Significance
How high levels of aggression are generated in any organism is poorly understood, especially the genetic basis. Analyses of a hyperaggressive line of fruit flies (Bullies) generated by a genetic selection approach revealed a loss of the aggressive phenotype when animals were reared at a lower temperature. This effect offered an opportunity to perform RNA-seq analyses searching for genetic differences specifically related to hyperaggression. The results showed a small number of gene differences of twofold or more in the Bullies; one is a member of a neutral amino acid family of transporters suggested to be important in glutamate and GABA neurotransmitter regulation. Lowering levels of this gene in Drosophila melanogaster partially duplicates the high-aggression phenotype.
Keywords: aggression, Drosophila, transmembrane neutral amino acid transporter
Abstract
By selection of winners of dyadic fights for 35 generations, we have generated a hyperaggressive Bully line of flies that almost always win fights against the parental wild-type Canton-S stock. Maintenance of the Bully phenotype is temperature dependent during development, with the phenotype lost when flies are reared at 19 °C. No similar effect is seen with the parent line. This difference allowed us to carry out RNA-seq experiments and identify a limited number of genes that are differentially expressed by twofold or greater in the Bullies; one of these was a putative transmembrane transporter, CG13646, which showed consistent and reproducible twofold down-regulation in Bullies. We examined the causal effect of this gene on the phenotype with a mutant line for CG13646, and with an RNAi approach. In all cases, reduction in expression of CG13646 by approximately half led to a hyperaggressive phenotype partially resembling that seen in the Bully flies. This gene is a member of a very interesting family of solute carrier proteins (SLCs), some of which have been suggested as being involved in glutamine/glutamate and GABA cycles of metabolism in excitatory and inhibitory nerve terminals in mammalian systems.
Aggression is an innate, complex social behavior, whose manifestation is continually modifiable by social context. Great heterogeneity exists in the display of aggressiveness among individuals in competition for desired resources, even within inbred populations of organisms. The root causes of this heterogeneity are relatively unknown, with interactions among genes, development, and environment commonly offered as underlying explanations. Moreover, how organisms transition from safer, lower levels of aggression that initiate conflicts, to potentially damaging, higher levels of aggression that end conflicts and establish rank orders of organisms, also is poorly understood. In this article, we begin to address several of these issues by comparing the genetic makeup of a highly aggressive line of flies called Bullies (selected by inbreeding) to a parental wild-type stock of Canton-S flies (1), and identifying candidate nervous system genes important in establishing higher levels of aggression.
In both vertebrate and invertebrate nervous systems, numerous genes of widely varying functions have been identified as important contributors to aggression (2–8), including small molecules, biogenic amines, and peptide transmitter-related genes, their receptors, and their second messenger targets (9–15); steroid hormones and their membrane and nuclear receptor targets (16, 17); transcription regulators (18); and key metabolic regulators (16, 19, 20). Many of these genes were identified using transgenic mutants directed at specific genes. Recent studies have taken a more unbiased approach and identified multiple new genes by comparing differential mRNA transcript abundance between selected lines of highly aggressive and less-aggressive animals (21).
In recent years, Drosophila has been established as a strong model system for the study of aggression displaying characteristic fighting patterns that can be examined by forward and reverse genetic approaches, and by a powerful genetic toolkit, including binary systems that allow gene expression to single-cell levels (22–24). After originally being described briefly in 1915 (25), aggression in Drosophila was further described in more detail by Jacobs (26) and Dow and von Schilcher (27), with the latter paper being the first to describe an escalated high-intensity boxing behavior. In 2002, Chen et al. (28) reported a dyadic fight setup that has since become a standardized aggression paradigm in Drosophila. As in vertebrate models, a majority of the genes identified through a candidate gene approach in studies of Drosophila aggression belong to the biogenic amine and peptide families. Thus, the amines serotonin (29, 30), dopamine (31), and octopamine (32) have been shown to modulate levels of aggression, along with the peptides tachykinin (33) and NPF (29). In addition, candidate gene approaches also have identified tailless, a fly ortholog of mouse nr2e1, and its corepressor, atrophin, as influencing aggressive behaviors (34). Unbiased screens have also been used to identify behavioral candidate loci in Drosophila by choosing extreme phenotypes in selection experiments (35, 36). In relation to aggression, similar approaches have been taken in recent years by several laboratories via selection of hyperaggressive flies (1, 37, 38). Transcriptome analysis of aggressive compared with less-aggressive lines generated lists of many genetic differences with little agreement between the results of different laboratories. One gene that was followed up on by several laboratories was a member of the cytochrome P450 gene family that is closely associated with sensory signal detection (37, 39). Even after such initial forward and reverse genetic screens, the genetic basis of aggression in Drosophila remains poorly understood.
In this study we took advantage of the well-characterized aggression phenotype displayed by the hyperaggressive Bully flies (1) and began a search for any underlying genetic roots. Using RNA-seq and quantitative RT-PCR (qRT-PCR), we identified six genes with consistent twofold or greater differential transcript abundance in Bully heads compared with the heads of nonselected parental Canton-S flies. One gene, CG13646, a putative neutral amino acid transmembrane transporter, showed consistent and significant down-regulation that was correlated with the hyperaggression phenotype. Down-regulation of CG13646 using a P-element insertion mutant and a pan-neuronally driven RNAi line, resulted in flies that showed higher-level aggression and that had a competitive advantage over control flies. These findings demonstrated that the expression level of CG13646 significantly influences the display of higher-level aggression in Drosophila.
Results
Hyperaggressive Phenotype in Bully Line Is Temperature Sensitive.
By selecting winners of fights for 35 generations, we produced a hyperaggressive line of flies called Bullies (1). These flies start fights sooner, retaliate more often, and almost always win fights against the parent Canton-S line (1). The higher levels of aggression observed compared with the Canton-S parent line include shortening of the latency to lunge; large increases in the display of lunging; and increases in both the numbers of boxing events and in the amount of time spent boxing (40). In recent genetic experiments with the Bully line, we accidentally discovered that maintenance of the Bully phenotype depended on the rearing temperature: at 25 °C the hyperaggression phenotype was maintained, whereas at 19 °C it was lost. Recognizing that temperature sensitivity might offer a possible avenue toward unraveling the genetic basis of the hyperaggression phenotype, we initiated studies in which we compared aggression-related behavioral metrics in adult male flies of the Bully and Canton-S stock lines, using animals that had been reared from egg to eclosion at either 19 °C or 25 °C.
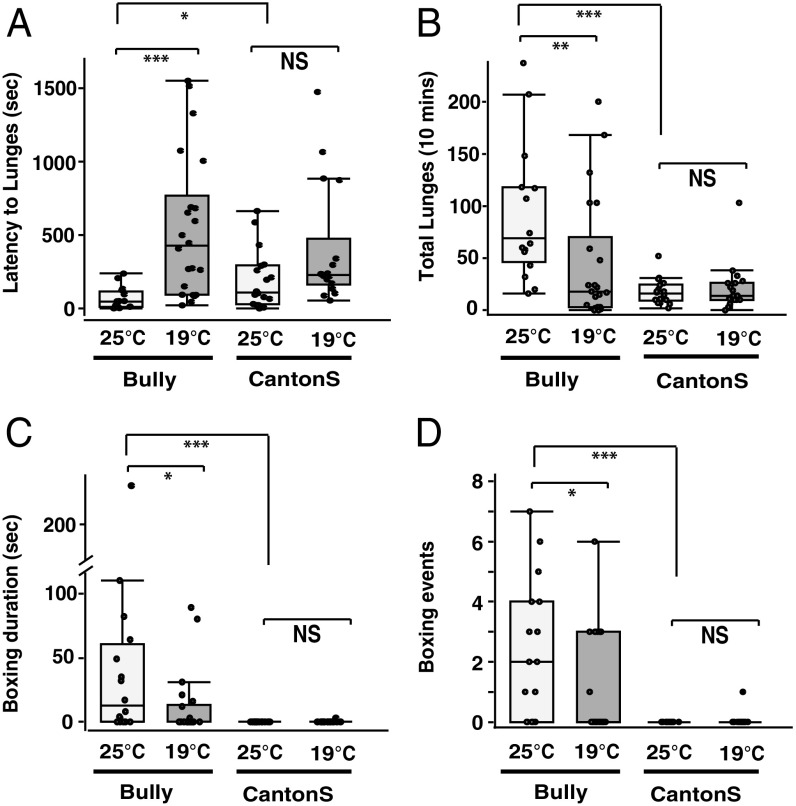
Bullies grown at 25 °C started lunging in fights significantly sooner than Bullies grown at 19 °C (Fig. 1A). No comparable differences in aggression levels were seen in the Canton-S parent flies reared at either temperature. The total numbers of lunges seen (Fig. 1B) were greatest in the Bully25 fights. Bully19 flies displayed smaller numbers of lunges that were not different from the Canton-S flies at either temperature. The amount of time spent boxing (Fig. 1C) and the numbers of boxing events (Fig. 1D) also showed similar changes: in all cases, Bully25 flies displayed higher instances of high-level aggression than Bully19 flies or than Canton-S flies at either temperature (Fig. 1). The above findings suggest that the effects of developmental temperature are selective for the Bully flies, reducing their higher-level aggression when flies are reared before eclosion at a lowered temperature.
Fig. 1.
Lowered developmental temperature reduces hyperaggression phenotype in Bullies. (A) A significant increase in latency to lunging was observed for male Bullies grown at 19 °C (Bully19) compared with those grown at 25 °C (Bully25) (Wilcoxon test, P < 0.001). Canton-S raised at 25 °C (Canton-S25) also showed a significant increase in latency compared with Bully25 (Wilcoxon test, P < 0.05). However, there was no significant difference in latency to lunge between Canton-S19 and Canton-S25 reared male flies (Wilcoxon test, P = 0.051). (B) Bully25 showed a significant increase in lunge numbers compared with Bully19 (Wilcoxon test, P = 0.003), Canton-S25 and Canton-S19 flies (Wilcoxon test, P < 0.001). (C) Boxing duration was significantly higher in Bully25 flies, compared with either Bully19 (Wilcoxon test, P = 0.02), or Canton-S flies raised at either temperature (Wilcoxon test, P < 0.0001). No significant difference in boxing duration was found in Canton-S reared at either temperature (Wilcoxon test, P = 0.34). (D) The numbers of Boxing events were significantly higher in Bully25 compared with all other groups (Bully19 to Bully25 Wilcoxon test, P = 0.03; Bully19 to Canton-S25 Wilcoxon test, P < 0.0001; and Canton-S19 to Canton-S25 Wilcoxon test, P = 0.34, n = 16–22).
Narrowing the Developmental Window of the Temperature-Sensitive Effect.
We next investigated whether a narrower developmental window could be found during which the hyperaggressive behavior displayed by adult Bully flies can be modulated. As holometabolous insects, Drosophila melanogaster undergo complete metamorphosis during pupal life. The adult brain that is formed during this stage is very different from the larval brain it replaces. To ask whether the temperature-sensitive period is before, during, or after metamorphosis, we reared flies through their embryonic and larval phases at 25 °C and shifted the temperature from 25 °C to 19 °C at two different points during the pupal stage [immediately after puparium formation (APF0) and APF48 for 2 d (48 h) later]. As controls, we included groups of flies that were maintained at 19 °C throughout development (Bully19) until eclosion, then shifted to 25 °C, and a second group that were reared completely at 25 °C (Bully25).
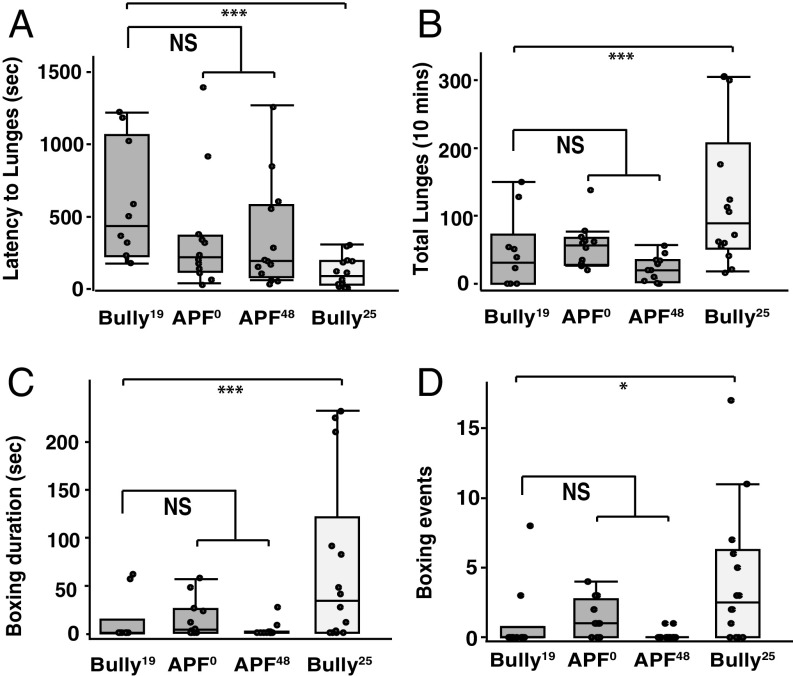
In both the APF0 and APF48 groups of flies, the hyperaggression phenotype was lost as completely as in the flies reared to eclosion at 19 °C (Bully19). Small trends were observed toward reduced latency to lunge in both APF groups, but these values were not significantly different from the Bully19 flies (Fig. 2A). The total numbers of lunges, boxing duration, and boxing events all were reduced in both APF groups to the levels found in the Bully19 group (Fig. 2 B–D): all of these values were lower than the Bully25 group. The results, therefore, suggested the existence of a short developmental window—after the second day of pupal life at 25 °C and before eclosion—that is temperature dependent and that can modulate adult higher-level aggression in Bullies. Efforts to find an even narrower developmental window of temperature sensitivity yielded variable results, possibly due to temperature affecting the rate of development, and the difficulty of exactly comparing developmental stages of flies reared at 19 °C and 25 °C. In these experiments (Fig. 2), the Bully25 group showed aggression levels similar to those seen in Fig. 1.
Fig. 2.
Hyperaggression is influenced by a temperature shift during a short preeclosion window. Several aspects of the Bully phenotype were examined when flies were grown at 19 °C or 25 °C throughout development or were shifted from 25 °C to 19 °C at different points during pupal life (shifts were made at APF0 or APF48). All flies were shifted to 25 °C after eclosion and until used in experiments. (A) The latency to the first lunge was longer when Bullies were reared at 19 °C (Bully19) and in the APF0 and APF48 groups compared with Bullies reared at 25 °C (Bully25) (APF0 Wilcoxon test, P = 0.08; APF48 Wilcoxon test, P = 0.067; Bully25 Wilcoxon test, P = 0.0004). (B) Bully25 flies showed higher numbers of lunges than those displayed by Bully19 flies (Wilcoxon P = 0.02) and by the APF0 and APF48 groups of flies. (C) Bully25 flies spent a greater amount of time boxing than all other groups (Wilcoxon test, P = 0.02), and (D) engaged in a higher number of boxing events than the Bully19 and the other two groups of flies (Wilcoxon test, P = 0.03).
Identification of a Small Group of Differentially Expressed Genes Associated with Higher-Level Aggression.
Because a temperature effect on hyperaggression was found only in Bully, and not in Canton-S flies, this offered a possible experimental avenue by which to search for gene expression differences that might relate selectively to the Bully phenotype; to explore this, we gathered heads from 4 different groups of socially isolated 6-d-old adult male flies: Bullies at 25 °C; Bullies at 19 °C; Canton-S at 25 °C; and Canton-S at 19 °C. We performed RNA-seq analyses on these groups and searched for differentially expressed genes (DEG) between the groups that were selectively related to the Bully phenotype.
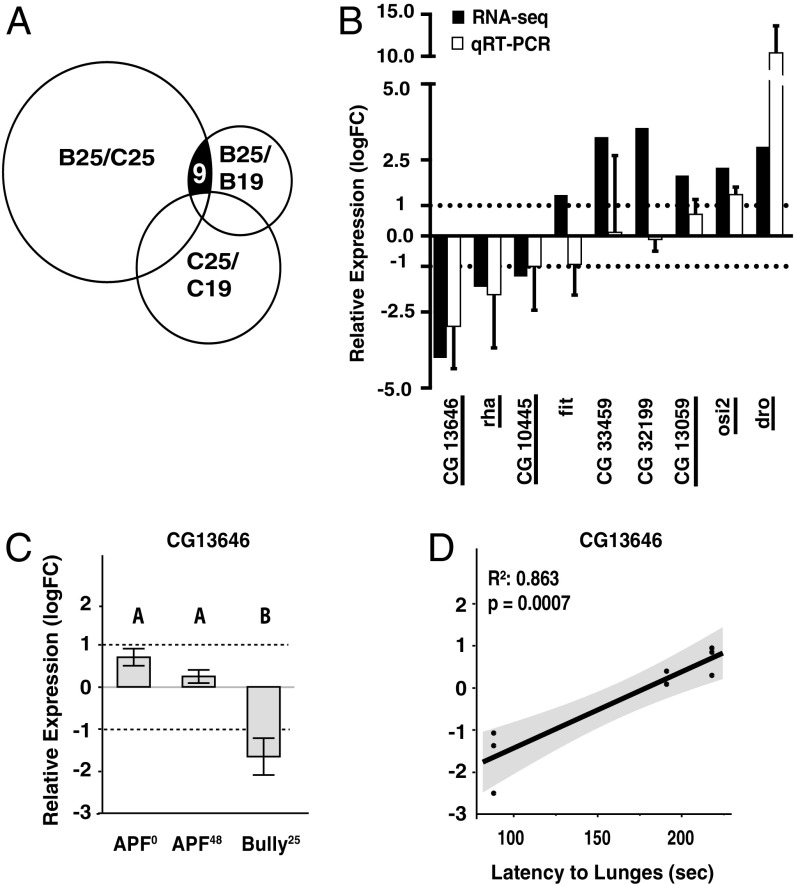
A comparison of RNA profiles of heads from Bully25 and Canton-S25 flies revealed 336 genes (Dataset S1) that were differentially expressed at a threshold of twofold or greater differences in gene expression levels. Next we attempted to separate gene differences that were selectively related to maintenance of the hyperaggression phenotype and were not related to the differences in genetic background. For this purpose, we examined gene expression differences in Bully25 and Bully19 fly heads and compared these to the differences seen in the initial group. This examination reduced the list of candidate genes from 336 to 11. Finally, to control for any additional genes that changed due to temperature alone, the Canton-S25 and Canton-S19 parent groups that showed no hyperaggression phenotype at either temperature were included as well, leaving only nine candidate genes (Fig. 3A). The genes involved were CG13646, rha, CG10445, fit, CG33459, CG32199, CG13059, osi2, and dro.
Fig. 3.
Identification of candidate genes correlated with hyperaggression using differential expression profiles. (A) Relative transcript abundance in fly heads was measured using RNA-seq. The genotypes compared for differential expression were as follows: Bully25 and Canton-S25; Bully25 and Bully19; and Canton-S25 and Canton-S19. Using FDR < 0.05 (adjusted P values) and minimum FC > 2, nine genes were identified that show consistent differential expression that was correlated with hyperaggression. (B) Relative expression of the nine candidate genes in the Bully25 groups were compared with Canton-S25. qRT-PCR confirmed six (underlined) of the nine genes as candidates for the higher-level aggression phenotype (three biological replicates per group). (C) The expression of CG13646 in Bully25 flies was significantly down-regulated compared with both Bully APF0 (Student’s t test P = 0.003), and Bully APF48 flies (Student’s t test P = 0.01). Log fold changes were calculated using CG13646 expression in Bully19 as reference. (D) Lunge latency was strongly related to CG13646 relative expression (linear regression analysis, lunge latency, R2 = 0.86, P = 0.0007).
We attempted to verify the expression levels of the nine candidate genes using qRT-PCR in an additional three independent samples per group. Six of the nine genes consistently showed expression level differences that were in the same direction and of similar magnitude to the values found in the RNA-seq analysis (underlined in Fig. 3B and Dataset S2). To further support the notion that the expression differences of the group of six were selectively related to the Bully phenotype, we again used qRT-PCR to measure levels of gene expression in RNA samples from heads of flies reared under the APF0, APF48, and Bully25 experimental protocols (Fig. 3C and Fig. S1). In this experiment, we anticipated that the largest gene differences would be seen in the Bully25 group. Among the six candidate genes, the most consistent results with the least variation were seen with two genes: CG13646 and osi2 (Fig. 3C and Fig. S1). Focusing on CG13646 (see below for selection of this gene), we asked whether several indicators of high-level aggression might correlate with the relative expression level of this gene. To explore this further, we constructed regression plots of transcript abundance in the RNA samples from the APF0, APF48, and Bully25 groups of flies vs. the median latency to the first lunge and the median numbers of lunges seen during 10 min after the first lunge (graph not shown). A strong positive correlation (R2 = 0.86; P < 0.0007) was shown only between the median lunge latency and the expression level of CG13646 (Fig. 3D).
Fig. S1.
The expression levels of candidate genes in Bully groups APF0, APF48, and Bully25 were compared relative to the Bully19 group. There were no significant differences in gene expression in rha (pAPF0 = 0.28; pAPF48 = 0.91), CG10445 (pAPF0 = 0.68; pAPF48 = 0.08), CG13059 (pAPF0 = 0.21; pAPF48 = 0.49), and dro (pAPF0 = 0.47; pAPF48 = 0.4) compared with Bully25 flies. osi2 showed significant up-regulation in Bully25 compared with APF0 (P = 0.007), but a similar effect was not observed compared with APF48 (P = 0.06). For all comparisons, Student’s t test was performed.
CG13646 Is a Putative Neutral Amino Acid Transporter Gene That Modulates Higher-Level Aggression.
To begin the analysis of possible functional roles of genes important in maintaining the Bully phenotype, we selected CG13646 for further examination. This gene is a postulated member of a large family of closely related amino acid transporter genes found in many species, including humans and Drosophila (41). Of particular interest in this transporter group is that family members have been suggested to serve important roles in neurons and glia relating to GABA, glutamate, and glutamine metabolism in nerve terminals (42, 43). Moreover, as noted above, reduced expression of CG13646 is well correlated with the developmental time window during which temperature influences the maintenance of the Bully phenotype (Fig. 3C).
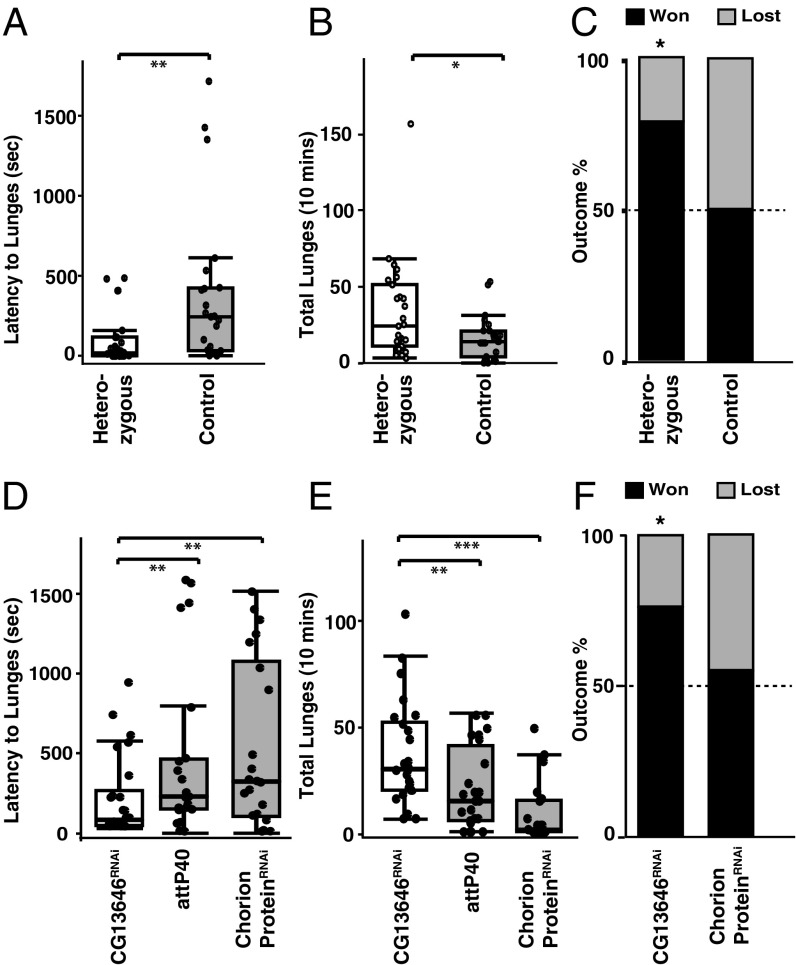
We took two different experimental approaches to ask whether direct manipulation of expression levels of this gene would alter the levels of aggression displayed by male flies during fights. First we found a P-element insertion mutant (CG13646e03587) that disrupted the coding region of CG13646. In the homozygous mutant of this line, no functional mRNA was found by qRT-PCR, confirming that it is a null mutant. Although the homozygous null mutants were viable, they displayed gross locomotion defects. Therefore, we used heterozygotes for behavioral assays. qRT-PCR showed that the transcript abundance was ∼60% of normal in heads from the heterozygotes. For the aggression assays, we paired flies in same genotype fights, and also carried out competitive advantage assays in which we fought mutants against control flies. In the same genotype fights, we observed a significant decrease in the latency to lunge, and a significant increase in total numbers of lunges in the experimental genotype (Fig. 4 A and B) compared with the corresponding controls without the P-element insertion. In the competitive advantage assay, experimental flies won significantly more of their fights than control flies (Fig. 4C). The second approach to altering CG13646 expression was to use RNAi knockdown of the gene. In these experiments we used an elavc155-Gal4 driver to express an RNAi transgene targeting CG13646. As controls for these experiments, we used a TRiP stock that contained no RNAi transgene (attP40 docking site only) for background effects, and an RNAi allele targeting a female-specific chorion protein as a control for the insertion site of the RNAi. Using qRT-PCR, we found that the transcript level of CG13646 in the heads of knockdown flies was ∼20% of that found in both control groups. No significant differences in transcript abundance were found in the control lines. Next, we performed aggression assays using same genotype pairings in one set of experiments and competition between the two different groups of RNAi knockdown flies (CG13646, and chorion protein) and genetic background controls in a second set. Once again, significant reductions in latency to lunge and increases in lunge numbers in CG13646 knockdown flies were seen compared with controls (Fig. 4 D and E). The experimental group also won significantly more fights against the background line. When the background line fought against the chorion RNAi line, however, the outcome was not significantly different from 50:50 (Fig. 4F). These findings show that down-regulation of CG13646 expression by two different routes leads to significant increases in several indices of higher-level aggression in Drosophila.
Fig. 4.
Down-regulation of CG13646 transcripts elevates level of aggression. (A) Heterozygous P-element mutants of CG13646 began fights significantly earlier (Wilcoxon test, P = 0.002) and (B) lunged significantly more (Wilcoxon test, P = 0.03) than control flies (n = 23–27). (C) Heterozygous mutants won significantly more fights (χ2 test, P = 0.001, n = 32) against wild-type Canton-S flies (first bar) than did control flies (second bar, χ2 test, P = 1.0, n = 28). (D) Flies with pan-neuronal knockdown using RNAi of CG13646 (elavc155-Gal4;UAS-dcr2;UAS-CG13646RNAi) began fights significantly earlier than both control lines (elavc155-Gal4;UAS-dcr2/+ and elavc155-Gal4;UAS-dcr2;UAS-chorion proteinRNAi) (Wilcoxon test, P < 0.005). (E) RNAi knockdown of CG13646 flies also lunge significantly more than attP40 (Wilcoxon test, P = 0.004) and chorion protein RNAi (Wilcoxon test, P < 0.001) control flies (n = 22–26). (F) Pan-neuronal knockdown of CG13646 yielded flies that were significantly more likely to win dyadic fights against attP40 control flies (elavc155-Gal4;UAS-dcr2/+) (χ2 test, P = 0.01, n = 21), whereas flies in the RNAi control group (elavc155-Gal4;UAS-dcr2;UAS-chorion proteinRNAi) had no significant advantage on the outcome of fights (χ2 test, P = 0.68, n = 24).
Discussion
Aggression is a complex behavior, triggered in varying social situations via a variety of sensory cues and likely processed via multiple circuitries to yield stereotypical sequences of patterned motor output that are used to defend or acquire desired resources. One can imagine many nodes in such processing in which alterations of gene expression could impact on the final outcome motor pathway. Moreover, the final output pathway itself involves sequences of motor pattern displays of differing intensity, with only the highest levels of these usually determining the outcome of bouts (28, 40, 44). In Drosophila, many genes have been identified that are differentially expressed between high- and low-aggression level male flies using positive and negative methods of screening by both candidate gene and genome-wide screening methods (37–39). The genetic screens have identified hundreds to thousands of genes that are differentially expressed in high- and low-aggression flies, and there has been little overlap in the genes detected using differing aggression assays by different laboratories. What then is the value of identifying single genes that are involved in the modulation of aggression as has been done in this study? One obvious advantage of such studies is that they can lead to the identification of neurons, neuron types, and circuitries directly or indirectly involved in the expression of higher levels of aggression by organisms. A case in point is in the identification of certain sensory neurons that are associated with one form of the family of cytochrome P450 proteins, Cyp6a20 [originally identified by Dierick and Greenspan (37) and then used in studies by Wang et al. (39)], identifying the gene, in this case, as coding for a protein found in nonneural support cells associated with pheromone-detecting sensory olfactory sensilla. In experiments comparing socially isolated highly aggressive flies with group-reared lower level-aggression fruit flies, Wang et al. (39) demonstrated that social experience increased activity of this protein, and proposed that this thereby decreased aggressivity, whereas social isolation had opposite effects. We too detected changes in the levels of genes of the P450 family of cytochromes (cyp6a2, cyp6a8, cyp6a14, cyp6a17, and cyp6a23) between Bullies and the control parent stocks, but these were less than twofold different in the Bully19 and Bully25 flies, so they were not included with the investigations presented here.
In these studies, we add a member of a different family of genes than those previously identified that is related to higher-level aggression. The gene CG13646 is significantly decreased in the hyperaggressive Bully25 flies compared with the parent and Bully19 lines of flies. This gene is a member of the evolutionarily conserved solute-linked carrier superfamily of proteins (SLCs) (41, 45). The SLCs are the second largest collection of distinct membrane proteins in the human genome consisting of close to 400 proteins, distributed in 46 families in humans: 42 closely related families are found in the Drosophila genome (41). CG13646 belongs to the SLC38 family of the SLC superfamily (46). In the human genome and in other mammalian species there are 11 members of this family, and they all are Na+-dependent amino acid transporters that are widely distributed in many tissues of the body, including the nervous system (45, 47). A recent publication has characterized the SLC38A8 member of that family in mouse brain and has demonstrated that this transporter is found in both inhibitory and excitatory neurons and that it has a preference for transporting glutamine, alanine, arginine, histidine, and aspartate into neurons via a Na+-dependent mechanism (48). Of particular interest is that this transporter is hypothesized to be important in a glutamine/glutamate (GABA) cycle in the brain (49), in which glutamine, the amidated form of glutamic acid, serves to regulate stores of glutamate and GABA in nerve terminals. Another recent publication suggests that the A6 member of the SLC38 family of proteins is selectively localized at synaptic membranes of glutamatergic excitatory neurons (47). Different isoforms of the SLC38 gene are highly expressed in different regions of the mammalian brain, where they likely serve different functions. For example, the A1 isoform is highly expressed in the thalamus, arcuate nucleus, and amygdala where it is postulated to be involved in aggression (45), whereas the A6 isoform is concentrated in the lateral hypothalamus where it is speculated to be involved in motivation and reward (42).
In the present studies we showed a consistent twofold decrease of CG13646 expression in the Bully line reared at 25 °C in comparison with Bullies reared at 19 °C and to the control Canton-S parent line reared at either temperature. At least five other genes also showed elevations or reductions of expression in the heads of Bully25 flies compared with all controls, and an even larger number showed smaller and more variable-level changes (Dataset S1). We selected CG13646 for further study due to the consistent down-regulation of its mRNA levels associated with higher-level aggression, and because it showed a strong and highly significant coefficient of determination with elevated aggression. Because members of SLC family are commonly found in synaptic regions where they have been postulated to serve in glutamine/glutamate (GABA) and glutamine/glutamate cycles of transmitter regulation in nerve terminals (50), we hypothesized that genes like CG13646 might be functioning at important nodes in pathways of behavioral regulation. We were delighted, therefore, to find that lowering levels of CG13646 to similar levels of expression as those seen in Bully25 fly heads, partially duplicated the aggression phenotype displayed by Bullies. Heterozygous mutants and RNAi knockdown of CG13646 both yielded progeny that showed higher levels of aggression measured by lunge latency and total numbers of lunges displayed during fights; these flies also were significantly more likely to win fights against their control counterparts. Down-regulation of this single gene, however, did not result in elevated levels of boxing, the highest level of aggression seen during fights in Drosophila. This result may not be surprising given the complex architecture of aggressive behavior, which involves internal arousal, sensory experience, decision-making, and a persistent elevated aggressive state (30, 32, 51, 52).
In summary, in the present studies, beginning with the Bully line of hyperaggressive flies previously generated in our laboratory, we have identified a member of a new and interesting category of genes that is tied selectively to the Bully phenotype. The gene CG13646 is a member of the SLC38 category of neutral amino acid transporters that have been proposed to be important in mammalian systems in glutamate and GABA neurotransmitter regulation. In the next stages of these studies, we will (i) examine the substrate specificity of the protein product of CG13646 to ask if it indeed is a glutamine-selective transporter and (ii) develop reagents that will allow us to localize the protein product in the Drosophila central nervous system.
Methods
Stocks and Fly Husbandry.
Flies were maintained on standard cornmeal medium on a 12-h light/dark cycle at 25 °C and ambient humidity. The following strains were used for this study: Canton-S, Canton-S Bully (1), elavc155-Gal4;UAS-dcr2, transgenic RNAi lines targeting the following genes: CG13646 (no. 53701), Chorion protein 36 (no. 57838), and the negative control without any RNAi transgene (no. 36304). For mutant studies, PBac{RB}CG13646e03587 (Exelixis Harvard Stock Center) and the corresponding background strain used for the insertion, w1118 (no. 6326), were crossed to Canton-S females to generate the heterozygous and control flies, respectively. For RNAi knockdown studies, elavc155-Gal4;UAS-dcr2 females were crossed to UAS-CG13646RNAi, UAS-chorion proteinRNAi, and the negative control (attP40 docking site only) to generate the corresponding lines for aggression assays.
For experiments involving different developmental temperatures, control flies were raised at 19 °C or 25 °C throughout development until eclosion (i.e., Bully19 or Bully25). A temperature shift from 25 °C to 19 °C was performed at two different points during pupal development (APF0 and APF48). After eclosion, flies were kept at 25 °C for 5–7 d before performing the aggression assays.
Aggression Assays.
Socially naive males were isolated at late pupal stage and housed individually in single glass vials (16 × 100 mm) containing ∼1.5 mL standard fly food. For flies reared at 19 °C, isolated pupae were kept at 19 °C until eclosion, and then transferred to 25 °C on the day after eclosion. Flies were aged for 5–7 d at 25 °C and ambient humidity before carrying out aggression assays. Aggression assays were always performed between pairs of males of the same genotype and experimental treatment unless otherwise specified. For winner–loser experiments, pairs of flies were briefly anesthetized with CO2 at least 24 h before the behavioral assays. Different colors of water-based acrylic paint were applied to the dorsal thorax of both flies to facilitate visual tracking of individual flies.
For the experiments involving different developmental temperatures, assays were carried out in 12-well polystyrene plates as previously described (53). For all other experiments, aggression assays were performed in a newly developed behavioral chamber that involves minimal handling of flies (44). Food cups with standard fly food and with a drop of yeast paste on top were used as an attractive resource in all fights. Lunge and boxing behavior was scored as previously described (28): latency to lunge was the time between the first encounter on the food cup (social interaction lasting at least 3 s) and the occurrence of first lunge on the food cup. Winner–loser outcomes were defined as previously described (1). All aggression assays were carried out with age- and size-matched males during the first 3 h of the daily light cycle. Each fight was videotaped for at least 40 min, and videos were analyzed manually.
RNA-Seq and Differential Gene Expression Analysis.
Two biological replicates of four groups of flies (CS19, CS25, Bully19, and Bully25) were used to carry out an RNA-seq analysis. For each group, 6-d-old socially isolated male flies were frozen in liquid nitrogen. Next, 90–100 frozen heads were collected per group and the samples were separated into two biological replicates. Total RNA extraction and the following RNA-seq analysis were performed at the Harvard Medical School Biopolymers Facility. Sequencing was performed on an Illumina HiSeq instrument, resulting in ∼30 million of 50-bp reads per sample. Sequencing reads were mapped to the Drosophila transcriptome (dm3 RefSeq annotation) using TopHat (54). Gene expression counts were calculated using GenomicFeatures (55). Calculation of expression values and differential expression analysis were performed using the edgeR package (56). The genes with false discovery rate (FDR) < 0.05 (adjusted P values) and fold change (FC) > 2 were considered to indicate significant up-regulation; FDR < 0.05 and FC < 0.5 were considered as significant down-regulation; FC between 0.5 and 2 was considered not significant. For the identification of aggression-related genes, DEGs that showed different expression directions in CS25 and Bully25 flies were considered aggression related, such as genes up-regulated in CS25 but suppressed in Bully25, or down-regulated in CS25 but up-regulated in Bully25. Similarly, DEG of different expression directions in Bully19 and Bully25 flies were considered aggression related, whereas DEGs between CS25 and CS19 were considered not aggression related.
Quantitative PCR.
Independent samples of fly heads (6-d-old socially naive males) were used to verify the RNA-seq results. Three samples of fly heads, each containing a pool of at least 30 individual heads, were collected from different groups and frozen in liquid nitrogen. Frozen heads were homogenized and total RNA was extracted using TRIzol (Life Technologies) following the protocol described by the manufacturer. cDNA from each sample was generated using the QuantiTect Reverse Transcription Kit (QIAGEN). Quantitative PCR were performed using the iQ SYBR Green Supermix and CRX96 System (Bio-Rad). The list of primers used in the qRT-PCR analysis is provided in Table S1. Three replicates were included for each sample and the results were normalized against the mRNA level of actin to determine the ΔCt (cycle threshold) values.
Table S1.
qRT-PCR primer pairs used to amplify regions of each of the nine preliminary candidate genes
| Gene | Direction | Primers |
| CG32199 | Fwd | ACCTCTGCGTTCCTTAAGTTAG |
| Rev | GGCTATGTCTCGCATTGATTTG | |
| CG33459 | Fwd | TGTGCCGGAAGTAGCAAATC |
| Rev | CCACTTGAGCGTGAATGTATCT | |
| dro | Fwd | GCTGCTTGCTTGCGTTT |
| Rev | TGAGTCAGGTGATCCTCGAT | |
| osi2 | Fwd | TCTCGCGTAAGAAGTTGAAGAA |
| Rev | AGCGATTCCAAGGATGAAGG | |
| CG10445 | Fwd | AGGTACAACGAGATCTACGAAAG |
| Rev | CAGCAGTAGCACCAGGATATAG | |
| CG13646 | Fwd | TCCTGTACGTGGTGGATTTG |
| Rev | GAGAGCAGGAAGGAGGTATAGA | |
| CG13059 | Fwd | CTTAAGACTGTGGTCTCCTCTG |
| Rev | CTGGGATCACGGTCTTGATAA | |
| fit | Fwd | GTCCCAAGGTCCAAGTCTAAG |
| Rev | CAGTCGCTTATCCACCTTGT | |
| rha | Fwd | CGTGGTCGAGGCGATAAACTT |
| Rev | CACACTCCCAGATACAGTTGC | |
| actin5c | Fwd | GCCCATCTACGAGGGTTATG |
| Rev | CGGTGGTGGTGAAAGAGTAA | |
| CG13646 (exon 2–3) | Fwd | ATTGTGGCGCTGTTGTGGTT |
| Rev | ACTGGAAGGCCAATATGCTG |
Supplementary Material
Acknowledgments
We thank Ayla Ergun and Ruslan Sadreyev for the analysis of RNA-seq data, and Severine Trannoy and other members of E.A.K.’s laboratory for discussions and comments. Preliminary studies on this work demonstrating the temperature sensitivity of the hyperaggression phenotype were performed by Jill Penn and Olga Alekseyenko. This work was supported by National Institute of General Medical Sciences Grants R01 GM099883, R01 GM074675, and R35 GM118137 (all to E.A.K.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE94293).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618354114/-/DCSupplemental.
References
- 1.Penn JKM, Zito MF, Kravitz EA. A single social defeat reduces aggression in a highly aggressive strain of Drosophila. Proc Natl Acad Sci USA. 2010;107(28):12682–12686. doi: 10.1073/pnas.1007016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roubertoux PL, et al. Attack behaviors in mice: From factorial structure to quantitative trait loci mapping. Eur J Pharmacol. 2005;526(1-3):172–185. doi: 10.1016/j.ejphar.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Arias M, et al. CB1 cannabinoid receptor-mediated aggressive behavior. Neuropharmacology. 2013;75:172–180. doi: 10.1016/j.neuropharm.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Schneider R, Hoffmann HJ, Schicknick H, Moutier R. Genetic analysis of isolation-induced aggression. I. Comparison between closely related inbred mouse strains. Behav Neural Biol. 1992;57(3):198–204. doi: 10.1016/0163-1047(92)90150-3. [DOI] [PubMed] [Google Scholar]
- 5.Sandnabba NK. Selective breeding for isolation-induced intermale aggression in mice: Associated responses and environmental influences. Behav Genet. 1996;26(5):477–488. doi: 10.1007/BF02359752. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi A, Miczek KA. Neurogenetics of aggressive behavior: Studies in rodents. Curr Top Behav Neurosci. 2014;17:3–44. doi: 10.1007/7854_2013_263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norton WHJ, et al. Modulation of Fgfr1a signaling in zebrafish reveals a genetic basis for the aggression-boldness syndrome. J Neurosci. 2011;31(39):13796–13807. doi: 10.1523/JNEUROSCI.2892-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teles MC, Dahlbom SJ, Winberg S, Oliveira RF. Social modulation of brain monoamine levels in zebrafish. Behav Brain Res. 2013;253:17–24. doi: 10.1016/j.bbr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Audero E, et al. Suppression of serotonin neuron firing increases aggression in mice. J Neurosci. 2013;33(20):8678–8688. doi: 10.1523/JNEUROSCI.2067-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saudou F, et al. Enhanced aggressive behavior in mice lacking 5-HT1B receptor. Science. 1994;265(5180):1875–1878. doi: 10.1126/science.8091214. [DOI] [PubMed] [Google Scholar]
- 11.Carvajal C, Dumont Y, Quirion R. Neuropeptide Y: Role in emotion and alcohol dependence. CNS Neurol Disord Drug Targets. 2006;5(2):181–195. doi: 10.2174/187152706776359592. [DOI] [PubMed] [Google Scholar]
- 12.Adamczyk A, et al. GluA3-deficiency in mice is associated with increased social and aggressive behavior and elevated dopamine in striatum. Behav Brain Res. 2012;229(1):265–272. doi: 10.1016/j.bbr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stork O, et al. Postnatal development of a GABA deficit and disturbance of neural functions in mice lacking GAD65. Brain Res. 2000;865(1):45–58. doi: 10.1016/s0006-8993(00)02206-x. [DOI] [PubMed] [Google Scholar]
- 14.Nelson RJ, et al. Behavioural abnormalities in male mice lacking neuronal nitric oxide synthase. Nature. 1995;378(6555):383–386. doi: 10.1038/378383a0. [DOI] [PubMed] [Google Scholar]
- 15.De Felipe C, et al. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature. 1998;392(6674):394–397. doi: 10.1038/32904. [DOI] [PubMed] [Google Scholar]
- 16.Toda K, Saibara T, Okada T, Onishi S, Shizuta Y. A loss of aggressive behaviour and its reinstatement by oestrogen in mice lacking the aromatase gene (Cyp19) J Endocrinol. 2001;168(2):217–220. doi: 10.1677/joe.0.1680217. [DOI] [PubMed] [Google Scholar]
- 17.Sallinen J, Haapalinna A, Viitamaa T, Kobilka BK, Scheinin M. Adrenergic alpha2C-receptors modulate the acoustic startle reflex, prepulse inhibition, and aggression in mice. J Neurosci. 1998;18(8):3035–3042. doi: 10.1523/JNEUROSCI.18-08-03035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young KA, et al. Fierce: A new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav Brain Res. 2002;132(2):145–158. doi: 10.1016/s0166-4328(01)00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gogos JA, et al. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci USA. 1998;95(17):9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto T, Honda S, Harada N. Alteration in sex-specific behaviors in male mice lacking the aromatase gene. Neuroendocrinology. 2003;77(6):416–424. doi: 10.1159/000071313. [DOI] [PubMed] [Google Scholar]
- 21.Malki K, et al. Transcriptome analysis of genes and gene networks involved in aggressive behavior in mouse and zebrafish. Am J Med Genet B Neuropsychiatr Genet. 2016;171(6):827–838. doi: 10.1002/ajmg.b.32451. [DOI] [PubMed] [Google Scholar]
- 22.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Lai S-L, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9(5):703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 24.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24(5):251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 25.Sturtevant HA. Experiments on sex recognition and the problem of sexual selection in Drosophila. J Anim Behav. 1915;5(5):351–366. [Google Scholar]
- 26.Jacobs ME. Influence of beta-alanine on mating and territorialism in Drosophila melanogaster. Behav Genet. 1978;8(6):487–502. doi: 10.1007/BF01067478. [DOI] [PubMed] [Google Scholar]
- 27.Dow MA, von Schilcher F. Aggression and mating success in Drosophila melanogaster. Nature. 1975;254(5500):511–512. doi: 10.1038/254511a0. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Lee AY, Bowens NM, Huber R, Kravitz EA. Fighting fruit flies: A model system for the study of aggression. Proc Natl Acad Sci USA. 2002;99(8):5664–5668. doi: 10.1073/pnas.082102599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dierick HA, Greenspan RJ. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet. 2007;39(5):678–682. doi: 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- 30.Alekseyenko OV, et al. Single serotonergic neurons that modulate aggression in Drosophila. Curr Biol. 2014;24(22):2700–2707. doi: 10.1016/j.cub.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alekseyenko OV, Chan Y-B, Li R, Kravitz EA. Single dopaminergic neurons that modulate aggression in Drosophila. Proc Natl Acad Sci USA. 2013;110(15):6151–6156. doi: 10.1073/pnas.1303446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Certel SJ, Savella MG, Schlegel DCF, Kravitz EA. Modulation of Drosophila male behavioral choice. Proc Natl Acad Sci USA. 2007;104(11):4706–4711. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asahina K, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156(1-2):221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis SM, Thomas AL, Nomie KJ, Huang L, Dierick HA. Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat Commun. 2014;5:3177. doi: 10.1038/ncomms4177. [DOI] [PubMed] [Google Scholar]
- 35.Hirsch J, Boudreau JC. Studies in experimental behavior genetics. I. The heritability of phototaxis in a population of Drosophila melanogaster. J Comp Physiol Psychol. 1958;51(6):647–651. doi: 10.1037/h0039498. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch J. Studies in experimental behavior genetics. II. Individual differences in geotaxis as a function of chromosome variations in synthesized Drosophila populations. J Comp Physiol Psychol. 1959;52(3):304–308. doi: 10.1037/h0043498. [DOI] [PubMed] [Google Scholar]
- 37.Dierick HA, Greenspan RJ. Molecular analysis of flies selected for aggressive behavior. Nat Genet. 2006;38(9):1023–1031. doi: 10.1038/ng1864. [DOI] [PubMed] [Google Scholar]
- 38.Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2006;2(9):e154. doi: 10.1371/journal.pgen.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci USA. 2008;105(15):5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsen SP, Chan Y-B, Huber R, Kravitz EA. Gender-selective patterns of aggressive behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101(33):12342–12347. doi: 10.1073/pnas.0404693101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Höglund PJ, Nordström KJV, Schiöth HB, Fredriksson R. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol Biol Evol. 2011;28(4):1531–1541. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L, Yee SW, Kim RB, Giacomini KM. SLC transporters as therapeutic targets: Emerging opportunities. Nat Rev Drug Discov. 2015;14(8):543–560. doi: 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Featherstone DE. Glial solute carrier transporters in Drosophila and mice. Glia. 2011;59(9):1351–1363. doi: 10.1002/glia.21085. [DOI] [PubMed] [Google Scholar]
- 44.Trannoy S, Chowdhury B, Kravitz EA. A new approach that eliminates handling for studying aggression and the “loser” effect in Drosophila melanogaster. J Vis Exp. 2015;(106):e53395. doi: 10.3791/53395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundberg BE, et al. The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. J Mol Neurosci. 2008;35(2):179–193. doi: 10.1007/s12031-008-9046-x. [DOI] [PubMed] [Google Scholar]
- 46.Romero-Calderón R, et al. A screen for neurotransmitter transporters expressed in the visual system of Drosophila melanogaster identifies three novel genes. Dev Neurobiol. 2007;67(5):550–569. doi: 10.1002/dneu.20342. [DOI] [PubMed] [Google Scholar]
- 47.Bagchi S, Baomar HA, Al-Walai S, Al-Sadi S, Fredriksson R. Histological analysis of SLC38A6 (SNAT6) expression in mouse brain shows selective expression in excitatory neurons with high expression in the synapses. PLoS One. 2014;9(4):e95438. doi: 10.1371/journal.pone.0095438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hägglund MGA, et al. Transport of L-glutamine, L-alanine, L-arginine and L-histidine by the neuron-specific Slc38a8 (SNAT8) in CNS. J Mol Biol. 2015;427(6 Pt B):1495–1512. doi: 10.1016/j.jmb.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Albrecht J, Sidoryk-Węgrzynowicz M, Zielińska M, Aschner M. Roles of glutamine in neurotransmission. Neuron Glia Biol. 2010;6(4):263–276. doi: 10.1017/S1740925X11000093. [DOI] [PubMed] [Google Scholar]
- 50.Schlessinger A, Khuri N, Giacomini KM, Sali A. Molecular modeling and ligand docking for solute carrier (SLC) transporters. Curr Top Med Chem. 2013;13(7):843–856. doi: 10.2174/1568026611313070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoopfer ED, Jung Y, Inagaki HK, Rubin GM, Anderson DJ. P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife. 2015;4:e11346. doi: 10.7554/eLife.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernández MP, Kravitz EA. Aggression and courtship in Drosophila: Pheromonal communication and sex recognition. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2013;199(11):1065–1076. doi: 10.1007/s00359-013-0851-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernández MP, et al. Pheromonal and behavioral cues trigger male-to-female aggression in Drosophila. PLoS Biol. 2010;8(11):e1000541. doi: 10.1371/journal.pbio.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.