that express receptors that engage type I or type II IFN are very low (SI Appendix, Fig. S3). Mouse ESC (62) and embryonal carcinoma cells (63), for example, do not respond to type I IFN. A second reason underpinning the susceptibility of ESC-generated trophoblast to ZIKV may be that the cells permit easy viral access. Flaviviruses enter the host cell by way of endosomes (36) in a process sometimes known as “apoptotic mimicry” (44). Although two factors implicated in ZIKV entry into skin cells, CD209 (DG-SIGN) and HAVCR1 (TIM-1) (37), appear not to be expressed in ESCd < 40 and ESCd > 70 (Fig. 1A), the genes for several others, including AXL and TYRO3, are (Fig. 1 A and B and SI Appendix, Fig. S2). Transcripts for the third member of the TAM trio, namely MERTK, are also expressed in the differentiated ESC. At d 4 of differentiation, the time the cultures were exposed to ZIKV, MERTK, like TYRO3, is most concentrated in areas positive for the α-subunit of hCG, CGA, which is a marker that predicts future STB (32, 64) (Fig. 2), and, as discussed above, STB seems to be the most immediate target for cellular destruction in infected colonies. It remains unclear why ESCu cells are less sensitive to ZIKVU than their differentiated counterparts. Possibly it is due to low expression of MERTK or some other entry factor. Nevertheless, it should be recognized that expression of these potential entry factors does not guarantee susceptibility of these tissues to any particular flavivirus, including ZIKV, although an inescapable conclusion from our experiments is that ESC-derived trophoblast is a better target for ZIKVU than PHTu and PHTd.
Unexpectedly, the Asian strain ZIKVC appeared to showed a lower predilection for the ESCd than ZIKVU (Figs. 5 and 6 and SI Appendix, Figs. S5 and S6). Of note, the ESCd infected with ZIKVC at MOI 0.27 displayed minimal cell lysis, even after 72 h, yet a majority of the cells contained virus (SI Appendix, Fig. S6). Production of infection-competent virus was also much lower with ZIKVC than with ZIKVU (Fig. 5 and SI Appendix, Fig. S5). Somewhat similar results were obtained with monolayers of JAr choriocarcinoma cells, although such cells were clearly much less susceptible than ESCd (SI Appendix, Fig. S7). Together, these results suggest that ESCd are vulnerable to both strains of ZIKV, although the Cambodian strain (ZIKVC) may infect less readily and cause less cell lysis. We have no ready explanation for this observation, although it is clearly of interest, as it is the Asian strains of ZIKV that have been linked to fetal and pediatric brain abnormalities. Others have noted similar differences in virulence between an African and Asian strain on human neural cells and astrocytes and suggested that African strains should not necessarily be dismissed as benign with regard to causing neurological impairment (15).
What, then, do the ESC-derived trophoblasts represent? They display a progression of differentiation over time, expressing markers of trophoblast stem cells soon after initiating differentiation, but, a few days later, after the stem cell signature genes have more or less been down-regulated, STB formation begins (29, 31, 44, 65, 66). By day 8, the transcriptome signature of the ESCd > 70 STB fraction is so different from that of PHTd, the placenta-derived STB fraction, that it is highly improbable that the two could be functional homologs (32). STB arises at two distinct stages of placental development. Whereas the PHTd corresponds to the surface layer that covers the exterior of placental villi, a very different type of STB arises at around the time the conceptus first establishes itself in the wall of the uterus (67). We have argued that the STB that forms from ESC by BAP treatment (ESCd > 70) represents this early syncytium (32). Unfortunately, this period of early pregnancy is poorly studied because of the lack of available human tissue. Most of what is known has come from older literature (68–70), archived samples (71), and from analogies drawn from histological studies on nonhuman primates (71–73). As the primate embryo implants, a penetrating mass of invasive STB forms in the implantation zone and soon surrounds the conceptus. Such a syncytium also forms in human conceptuses cultured for 13 d or 14 d in vitro, when it forms the periphery of a layer of trophoblast encircling the epiblast (74, 75). It has been inferred that this syncytial mass, as it advances within the uterine wall, hollows out lacunae, which become filled with fluid from breached uterine glands and maternal blood vessels and likely provision the conceptus (67, 76–78). Within a few days, however, columns of CTB penetrate the STB to form primary villi, which eventually branch, acquire cores of blood vessels and connective tissue, and form the placenta proper.
Clearly, a major limitation to our study is that we cannot be certain that the model represents this primitive trophoblast. Nor have we tested whether or not ESC-derived trophoblast can be infected with other flaviviruses, which might be expected to use similar mechanisms of entry into the cells. Nonetheless, if our hypothesis is correct and the ESCd < 40 and ESCd > 70 represent the trophoblast encompassing the implanting conceptus, this tissue may be at particular risk for infection because it is exposed to maternal blood and fluids. It also may lack a responsive innate immune system that would protect against viral replication and release. Accordingly, we suggest that the developing fetus could be most vulnerable to infection by ZIKV and certain other pathogens, including rubella virus, cytomegalovirus, and herpes simplex virus (59), during a relatively narrow window within the first trimester of pregnancy before a protective zone of more resilient villous trophoblast has become established, yet when organogenesis is being initiated. One speculative explanation for the ostensibly more benign effects of an African strain of ZIKV, such as ZIKVU, on fetal development is that such viruses are so destructive to the primitive trophoblast surrounding the embryo that any pregnancy in its early stages would be terminated, possibly without a significant extension of the mother’s menstrual cycle. On the other hand, infection with an Asian strain may be less destructive to the early placenta and allow the pregnancy to continue and fetal infection to become established.
Materials and Methods
Human ESC Culture and Differentiation.
Human ESC (H1, WA01) were cultured in six-well tissue culture plates (Thermo Scientific) coated with Matrigel (BD Bioscience) under an atmosphere of 5% (vol/vol) CO2/air at 37 °C in mTeSR1 medium (STEMCELL Technologies). Cells were passaged every 5–6 d. The method for trophoblast differentiation has been described elsewhere (29). Briefly, on the day after passaging onto Matrigel-coated dishes at 1.2 × 104 cells/cm2, the culture medium was changed to DME/F12 medium (Thermo Scientific) with knock-out serum replacement (KOSR, Invitrogen) that had been conditioned by mouse embryonic fibroblasts (MEF) and supplemented with FGF2 (4 ng/mL). After 24 h, the conditioned medium was replaced with daily changes of nonconditioned DME/F12/KOSR medium lacking FGF2, but containing BMP4 (10 ng/mL), A83-01 (1 μM), and PD173074 (0.1 μM) (BAP treatment) for up to 8 d. Control cultures (ESCu) were maintained in conditioned medium containing 4 ng/mL FGF2.
Cell Separation on Strainers.
These procedures have been described elsewhere (32). In brief, the colonies were dissociated by using Gentle Cell Dissociation Reagent (STEMCELL Technologies), and the larger STB sheets (ESCd > 70) were collected by passing the suspension through a nylon strainer designed to retain objects >70 μm across (Fisher Scientific). The <40-μm fraction (ESCd < 40), which consists largely of mononucleated cell types, was the cell fraction able to pass through 40-μm cell strainers. The subsequent RNAseq analysis was performed on RNA from ESCd < 40 and ESCd > 70 isolated from BAP-treated H1 ESC in three separate experiments, each performed over a period of 6 wk. In each instance, ESCu cultured in parallel in the presence of FGF2 but without BAP exposure served as controls.
Derivation of PHTu and PHTd.
Placental tissue samples were collected by the Obstetrical Specimen Procurement Unit at Magee-Womens Hospital of the University of Pittsburgh Medical Center. Collection was conducted under an approved exempt protocol by the Human Research Protection Office of the University of Pittsburgh. Patients provided written consent for the use of de-identified, discarded tissues for research upon admittance to the hospital.
Primary villous CTB were derived and cultured according to published procedures (79–81) from three human placentas (one female and two male). Multiple primary cultures were established from each placenta at a density of 3.5 × 105 cells/cm2 in DMEM supplemented with 10% (vol/vol) FBS and antibiotics under a 5% (vol/vol) CO2/air atmosphere at 37 °C. Triplicate cultures from each placenta were harvested at 9 h (PHTu) before syncytium formation and subsequently at 48 h (PHTd) when syncytium formation had occurred. Total RNA was extracted from each sample (3 × 3 at 9 h and 48 h, respectively) to provide a total of 18 samples for RNAseq analysis.
RNAseq Analyses.
RNA was obtained from each size-fractioned sample of cells from ESCu that had been BAP-treated for 8 d and from untreated ESCu controls cultured in parallel (32). The quantitation and quality control of RNA from ESCd < 40 and ESCd > 70 and from the 18 samples derived from the primary PHTu and PHTd was performed on a Fragment Analyzer (Advanced Analytical), and cDNA libraries were constructed by standard methods (Illumina TruSeq mRNA stranded kit) with index adapters (Illumina TruSeq indexes). DNA was then sequenced as single-end, 50 base-length reads on an Illumina HiSEq. 2500 instrument (Illumina Inc.) (Dataset S2). The data have been deposited in the Gene Expression Omnibus (GEO) database (GSE73017). Briefly, the data were preprocessed with the read trimming and cropping tool Trimmomatic v.0.30 (83) and the command line parameters: −phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:20. RNAseq reads were mapped to the reference genome Ensembl GRCh37/hg19 with the sequence-aligning tool Bowtie v.2.2.3 (84) and the splice junction mapper for RNAseq reads TopHat v.2.0.12 (85) with the command line parameters: −no-coverage-search–b2-sensitive (Dataset S2). Uniquely mapped reads were counted with htseq-count v.0.6.1p1 and GTF transcriptome annotation file downloaded from GEO accession GSE57049 (86). Gene expression levels were calculated in FPKM, considering gene length as a sum of all exonic nonoverlapping sequences of all isoforms of a given gene. Tests for differential expression (q-value < 0.01, fold-change > 4; Benjamini–Hochberg P value adjustment method) were done by using DESeq within Spotfire v.6.5.3.25 (TIBCO Software). Networks of related enriched GO terms were created and analyzed with Metascape (87).
Public RNAseq Datasets.
Publicly available RNAseq datasets for normal chorionic villi samples (12.7–14.6 wk of gestation, i.e., late first–early second trimester) from ref. 41 were obtained from GEO database accession GSE42142 (RNAseq samples N5, N7, N8, N9, N10). RNAseq datasets for H1 ESC-derived trophoblast-like cells from the Roadmap Epigenomics Project (40) were downloaded from GEO database accessions GSM915320 and GSM915321. RNAseq reads from human NPC were obtained from GEO entry GSE78711 (10). RNAseq data for amnion, chorion, and decidua samples were retrieved from Sequence Read Archive (SRA) accession SRP017583 (42). Data for PHTd cells (SRA accession SRP072501) were from Bayer et al. (24).
Propagation of ZIKV in Vero Cells.
Vero cells (ATCC; CCL-81) were seeded into T25 flasks (0.5 × 106 cells/mL). At 3 d (confluent density ∼3 × 106 cells/mL) the monolayer was infected with either the ZIKVU (strain MR-766 Uganda, African Lineage, GenBank accession HQ234498.1) or with ZIKVC (strain FSS13025 Cambodia, Asian Lineage, GenBank accession KU955593.1) (51) at an MOI of 0.01. Flasks were incubated at 37 °C for 1 h with gentle rocking. After adding 5 mL additional medium [DMEM, Mediatech, supplemented with 10% (vol/vol) FBS], cultures were maintained for a further 72 h. ZIKV stocks were generated by collecting the medium at 72 h postinfection, when typically 40–50% of cells showed cytopathic effects. Virus stocks were maintained at −80 °C until used.
Tissue Culture Infectious Dose50 Assays.
DMEM (90 µL) supplemented with 10% (vol/vol) FBS was added to each well of a 96-well culture plate. Virus-containing samples (either ZIKVU or ZIKVC) in 10 µL of medium were added in triplicate to the top row of wells in the plate. Virus samples were then serially diluted in 10-fold steps in successive rows of wells. Vero cells from a confluent T25 flask were dissociated with trypsin and resuspended in 11 mL DMEM/10% (vol/vol) FBS. Cell suspension (0.1 mL) was then added to each well of the 96-well plate. At day 7, medium was removed from each well, and the cells were stained with 0.1 mL of crystal violet solution [0.2% wt/vol crystal violet in an aqueous solution containing 10% (vol/vol) formaldehyde and 20% (vol/vol) ethanol] for 10–15 min. After washing with tap water, titer was calculated based on 50% endpoints by using the Reed and Muench algorithm and expressed as log 10 TCID50/mL (88).
Plaque Assay to Analyze ZIKVC Titers.
Vero 81 cells were plated at the density of 5 × 104 cells/well in a 24-well plate and cultured for 3 d at 37 °C under air/5% (vol/vol) CO2. After removing the medium, viral supernatant (150 μL) was added to each well. The culture plates were rocked every 15 min for 1 h before layering 1 mL of plaque assay agarose medium [1× Medium 199 (Sigma-Aldrich), 10% (vol/vol) FBS, 4% (wt/vol) NaHCO3, 0.5% MEM vitamins, 0.5% MEM amino acids (Mediatech Inc.)] above the cells. After the agarose solidified, the plates were turned upside down and maintained for 5 d at 37 °C in an atmosphere of air/5% (vol/vol) CO2. As a substrate, MTT solution [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; 0.15 mL; 0.5%] was added to the wells, and the plates were incubated overnight. The plaques were counted the following day.
Immunofluorescent staining and Western blotting were conducted as described before (28, 29). Primary and secondary antibodies used in Western blotting and immunostainings are summarized SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank D. F. Reith for administrative assistance and Dr. Alexander Jurkevich and the University of Missouri Molecular Cytology Core for assistance with the confocal imaging. Zika virus stocks were prepared and maintained in the virology laboratory of the Laboratory for Infectious Disease Research at the University of Missouri. This study was supported by by National Institutes of Health (NIH) Grant R01HD077108 (to T.E. and D.J.S.) and NIH Grant R01HD067759 (to R.M.R.); by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) Grant 2014/03620-2 (to S.V.-A.); and by Fellowship 2010/51152-7 from FAPESP (to D.Y.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus database (accession no. GSE73017).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616097114/-/DCSupplemental.
References
- 1.Doran KS, Banerjee A, Disson O, Lecuit M. Concepts and mechanisms: Crossing host barriers. Cold Spring Harb Perspect Med. 2013;3(7):a010090. doi: 10.1101/cshperspect.a010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardenas I, et al. Viral infection of the placenta leads to fetal inflammation and sensitization to bacterial products predisposing to preterm labor. J Immunol. 2010;185(2):1248–1257. doi: 10.4049/jimmunol.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delorme-Axford E, Sadovsky Y, Coyne CB. The placenta as a barrier to viral infections. Ann Rev Virol. 2014;1(1):133–146. doi: 10.1146/annurev-virology-031413-085524. [DOI] [PubMed] [Google Scholar]
- 4.Adibi JJ, Marques ET, Jr, Cartus A, Beigi RH. Teratogenic effects of the Zika virus and the role of the placenta. Lancet. 2016;387(10027):1587–1590. doi: 10.1016/S0140-6736(16)00650-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasil P, et al. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med. 2016;375(24):2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira Melo AS, et al. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: Tip of the iceberg? Ultrasound Obstet Gynecol. 2016;47(1):6–7. doi: 10.1002/uog.15831. [DOI] [PubMed] [Google Scholar]
- 7.Campos GS, Bandeira AC, Sardi SI. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21(10):1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mlakar J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374(10):951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 9.Cugola FR, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534(7606):267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang H, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18(5):587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 12.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang J, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19(2):258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19(5):672. doi: 10.1016/j.stem.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Simonin Y, et al. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine. 2016;12:161–169. doi: 10.1016/j.ebiom.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soares de Souza A, et al. Fetal infection by Zika virus in the third trimester: Report of 2 cases. Clin Infect Dis. 2016;63(12):1622–1625. doi: 10.1093/cid/ciw613. [DOI] [PubMed] [Google Scholar]
- 17.Honein MA, et al. US Zika Pregnancy Registry Collaboration Birth defects among fetuses and infants of US women with evidence of possible Zika virus infection during pregnancy. JAMA. 2017;317(1):59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 18.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17(9):1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priyamvada L, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci USA. 2016;113(28):7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simister NE. Placental transport of immunoglobulin G. Vaccine. 2003;21(24):3365–3369. doi: 10.1016/s0264-410x(03)00334-7. [DOI] [PubMed] [Google Scholar]
- 21.Cauchemez S, et al. Association between Zika virus and microcephaly in French Polynesia, 2013-15: A retrospective study. Lancet. 2016;387(10033):2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson MA, Mier-y-Teran-Romero L, Reefhuis J, Gilboa SM, Hills SL. Zika and the risk of microcephaly. N Engl J Med. 2016;375(1):1–4. doi: 10.1056/NEJMp1605367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delorme-Axford E, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA. 2013;110(29):12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayer A, et al. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016;19(5):705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazear HM, et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19(5):720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi SL, et al. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg. 2016;94(6):1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miner JJ, et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell. 2016;165(5):1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yockey LJ, et al. Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell. 2016;166(5):1247–1256 e1244. doi: 10.1016/j.cell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amita M, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110(13):E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz LC, et al. Human embryonic stem cells as models for trophoblast differentiation. Placenta. 2008;29(Suppl A):S10–S16. doi: 10.1016/j.placenta.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, et al. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci USA. 2015;112(18):E2337–E2346. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabe S, et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA. 2016;113(19):E2598–E2607. doi: 10.1073/pnas.1601630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CQ, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 2016;6(2):257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod. 2011;84(3):412–421. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller-Tank S, Maury W. Phosphatidylserine receptors: Enhancers of enveloped virus entry and infection. Virology. 2014;468–470:565–580. doi: 10.1016/j.virol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera-Lecoin M, Meertens L, Carnec X, Amara A. Flavivirus entry receptors: An update. Viruses. 2013;6(1):69–88. doi: 10.3390/v6010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamel R, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;89(17):8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowakowski TJ, et al. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18(5):591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. Flavivirus cell entry and membrane fusion. Viruses. 2011;3(2):160–171. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin S, et al. Global DNA hypermethylation in Down syndrome placenta. PLoS Genet. 2013;9(6):e1003515. doi: 10.1371/journal.pgen.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, et al. Transcriptome landscape of the human placenta. BMC Genomics. 2012;13:115. doi: 10.1186/1471-2164-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8(5):327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharyya S, et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 2013;14(2):136–147. doi: 10.1016/j.chom.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colpitts CC, Schang LM. A small molecule inhibits virion attachment to heparan sulfate- or sialic acid-containing glycans. J Virol. 2014;88(14):7806–7817. doi: 10.1128/JVI.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klimyte EM, Smith SE, Oreste P, Lembo D, Dutch RE. Inhibition of human metapneumovirus binding to heparan sulfate blocks infection in human lung cells and airway tissues. J Virol. 2016;90(20):9237–9250. doi: 10.1128/JVI.01362-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neal JW. Flaviviruses are neurotropic, but how do they invade the CNS? J Infect. 2014;69(3):203–215. doi: 10.1016/j.jinf.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Connell BJ, Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: From attachment to entry inhibition. Front Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talarico LB, et al. The antiviral activity of sulfated polysaccharides against dengue virus is dependent on virus serotype and host cell. Antiviral Res. 2005;66(2-3):103–110. doi: 10.1016/j.antiviral.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46(5):509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 51.Haddow AD, et al. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Negl Trop Dis. 2012;6(2):e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borden EC, et al. Interferons at age 50: Past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Surasombatpattana P, et al. Dengue virus replication in infected human keratinocytes leads to activation of antiviral innate immune responses. Infect Genet Evol. 2011;11(7):1664–1673. doi: 10.1016/j.meegid.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15(7):429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 55.Cho H, et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med. 2013;19(4):458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donker RB, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18(8):417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabata T, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe. 2016;20(2):155–166. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aldo P, et al. HSV-2 enhances ZIKV infection of the placenta and induces apoptosis in first-trimester trophoblast cells. Am J Reprod Immunol. 2016;76(5):348–357. doi: 10.1111/aji.12578. [DOI] [PubMed] [Google Scholar]
- 59.Coyne CB, Lazear HM. Zika virus: Reigniting the TORCH. Nat Rev Microbiol. 2016;14(11):707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 60.Jurado KA, et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight. 2016;1(13):e88461. doi: 10.1172/jci.insight.88461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quicke KM, et al. Zika virus infects human placental macrophages. Cell Host Microbe. 2016;20(1):83–90. doi: 10.1016/j.chom.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, et al. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem. 2013;288(22):15926–15936. doi: 10.1074/jbc.M112.421438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harada H, et al. Absence of the type I IFN system in EC cells: Transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63(2):303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 64.Hoshina M, Boothby M, Boime I. Cytological localization of chorionic gonadotropin alpha and placental lactogen mRNAs during development of the human placenta. J Cell Biol. 1982;93(1):190–198. doi: 10.1083/jcb.93.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horii M, et al. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA. 2016;113(27):E3882–E3891. doi: 10.1073/pnas.1604747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchand M, et al. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod. 2011;84(6):1258–1271. doi: 10.1095/biolreprod.110.086413. [DOI] [PubMed] [Google Scholar]
- 67.James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta. 2012;33(5):327–334. doi: 10.1016/j.placenta.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 68.Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. Am J Anat. 1956;98(3):435–493. doi: 10.1002/aja.1000980306. [DOI] [PubMed] [Google Scholar]
- 69.Boyd JD, Hamilton WJ. The Human Placenta. Heffer & Sons; Cambridge, UK: 1970. [Google Scholar]
- 70.Amoroso E. Placentation. In: Parkes A, editor. Marshall’s Physiology of Reproduction. Vol 2. Little Brown & Co.; Boston: 1952. pp. 127–311. [Google Scholar]
- 71.Enders AC. Trophoblast differentiation during the transition from trophoblastic plate to lacunar stage of implantation in the rhesus monkey and human. Am J Anat. 1989;186(1):85–98. doi: 10.1002/aja.1001860107. [DOI] [PubMed] [Google Scholar]
- 72.Enders AC, King BF. Early stages of trophoblastic invasion of the maternal vascular system during implantation in the macaque and baboon. Am J Anat. 1991;192(4):329–346. doi: 10.1002/aja.1001920403. [DOI] [PubMed] [Google Scholar]
- 73.Enders AC, Lantz KC, Peterson PE, Hendrickx AG. From blastocyst to placenta: The morphology of implantation in the baboon. Hum Reprod Update. 1997;3(6):561–573. doi: 10.1093/humupd/3.6.561. [DOI] [PubMed] [Google Scholar]
- 74.Shahbazi MN, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18(6):700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deglincerti A, et al. Self-organization of the in vitro attached human embryo. Nature. 2016;533(7602):251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 76.Hustin J, Schaaps JP. Echographic [corrected] and anatomic studies of the maternotrophoblastic border during the first trimester of pregnancy. Am J Obstet Gynecol. 1987;157(1):162–168. doi: 10.1016/s0002-9378(87)80371-x. [DOI] [PubMed] [Google Scholar]
- 77.Foidart JM, Hustin J, Dubois M, Schaaps JP. The human placenta becomes haemochorial at the 13th week of pregnancy. Int J Dev Biol. 1992;36(3):451–453. [PubMed] [Google Scholar]
- 78.Burton GJ, Jauniaux E, Watson AL. Maternal arterial connections to the placental intervillous space during the first trimester of human pregnancy: The Boyd collection revisited. Am J Obstet Gynecol. 1999;181(3):718–724. doi: 10.1016/s0002-9378(99)70518-1. [DOI] [PubMed] [Google Scholar]
- 79.Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF., III Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118(4):1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- 80.Nelson DM, Johnson RD, Smith SD, Anteby EY, Sadovsky Y. Hypoxia limits differentiation and up-regulates expression and activity of prostaglandin H synthase 2 in cultured trophoblast from term human placenta. Am J Obstet Gynecol. 1999;180(4):896–902. doi: 10.1016/s0002-9378(99)70661-7. [DOI] [PubMed] [Google Scholar]
- 81.Schaiff WT, et al. Peroxisome proliferator-activated receptor-gamma and retinoid X receptor signaling regulate fatty acid uptake by primary human placental trophoblasts. J Clin Endocrinol Metab. 2005;90(7):4267–4275. doi: 10.1210/jc.2004-2265. [DOI] [PubMed] [Google Scholar]
- 82.Yunusov D, et al. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci Rep. 2016;6:32753. doi: 10.1038/srep32753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cabili MN, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tripathi S, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li MI, Wong PS, Ng LC, Tan CH. Oral susceptibility of Singapore Aedes (Stegomyia) aegypti (Linnaeus) to Zika virus. PLoS Negl Trop Dis. 2012;6(8):e1792. doi: 10.1371/journal.pntd.0001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.