Abstract
Cell migration is a complex and multi-step process involved in homeostasis maintenance, morphogenesis and disease development, such as cancer metastasis. Modeling cell migration and the relevant cytoskeleton dynamics has profound implications for studying fundamental development and disease diagnosis. This review focuses on some recent models of both cell migration and migration-related cytoskeleton dynamics, addressing issues such as the difference between amoeboid and mesenchymal migration modes, and between single-cell migration and collective cell migration. There view also highlights the computational integration among variable external cues, especially the biochemical and mechanical signaling that affects cell migration. Finally, we aim to identify the gaps in our current knowledge and potential strategies to develop integrated modeling-experimental frameworks for multi-scale behavior integrating gene expression, cell signaling, mechanics and multi-cellular dynamics.
Graphical abstract
Introduction
Cell migration, both at the single-cell and at the collective cell level, plays a significant role in regulating living system behavior. It is a complex process involved in development, tissue formation and engineering, cancer and metastasis.1,2 From a mechanistic stand point, it is argued that individual cell migration has three key components: protrusion, contractility, and adhesion, for migration in different environments.3,4 Additionally, single cell migration has been classified into two types,5 namely amoeboid and mesenchymal migration. Amoeboid migration commonly refers to the faster movement of rounder cells that lack mature focal adhesion and stress fibers, while cells with high cell-matrix adhesion and high cytoskeleton contractility undergo the fibroblast-like mesenchymal migration.6 There are also several modes of collective cell migration, which are observed in vivo and differ from single migration modes.5 Cell migration is a result of a balance between cell-environment interaction and intracellular cytoskeleton dynamics. Seeded on a two-dimension (2D) substrate, cells have been shown to migrate towards a stiffer environment, a process that is termed as durotaxis.7 Cells in three-dimension (3D) matrices also tends to move faster under stiff environment, however, with a certain limit over which the cell speed will be hindered due to the matrix resistance force.8–15 Mechanical inputs also play a role in migration, for example, the increased collagen crosslink and extracellular matrix (ECM) stiffening, cooperating with oncogenes such as ErbB2 can promote the invasiveness of mammary epithelial cells,10,16,17 even with the absence of cellular and soluble tissue and systemic factors.16 Other than mechanical inputs, chemotaxis, due to growth factors, is important in modulating both 2D and 3D cell migration.18,19 These and other studies have shown that environmental factors, both chemical and mechanical, are key to developing a comprehensive understanding of single and collective migratory behavior. Observations of these cellular behaviors in experiments inspire computational models aiming to unveil underlying mechanisms that are hard to test directly due to the complexity or to bridge current gaps in our current understanding. A key aim of this review is to understand the current computational models available, and their strengths and weaknesses, in describing both the environment and the migratory behavior at a single and collective level.
With the improved computational power, quantitative cell tracking techniques for cell migration have received an augment as well. Recent studies have used several trackers for migration and cellular reorganization including the nucleus,20 Cytotrackers,21 single-particle tracking systems,20,22–24 the particle image velocimetry (PIV)25 and additional methods such as optical flow tracking.26 Utilization of these tools have made it possible not only to track cellular motion velocity but also directional migration persistence,27 mean squared displacement (MSD),28 the velocity correlation,25 the cell-pair separation distance20 and so on. Nonetheless, more quantified variables are needed to quantify and distinguish different patterns of migration modes in both single-cell and collective cell migration, such as transition between amoeboid and mesenchymal migration, cellular leadership in collective behavior.29,30 Additionally, not all tools are applicable in native like three-dimensional environments. This review aims to address both the recent developments and future opportunities in integrated modeling-experimental frameworks to understand single and collective cellular behavior in complex environments.
Cellular computational models in cell migration
To investigate microenvironment effects on cell migration, several models have been proposed to quantitatively predict the migration speed due to different external cues for both single-cell and collective cell migration. There include, among others, force balancing models to predict the migration speed for the single-cell migration10,31, and also the energy based model to predict the collective cell migration for wound healing process, tumor cell invasion and angiogenesis.
Force balancing models in cell migration
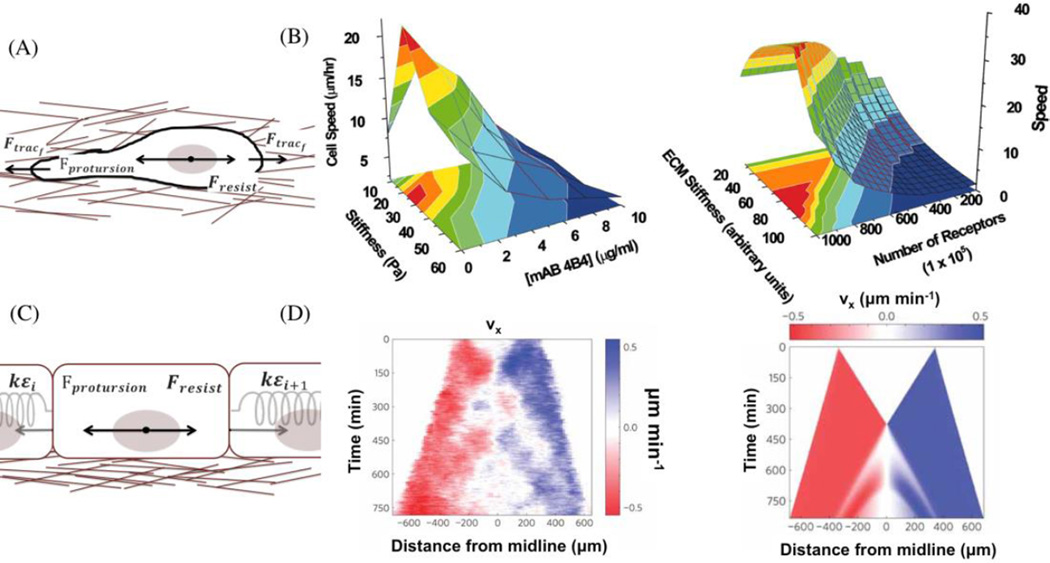
The mechanical interventions due to external stimuli such as changes in physical, chemical or mechanical properties of ECM are intuitively characterized with force balancing models. In 3D single-cell migration, the balance among the cell protrusion force, the cell traction force from both the cell front and back due to cell-ECM interactions, and the resistive force is used to study cell speed change due to different ECM stiffness, density, cell integrin amounts, and matrix metalloproteinase (MMP) expressions (Fig 1A).8,10,31
Fig 1. Force balancing models of cell migration.
(A) A force balancing model of individual cell migration. (B) The later experimental validation (left) is consistent with the computational predictions (right), which successfully predicted the cell migration speed dependence on both the integrin concentration and the matrix stiffness. Iamges are obtained from Zaman et al.10 (Copyright (2006) National Academy of Sciences)(C) A force balancing model of collective monolayer cell migration. (D) This model successfully explained the mechanical wave-like cell migration and stress patterns, which are due to the reinforcement and fluidization of cytoskeleton. The experimental measurement is on the left and the numerical simulation is on the right using the force balancing model. Images are obtained from Serra-Picamal et al.34 (Adapted by permission from Macmillan Publishers Ltd: Nat. Phys. (Ref 34), copyright (2012))
Here, cell migration speed ν is determined by solving the Fresistive, given the ECM stiffness Emol, the ECM ligand density LD, the cell-expressing integrin concentration ITG and MMP density. Fprotusion is the cell protrusion force in 3D that is simulated by a vector with constant magnitude but random direction. The traction forces and the resistive force can be mathematically represented as
Here f(Emol) is a linear function of Emol until Emol reaches a threshold and then this function plateaus. The ligand density LDf(MMP) is linearly dependent on MMP-mediated matrix degradation.8 R and η are the cell radius and the effective viscosity of the matrix material. This single-cell force balancing model not only predicts the biphasic dependence of cell speed on ECM density in 3D, but also shifting of such dependence due to changes in integrin-mediated adhesions in vitro (Fig 1B).10 In this experimental validation single cell speed is measured using a live cell membrane dye and IMARIS (Bitplane, St. Paul, MN) spots/isosurface tracking routines. Here this tool offers major advantages in single particle tracking in 3D and being automated, though like most single particle tracking algorithm it does not deal with cell division/apoptosis. Excluding the matric resistance force enables this model to predict cell migrations in 2D as well.31,32 Overall, this model is significant because it qualitatively predicts the cellular behavior due to variations in ECM properties in 3D, and unveils the underlying mechanism leading to these variations, though it doesn’t capture the changes in local stiffness or morphology change of the gel due to cell-ECM interactions.33
Another force balancing model focused on characterizing the collective 1D or semi-2D cell migration (average on one direction) considers cell-cell stress and cell elastic properties, in addition to the cell traction force, cell protrusion force, and resistive force34 (Fig 1C).
Here Fprotusion is the cell protrusion force, or the cell self-propelled force in collective migration, ε is the neighbor cell deformation strain, i and i + 1 denotes different neighbor cells, and k is the cell elastic constant. Both the cell-cell stress and cell migration velocity are the outputs of this model. Results from the model showed that only with cell reinforcement (stiffening) and followed soon by a fluidization (softening) when the strain is above a threshold, can the cells have a mechanical wave pattern of migration speed and cell-cell stress observed in experiments.34 Without such a quantitative model, it would be difficult to understand this wave pattern and to further link it to the cytoskeleton reinforcement/fluidization. Here cell speed in a 2D monolayer is measured using the particle image velocimetry (PIV) algorithm. This algorithm offers major advantages in measuring collective cell migration in 2D monolayer using phase-contract images, and do not require any fluorescence staining. Although it does not track motions of each cell but rather identifies average movements of subregions in an image.26 A possible factor that this collective cell migration force balancing model can be extended to capture directly is the cell-cell adhesion,35,36 which is not always the same as the cell-cell stress.
These force-balance models are mainly deterministic models. The advantage for these types of models is the computational efficiency, since one can directly solve the cell speed in a linear manner. One of the limitations is that when considering chemically relevant effects, or factors that cannot be explicitly expressed in terms of force, this model is unable to capture the chemical complexity. That said, studies have used force-like terms to model the chemotaxis in cell migration.37 This force-like term linearly depends on the cell sensitivity to the chemical gradient. Force balancing models can also be carried out in a stochastic fashion using the Langevin equation with a stochastic noise term.37,38 Overall, force balancing models are an approximation of equations of motions, in which usually the inertia terms are neglected due to the relative high friction between the cells and their environment for high computational efficiency.
Energy based models in cell migration
To address factors that are chemically relevant or cannot be explicitly expressed in terms of forces, the cellular Potts model39 with energy minimization and Monte Carlo steps offers a major advantage. Its off-lattice alternative, the vertex model, is also well used in morphogenesis and patterning.40,41 The factors affecting cell migration are evaluated in terms of Hamiltonian or energy form.42–45
Here the adhesion energy term Eadhesion, which was developed from Steinberg’s Differential Adhesion Hypothesis (DAH),39,46 has two parts: the adhesion among cells, and between cell and ECM or medium.41 σ is the adhesion energy per unit area, and A is the adhesion area for cell-cell or cell-ECM contacts. Econtinuity is the conservation of cell volume V in 3D or surface area in 2D due to cell elasticity and/or contractility,41,47 and is usually a quadratic function of the change ratio. Egrowth captures favorable energy change due to proliferation via the doubling cell volume and surface area.43 In some models however, the effect of proliferation is set within the Econtinuity. The chemotaxis energy term Echemotaxis is simulated to be linearly dependent on the growth factor concentration gradient ∇GF.43 The probability of accepting each Monte Carlo step is given by43
The cellular Potts model has addressed multiple cell migration predictions that can be validated by experiments. One of the successes is in predicting endothelial cell migration in angiogenesis.48 Shamloo et al48 first used an in silico collagen fibrils model to estimate the corresponding matrix density index used in their model via matching collagen area in microscopic and in silico images under different densities. We also have used an in silico collagen fibers model to mimic microscopic collagen images in vitro.33 With this corresponding matrix density index obtained, the angiogenesis process, that is the migration of endothelial cells, is simulated mimicking different collagen density effect. Shamloo et al48 confirmed their previous model predictions43 on endothelial sprout formation morphology under different ECM matrix. Recent studies also consider the effect of MMP and the nucleus compartments in cell migration using cellular Potts models. These models have been able to predict that cell migratory ability in subcellular channels closely depends on proteolytic machinery, and the cancer cell can achieve a sustained locomotion by either a pericellular proteolysis or by deforming its nucleus in a subnuclear track.42,49 A limitation for these Monte Carlo based energy minimization models is that they can only simulate a relatively small number of cell populations due to the heavy computational task.
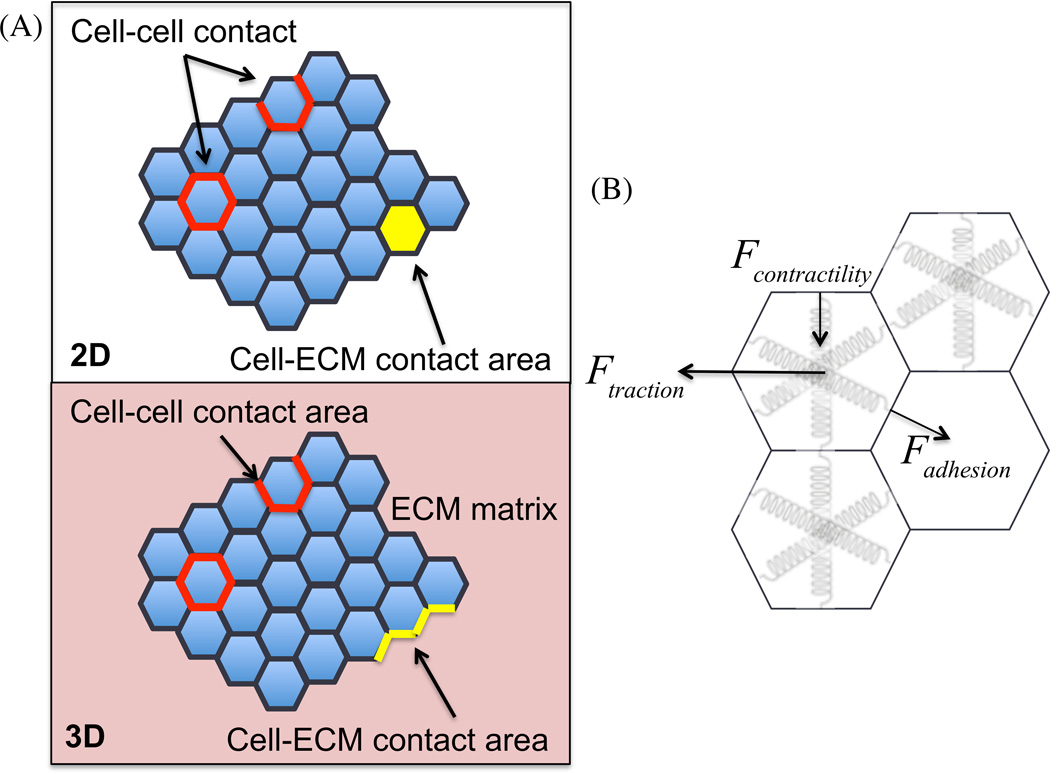
In comparison, the vertex model using the similar form of energy term is solved as an optimization problem. It tracks the movement of vertices during the optimization process. The gradient descent method can help to find the local minima state of the system energy. The steady state of 2D or 3D vertex model (Fig 2B) is close to the force balancing model result. For example, both approaches lead to the equilibrium between the contractility and the cell-cell adhesive force,41 and so does the Cellular Potts model. However, to fully convert the results between these models, vertex version of force balancing model (Fig 2C), or more generally, an appropriate conversion between the energy based model and the force balancing model is needed.
Fig 2. The vertex model and its potential force balancing correspondence.
(A) The vertex model for collective cell migration in 2D and 3D, with the difference in cell-ECM contact area part and whether to include the elasticity of ECM matrix when cells invading into the matrix. (B) An appropriate analog between the energy based model and force balancing model is needed.
This is a challenging problem, but a significant one. With recent advancements in stress and force measurement techniques, such as cell traction microscopy and cell stress microscopy, the mechanical properties can be measured directly during collective cell migration. A force-based model will have an advantage of direct correspondence with these mechanical measurements, and it can be directly and easily tested, in comparison with the energy based model. During collective cell migration, there exists largely motility, mechanical, molecular signaling heterogeneities within monolayers and cell clusters. The next step of unveiling the collective cell migration patterns is to explain the source and predict quantitatively these heterogeneities.
In addition to the models discussed above, there are also stochastic models on collective cell migration, especially focused on angiogenesis involving tip cell migration, sprouting and anastomosis.50 Maclaw et al. developed an evolutionary model of tumor progression regarding cell growth and migration, suggesting that targeting short-range cellular migratory activity could have marked effects on tumor growth rates.51 There is also a phase-field model, which does not identify cell boundaries and studies cell migration in a relatively deterministic manner.52 Additionally cell migration models have been developed that capture how migration affects ECM fibrous structure.33 There is an opportunity that it can be combined with recent additional ECM fibrous models53,54 focusing on microenvironment heterogeneity in cell migration.
Signaling pathways in cell migration
Cell signaling plays a central role in single and collective migration. The following section will review recent advancements in modeling signaling during migration. Due to a greater number of models focused on mesenchymal migration, and more data and literature focusing on these modes, the next section will have a greater discussion of mesenchymal modes, compared to amoeboid studies.
Signaling pathways in mesenchymal migration
As previously mentioned, modes of single-cell migration are divided into two broad categories, namely mesenchymal and amoeboid migratory modes. The mesenchymal migration relies mainly on high adhesion and form mature focal adhesion.
In mesenchymal modes, cell migration involves formation of cell-ECM adhesions at the leading edge of protrusions and disassembly of adhesions, mainly at cell rear.55,56 Integrins bind to the ECM and initially form small, transient matrix contact structures, called nascent adhesions. Proteins are recruited to bind with integrin cytoplasmic domains, such as paxillin.57,58 During adhesion maturation to focal adhesion, FAK and Src are activated by phosphorylation. FAK activity can be enhanced by the collagen crosslinking and ECM stiffening and further drive invasion of tumor cells.16,59 Importantly, it has been found recently that for cells in 3D matrices, focal adhesion proteins such as FAK do not form aggregates but are more diffused throughout the cytoplasm, though they still modulate cell motility by affecting protrusion activity and matrix deformation.60 Adhesion-relevant kinases regulate Rho-family GTPases, such as Rac and RhoA.61,62 FAK has been shown to be able to activate63–65 or inhibit RhoA activity.66,67 Though with these opposing effects, there is an apparent global increase in FAK and RhoA activity in stiffer 3D ECM environment.59 The increased collagen crosslink and ECM stiffening also promote 3D cell migration via up-regulating growth factor dependent PI3K activity.16,68 Following EGF stimulation, protein complexes containing Src, F-actin, mDia1 and Dia-interacting protein 1 stimulate Rac1.66
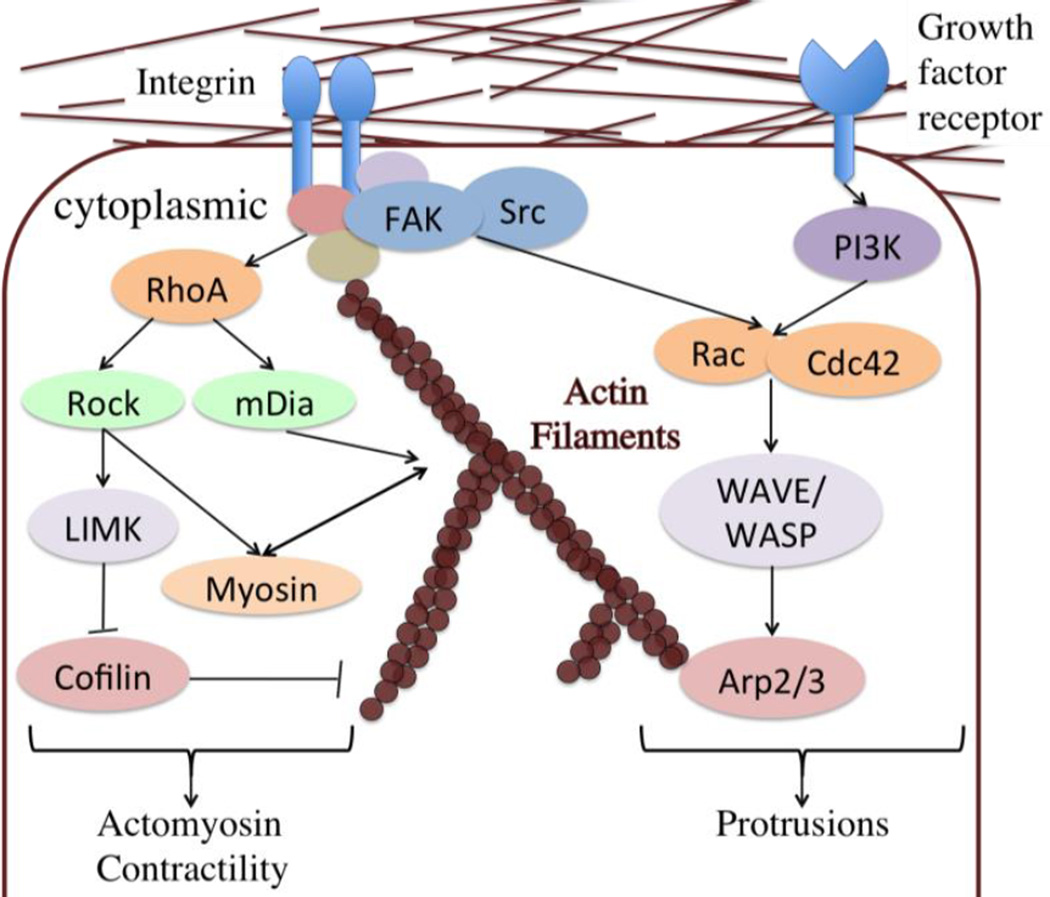
Cell protrusions at leading edge in migration process involves actin polymerization, while at cell rear there are cell contractions.56 The cytoskeleton activities are coordinated with formation/disassembly of cell-ECM adhesions. Rho-family GTPases are molecular ‘switches’ within cells bridging adhesion activities and cytoskeleton dynamics by controlling the formation and disassembly of actin cytoskeletal structures (Fig 3). RhoA-GTP can activate formin (mDia)-based actin nucleation69,70 and ROCK dependent myosin II contractility71. mDia stimulates the formation of stable microtubules that are capped and oriented towards the wound edge to promote cell migration.72,73 Rac stimulates branching and polymerization of actin filaments by the Arp2/3 complex via the activation of WAVE,74 and it promotes the lamellipodia formation in cell migration.75,76 Cdc42 activity is also prominent at the tip of the leading edge and regulates the migration direction by regulating the cell polarization.77,78 WASP, which is autoinhibited, can be activated by binding with Cdc42 and further activates Arp2/3.79 Conventionally, Rac promotes membrane protrusions at the leading edge, while RhoA is active in the cell rear and regulates its contractility in 2D cell migration.80,81 This is most likely due to the antagonism between Rac and RhoA at the leading edge.82,83 However, it has also been found that in randomly migrating cells, RhoA activity is high in a sharp band directly at the edge of protrusions.84
Fig 3. The signaling pathways of cytoskeleton dynamics in cell migration.
Here is a signaling pathway map that represents how external cues regulate cytoskeleton dynamics, and further induce directional cell migration via these downstream signaling molecules. Several modeling work of these signaling pathways have been built upon these interactions and crosstalks, and they further predict or explain the integrated effects of signaling molecules in affecting cell migration.
Signaling pathways in amoeboid migration
In amoeboid cell migration mode, cells migrate in a largely rounded shape, which results from a high cell tension relative to cell adhesions.85 In cells that undergo amoeboid migration, actin filaments assemble into a fibrous cortex that is absent from the cell front, but become progressively denser toward the cell rear, and so does myosin.86 With myosin II-dependent contraction at the cell rear, cells are even able to squeeze their nuclei through a narrow gap.3 High cell tension, that is, high cortical actomyosin contractility is due to high Rho and low Rac activities.85 Relying on increased activity of the RhoA pathway, formin-based actin nucleation and myosin II contractility are activated, which meanwhile promotes a retrograde actin flow during migration process.87 A retrograde flow is a translocation and recycling at proximal sites due to actin filament severing and/or disassembly following assembly at nucleation sites.88 The cell-scale retrograde flow of actin and myosin II is the driving force for the stable bleb mode in amoeboid migration. The pseudopods mode of amoeboid migration, which has protrusion due local actin assembly at the leading edge, only has the retrograde flow in the front.4
Manipulating cytoskeleton regulators, such as Rac,74 Rho89 and Rock,86 cell-ECM adhesion proteins,86 can make cells to switch migration modes between the mesenchymal and amoeboid migration. Adding lysophosphatidic (LPA), a serum phospholipid known to activate cortical contractility via the Rho/Rock pathway, also induces amoeboid migration.90 After LPA addition, myosin II rapidly redistributed to the cell cortex. The cortical accumulations of myosin II and the size of bleb are correlated with LPA addition. And a gradient of LPA can induce an asymmetric contraction of the cortical cytoskeleton to further influence cell migration. Both the mesenchymal and amoeboid single-cell migration requires a high myosin activity for mechanotransduction and great amount actin assemblies. However, other than the integrin or the external environment regulations, what kinds of direct molecular actin regulators lead to the difference between the cortical actin assemblies and the stress fiber assemblies? Do the stress fiber contractility and the cortical actomyosin contractility depend on the same cascade of myosin II signaling? These are the questions in need to be answered in unveiling the regulation of the single-cell migration signaling.
Signaling pathways in collective cell migration
Though the collective cell migration is ubiquitous in biology, detailed mechanistic and quantitative understanding linking molecular events with collective behavior is lacking compared to single-cell migration due to its complexity. When studying collective cell behavior, velocity correlation is sometimes used,25,30 to understand collective cellular motion. This correlation is a coefficient of coordination in collective cell migration in addition to cell speed and directional persistence. The tumor suppressor, merlin, plays an important role in coordinating cells interacting with Rac1 in monolayer epithelial collective migration.25 Also results from experiments show that collective migration has largely related to the intercellular stress.34 Within a confluent monolayer cells tend to move along directions with minimal intercellular shear stress, so that cell-cell junctions along these directions carry only minimal shear stresses.91 In the adherens junctions of epithelial cells, the extracellular domain of E-cadherin interact with cadherin protein on other cells (trans-binding) in a calcium-dependent way,92 while the cytoplasmic domain of E-cadherin interacts with p120 catenin, β-catenin and α-catenin, which links with the actin cytoskeleton.93 P-cadherin, whose expression is highly associated with undifferentiated cells in normal adult epithelial tissues and breast cancer, has the similar cadherin-catenin complex structure and actin cytoskeleton connection in cells.94 Results from the experiment show that Wnt slows down collective cell migration, which involves with the interaction with β-catenin.30 Substrate stiffness not only regulates cellular migration at a single cell level, but also at a collective level. The stiffer substrate has resulted in higher migration speed and persistence at the edge, which emerge with a higher myosin-mediated contractility.20 Cell coordination relies on a balance between cadherin complex mediated cell-cell adhesion and myosin-mediated contractility during collective motion. Epithelial polarity is another important factor regulating the cooperativity during cell migration, though to our knowledge researchers have not fully studied how the balance of the contractility and cell-cell adhesion interacts with this polarity in this directionality information synchronization. With current molecular assays in collective cell migration, the challenges stem from the investigation of downstream cascade signaling effects and quantification of patterns of collective cell migration in various environments. Thus, the computational models have the ability to provide novel insights to understand these questions.
Modeling signaling in cell migration
In the previous section we mainly reviewed the signaling pathways during cell migration, at the single and collective level. In the following section, we will turn our attention to additional comprehensive modeling work that integrates cell signaling and migration, with an eye towards the future direction and opportunities for computational modeling.
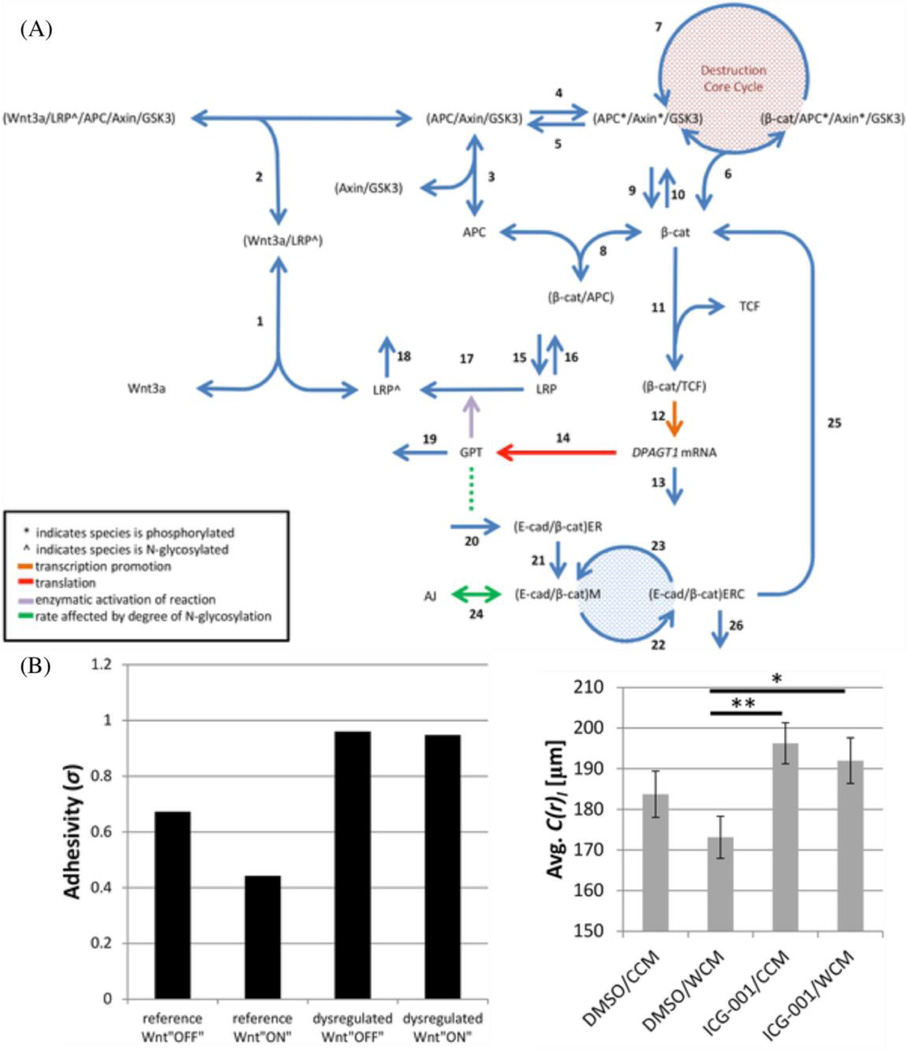
It is of fundamental importance to understand the molecular programmes that regulate cell migration in an integrated modeling-experimental framework. We recently have established an integrated signaling model of N-glycosylation metabolic pathway, Wnt/β-catenin signaling pathway, and cadherin-mediated adhesion in affecting cell migrations (Fig. 4A).30 A fundamental, ER glycosylation enzyme, GPT, is a Wnt/β-catenin transcriptional target that can promote the N-glycosylation of E-cadherin. This glycosylation, in turn, limits the adhesivity of cadherin ectodomains, which, in turn, leads to enhanced β-catenin signaling. This consists of multiple signaling pathways and feedback/feedforward pathways with extensive complexity. With the integrated modeling-experimental framework, we can reduce this complexity and unveil underlying molecular mechanisms regulating cell migration. In this model, binding and dissociation processes are described as linear product of the kinetic rate and the corresponding molecule concentrations: kiXY and k−i(X/Y). Here X and Y are the free concentrations of the binding partners, (X/Y) the concentration of the complex in which X and Y binds together, and ki and k−i the association and dissociation rates. Syntheses of proteins are described by constant rates (νi). Phosphorylation and dephosphorylation processes are described as kiX and kiX*, in which phosphorylated molecules are labeled with *. For example,
Fig 4. The integrated signaling pathway of N-glycosylation, Wnt/β-catenin and E-cadherin/β-catenin.
The signaling pathway map (A) that integrates model of N-glycosylation, Wnt/β-catenin and E-cadherin/β-catenin represents how signaling molecules interact in a kinetic network. The quantitative predictions of the model (B: left) are validated in experiments of cell migration (B: right). (Copyright 2016 PLoS Comput. Biol.30 :https://creativecommons.org/licenses/by/4.0/)
Here the dynamics of active β-catenin is solved based on the pathway processes that it involves in. The parameters are either taken from the direct measurement of the binding/unbinding rate and the phosphorylation/dephosphorylation rate, or estimated from known cellular responses. And the adhesivity of adhesion junctions is solved by the concentration of the adhesion junctions, the concentration of the (E-caderin/β-catenin) complex in the membrane, and the N-glycosylation degree of E-cadherin in the membrane (σM). We also carried out the experimental validation for our signaling pathway model. We not only measured the molecular amount change under the addition of Wnt3a and the β-catenin/TCF binding inhibitor, but also quantify the collective migration pattern change under these conditions in wound assay. The average speed, the movement angle orientations that indicate the coordination and the persistence, and more importantly, the correlation length are calculated based on the PIV measurement, which was previously discussed and offers an advantage in tracking motions in a monolayer. The correlation length quantifies the length scale within which cells have coordination in their motions,25 and the adhesivity predicted in our model has a good agreement with this intercellular coordination measurement in wound assay experiment (Figure 4B). To the authors’ knowledge, this is one of the few successful studies investigating multiple signaling pathways in influencing cell migration in an integrated modeling-experimental framework.
Over the years, a number of additional computational studies have modeled dynamics of signaling molecules to understand cell migration. The enormous complexity of the cell, combined with limited spatial and temporal data at the molecular level and prohibitive cost of detailed molecular modeling, result in models that cannot capture the dynamics of all the key molecules in complex environments. The molecules selected in the computational model are therefore often representative of the questions the researchers want to ask. For example, some models focus on how Rho-family GTPases sense the ECM properties characterizing the adhesion related kinases,95 other models have focused on rate of protrusion96,97 and polymerization98 in cell migration using cytoskeletal dynamics. However, most such computational models due to computational complexity and lack of detailed mechanistic data, have focused on single cells and have occasionally incorporated ECM,33,99,100 suggesting that integrated modeling of cellular interior, cell-cell and cell-ECM interactions continues to be an elusive goal. To this end, one of the most important processes in designing a multi-scale model is the “bridge”. The challenge in creating this bridge is about how to transfer simulation information/results between multiple scales. There have been some efforts, for example, the previous force balancing model inside the cell can potentially coupled with the previous force balancing model of cell migration. Similarly, the signaling pathway of Hippo95,101 can be combined in the energy based model and characterizing how the cell-cell stress difference can lead to different pattern of collective cell migration.
Yet, the broad based frameworks connecting scales and providing detailed and valuable information at multiple scales continues to be a major challenge.
Outlook
With increasing computational power in the past twenty years, significant effort has been made to study cell migration in complex and clinically relevant environments. Combined with new developments in biomedical engineering, new tools are being developed to effectively characterize cell migration and the associated mechanisms, such as tracking individuals within cell cohorts embedded in three dimensional collagen scaffolding29 and 3D cell traction microscopy.102 With the help of these quantification tools, corresponding numerical modeling work has also shed light on 3D migratory mechanisms in disease understanding. Availability of this data is revolutionizing modeling strategies and approaches to understand cellular behavior in complex environments and possibly identify new therapeutic targets. Finally, with the development of genome-scale analyses, patient specific and precision medicine strategies will also benefit from integrated experimental and computational models to understand how cell migration alterations may influence clinical outcomes,103 Thus while new challenges in understanding cell migration behavior remain, the opportunities to develop new, powerful and potent therapeutic regimens, and create new platforms for multi-scale investigation provide exciting possibilities for fundamental and applied understanding of cellular form, fate and function.
Acknowledgments
The authors acknowledge the research support from the National Institutes of Health (grant Nos. U01-CA177799, U01-CA202123-01, andP01HL120839) and the National Science Foundation (grant No. DMR-1206635) for the work.
Reference
- 1.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev. Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 3.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 4.Welch MD. Cell Migration, Freshly Squeezed. Cell. 2015;160:581–582. doi: 10.1016/j.cell.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 5.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Friedl P, Wolf K. Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo C-M, Wang H-B, Dembo M, Wang Y. Cell Movement Is Guided by the Rigidity of the Substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harjanto D, Zaman MH. Computational study of proteolysis-driven single cell migration in a three-dimensional matrix. Ann. Biomed. Eng. 2010;38:1815–1825. doi: 10.1007/s10439-010-9970-0. [DOI] [PubMed] [Google Scholar]
- 9.Zaman MH, Matsudaira P, Lauffenburger DA. Understanding effects of matrix protease and matrix organization on directional persistence and translational speed in three-dimensional cell migration. Ann. Biomed. Eng. 2007;35:91–100. doi: 10.1007/s10439-006-9205-6. [DOI] [PubMed] [Google Scholar]
- 10.Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak M, Spill F, Kamm RD, Zaman MH. Single-Cell Migration in Complex Microenvironments: Mechanics and Signaling Dynamics. J. Biomech. Eng. 2016 doi: 10.1115/1.4032188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto Y, Prudhomme S, Zaman MH. Modeling of adhesion, protrusion, and contraction coordination for cell migration simulations. J. Math. Biol. 2014;68:267–302. doi: 10.1007/s00285-012-0634-6. [DOI] [PubMed] [Google Scholar]
- 13.Maffei JS, Srivastava J, Fallica B, Zaman MH. Combinative in vitro studies and computational model to predict 3D cell migration response to drug insult. Integr. Biol. (Camb) 2014;6:957–972. doi: 10.1039/c4ib00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepzelter D, Zaman MH. Modeling persistence in mesenchymal cell motility using explicit fibers. Langmuir. 2014;30:5506–5509. doi: 10.1021/la404832t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vargas DA, Bates O, Zaman MH. Computational model to probe cellular mechanics during epithelial- mesenchymal transition. Cells Tissues Organs. 2013;197:435–444. doi: 10.1159/000348415. [DOI] [PubMed] [Google Scholar]
- 16.Levental KR, et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spill F, Reynolds DS, Kamm RD, Zaman MH. Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 2016;40:41–48. doi: 10.1016/j.copbio.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lü D, et al. Asymmetric Migration of Human Keratinocytes under Mechanical Stretch and Cocultured Fibroblasts in a Wound Repair Model. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat. Rev. Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 2012;199:545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdan P, Deasy BM, Gharaibeh B, Roehrs T, Marculescu R. Heterogeneous structure of stem cells dynamics: statistical models and quantitative predictions. Sci. Rep. 2014;4:4826. doi: 10.1038/srep04826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mak M, Kamm RD, Zaman MH. Impact of Dimensionality and Network Disruption on Microrheology of Cancer Cells in 3D Environments. PLoS Comput Biol. 2014;10:1–12. doi: 10.1371/journal.pcbi.1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker EL, Srivastava J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell migration: Integrated roles of matrix mechanics and transforming potential. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker EL, Bonnecaze RT, Zaman MH. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys. J. 2009;97:1013–1021. doi: 10.1016/j.bpj.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das T, et al. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol. 2015;17 doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 26.Vig DK, Hamby AE, Wolgemuth CW. On the Quantification of Cellular Velocity Fields. Biophys. J. 2016;110:1469–1475. doi: 10.1016/j.bpj.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novikova EA, Raab M, Discher DE, Storm C. Persistence-driven durotaxis: Generic, directed motility in rigidity gradients. ArXiv e-prints. 2015 doi: 10.1103/PhysRevLett.118.078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi D, Yang X, Marchetti MC, Manning ML. Motility-driven glass and jamming transitions in biological tissues. arXiv Prepr. 2015 doi: 10.1103/PhysRevX.6.021011. at < http://arxiv.org/abs/1509.06578>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma Y, et al. Collective Motion of Mammalian Cell Cohorts in 3D. Integr. Biol. 2015;7:1526–1533. doi: 10.1039/c5ib00208g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vargas DA, Sun M, Kukuruzinska MA, Zaman MH. The Integrated Role of Wnt/β-catenin, N-glycosylation, and E-cadherin-mediated Adhesion in Network Dynamics. PLoS Comput. Biol. 2016;12:1–26. doi: 10.1371/journal.pcbi.1005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaman MH, Kamm RD, Matsudaira P, Lauffenburger Da. Computational model for cell migration in three-dimensional matrices. Biophys. J. 2005;89:1389–1397. doi: 10.1529/biophysj.105.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asthagiri a, Nelson CM, Horwitz aF, Lauffenburger Da. Quantitative relationship among integrin ligand-binding, adhesion, and signaling via focal adhesion kinase and extracellular signal-regulated kinase 2. J. Biol. Chem. 1999;274:27119–27127. doi: 10.1074/jbc.274.38.27119. [DOI] [PubMed] [Google Scholar]
- 33.Sun M, Bloom AB, Zaman MH. Rapid quantification of 3D collagen fiber alignment and fiber intersection correlations with high sensitivity. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0131814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serra-Picamal X, et al. Mechanical waves during tissue expansion. Nat. Phys. 2012;8:628–634. [Google Scholar]
- 35.Shih W, Yamada S. N-cadherin-mediated cell-cell adhesion promotes cell migration in a three-dimensional matrix. J. Cell Sci. 2012;125:3661–3670. doi: 10.1242/jcs.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber GF, Bjerke Ma, DeSimone DW. A Mechanoresponsive Cadherin-Keratin Complex Directs Polarized Protrusive Behavior and Collective Cell Migration. Dev. Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramis-Conde I, Drasdo D, Anderson ARA, Chaplain MAJ. Modeling the Influence of the E-Cadherin-β-Catenin Pathway in Cancer Cell Invasion: A Multiscale Approach. Biophys. J. 2008;95:155–165. doi: 10.1529/biophysj.107.114678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spill F, Guerrero P, Alarcon T, Maini PK, Byrne H. Hybrid approaches for multiple-species stochastic reaction-diffusion models. J. Comput. Phys. 2015;299:429–445. doi: 10.1016/j.jcp.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graner F, Glazier Ja. Simulation of Biological Cell Sorting Using a Two-Dimensional Extended Potts Model. Phys. Rev. Lett. 1992;69:2013–2016. doi: 10.1103/PhysRevLett.69.2013. [DOI] [PubMed] [Google Scholar]
- 40.Bi D, Lopez JH, Schwarz JM, Manning ML. A density-independent glass transition in biological tissues. arxiv. 2014:1–5. [Google Scholar]
- 41.Farhadifar R, Röper JC, Aigouy B, Eaton S, Jülicher F. The Influence of Cell Mechanics, Cell-Cell Interactions, and Proliferation on Epithelial Packing. Curr. Biol. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 42.Scianna M, Preziosi L. A cellular Potts model for the MMP-dependent and -independent cancer cell migration in matrix microtracks of different dimensions. Comput. Mech. 2013:1–13. [Google Scholar]
- 43.Bauer AL, Jackson TL, Jiang Y. Topography of extracellular matrix mediates vascular morphogenesis and migration speeds in angiogenesis. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Valle SY, Hyman JM, Chitnis N. Mathematical Models of Mixing Patterns Between Age Groups for Predicting the Spread of Infectious Diseases. Math. Biosci. Eng. 2013;20:1–20. doi: 10.3934/mbe.2013.10.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner S, Sherratt Ja. Intercellular adhesion and cancer invasion: a discrete simulation using the extended Potts model. J. Theor. Biol. 2002;216:85–100. doi: 10.1006/jtbi.2001.2522. [DOI] [PubMed] [Google Scholar]
- 46.Steinberg MS. Does differential adhesion govern self-assembly processes in histogenesis? Equilibrium configurations and the emergence of a hierarchy among populations of embryonic cells. J. Exp. Zool. 1970;173:395–433. doi: 10.1002/jez.1401730406. [DOI] [PubMed] [Google Scholar]
- 47.Bazellières E, et al. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shamloo A, Mohammadaliha N, Heilshorn SC, Bauer AL. A Comparative Study of Collagen Matrix Density Effect on Endothelial Sprout Formation Using Experimental and Computational Approaches. Ann. Biomed. Eng. 2016;44:929–941. doi: 10.1007/s10439-015-1416-2. [DOI] [PubMed] [Google Scholar]
- 49.Krause M, Wolf K. Cancer cell migration in 3d tissue: Negotiating space by proteolysis and nuclear deformability. Cell Adhes. Migr. 2015;9:357–366. doi: 10.1080/19336918.2015.1061173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spill F, Guerrero P, Alarcon T, Maini PK, Byrne HM. Mesoscopic and continuum modelling of angiogenesis. J. Math. Biol. 2014:1–48. doi: 10.1007/s00285-014-0771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Travasso RDM, Poiré EC, Castro M, Rodrguez-Manzaneque JC, Hernández-Machado a. Tumor angiogenesis and vascular patterning: A mathematical model. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0019989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Abhilash AS, Chen CS, Wells RG, Shenoy VB. Long-Range Force Transmission in Fibrous Matrices Enabled by Tension-Driven Alignment of Fibers. Biophys. J. 2014;107:2592–2603. doi: 10.1016/j.bpj.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abhilash AS, Baker BM, Trappmann B, Chen CS, Shenoy VB. Remodeling of Fibrous Extracellular Matrices by Contractile Cells: Predictions from Discrete Fiber Network Simulations. Biophys. J. 2014;107:1829–1840. doi: 10.1016/j.bpj.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb DJ, et al. FAK–Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 56.Sheetz MP, Felsenfeld D, Galbraith CG, Choquet D. Cell migration as a five-step cycle. Biochem. Soc. Symp. 1999;65:233–243. [PubMed] [Google Scholar]
- 57.Deakin NO, Turner CE. Paxillin comes of age. J. Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Z, Lambacher A, Fassler R. Nascent adhesions: From fluctuations to a hierarchical organization. Curr. Biol. 2014;24:R801–R803. doi: 10.1016/j.cub.2014.07.061. [DOI] [PubMed] [Google Scholar]
- 59.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraley SI, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim. Biophys. Acta - Mol. Cell Res. 2004;1692:63–76. doi: 10.1016/j.bbamcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Huveneers S, Danen EHJ. Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 63.Rico B, et al. Control of axonal branching and synapse formation by focal adhesion kinase. Nat. Neurosci. 2004;7:1059–1069. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhai J, et al. Direct Interaction of Focal Adhesion Kinase with p190RhoGEF. J. Biol. Chem. 2003;278:24865–24873. doi: 10.1074/jbc.M302381200. [DOI] [PubMed] [Google Scholar]
- 65.Guilluy C, et al. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng W, et al. DIP (mDia interacting protein) is a key molecule regulating Rho and Rac in a Src-dependent manner. EMBO J. 2004;23:760–771. doi: 10.1038/sj.emboj.7600095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Playford MP, Vadali K, Cai X, Burridge K, Schaller MD. Adhesion Kinase regulates cell–cell contact formation in epithelial cells via modulation of Rho. Exp. Cell Res. 2008;314:3187–3197. doi: 10.1016/j.yexcr.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloom AB, Zaman MH. Influence of the microenvironment on cell fate determination and migration. Physiol. Genomics. 2014;46:309–314. doi: 10.1152/physiolgenomics.00170.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakamoto S, et al. Liprin-controls stress fiber formation by binding to mDia and regulating its membrane localization. J. Cell Sci. 2012;125:108–120. doi: 10.1242/jcs.087411. [DOI] [PubMed] [Google Scholar]
- 70.Loria PM, Duke A, Rand JB, Hobert O. The Mouse Formin mDia1 Is a Potent Actin Nucleation Factor Regulated by Autoinhibition. Curr. Biol. 2003;13:1317–1323. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 71.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 72.Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 73.Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized Stabilization of Microtubules by Integrin- and FAK-Facilitated Rho Signaling. Sci. 2004;303:836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 74.Sanz-Moreno V, et al. Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 75.Marignani PA, Carpenter CL. Vav2 is required for cell spreading. J. Cell Biol. 2001;154:177–186. doi: 10.1083/jcb.200103134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Obergfell A, et al. Coordinate interactions of Csk, Src, and Syk kinases with αIIbβ3 initiate integrin signaling to the cytoskeleton. J. Cell Biol. 2002;157:265–275. doi: 10.1083/jcb.200112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Butty A, et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stowers L, Yelon D, Berg LJ, Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miki H, Takenawa T. Regulation of Actin Dynamics by WASP Family Proteins. J. Biochem. 2003;134:309–313. doi: 10.1093/jb/mvg146. [DOI] [PubMed] [Google Scholar]
- 80.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 81.Burridge K, Wennerberg K. Rho and Rac Take Center Stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 82.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac Downregulates Rho Activity: Reciprocal Balance between Both Gtpases Determines Cellular Morphology and Migratory Behavior. J. Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol. 2003;5:236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 84.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 85.Sanz-Moreno V, Marshall CJ. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 2010;22:690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y-J, et al. Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 87.Kardash E, et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol. 2010;12:47–53. doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- 88.Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin Drives Retrograde F-Actin Flow in Neuronal Growth Cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 89.Sahai E, Garcia-Medina R, Pouysségur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J. Cell Biol. 2007;176:35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruprecht V, et al. Cortical Contractility Triggers a Stochastic Switch to Fast Amoeboid Cell Motility. Cell. 2015;160:673–685. doi: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Notbohm J, et al. Cellular Contraction and Polarization Drive Collective Cellular Motion. Biophys. J. 2016;110:2729–2738. doi: 10.1016/j.bpj.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 93.Nelson WJ, Nusse R. Convergence of Wnt, {β}-Catenin, and Cadherin Pathways. Science (80-.) 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albergaria A, et al. P-cadherin role in normal breast development and cancer. International Journal of Developmental Biology. 2011;55:811–822. doi: 10.1387/ijdb.113382aa. [DOI] [PubMed] [Google Scholar]
- 95.Sun M, Spill F, Zaman MH. A Computational Model of YAP/TAZ Mechanosensing. Biophys. J. 2016;110:2540–2550. doi: 10.1016/j.bpj.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rubinstein B, Jacobson K, Mogilner A. Multiscale Two-Dimensional Modeling of a Motile Simple-Shaped Cell. Multiscale Model. Simul. 2005;3:413–439. doi: 10.1137/04060370X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stéphanou A, Mylona E, Chaplain M, Tracqui P. A computational model of cell migration coupling the growth of focal adhesions with oscillatory cell protrusions. J. Theor. Biol. 2008;253:701–716. doi: 10.1016/j.jtbi.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 98.Spill F, Andasari V, Mak M, Kamm RD, Zaman MH. Effects of 3D Geometries on Cellular Gradient Sensing and Polarization. 2015 doi: 10.1088/1478-3975/13/3/036008. at < http://arxiv.org/abs/1508.02786>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee B, et al. A Three-Dimensional Computational Model of Collagen Network Mechanics. PLoS One. 2014;9:e111896. doi: 10.1371/journal.pone.0111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steinwachs J, et al. Three-dimensional force microscopy of cells in biopolymer networks. Nat. Methods. 2015 doi: 10.1038/nmeth.3685. advance on. [DOI] [PubMed] [Google Scholar]
- 103.Collins FS, Varmus H. A New Initiative on Precision Medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]