Pectin methylesterase inhibitors control PME activity to hinder pectin degradation as part of the plant immune response.
Abstract
Infection by necrotrophs is a complex process that starts with the breakdown of the cell wall (CW) matrix initiated by CW-degrading enzymes and results in an extensive tissue maceration. Plants exploit induced defense mechanisms based on biochemical modification of the CW components to protect themselves from enzymatic degradation. The pectin matrix is the main CW target of Botrytis cinerea, and pectin methylesterification status is strongly altered in response to infection. The methylesterification of pectin is controlled mainly by pectin methylesterases (PMEs), whose activity is posttranscriptionally regulated by endogenous protein inhibitors (PMEIs). Here, AtPMEI10, AtPMEI11, and AtPMEI12 are identified as functional PMEIs induced in Arabidopsis (Arabidopsis thaliana) during B. cinerea infection. AtPMEI expression is strictly regulated by jasmonic acid and ethylene signaling, while only AtPMEI11 expression is controlled by PME-related damage-associated molecular patterns, such as oligogalacturonides and methanol. The decrease of pectin methylesterification during infection is higher and the immunity to B. cinerea is compromised in pmei10, pmei11, and pmei12 mutants with respect to the control plants. A higher stimulation of the fungal oxalic acid biosynthetic pathway also can contribute to the higher susceptibility of pmei mutants. The lack of PMEI expression does not affect hemicellulose strengthening, callose deposition, and the synthesis of structural defense proteins, proposed as CW-remodeling mechanisms exploited by Arabidopsis to resist CW degradation upon B. cinerea infection. We show that PME activity and pectin methylesterification are dynamically modulated by PMEIs during B. cinerea infection. Our findings point to AtPMEI10, AtPMEI11, and AtPMEI12 as mediators of CW integrity maintenance in plant immunity.
The cell wall (CW) is the primary interface where most plant-microbe interactions occur and the main physical and molecular line of defense evolved by plants to restrict pathogen penetration and infection spreading (Malinovsky et al., 2014). Some of the most devastating plant diseases are caused by necrotrophic fungi. Necrotrophic infection is a complex process mediated by numerous extracellular enzymes, proteins, and metabolites. CW-degrading enzymes perturb CW integrity, facilitating penetration into the host surface, while toxins, oxalic acid (OA), and reactive oxygen species may contribute to killing of the host cells (Laluk and Mengiste, 2010; King et al., 2011; Nakajima and Akutsu, 2014). The plant susceptibility to necrothrophs and the efficiency of CW degradation are largely affected by CW composition and structure (Lionetti et al., 2010; Francocci et al., 2013; Bellincampi et al., 2014). CWs of Arabidopsis (Arabidopsis thaliana) leaves consist mainly of cellulose fibers encased in a network of hemicellulose and embedded in a pectin matrix (Zablackis et al., 1995). Hemicelluloses include xyloglucan containing a (1,4)-β-linked glucan backbone substituted with (1,6)-α-linked xylosyl residues or side chains of xylosyl, galactosyl, and fucosyl residues. Low percentages of glucuronoarabinoxylans, mannans, and glucomannans also are present in CW of Arabidopsis leaves. Pectins are a complex group of polysaccharides composed of homogalacturonan (HG), rhamnogalacturonan I (RGI), rhamnogalacturonan II (RGII), and xylogalacturonan. HG, a linear polymer of (1,4)-α-linked GalA residues, is the prevalent component of leaf CW pectins (Zablackis et al., 1995) and is critical for tissue integrity, wall plasticity, and cell adhesion (Lionetti et al., 2014a). RGI consists of a backbone of alternating disaccharides, (1,4)-α-d-GalA-(1,2)-α-l-Rha with (1,4)-β-galactan, branched arabinan, or arabinogalactan side chains and constitute 20% to 25% of pectin of primary CW of dicots (Mohnen, 2008). RGII is a highly substituted galacturonan with side chains containing Ara, Rha, Gal, Xyl, or Fuc residues (Mohnen, 2008; Harholt et al., 2010). Xylogalacturonan is formed by a linear (1,4)-α-GalA backbone substituted with d-Xyl residues. RGII and xylogalacturonan represent a minor component and account of about 10% of leaf CW pectin (Zandleven et al., 2007; Mohnen, 2008).
During pathogen attack, plants sense pathogens and exploit constitutive and/or induced defense mechanisms based on biochemical modification of the CW components (Williamson et al., 2007; Raiola et al., 2011; Bellincampi et al., 2014; Blümke et al., 2015). Upon cuticle breaking, the adhesive pectin matrix is an early target for fungal necrotrophs (van Kan, 2006). Botrytis cinerea is considered one of the most important necrotrophic pathogens, mainly due to its large host range and its ability to produce severe damage, both preharvest and postharvest (Dean et al., 2012). Analysis of the B. cinerea genome indicates the presence of 118 genes associated with plant CW degradation (Amselem et al., 2011), including a large array of pectinases such as polygalacturonases and pectate lyases (Blanco-Ulate et al., 2014). Pectins are synthesized in the Golgi and secreted into the CW in a highly methylesterified form (Harholt et al., 2010; Kim et al., 2015). Methylesterified HG is demethylesterified after biosynthesis by plant pectin methylesterases (PMEs; EC 3.1.1.11; Pfam 01095; CE8; www.cazy.org), which release protons and methanol (MeOH) in the apoplast. PMEs belong to a large multigene family (67 putative isoforms in Arabidopsis) whose members display different modes of action and produce HG with different distribution and degree of methylesters in a locally regulated manner, not yet completely understood (Wang et al., 2013). PME activity can be tightly regulated by PME inhibitors (PMEIs) identified in numerous plant species (Sénéchal et al., 2014; Lionetti et al., 2015c). PMEIs belong to the large multigene family PF04043 (http://pfam.xfam.org/family/PF04043; 69 genes in Arabidopsis), which also includes the structurally related invertase inhibitors (INHs).
The methylesterification status of pectin affects plant resistance to diseases (Lionetti et al., 2012; Bellincampi et al., 2014). In several plant-microbe interactions, a high level of pectin methylesterification correlated with an increased resistance to pathogens. This feature was associated with the low susceptibility of high methylesterified pectin to pectic enzymes, virulence factors with active roles in pathogenesis (Herron et al., 2000; Wydra and Berl, 2006; Lionetti et al., 2012). Biotechnological approaches were used to increase the basal level of pectin methylesterification, aiming at engineering a pectin substrate less prone to degradation by fungal pectinases. These strategies were based mainly on reducing PME activity through the constitutive expression of development-related PMEIs. PMEI overexpression in different plant species reduced their susceptibility to fungal, bacterial, and viral pathogens (Lionetti et al., 2007, 2014b, 2015b; An et al., 2008; Raiola et al., 2011). In particular, plants overexpressing AtPMEI1 or AtPMEI2 (Hothorn et al., 2004; Raiola et al., 2004; Wolf et al., 2009; De Caroli et al., 2011) showed a lower level of PME activity, a higher degree of methylesterification (DME) of pectin, and a reduced susceptibility to B. cinerea and Pectobacterium carotovorum (Lionetti et al., 2007; Raiola et al., 2011). The reduced susceptibility to fungal diseases also was observed in wheat (Triticum aestivum) plants overexpressing AdPMEI from kiwi (Actinidia deliciosa; Camardella et al., 2000; Giovane et al., 2004; Di Matteo et al., 2005; Lionetti et al., 2014b), indicating that the inhibition of PME activity also improves plant resistance in low-pectin-containing monocot species (Vogel, 2008; Volpi et al., 2011).
Despite this evidence, the current knowledge of a possible regulation of pectin methylesterification during disease as well as of the molecular mechanisms involved in this regulation remains scarce. Emerging clues indicate that PME activity and the level of pectin methylesterification are precisely and temporally regulated during the course of a plant disease. A steady increase in plant PME activity resulted in a reduced level of pectin methylesterification, which was observed in Arabidopsis during infection by different pathogens (Bethke et al., 2014; Lionetti, 2015). Genome-wide transcriptional profiling suggests that specific Arabidopsis PMEs and PMEIs are expressed in response to pathogens and strictly dependent on the pathogen lifestyle (Lionetti et al., 2012; Windram et al., 2012). However, very few PME and PMEI isoforms have been identified to date that are involved in disease resistance, and their exact role during disease remains undetermined. AtPME3 is induced in Arabidopsis when challenged with either B. cinerea or P. carotovorum, and the pme3 mutant is more resistant to these necrotrophic pathogens (Raiola et al., 2011). TaPME1 has been considered as a candidate gene involved in wheat susceptibility against the necrotroph Fusarium graminearum (Lionetti et al., 2015a). A recent comprehensive analysis of the PME gene family in wheat indicated the possible involvement of specific TaPMEs in wheat defense against F. graminearum (Zega and D’Ovidio, 2016). Specific Arabidopsis PMEs contribute to immunity against the hemibiotrophic bacterial pathogen Pseudomonas syringae (Bethke et al., 2014). However, there is no evidence that the same genes are important against the necrotrophic fungus Alternaria brassicicola.
Accumulating evidence suggests that immune hormones regulate pectin methylesterification during plant-pathogen interactions (Nafisi et al., 2015). Jasmonic acid (JA) modulates the pectin DME in potato (Solanum tuberosum) to protect pectin degradation by pectate lyase produced by Dickeya dadantii (Taurino et al., 2014). PME activity is triggered through a JA-dependent pathway when Arabidopsis is challenged with P. syringae or A. brassicicola (Bethke et al., 2014). PME activity was proposed to be involved in the release and perception of defense signals during infection. PME activity is required for the production of demethylesterified oligogalacturonides, damage-associated molecular patterns (DAMPs), released upon partial degradation of HG by fungal pectinases (Ferrari et al., 2013). The ectopic expression of the strawberry (Fragaria × ananassa) gene FaPE1 in Fragaria vesca resulted in the generation of bioactive oligogalacturonides (OGs) responsible for the constitutive activation of defense responses and plant resistance to B. cinerea (Osorio et al., 2008, 2011). OGs are perceived in Arabidopsis by the receptor WALL-ASSOCIATED KINASE1 (WAK1) to activate plant immune responses (Brutus et al., 2010). WAK2 requires AtPME3-induced pectin demethylesterification to activate OG-dependent stress responses in Arabidopsis (Kohorn et al., 2014). Furthermore, PMEs release MeOH, a DAMP-like alarm signal (Hann et al., 2014), which alerts adjacent noninfected tissues or neighboring plants (Dorokhov et al., 2012; Komarova et al., 2014). The molecular mechanisms behind OG/MeOH release and signaling during pathogen infection are still largely unknown.
In this work, we advance the understanding about CW remodeling during necrotrophy. We sought to unravel the mechanisms regulating pectin methylesterification during pathogen infection. We identified AtPMEI10, AtPMEI11, and AtPMEI12 as functional PME inhibitors induced upon B. cinerea attack. AtPMEI10, AtPMEI11, and AtPMEI12 are regulated by immune hormonal pathways and by DAMPS such as OGs and MeOH. We isolated pmei mutants, which showed an impaired PMEI expression and a compromised fungal resistance. Our findings point to AtPMEI10, AtPMEI11, and AtPMEI12 as mediators of CW integrity maintenance in plant immunity.
RESULTS
The Expression of Specific PMEI Isoforms Is Altered during B. cinerea Infection
We attempted to understand if PMEIs are exploited by plants to regulate pectin methylesterification during disease. Analyzing publicly available microarray data (AbuQamar et al., 2006; Lionetti et al., 2012; Windram et al., 2012), we selected eight Arabidopsis genes, predicted to belong to the plant INH/PMEI family, which showed altered expression during B. cinerea infection. A homology tree, generated with the selected PMEIs and the INH/PMEIs characterized so far in dicotyledonous plants, identified three independent groups (Fig. 1). Group 1 included the functional AdPMEI expressed in kiwi fruits (Balestrieri et al., 1990; Giovane et al., 1995), the pollen-specific AtPMEI1 and AtPMEI2 (Raiola et al., 2004), and At5g46960 (henceforth AtPMEI12). The same group also contained SolyPMEI (Reca et al., 2012) and VvPMEI1 (Lionetti et al., 2015c), two PMEIs from tomato (Solanum lycopersicum) and grapevine (Vitis vinifera), respectively, both involved in fruit development. Group 2 comprised At1g62760 (henceforth AtPMEI10) and At3g47380 (henceforth AtPMEI11), both clustering with CaPMEI1, a pepper (Capsicum annuum) inhibitor involved in plant resistance to biotic and abiotic stresses (An et al., 2008). At5g51520 also was present in the same group with a higher similarity to AtPMEI4, an inhibitor expressed in Arabidopsis roots (Sénéchal et al., 2015). Group 2 also included AtPMEI3, expressed in apical meristems and affecting primordia formation (Peaucelle et al., 2008), and AtPMEI6, involved in seed maturation and germination (Saez-Aguayo et al., 2013). In group 3, At3g17140 clustered with the biochemically characterized INHs, suggesting a possible INH nature for this protein. Also in the same group, although with a low amino acid identity, is AtPMEI5, previously involved in seed germination (Müller et al., 2013). BoPMEI, involved in pollen tube growth of Brassica oleracea (Zhang et al., 2010), At4g15750, and At5g50040 (not yet characterized) showed low levels of amino acid identity with the other proteins analyzed.
Figure 1.
Homology tree of selected PMEI isoforms. The percentage amino acid identity (numbers at branch points) was evaluated between the putative Arabidopsis INH/PMEI isoforms with an altered expression during B. cinerea infection and PMEIs and INHs characterized so far. Multiple sequence alignment was performed using the DNAman software package (Lynnon Biosoft). The defense-related AtPMEI10, AtPMEI11, and AtPMEI12 selected in this study for further characterization are highlighted in boldface.
PMEIs and INHs can be distinguished on the basis of specific features in their amino acid sequences (Di Matteo et al., 2005). With the aim of identifying the nature of the eight selected genes as PMEIs or INHs, we searched for the conserved amino acid domains characterizing the two types of inhibitors (Supplemental Fig. S1). All selected PMEIs presented the four Cys residues typically engaged in the formation of two disulfide bridges and required to stabilize their four-helical-bundle structure. Excluding At3g17140, At5g46960, and At5g50040, the putative PMEIs also showed a conserved Thr residue required to strengthen the PMEI-PME interactions at apoplastic pH. A typical SAA motif, contributing to the PMEI-PME complex formation (Di Matteo et al., 2005), also was shared by putative PMEIs, except for At3g17140 and At4g15750. In At3g17140, the presence of a typical PKFAE motif, critical for invertase-INH interaction (Hothorn et al., 2010), suggests that the protein is an INH; therefore, it was not investigated further.
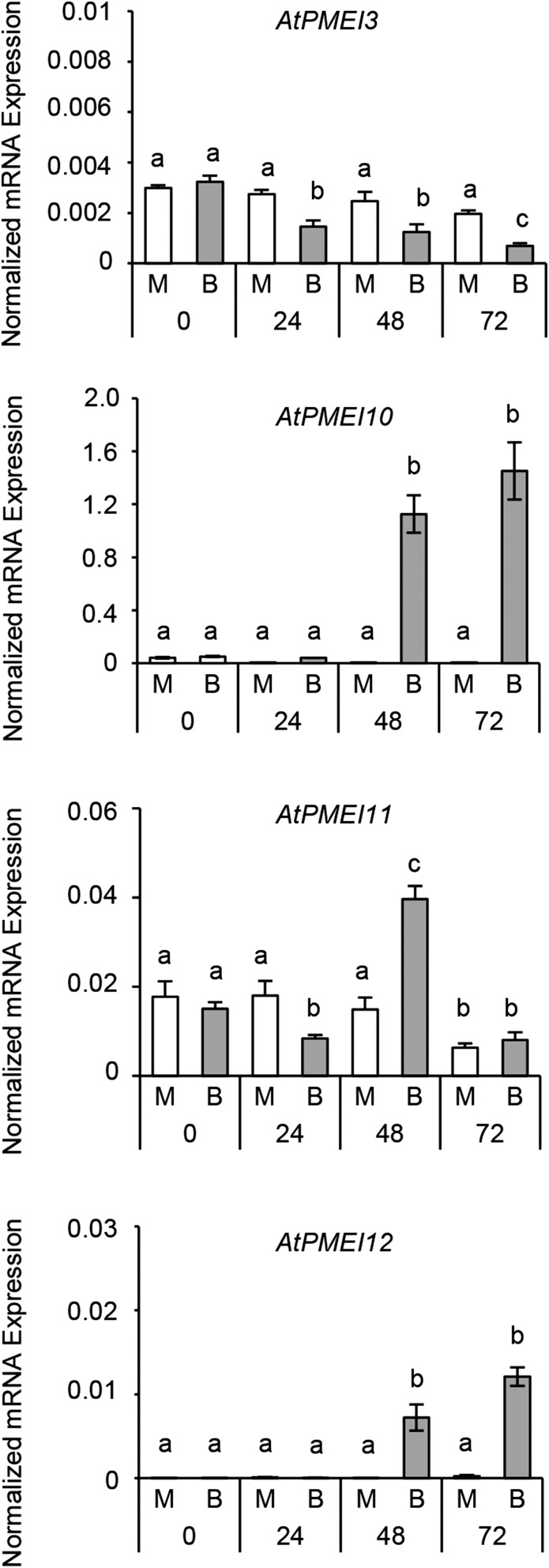
The level of expression of the selected genes was analyzed by reverse transcription (RT)-PCR at 0, 24, 48, and 72 h post B. cinerea infection. Different from what was reported in the microarray data, our analysis did not reveal an altered expression for At5g51520, At5g50040, and At4g15750, which were not further investigated. An altered pattern of expression of AtPMEI3 (Peaucelle et al., 2008), AtPMEI10, AtPMEI11, and AtPMEI12 was detected (Fig. 2). In particular, AtPMEI3 showed an early down-regulation at 24 h post inoculation (hpi), which proceeded during the course of infection. AtPMEI11 was rapidly and significantly repressed at 24 hpi, subsequently induced at 48 hpi, and then declined at 72 hpi. AtPMEI10 and AtPMEI12 were significantly induced at 48 hpi and further increased at 72 hpi. These results indicate that PMEI members have different levels and timings of expression in response to B. cinerea. Therefore, we decided to further study only AtPMEI10, AtPMEI11, and AtPMEI12, which were induced during B. cinerea infection.
Figure 2.
Levels and kinetics of AtPMEI3, AtPMEI10, AtPMEI11, and AtPMEI12 expression in Arabidopsis leaves during B. cinerea infection. The expression of different PMEI genes in infected leaves of 6-week-old Arabidopsis wild-type plants was analyzed by quantitative PCR (qPCR) at 24, 48, and 72 hpi. The expression levels were normalized to UBIQUITIN5 (UBQ5) expression. Values are means ± sd (n = 3). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.05). The experiments were repeated three times with similar results. B, B. cinerea-inoculated leaves; M, mock-inoculated leaves.
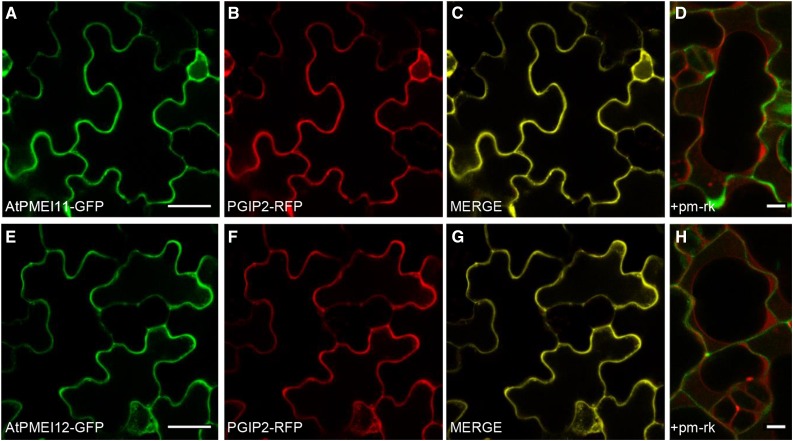
AtPMEI10 and AtPMEI11 share about 32% identity at the amino acid level with each other and about 13% identity with AtPMEI12 (Supplemental Fig. S2). All three PMEIs showed the presence of an N-terminal signal peptide for targeting to the secretory pathway. Interestingly, AtPMEI10 stands out for the presence, after the signal peptide and upstream of the PMEI domain, of a 115-amino acid region enriched in Ser/Pro-rich repeat (SPRR) modular domains, to our knowledge never described previously in PMEIs (Supplemental Figs. S2 and S3). The SPRR region is delimited by two SSSS motifs and is formed mainly by contiguous and repeated SL(S)SPS(L)-SP(A)P regions including two interspersed repetitions of four Pro residues. Prediction analysis indicated an extracellular localization for the three PMEIs (http://abi.inf.uni-tuebingen.de/Services/MultiLoc2). We attempted to investigate the in vivo subcellular localization of the three new inhibitors. None of the three inhibitors were predicted to show the C-terminal ω site common to glycosylphosphatidylinositol-anchored proteins (predGPI software; http://gpcr2.biocomp.unibo.it/predgpi/) and found previously for AtPMEI1 (De Caroli et al., 2011). Consequently, we decided to link GFP to the C terminus of the proteins. AtPMEI11-GFP and AtPMEI12-GFP constructs under the control of the 35S promoter were expressed transiently in Arabidopsis cotyledon epidermal cells using the FAST technique (Li et al., 2009). The Phaseolus vulgaris polygalacturonase-inhibiting protein2 fused to red fluorescent protein (PGIP2-RFP) was coexpressed as a CW marker (Fig. 3, B and F) and the plasma membrane aquaporin fused to mCherry (pm-rk) as a marker of the plasma membrane (Fig. 3, D and H; Nelson et al., 2007; De Caroli et al., 2011, 2015). At 36 h post transformation, the CW was fluorescently labeled by both AtPMEI11-GFP and AtPMEI12-GFP protein fusions (Fig. 3), as indicated by (1) the colocalization of both PMEI11-GFP (Fig. 3A) or PMEI12-GFP (Fig. 3E) with PGIP2-RFP (Fig. 3, C and G), and (2) the absence of colocalization of PMEI11-GFP or PMEI12-GFP chimeras with pm-rk in plasmolyzed cells (Fig. 3, D and H). These results confirm that AtPMEI11 and AtPMEI12 are correctly secreted into the apoplast. The consistent efforts spent on the construction of the AtPMEI10-GFP fusion were unsuccessful.
Figure 3.
Subcellular localization of AtPMEI11-GFP and AtPMEI12-GFP proteins. Confocal microscopy images of cotyledon epidermal cells of Arabidopsis 7-d-old-seedlings show the coexpression of AtPMEI11-GFP (A) or AtPMEI12-GFP (E) with PGIP2-RFP (B and F) and their colocalization in the apoplast (C and G) at 36 h post transformation. Plasmolyzed cells (1 m NaCl) expressing AtPMEI11-GFP (D) or AtPMEI12-GFP (H) and the plasma membrane marker pm-rk show the green fluorescent CW and the retracted red fluorescent plasma membrane. Bars = 20 μm (A–C and E–G) and 5 μm (D and H).
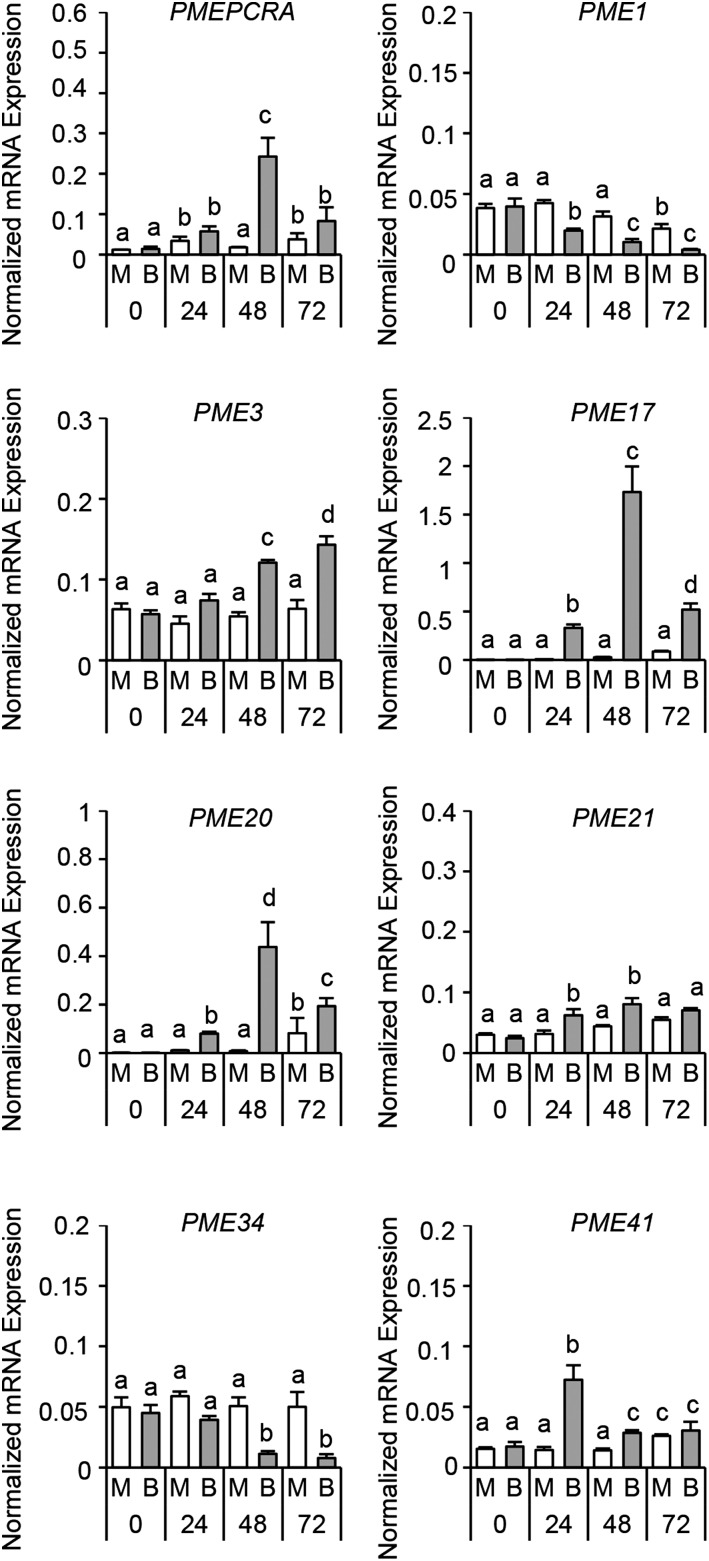
We attempted to explore possible PMEs expressed during B. cinerea infection and possibly recognized by the selected PMEIs. Twelve Arabidopsis PMEs exhibiting an altered expression upon B. cinerea infection were selected by exploiting publicly available microarray data (AbuQamar et al., 2006; Lionetti et al., 2012; Windram et al., 2012). We challenged Arabidopsis leaves with B. cinerea and quantified the expression level of the selected PMEs at 0, 24, 48, and 72 hpi. METHYLESTERASE PCR A (PMEPCRA), PME1, PME3, PME17, PME20, PME21, PME34, and PME41 members exhibited significantly altered timing and level of expression during B. cinerea infection, while the expression of PME4, PME6, PME7, and PME8 was not altered under our experimental conditions (Fig. 4). The higher level of expression was detected for PME17 and PME20. AtPMEPCRA, PME17, PME20, and PME21 expression started at 24 hpi, showed peaks at 48 hpi, and declined at 72 hpi. A steady increase of PME3 expression was observed during infection, consistent with previous results (Raiola et al., 2011). PME41 showed a peak of expression at 24 hpi and declined at 48 hpi. In contrast, PME1 and PME34 were repressed during the course of infection.
Figure 4.
Specific Arabidopsis PMEs show altered expression during B. cinerea infection. The expression of different PME isoforms in infected leaves of 6-week-old Arabidopsis wild-type plants was analyzed by qPCR at 0, 24, 48, and 72 h post infection. The expression levels were normalized to UBQ5 expression. The values are means ± sd (n = 3). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.01). The experiments were repeated three times with similar results. B, B. cinerea-inoculated leaves; M, mock-inoculated leaves.
AtPMEI10, AtPMEI11, and AtPMEI12 Expression Is Controlled by Plant Immune Signaling
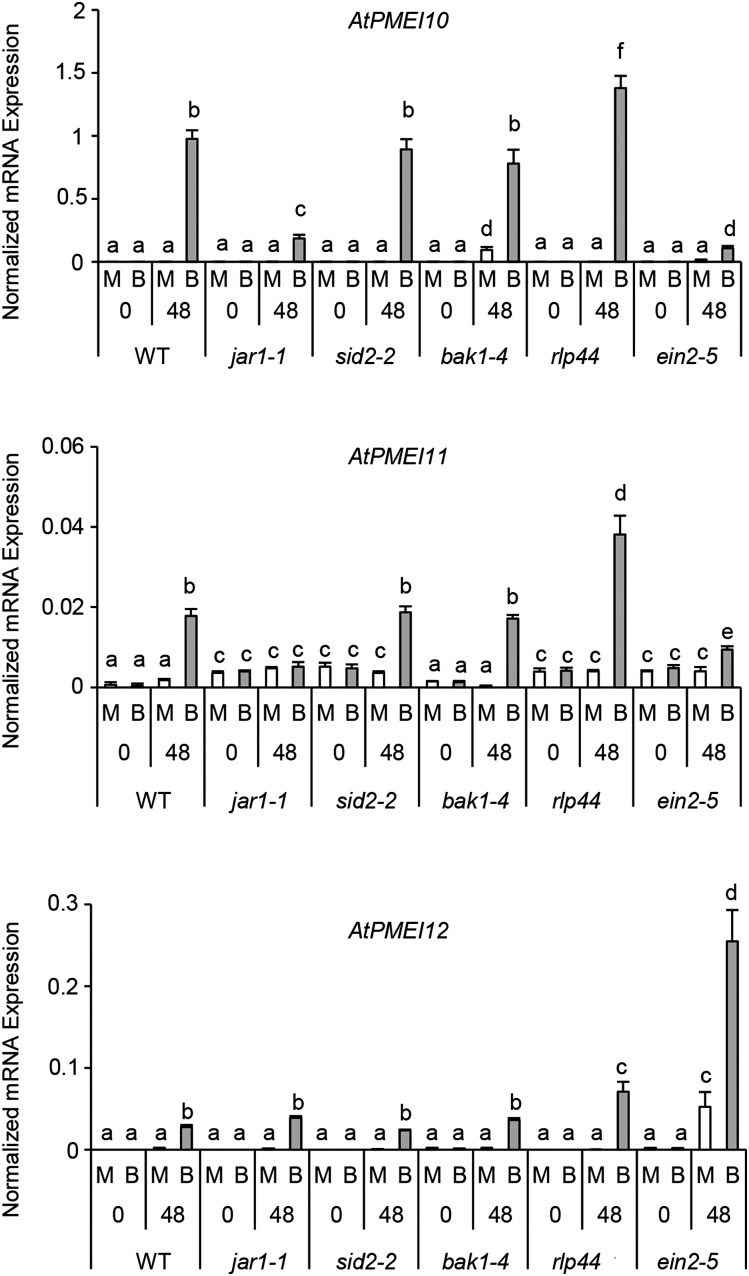
We evaluated the possibility that PMEI expression is exploited by Arabidopsis as part of the immune response against B. cinerea. The expression of AtPMEI10, AtPMEI11, and AtPMEI12 was determined in mutants defective in defense hormone signaling or biosynthetic pathways after infection with B. cinerea (Table I; Fig. 5). The induction of AtPMEI10 was reduced significantly in jar1-1 plants with respect to wild-type plants and almost completely abolished in ein2-5 plants. The same mutant also showed a lower induction of AtPMEI11 expression. Instead, AtPMEI12 induction was not affected in jar1-1 while it was significantly higher in ein2-5 mutants. These results indicate that JA and ethylene (ET) positively contribute to AtPMEI10 and AtPMEI11 induction against B. cinerea and that ET could exert a negative regulation of AtPMEI12. No differences were observed in sid2-2 and bak1-4 mutants with respect to the wild type, indicating that salicylic acid (SA) and BAK1 are not required for the induction of the inhibitors during B. cinerea attack. Interestingly, the induction of expression of all inhibitors also was higher in infected rlp44 plants with respect to the wild type, indicating that the signaling triggered by this receptor could negatively regulate PMEI expression. However, the high PMEI induction in rlp44 and the high expression of AtPMEI12 in ein2-5 also could be influenced by the high susceptibility of these mutants to the pathogen (Supplemental Fig. S4).
Table I. Arabidopsis mutants defective in immune hormone signaling used in this study.
| Name | Description | Gene Function | Literature |
|---|---|---|---|
| jar1-1 | Jasmonate-resistant1 | Defective in JA signaling | Staswick et al. (1992) |
| sid2-2 | SA induction-deficient mutant | Defective in SA biosynthesis | Nawrath and Métraux (1999) |
| bak1-4 | BRI1-associated receptor kinase | Defective in brassinosteroid perception | Kemmerling et al. (2007) |
| rlp44 | Receptor-like protein44 | Defective in the activation of brassinosteroid signaling triggered by alteration of methylesterification | Wolf et al. (2014) |
| ein2-5 | ET-insensitive | Defective in ET signaling | Alonso et al. (1999) |
Figure 5.
Plant immunity hormones regulate the expression of disease-related PMEIs. AtPMEI10, AtPMEI11, and AtPMEI12 expression was analyzed by qPCR at 0 and 48 hpi in B. cinerea- or mock-inoculated leaves of 6-week-old Arabidopsis wild-type (WT) plants. The expression levels were normalized to UBQ5 expression. The results represent means ± sd (n = 3). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.01). The experiments were repeated three times with similar results. B, B. cinerea-inoculated leaves; M, mock-inoculated leaves.
The 1,000 bp upstream of the transcriptional start site of the identified PMEIs was analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/) for the presence of putative cis-acting DNA elements with regulatory functions in plant immunity (Supplemental Fig. S5). All PMEIs hold the AWTTCAAA motif, an ethylene-responsive element (Tapia et al., 2005), and AACGTG, a T/G box element involved in JA gene induction (Boter et al., 2004). AtPMEI10 and AtPMEI12 promoters contain CGTG(T/C), a brassinosteroid response element (He et al., 2005). In addition, the AtPMEI11 and AtPMEI12 promoters incorporate TGTCA, a binding site of different transcription factors whose expression is associated with the resistance response in rice (Oryza sativa; Boyle and Brisson, 2001; Luo et al., 2005). Highly enclosed in all PMEI promoters is the W box motif, (T)TGAC(C/T), that is also found in the NPR1 promoter of Arabidopsis and was shown previously as an elicitor-responsive element recognized by different WRKY transcription factors (Eulgem et al., 1999; Chen and Chen, 2002). The AtPMEI11 promoter contains the Myb1 element, GTTAGTT, recognized by MYB transcription factors and found in well-characterized hypersensitive response-related genes (Kranz et al., 1998; Daniel et al., 1999; Pontier et al., 2001). We next investigated whether PME-related DAMPs can impact the expression of disease-related PMEIs. Leaves from wild-type plants were infiltrated with OGs, MeOH, or water, and the expression of AtPMEI10, AtPMEI11, and AtPMEI12 was evaluated at 1.5 h post infiltration. AtPMEI11 expression was induced after treatment with OGs and repressed by MeOH (Fig. 6). The expression of AtPMEI10 and AtPMEI12 was not altered by DAMP treatment. These results indicate that a posttranscriptional regulation of PME activity by AtPMEI11 can be mediated by PME-related DAMPs. Collectively, our findings point to a regulation of PME activity by PMEIs as part of the Arabidopsis immune response to B. cinerea.
Figure 6.
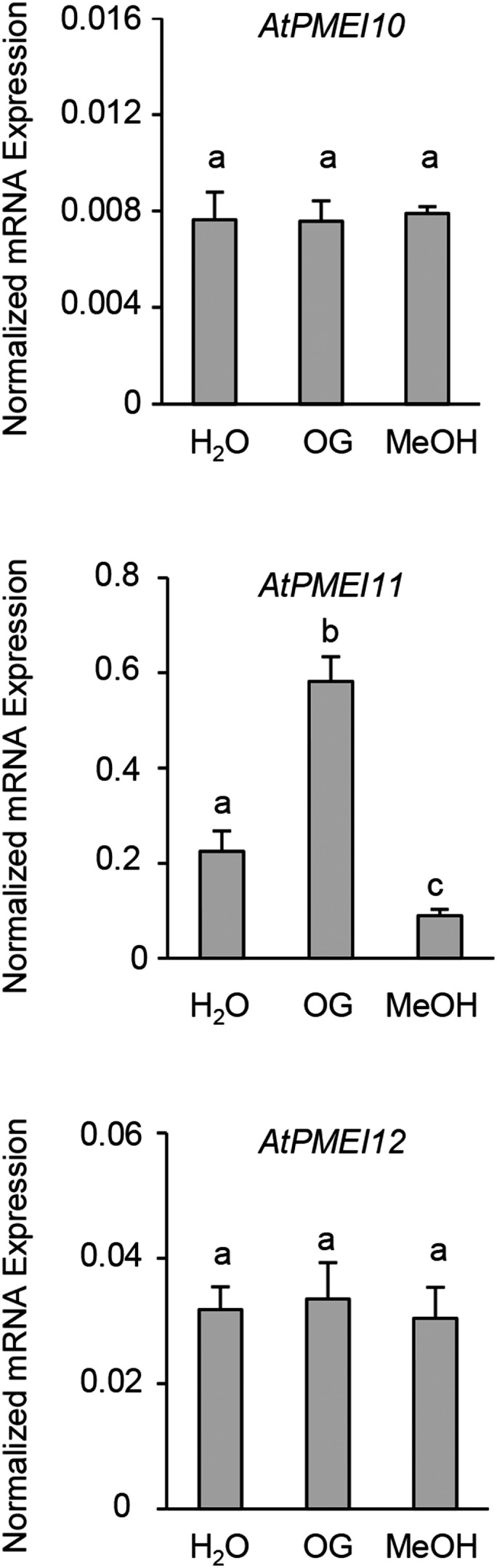
DAMPs regulate AtPMEI11 expression. The expression of AtPMEI10, AtPMEI11, and AtPMEI12 was analyzed by qPCR at 1.5 h post infiltration of leaves of 6-week-old Arabidopsis wild-type plants with OGs (50 μg mL−1), MeOH (0.1%, v/v), or water as a control. The expression levels were normalized to UBQ5 expression. Results represent means ± sd (n = 3). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.01). The experiments were repeated three times with similar results.
AtPMEI10, AtPMEI11, and AtPMEI12 Contribute to Arabidopsis Resistance against B. cinerea
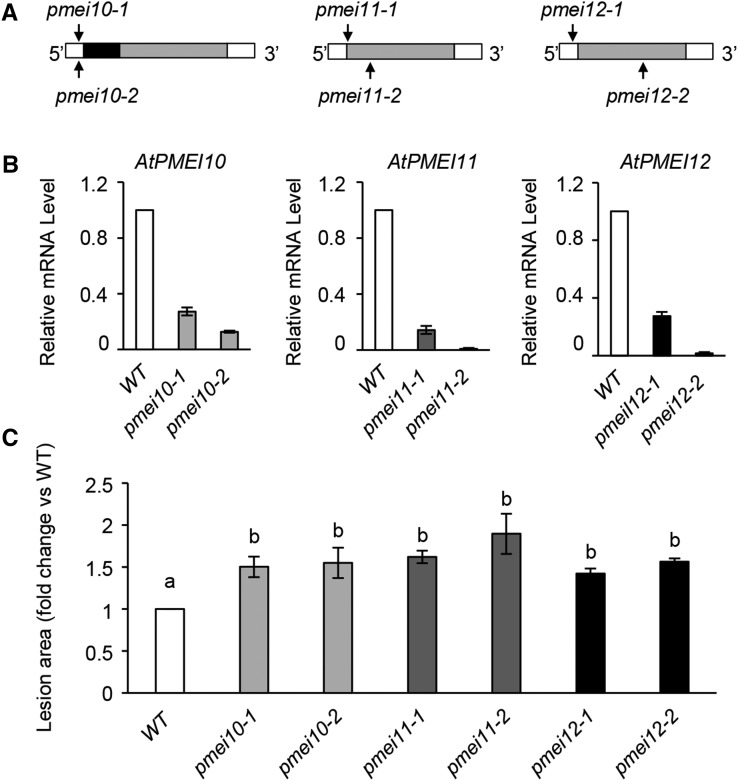
A reverse genetic approach was used to elucidate the role of the PMEIs against B. cinerea. Arabidopsis T-DNA and transposon insertional PMEI mutants were selected (Fig. 7A). Two independent mutant lines for each AtPMEI were characterized: SALK_007859C (henceforth pmei10-1 plants) and SALK_072421 (henceforth pmei10-2 plants), both with a T-DNA insertion located in the 5′ UTR of AtPMEI10; SALK_015169c (henceforth pmei11-1 plants) and GT_5_108240 (henceforth pmei11-2 plants), with a T-DNA/transposon insertion located in the starting codon and in the exon of AtPMEI11, respectively; and SALK_108076c (henceforth pmei12-1 plants) and GT_5_108791 (henceforth pmei12-2 plants), with a T-DNA/transposon insertion located in the 5′ UTR and in the exon of AtPMEI12, respectively. We quantified the AtPMEI10, AtPMEI11, and AtPMEI12 transcript abundance by RT-PCR in wild-type and mutant plants challenged with B. cinerea. The induction of the three inhibitors was significantly reduced in pmei10, pmei11, and pmei12 mutants during disease (Fig. 7B).
Figure 7.
Arabidopsis pmei10, pmei11, and pmei12 mutants are more susceptible to B. cinerea. A, Schematic representation of PMEI gene structures. The localizations of T-DNA and transposon insertions in the genomic DNA sequences are shown (black arrows). The 5′ untranslated region (UTR) and 3′ UTR are represented in white, and exons are represented in gray. The SPRR domain is represented as a black block in PMEI10. B, The expression of PMEI genes was analyzed by qPCR using cDNA from leaves of 6-week-old wild-type (WT) and mutant plants at 48 hpi with B. cinerea. The expression levels were normalized to UBQ5 expression. The relative mRNA levels are represented as the ratio between gene expression in the mutants and that in the wild type. Results represent means ± sd of three independent experiments (n = 3). C, Quantification of lesion areas produced by the spreading of the fungus at 48 hpi. The values are means ± sd of three independent experiments (n = 30). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.05).
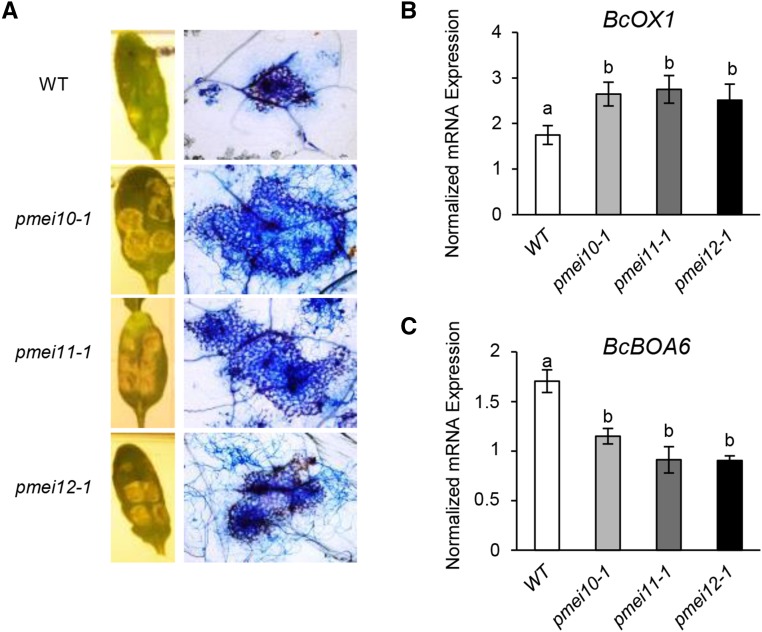
We next evaluated the susceptibility of pmei mutants to B. cinerea. Compared with the wild type, the local symptoms of the fungus were significantly higher in pmei10-1, pmei10-2, pmei11-1, pmei11-2, pmei12-1, and pmei12-2 plants, indicating that each inhibitor contributes to the resistance against the necrotroph (Fig. 7C). Since the two mutant lines for each gene showed a similar susceptibility phenotype against B. cinerea, we used pmei10-1, pmei11-1, and pmei12-1 mutants for further analysis. A greater development of B. cinerea mycelium around the inoculation sites was detected in leaf tissue of pmei mutants with respect to wild-type plants using Trypan Blue staining (Fig. 8A). We next tested if the increased susceptibility observed in pmei mutants could be due to a higher production of fungal toxic compounds, such as OA and botcinic acid. We analyzed the expression of BcOX1, encoding an oxaloacetate hydrolase, and of BcBOA6, encoding a polychetide synthase, key enzymes involved in the biosynthesis of OA and botcinic acid, respectively (Han et al., 2007; Dalmais et al., 2011; Schumacher et al., 2012). The induction of BcOX1 is higher, while the induction of BcBOA6 is significantly lower, in all pmei mutants with respect to the wild type (Fig. 8, B and C). These results indicate that a stimulated biosynthesis of the fungal OA can contribute to the higher susceptibility of pmei mutants.
Figure 8.
AtPMEI10, AtPMEI11, and AtPMEI12 expression alters B. cinerea colonization and mycotoxin synthesis. A, Photographs showing lesion areas produced by B. cinerea on leaves of 6-week-old Arabidopsis wild-type (WT) and pmei mutant plants (left) and microphotographs showing B. cinerea colonization revealed by Trypan Blue staining (right). B and C, The expression of BcOX1 (B) and BcBOA6 (C) genes was analyzed by qPCR at 48 hpi. Expression levels were normalized to BcTUBULIN expression. The results represent means ± sd (n = 3). Different letters indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.05). The experiments were repeated three times with similar results.
To determine if the increased susceptibility of pmei mutants could be due to an impaired ability to induce defense responses, we measured callose deposition and hydrogen peroxide (H2O2) accumulation after B. cinerea infection. Both defense responses were not compromised in leaves of pmei mutants with respect to the wild type (Supplemental Fig. S6). pmei mutants did not show any evident defects in rosette leaves, floral stems, flowers, seeds, and all developmental stages in which the inhibitors are significantly expressed (Supplemental Figs. S7 and S8).
AtPMEI10, AtPMEI11, and AtPMEI12 Control PME Activity and Pectin Methylesterification during B. cinerea Infection
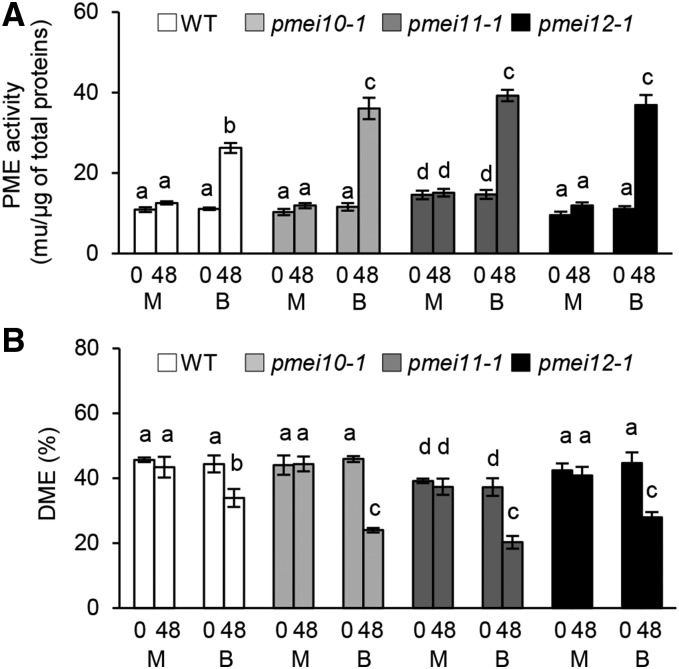
After establishing that AtPMEI10, AtPMEI11, and AtPMEI12 play a role in immunity, we evaluated their possible contribution to the control of PME activity and pectin methylesterification during B. cinerea infection. The level of PME activity was monitored in wild-type, pmei10-1, pmei11-1, and pmei12-1 plants after B. cinerea infection using histochemical and biochemical assays (Lionetti, 2015). A higher increase of pathogen-induced PME activity was observed in mutant plants with respect to the wild type (Fig. 9A; Supplemental Figs. S9 and S10). This indicates that AtPMEI10, AtPMEI11, and AtPMEI12 are functional PMEIs and are able to control the induction of PME activity observed during disease. No significant differences were observed in PME activity of untreated wild-type, pmei10-1, and pmei12-1 plants. Instead, a higher basal level of PME activity was detected in the pmei11-1 mutant compared with the wild type (Fig. 9A). Most likely, AtPMEI11 also modulates the PME activity resident in leaf tissue. B. cinerea is known to produce PMEs during infection (Valette-Collet et al., 2003; Kars et al., 2005). We attempted to understand if the PME activity controlled by the identified PMEIs was derived by plant and/or by fungus. To this purpose, the recombinant AtPMEI1, purified to homogeneity and unable to inhibit B. cinerea PMEs (Lionetti, 2015), was added to protein extracts isolated from infected leaves of wild-type and pmei plants at 48 hpi. The strong inhibition of PME activity by AtPMEI1 in both the wild type and pmei mutants indicated that the enzymatic activity targeted by defense-related PMEIs was mainly plant derived (Supplemental Fig. S10).
Figure 9.
AtPMEI10, AtPMEI11, and AtPMEI12 regulate PME activity and the level of pectin methylesterification during disease. Leaves of 6-week-old Arabidopsis wild-type (WT) and pmei mutant plants were inoculated with B. cinerea and mock treatment, and PME activity (A) and the DME of pectin (B) were quantified at 0 and 48 hpi. The results represent means ± sd (n = 3). Different letters on the bars indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.05). The experiment was repeated three times with similar results. B, B. cinerea-inoculated leaves; M, mock-inoculated leaves.
In order to understand the role of the identified PMEIs in the modulation of pectin methylesterification after infection, we started to monitor the DME in wild-type leaves at 24, 48, and 72 h post B. cinerea infection. The DME was reduced significantly up to 48 hpi, reaching a level of 80% reduction compared with a mock control (Supplemental Fig. S11), and intriguingly, no further reduction of DME was found at a later stage of infection. The possible contribution of PMEI expression on the level of pectin methylesterification was explored 48 h after B. cinerea infection by comparing the DME in the wild type and pmei10-1, pmei11-1, and pmei12-1 mutants. A significantly higher reduction of DME was observed in pmei10-1, pmei11-1, and pmei12-1 mutants compared with the wild type (Fig. 9B). The DME of uninfected pmei11-1 was lower with respect to the other genotypes, consistent with its higher PME activity in the leaves.
AtPMEI10, AtPMEI11, and AtPMEI12 Control CW Integrity against B. cinerea
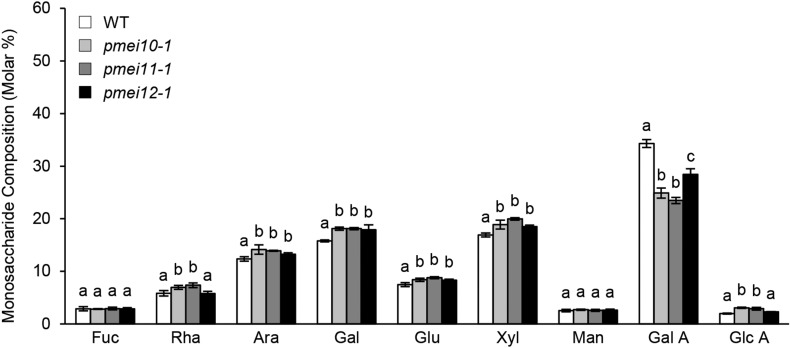
To study the role of the selected PMEIs in CW remodeling, it was useful first to clarify the dynamics of CW changes in wild-type plants at different times during B. cinerea infection (Supplemental Fig. S12). The amount of cellulose-derived Glc was reduced significantly at 24 hpi and decreased to a higher extent at 48 hpi, indicating a massive degradation of the polymer by B. cinerea cellulases. At the latter stage of infection (72 hpi), the level of Glc was not significantly different from that observed at 48 hpi (Supplemental Fig. S12A). The monosaccharide composition indicates a progressive reduction of GalA content at 24 and 48 hpi, confirming the pectinolytic nature of B. cinerea (Supplemental Fig. S12B). Interestingly, no further reduction of GalA content was detected later at 72 hpi. The lack of reduction of cellulose-derived Glc and GalA content at the latter stage of infection could be related to the lower increment of the lesion area detected between 48 and 72 hpi (0.4-fold change) with respect to that observed between 24 and 48 hpi (2.3-fold change; Supplemental Fig. S13). A significant increase in the relative amounts of all other monosaccharides was observed (Supplemental Fig. S12B), indicating a possible CW biosynthetic compensatory effect in response to cellulose and pectin degradation. No significant differences were observed in mock-inoculated leaves at the different time points (Supplemental Figs. S12A and S14). The contribution of PMEIs in the degradation and remodeling of CW polysaccharides was investigated by analyzing the monosaccharide composition of pmei10, pmei11, and pmei12 mutants in comparison with the wild type at 48 h after B. cinerea infection. A greater reduction of GalA level was detected in the CW of all pmei mutants with respect to the wild type, with a minor decrease in pmei12 plants (Fig. 10). This result indicates that a higher pectin degradation occurs if AtPMEI10, AtPMEI11, or AtPMEI12 expression is reduced. A higher induction in the relative amounts of Ara, Gal, Glu, and Xyl was observed in all pmei mutants with respect to the wild type, while increases of Rha and GlcA were found only in pmei10 and pmei11 plants. The monosaccharide composition of the noncellulosic polysaccharides extracted from mock-inoculated leaves of wild-type and pmei10 and pmei12 plants was similar (Supplemental Fig. S15). Instead, pmei11 CW had a reduced GalA content and an increased content of Ara, Gal, Glu, and Xyl. Since pmei11 has a basal higher PME activity and a basal low DME with respect to the wild type (Fig. 9), its pectin could be particularly sensitive to endogenous pectinases.
Figure 10.
pmei10, pmei11, and pmei12 mutants showed a higher pectin degradation with respect to the wild type (WT) during B. cinerea infection. The monosaccharide composition of matrix CW polysaccharides was monitored in leaves of 6-week-old Arabidopsis wild-type and pmei mutant plants challenged with B. cinerea at 48 hpi. The molar percentages of Fuc, Rha, Ara, Gal, Glc (Glu), Xyl, Man, GalA, and GlcA released after 2 m trifluoracetic acid (TFA) hydrolysis were quantified by high-performance anion-exchange chromatography with pulsed amperometric detection. Results represent means ± sd (n = 3). Different letters for each monosaccharide indicate data sets significantly different according to ANOVA followed by Tukey’s test (P < 0.05). The experiments were repeated three times with similar results.
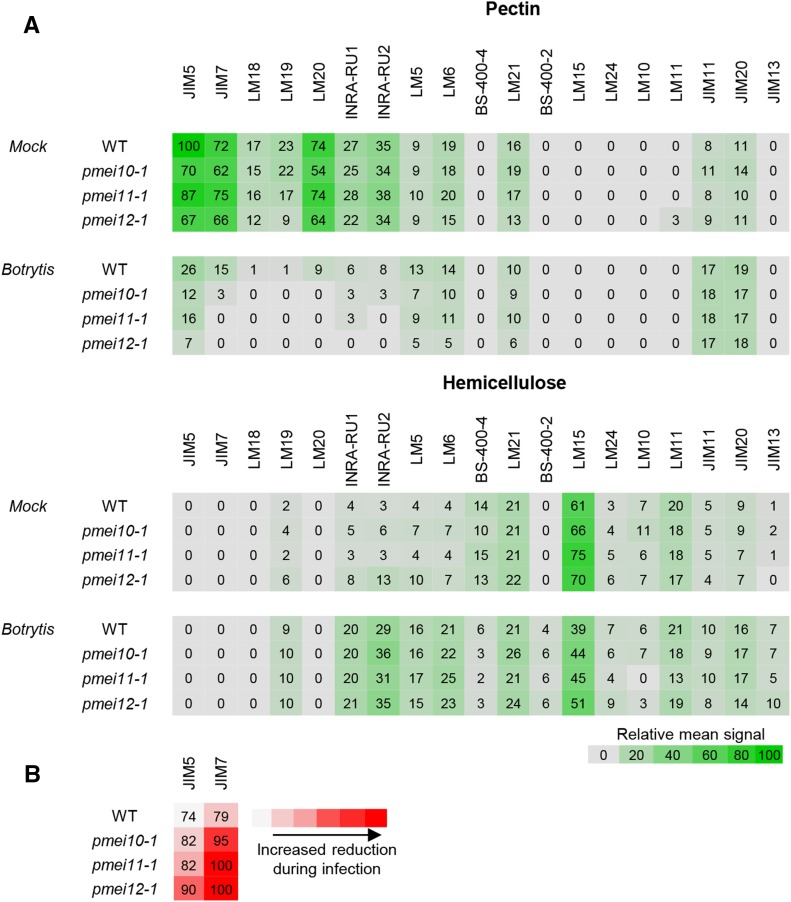
Afterward, we explored the impact of AtPMEI10, AtPMEI11, and AtPMEI12 expression on the dynamic of CW polysaccharide structures and proteins during infection. We performed a carbohydrate microarray polymer profiling (CoMPP; Moller et al., 2007, 2008) on pectin- and hemicellulose-enriched fractions isolated from CW of B. cinerea-infected and mock-inoculated wild-type and pmei leaves at 48 hpi. A set of monoclonal antibodies (mAbs) specific for CW glycan epitopes was used (Table II). A heat map showing the mean CoMPP signals obtained for all the mAbs is shown in Figure 11A. Specific modifications of CW structure and components occurred in Arabidopsis wild-type leaves when challenged with B. cinerea. The analysis indicates pectin as the substrate most significantly altered during B. cinerea infection. All epitopes relative to different degrees of HG methylesterification were reduced in the pectin fractions of infected wild-type and pmei plants, although to different extents. Interestingly, pmei mutants, in comparison with wild-type plants, showed a greater reduction in JIM7 signal, specific for high methylesterified pectins with respect to JIM5, recognizing pectin with a low DME (Fig. 11B). In contrast, a similar increase of unesterified pectin recognized by LM19 was detected in hemicellulose fractions of infected CWs in all plants analyzed. In pectin fractions of wild-type infected leaves, RGI backbone epitopes (recognized by mAbs INRA-RU1 and INRA-RU2) decreased at a higher extent with respect to epitopes relative to the RGI side chain (recognized by mAbs LM5 and LM6). These reductions were slightly higher in pmei mutants with respect to the wild type. In hemicellulose fractions, all RGI epitopes increased to the same extent in wild-type and pmei plants. The increase of different pectic epitopes in the hemicellulose fraction could reflect the induction of linkage between pectin and hemicelluloses as potential mechanisms to defend CW against the action of the degradative fungal enzymes. Signals of LM15 and BS-400-4, specific for nongalactosylated xyloglucan and galactomannans, respectively, were reduced in the hemicellulose of infected wild-type and mutant leaves. Instead, the signals of LM24, relative to galactosylated xyloglucan epitopes, and LM21, specific for galactomannans/glucomannans, seem not to be affected during infection. These results suggest that the addition of specific substitutions could represent a strengthening mechanism to protect xyloglucan and mannans from enzymatic degradation during disease, as already proposed during development (Peña et al., 2004). The appearance of signals of BS-400-2, specific for β-(1,3)-d-glucan, in infected CWs indicates that both the wild type and mutants accumulated callose against B. cinerea. The signals of JIM11 and JIM20, both recognizing extensin epitopes, and of JIM13, specific for arabinogalactan proteins, increased to similar extents in all infected genotypes. The increase of extensins and arabinogalactan proteins could be exploited by plants to increase CW polysaccharide assembly and to reduce the accessibility of fungal CW-degrading enzymes (Rashid, 2016).
Table II. mAbs used for CoMPP.
AGP, Arabinogalactan protein; XXXG, oligosaccharide motif consisting of three Xyl-substituted (X) residues and one unsubstituted (G) glucosyl residue. L in XLLG indicates Gal linkage to Xyl.
| mAb | Detected Epitope |
|---|---|
| JIM5 | Partially methylesterified/unesterified HG |
| JIM7 | Partially methylesterified/methylesterified HG |
| LM18 | Partially methylesterified HG |
| LM19 | Unesterified HG |
| LM20 | Methylesterified HG |
| INRA-RU1 | Backbone of RGI |
| INRA-RU2 | Backbone of RGI |
| LM5 | (1→4)-β-d-Galactan |
| LM6 | (1→5)-α-l-Arabinan |
| BS-400-4 | (1→4)-β-d-(Galacto)mannan |
| LM21 | (1→4)-β-d-(Galacto)(gluco)mannan |
| BS-400-2 | (1→3)-β-d-Glucan |
| LM15 | Xyloglucan (XXXG motif) |
| LM24 | Xyloglucan (XLLG motif) |
| LM10 | (1→4)-β-d-Xylan |
| LM11 | (1→4)-β-d-Xylan/arabinoxylan |
| JIM11 | Extensin |
| JIM20 | Extensin |
| JIM4 | AGP |
| JIM13 | AGP |
Figure 11.
Changes in Arabidopsis CW structures of wild-type (WT) and pmei mutant plants during B. cinerea infection analyzed by CoMPP. A, Heat map showing the relative abundance of CW glycans recognized by different monoclonal antibodies (top) on pectin-enriched (diaminocyclohexanetetraacetic acid [CDTA]) and hemicellulose-enriched (NaOH) fractions extracted from CW prepared from B. cinerea-infected and mock-inoculated leaves of 6-week-old Arabidopsis wild-type and pmei10-1, pmei11-1 and pmei12-1 plants at 48 h post infection. The distribution of CW polysaccharide epitopes is represented as a heat map where mean CoMPP spot signals (numbered values) are correlated to color intensity. The highest mean signal value in the data set was set to 100, and all other values were adjusted accordingly. B, Heat map reporting percentages of the reduction of CoMPP signals related to antibodies recognizing HG methylesterification in the CDTA fraction of B. cinerea-infected leaves with respect to mock-inoculated leaves.
DISCUSSION
In this work, we argued about the possibility that plants can modulate PME activity by expressing specific defense-related PMEIs as a physiological response to pathogens. We selected AtPMEI10, AtPMEI11, and AtPMEI12 as possible pathogenesis-related PMEIs. These inhibitors showed a sequence homology and the presence of structural motifs typical of functional PMEIs. Interestingly, AtPMEI10 shows an SPRR/PMEI structure not described previously in this class of CW proteins. Similar domains are present in CW structural proteins such as extensins (Lamport et al., 2011). Interestingly, chimeric Leu-rich repeat/extensin CW proteins with roles in plant development and defense were reported (Jones and Jones, 1997; Baumberger et al., 2001; Draeger et al., 2015). As described for Leu-rich repeat/extensin CW protein chimeras (Ringli, 2010), the SPRR region in SPRR/PMEI could be required to anchor PMEI10 within the CW during infection. We demonstrated that AtPMEI11 and AtPMEI12 are correctly secreted into the apoplast, as also reported for other characterized PMEIs (Röckel et al., 2008; Hong et al., 2010; Zhang et al., 2010; De Caroli et al., 2011; Vandevenne et al., 2011; Reca et al., 2012). Further experiments are needed to confirm the predicted extracellular localization of AtPMEI10. The different levels and timing of expression in response to B. cinerea suggest that AtPMEI10, AtPMEI11, and AtPMEI12 are not functionally redundant and could play different roles during disease. AtPMEI11 is expressed in uninfected Arabidopsis leaves and repressed early during infection. At early stages of pathogen attack, the AtPMEI11 down-regulation as well as the absence of induction of the other inhibitors could favor the effects mediated by pathogen-induced PMEs, such as the possible release of OGs and MeOH. Later during infection, AtPMEI10, AtPMEI11, and AtPMEI12 are significantly induced. Microarray experiments showed that AtPMEI10, AtPMEI11, and AtPMEI12 also are induced by P. syringae, an hemibiotrophic bacterial pathogen, but not by Erysiphe orontii, a fungal biotroph (Supplemental Fig. S16; www.geneinvestigator.ethz.ch/at/). Overall, this evidence suggests that the new AtPMEIs are involved in immunity against pathogens, including the necrotrophy in their lifestyle. Intriguingly, the three PMEI members are highly expressed in senescing leaves, a developmental stage in which CW degradation occurs (Supplemental Fig. S8; Patro et al., 2014), suggesting that the expression of these PMEIs is activated in response to the loss of CW integrity.
Meta-analyses indicated that plant PMEs are differentially expressed in Arabidopsis in response to B. cinerea (Lionetti et al., 2012; Windram et al., 2012); however, information about the kinetics of expression of these genes remains scarce. We found that six PMEs were induced significantly while two isoforms were repressed during infection. The rapid and efficient activation of several PME isoforms (PME17, PME20, PME21, and, in particular PME41) indicates that plants employ an expensive metabolic response early against B. cinerea, most likely to favor the release of OGs and MeOH and trigger immunity against the pathogen. On the other hand, the steady down-regulation of PME members (PME1 and PME34) may represent, together with PMEI induction, a defense mechanism to limit pectin demethylesterification and degradation during infection. Consistently, TaPME1 expression is repressed in a resistant wheat genotype but induced in a susceptible accession during F. graminearum infection (Lionetti et al., 2015a). On the basis of PME/PMEI coexpression, we suggest PME3, PMEPCRA, PME17, PME20, and PME21 as putative ligands of AtPMEI10, AtPMEI11, and AtPMEI12. Future efforts will be devoted to study the molecular bases of PME-PMEI interactions during pathogenesis.
It is known that JA and ET signaling plays a strong role in the immunity of Arabidopsis to the necrotrophs (Thomma et al., 2001; Laluk and Mengiste, 2010). Our findings uncover the involvement of JA and ET immune signaling pathways in the activation of AtPMEI10 and AtPMEI11 in response to B. cinerea. A similar regulation also was proposed for the defense proteins PDF1.2, β-Chi, and Thi2.2 (Epple et al., 1995; Penninckx et al., 1998; Bari and Jones, 2009). It is also known that JA- and ET-dependent defense responses are triggered when the integrity of the plant CW is perturbed (Ellis and Turner, 2001; Ellis et al., 2002; Zhong et al., 2002). AtPMEI10 and AtPMEI11 could be part of a plant defense machinery against necrotrophs activated by both JA and ET aimed to maintain CW integrity, preventing excessive damage to host tissues (Overmyer et al., 2003). In contrast, the expression of AtPMEI12 is not affected by the alteration of JA signaling while the gene is strongly induced in the ein2-5 mutant, suggesting that ET signaling could contribute to the repression of the inhibitor during infection. The higher susceptibility of bak1-4 mutants to B. cinerea (Supplemental Fig. S4) indicates that, although BAK1 is involved in disease resistance against B. cinerea, as demonstrated previously for hemibiotrophs and biotrophs (Roux et al., 2011), it seems not to be required for AtPMEI activation. Our results exclude a possible role of the SA-mediated pathway in AtPMEI activation during defense, consistent with the major role of SA in Arabidopsis-biotroph interactions (Thomma et al., 2001; Nafisi et al., 2015). The ability of Arabidopsis to orchestrate the activation of specific AtPMEI isoforms during disease indicates the requirement of a fine modulation of PME activity during the course of infection.
PMEs release MeOH, a DAMP-like alarm signal, to alert adjacent noninfected tissues or neighboring plants (Dorokhov et al., 2012; Komarova et al., 2014). MeOH treatment can alter plant signaling responses to different DAMPs and microbe-associated molecular patterns (Hann et al., 2014). We revealed that AtPMEI11 expression is repressed by MeOH. This effect could be related to the down-regulation of AtPMEI11 observed at early stages of infection. This evidence, together with previous observations indicating that specific PMEs are MeOH-induced genes, support a role of MeOH as a danger signal to amplify PME-related immunity (Downie et al., 2004; Dorokhov et al., 2012; Komarova et al., 2014). RLP44 senses the alteration of pectin methylesterification during growth and abiotic stresses (Wolf et al., 2014). However, a possible role of RLP44 in response to pathogens has not yet been explored. We found that rlp44 mutants showed a higher induction of AtPMEI10, AtPMEI11, and AtPMEI12 and was more susceptible to B. cinerea with respect to the wild type (Supplemental Fig. S4). It is possible that RLP44 perceives the alterations of pectin methylesterification induced during infection and, by controlling PMEI expression, could boost PME-related immunity in concert with MeOH. OGs are able to elicit defense responses, including the accumulation of reactive oxygen species and pathogenesis-related proteins, and to protect plants against pathogen infection (Ferrari et al., 2013). We demonstrated that OG specifically induced AtPMEI11 expression. AtPMEI11 expression also is triggered by a number of elicitors/effectors (Supplemental Figs. S16 and S17). For instance, AtPMEI11 is affected by another DAMP, Pep2 (Huffaker et al., 2006; Supplemental Fig. S17), by different pathogen-associated molecular patterns (PAMPs), such as elf18 (Kunze et al., 2004), flg22 (Felix et al., 1999), LPS (Zeidler et al., 2004), and GST-NPP1 (Fellbrich et al., 2002), and by the effector HrpZ (Alfano et al., 1996; Supplemental Figs. S16 and S17). This evidence indicates that AtPMEI11, and consequently PME activity, can be regulated precisely by DAMPs, PAMPs, and effector-triggered immunity.
We isolated pmei10-1, pmei10-2, pmei11-1, pmei11-2, pmei12-1, and pmei12-2 mutants that, when infected with B. cinerea, exhibited a compromised resistance to the fungus. The higher susceptibility observed in pmei mutants was related to higher mycelium growth. We found a higher induction of BcOX1 in all pmei mutants with respect to wild-type plants. OA may be a cofactor in pathogenesis, acting in synergy with endopolygalacturonases during tissue maceration (ten Have et al., 2002). OA can cause hydration and swelling of pectin, favoring its degradation by endopolygalacturonases. Moreover, by sequestering the Ca2+ ions, OA may perturb the integrity of the pectic structure (Mansfield and Richardson, 1981). Through the expression of PMEIs, plants could engineer a substrate less enriched in Ca2+ ions and, thus, less susceptible to OA-mediated CW damage. BcBOA6 is expressed at lower levels in pmei mutants with respect to the wild type. These results indicate that pectin methylesterification can affect the production of fungal toxic compounds and suggest that B. cinerea selects specific toxins based on the different CW substrate to be degraded during infection. pmei mutants showed a higher increase of PME activity upon infection, indicating that AtPMEI10, AtPMEI11, and AtPMEI12 are able to control plant PME activity induced during disease. The low level of BcPME1 and BcPME2 expression at 48 hpi (Supplemental Fig. S18), the strong inhibition of B. cinerea-induced PME activity by AtPMEI1 (Supplemental Fig. S10), together with the evidence that PMEIs typically recognize plant PMEs (Raiola et al., 2004; Di Matteo et al., 2005; Reca et al., 2012; Lionetti et al., 2015c) support our conclusion. However, further research is needed to evaluate if the identified PMEIs also are able to inhibit fungal enzymatic activity.
The compositional profiling of CW, extracted from infected Arabidopsis wild-type leaves, revealed an extensive degradation of cellulose and HG and a reduction of DME, which intriguingly stops at the latter stages of infection. This physiological dynamics suggests that, when pectin demethylesterification reaches a dangerous level, it could be limited to protect CW by the fungus-degrading machinery later during infection. It is noteworthy that AtPMEI induction fits well with the block of pectin demethylesterification and CW degradation observed at late stages of infection. All pmei mutants exhibited a higher reduction of HG methylesterification and a greater degradation of HG with respect to the wild type. CoMPP indicated that pmei mutants showed a greater reduction of highly methylesterified pectins with respect to pectin with a low DME and a higher degradation of HG and RGI with respect to wild-type plants. The lack of PMEI expression does not affect hemicellulose strengthening, callose deposition, and the synthesis of structural defense proteins, here detected as CW-remodeling mechanisms induced against B. cinerea. Overall, our evidence indicates that AtPMEI10, AtPMEI11, and AtPMEI12 are exploited by Arabidopsis to limit demethylesterification and to protect pectin degradation and CW integrity in response to the pathogen. During infection, the increased amount of some monosaccharides observed in the wild type, and to a greater extent in all pmei mutants, could reflect the activation of a CW biosynthetic compensatory effect dependent on the extent of CW degradation. Intriguingly, this can be a CW-remodeling mechanism induced as a plant defense response against fungal OA production. The evidence that the toxin DON from the necrotrophic pathogenic fungus F. graminearum was able to alter the monosaccharide composition of Brachypodium distachyon CW supports this last hypothesis (Blümke et al., 2015).
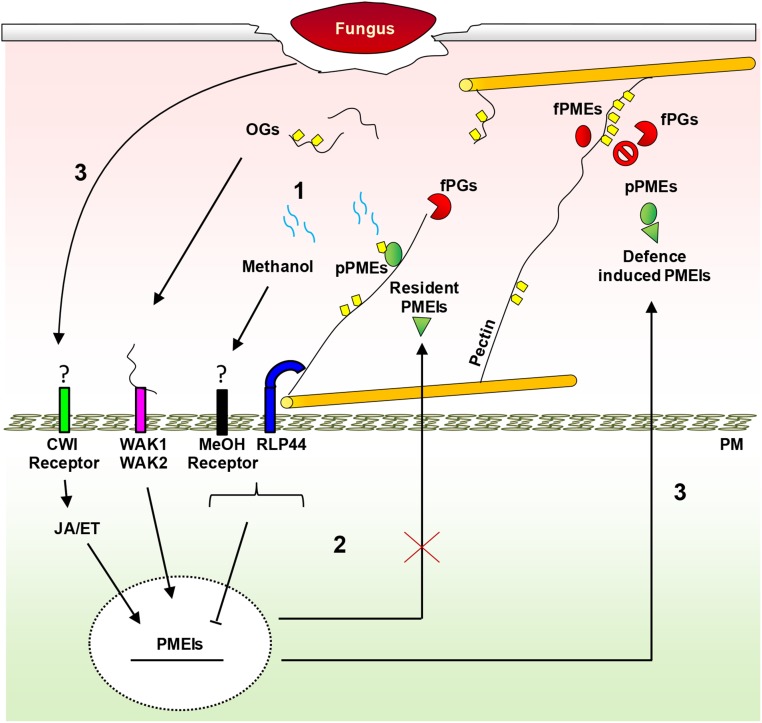
The PMEI family appeared evolutionarily later than the PME genes (Wang et al., 2013). It is possible that plants have evolved specific PMEIs to control PME activity as a result of plant-pathogen coevolution (Jones and Dangl, 2006). In Figure 12, a dynamic model shows the possible roles of PMEIs in plant immunity against necrotrophs. In early phases of pathogen infection, plants induce PMEs to release MeOH and OGs, danger signals sensed by specific receptors to activate immune responses. By a feedback loop, MeOH and RLP44 receptor down-regulate resident PMEIs, possibly to favor a rapid and efficient local production of PME-related DAMPs. However, necrotrophic pathogens have evolved the ability to efficiently degrade low methylesterified pectin. At later stages of infection, plants defend CW integrity by triggering a signaling mediated by OG, JA, and ET. This leads to the induction of specific PMEIs able to lock the decrease of pectin methylesterification, protecting CW by further enzymatic degradations and limiting pathogen diffusion. This study brings new insights to molecular mechanisms of the regulation of PME activity during infection and the dynamic role of pectin methylesterification in plant immunity against necrotrophs. We highlight pectin methylesterification as a biochemical determinant of plant immunity and indicate AtPMEI10, AtPMEI11, and AtPMEI12 as new targets for crop breeding approaches to improve plant resistance to disease.
Figure 12.
Dynamic model showing the possible roles of PMEIs in plant immunity against necrotrophs. Step 1, In the early phases of pathogen infection, PME is required to release MeOH and OG danger signals, sensed by specific receptors to activate immunity. Step 2, In a feedback loop, MeOH and RLP44 down-regulate the resident pathogen-related AtPMEI expression to favor a rapid and efficient elicitation of defense responses by PME-related DAMPs. Pathogens have evolved the ability to efficiently degrade low-methylesterified pectin produced by PME activity. Step 3, At later stages of infection, plants defend CW integrity (CWI) by triggering a signaling mediated by OGs, JA, and ET. This leads to the induction of specific PMEIs able to lock the decrease of pectin methylesterification, protecting CW by further enzymatic degradation and limiting fungal diffusion. fPGs, Fungal polygalacturonases; fPMEs, fungal PMEs; PM, plasma membrane; pPMEs, plant PMEs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a controlled-environment chamber maintained at 22°C and 70% relative humidity with a 12-h/12-h day/night cycle (photosynthetically active radiation level of 100 µmol m−2 s−1). Germplasm used includes jar1-1 (Staswick et al., 1992), bak1-4 (Kemmerling et al., 2007), rlp44 (Wolf et al., 2014), ein2-5 (Alonso et al., 1999), and sid2-2 (Nawrath and Métraux, 1999). Mutants characterized in this study are as follows: SALK_007859C (pmei10-1 plants), SALK_072421 (pmei10-2 plants), SALK_015169c (pmei11-1 plants), GT_5_108240 (pmei11-2 plants), SALK_108076c (pmei12-1 plants), and GT_5_108791 (pmei12-2 plants). All SALK_c lines belong to the SALK Homozygote T-DNA Collection (http://signal.salk.edu/cgi-bin/homozygotes.cgi; Alonso et al., 2003) and were obtained from the Nottingham Arabidopsis Stock Centre. The homozygosity of the mutants was confirmed by PCR-based genotyping.
Cloning of AtPMEI11 and AtPMEI12 and Plasmid Construction and Agrobacterium tumefaciens Transient Transformation of Arabidopsis Leaf Cotyledons
The intronless regions of AtPMEI11 and AtPMEI12, encoding the predicted protein, were amplified by PCR from genomic DNA (50 ng) with ExTaq polymerase (Takara) and then amplified to introduce the restriction sites BamHI and NheI in each gene using primers reported in Supplemental Table S3. The BamHI/NheI fragments were inserted into a GFP-containing vector (De Caroli et al., 2015). All constructs were inserted as BamHI/SacI fragments into the plant binary vector (Haseloff et al., 1997) for A. tumefaciens-mediated expression in Arabidopsis seedlings. The cloned genes and the GFP constructs were checked by sequencing (Eurofins Genomics). The GFP constructs AtPMEI11-GFP, AtPMEI12-GFP, pm-rk, and PGIP2-RFP (De Caroli et al., 2011, 2015) were introduced into A. tumefaciens (strain LBA4404) and used for transient gene expression and coexpression of the chimeras in Arabidopsis seedling cotyledons (Li et al., 2009).
Confocal Laser Scanning Microscopy
Observations were made using a confocal laser microscope (LSM 710; Zeiss) and Zen Software. To detect GFP fluorescence, a 488-nm argon ion laser line was used, and the emission was recorded with a 505- to 530-nm filter set; RFP was detected with a 560- to 615-nm filter set after helium-neon laser excitation at 543 nm, while chlorophyll epifluorescence was detected with the filter set for tetramethylrhodamine isothiocyanate (greater than 650 nm) and eliminated. The power of each laser line, the gain, and the offset were identical for each experiment so that the images were comparable. Appropriate controls were performed to exclude the possibility of cross talk between the two fluorochromes before image acquisition.
Arabidopsis Infection with Botrytis cinerea
B. cinerea strain SF1 (Lionetti et al., 2007) was grown for 15 d on potato dextrose agar at 39 g L–1 and 23°C with a 16-h-light/8-h-dark photoperiod before conidia collection. Conidia were harvested by washing the surface of the mycelium with 10 mL of sterile distilled water. Conidia suspensions were filtered to remove residual mycelium, and the spore concentration was determined using a Thoma chamber. To synchronize the germination, 2 × 105 mL of conidia was incubated in potato dextrose broth at 24 g L−1 at room temperature for 3 h. Fully developed leaves were detached from 6-week-old Arabidopsis plants. The detached leaves were placed in square petri dishes with petioles embedded in 0.8% (w/v) agar. Six droplets of conidia suspension (5 µL each) were placed on the surface of each leaf. Mock inoculation was performed using potato dextrose broth. Leaves were incubated at 24°C with a 16-h-light/8-h-dark photoperiod. The lesion size produced by B. cinerea was evaluated as an indicator of susceptibility to the fungus (Mengiste et al., 2003; Denby et al., 2004).
Determination of PME Activity
The quantification of PME activity was performed using the PECTOPLATE assay as described previously (Lionetti, 2015). Extractions of total proteins were obtained by homogenizing uninfected and infected leaves of 6-week-old Arabidopsis plants in the presence of 1 m NaCl, 12.5 mm citric acid, 50 mm Na2HPO4, 0.02% (w/v) sodium azide, and 1:100 (v/v) protease inhibitor (P8849; Sigma-Aldrich), pH 6.5 (2 mL of extraction buffer per g of tissue). The homogenates were shaken for 1.5 h at 4°C and centrifuged at 14,000g for 15 min, and the supernatant was collected. Protein concentration was determined in supernatants using the Bradford protein assay method (Bradford reagent; Sigma-Aldrich) and bovine serum albumin as a standard (Bradford, 1976). Equal amounts of protein samples (2 µg of total proteins in 20 μL) were loaded in each well of the PECTOPLATE. Plates were incubated at 30°C for 16 h and stained with 0.05% (w/v) Ruthenium Red (R2751; Sigma-Aldrich) for 30 min. The plates were destained by several washes with water, and the area of the fuchsia-stained haloes, resulting from the demethylesterification of pectin, was measured with ImageJ software (Abramoff et al., 2004). Known amounts of commercially available PME from orange (Citrus spp.) peel (P5400; Sigma-Aldrich) was used in the PECTOPLATE to generate a standard curve used to calculate PME activity in the protein extracts.
Elicitor Treatments, Gene Expression Analysis, and Mutant Genotyping
Leaves of 6-week-old Arabidopsis plants were infiltrated with OGs (50 μg mL−1), MeOH (0.1%, v/v), or water as a control using a needleless syringe. At 1.5 hpi, leaves were collected and processed for RNA extraction. In all experiments, leaves were frozen in liquid nitrogen and homogenized using a mixer mill (MM301; Retsch) and inox beads (5 mm diameter) for about 1 min at 30 Hz, and total RNA was extracted with Isol-RNA Lysis Reagent (5′-Prime) according to the manufacturer’s instructions. RNA was treated with RQ1 DNase (Promega), and first-strand cDNA was synthesized using ImProm-II reverse transcriptase (Promega). qPCR analysis was performed using the CFX96 Real-Time System (Bio-Rad). One microliter of cDNA (corresponding to 50 ng of total RNA) was amplified in 30 μL of reaction mix containing 1× Go Taq qPCR Master Mix (Promega) and 0.4 μm of each primer. Primer sequences are shown in Supplemental Table S1. Expression levels of each gene, relative to the UBQ5 gene, were determined using a modification of the method of Pfaffl (2001). Meta-analysis of publicly available microarray data was performed using the Genevestigator database (https://www.genevestigator.com/gv/plant.jsp; Zimmermann et al., 2004).
Genomic DNA was extracted from rosette leaves of 6-week-old Arabidopsis wild-type plants as described previously (Edwards et al., 1991) and subjected to a PCR-based screening using primer pairs described in Supplemental Table S2. Go-Taq DNA polymerase (Promega) was used at 0.5 units per 30-µL assay. Amplification was carried out in the presence of deoxyribonucleotide triphosphates (0.5 mm), and specific primers (300 nm) in the buffer were provided by the supplier. Conditions for amplification were as follows: 94°C for 3 min; 30 cycles of amplification at 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s; and a final extension of 72°C for 1 min. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Determination of H2O2 Accumulation, Callose Deposition, and Mycelium Growth
Arabidopsis leaves were assayed for H2O2 accumulation employing 3,3′-diaminobenzidine staining as described (Reem et al., 2016). For the determination of callose deposition in B. cinerea-infected and mock-inoculated leaves from 6-week-old Arabidopsis, plants were cleared with 100% (v/v) ethanol. Leaves were fixed in an acetic acid:ethanol (1:3) solution for 2 h, sequentially incubated for 15 min in 75% (v/v) ethanol, in 50% (v/v) ethanol, and in 150 mm phosphate buffer, pH 8, and then stained for 2 h at 25°C in 150 mm phosphate buffer, pH 8, containing 0.01% (w/v) Aniline Blue. After staining, leaves were mounted in 50% (v/v) glycerol and examined with a UV2A filter (excitation wavelength = 330–380 nm) using the epifluorescence microscope Nikon Eclipse E200. Photographs were taken with a Nikon Digital Sight DS-Fi1c camera. For the detection of B. cinerea hyphae in infected plant tissues, detached Arabidopsis leaves were washed for 1 h with gentle agitation in absolute ethanol at 60°C to remove chlorophyll. Thereafter, leaves were incubated for 30 min in lactophenol Trypan Blue solution (water:glycerol:lactic acid [1:1:1] + 10 µL of Trypan Blue solution [25 mg mL−1; Sigma-Aldrich]). Finally, stained leaves were incubated for 20 min with gentle agitation in a destained solution (water:glycerol:lactic acid [1:1:1]) and transferred into 50% (v/v) glycerol solution for microscopy.
Extraction of Alcohol-Insoluble Residue
Uninfected, mock-inoculated, and B. cinerea-infected leaves of 6-week-old Arabidopsis plants were collected in screw-cap tubes, frozen in liquid nitrogen, and homogenized for 1 min at 30 Hz using a mixer mill (MM301; Retsch) and inox beads (5 mm diameter). Milled tissue was washed twice in prewarmed (70°C) 70% (v/v) ethanol, vortexed, and pelleted by centrifugation at 14,000g for 10 min. The pellet was suspended twice with a chloroform:MeOH mixture (1:1, v/v), shaken for 30 min at room temperature, and centrifuged at 14,000g for 10 min. Samples were pelleted by centrifugation at 14,000g for 10 min. Pellets were resuspended twice in 1 mL of 80% (v/v) acetone and spun at 14,000g for 5 min. Supernatants were discarded, and the pellet containing the alcohol-insoluble residue (AIR) was dried overnight at room temperature under a chemical hood. Starch was removed by treating the AIR with porcine type IA α-amylase (100 units g−1 AIR; product no. A4268; Sigma-Aldrich) in 100 mm potassium phosphate buffer, pH 7.5, 5 mm NaCl, and 0.02% (w/v) NaN3 for 24 h at 37°C. The suspension was centrifuged at 14,000g for 20 min, and the pellet was then washed with distilled water and 80% (v/v) acetone.
Determination of DME and Monosaccharide Composition of CW
Destarched AIR (2 mg) was saponified by suspending it in 30 μL of water and 10 μL of 1 m NaOH. The solution was incubated at room temperature for 1 h and neutralized afterward with 10 μL of HCl. After centrifugation at 14,000g for 10 min, aliquots of the supernatant (10 and 20 μL) were loaded on 96-well microtiter plates (cod.9018; Costar), and the volume was adjusted to 50 μL. Alcohol oxidase (50 μL) was added to each well (0.03 units in 0.1 m sodium phosphate, pH 7.5; Sigma-Aldrich), and this mixture was incubated at room temperature for 15 min on a shaker. Thereafter, 100 µL of a mixture containing 0.02 m 2,4-pentanedione in 2 m ammonium acetate and 0.05 m acetic acid was added to each well. After 10 min of incubation at 68°C, samples were cooled on ice, and the absorbance was measured at 412 nm in a microplate reader (ETI-System reader; Sorin Biomedica Cardio). The MeOH amount was estimated as described previously (Klavons and Bennett, 1986). The DME was expressed as MeOH-to-uronic acid molar ratio (%).
Two milligrams of saponified AIR samples was incubated in 200 µL of 2 m TFA at 121°C. After 1 h, 200 µL of isopropanol was added, and the mixtures were evaporated at 40°C with a stream of N2 gas. This step was repeated twice, and the samples were dried at room temperature overnight. The TFA-hydrolyzed monosaccharides were suspended in 200 µL of water. The monosaccharide composition of destarched and TFA-hydrolyzed AIR was determined by high-performance anion-exchange chromatography with pulsed amperometric detection using a PA20 column (Dionex). Peaks were identified and quantified by comparison with a standard mixture of Rha, Ara, Fuc, Gal, Xyl, Man, GalA, and GlcA (Sigma-Aldrich). The crystalline cellulose was determined as described previously (Updegraff, 1969). The cellulose-derived Glc content in destarched AIR was determined by an anthrone colorimetric assay (Scott and Melvin, 1953) with Glc (G8270; Sigma-Aldrich) as a standard.
CoMPP Analysis of CW Material
For the CoMPP analysis, a pool of 100 B. cinerea-infected and mock-inoculated leaves isolated from at least 10 different Arabidopsis plants was used for AIR extraction for each experiment. The chemical fractionation process of AIR material (10 mg) was performed in two extraction steps: first, with 50 mm diaminocyclohexanetetraacetic acid for predominant pectin extraction; then, with 4 m NaOH in 0.1% (w/v) NaBH4, and the samples were spotted accordingly (Moller et al., 2007, 2008) using a microarray robot (Sprint; Arrayjet). Once printed, arrays were blocked with phosphate-buffered saline (PBS) containing 5% (w/v) low-fat milk powder (MPBS). Arrays were washed with PBS and probed with antibodies (PlantProbes; Leeds University) in 5% (w/v) MPBS. Subsequently, arrays were washed in PBS and incubated with anti-rat secondary antibody conjugated to alkaline phosphatase (Sigma-Aldrich) in 5% (w/v) MPBS (1:5,000). Arrays were developed in a solution containing 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium in alkaline phosphatase buffer (100 mm NaCl, 5 mm MgCl2, and 100 mm diethanolamine, pH 9.5). Developed microarrays were scanned at 2,400 dpi (CanoScan 8800 F; Canon) and converted to TIFFs. Antibody signals were measured using appropriate software (Array-Pro Analyzer 6.3; Media Cybernetics). The highest mean spot signal in the data set was assigned a value of 100%, and all other signals were adjusted accordingly (Moller et al., 2007, 2008).
Accession Numbers
Details regarding the sequences of the genes analyzed in this study can be obtained with the following accession numbers: AtPMEI1 (At1g48020), AtPMEI2 (At3g17220), AtPMEI3 (At5g20740), AtPMEI4 (At4g25250), AtPMEI5 (At2g31430), AtPMEI6 (At2g47670) AtPMEI10 (At1g62760), AtPMEI11 (At3g47380), AtPMEI12 (At5g46960), PMEPCRA (At1g11580), PME1 (At1g53840), PME3 (At3g14310), PME4 (At3g14300), PME6 (At4g33230), PME7 (At5g07420), PME8 (At3g60730), PME17 (At2g45220), PME20 (At2g47550), PME21 (At3g05610), PME34 (At3g49220), PME41 (At4g02330), JAR1 (At2g46370), ABA2 (At1g52340), BAK1 (At4g33430), RLP44 (AT3G49750), EIN2 (At5g03280), SID2 (At1g74710), UBQ5 (At3g62250), CaPMEI (ABG47806), AdPMEI (P83326), BoPMEI (AAZ20131), SolyPMEI (SGN-U601352), VvPMEI1 (XP_010660323), AtC/VIF1 (AT1G47960), NtCIF (CAA73333), NtVIF (CAA73334), SolyCIF (SGN-U317539), and SolyVIF (SGN-U567308)
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of amino acid sequences of putative Arabidopsis invertase/PMEI isoforms with an altered expression during B. cinerea infection.
Supplemental Figure S2. Alignment of amino acidic sequences of AtPMEI10, AtPMEI11, and AtPMEI12.
Supplemental Figure S3. Nucleotide and amino acid sequences of AtPMEI10.
Supplemental Figure S4. Disease symptoms produced by B. cinerea infection in immune signaling mutants.
Supplemental Figure S5. Analysis of cis-acting DNA elements with regulatory functions in plant immunity in the 5′ flanking regions of AtPMEI10, AtPMEI11, and AtPMEI12.
Supplemental Figure S6. Arabidopsis pmei10-1, pmei11-1, and pmei12-1 mutants are not defective in H2O2 and callose accumulation against B. cinerea.
Supplemental Figure S7. Representative images illustrating the morphology of different tissues of pmei10-1, pmei11-1, and pmei12-1 mutants and wild-type plants.
Supplemental Figure S8. AtPMEI10, AtPMEI11, and AtPMEI12 gene expression in different tissues and developmental stages of Arabidopsis.
Supplemental Figure S9. Histochemical evidence indicating the control of PME activity by AtPMEI10, AtPMEI11, and AtPMEI12 during B. cinerea infection.
Supplemental Figure S10. Effect of exogenous addition of recombinant AtPMEI1 on PME activity at 48 hpi.
Supplemental Figure S11. Dynamic modifications of DME in Arabidopsis leaves during B. cinerea infection.
Supplemental Figure S12. Dynamic modifications of CW in Arabidopsis leaves during B. cinerea infection.
Supplemental Figure S13. Evaluation of Arabidopsis susceptibility during B. cinerea infection.
Supplemental Figure S14. Monosaccharide composition at different time points of CW of mock-inoculated leaves of Arabidopsis wild-type plants.
Supplemental Figure S15. Monosaccharide composition of CW of mock-inoculated leaves of Arabidopsis wild-type and pmei mutant plants.
Supplemental Figure S16. AtPMEI10, AtPMEI11, and AtPMEI12 gene expression during infection of Arabidopsis with different pathogens and following treatment with several elicitors.
Supplemental Figure S17. Expression patterns of AtPMEI10, AtPMEI11, and AtPMEI12 mRNA in Arabidopsis in response to different PAMPs, effectors, and DAMPs.
Supplemental Figure S18. Levels and kinetics of expression of B. cinerea PMEs during infection.
Supplemental Table S1. Primers used for RT-PCR.
Supplemental Table S2. Primers used for mutant genotyping.
Supplemental Table S3. Primers used for AtPMEI11 and AtPMEI12 cloning and plasmid construction.
Supplementary Material
Acknowledgments
We thank Alessandra Riccitelli for technical assistance.
Glossary
- CW
cell wall
- HG
homogalacturonan
- RGI
rhamnogalacturonan I
- RGII
rhamnogalacturonan II
- MeOH
methanol
- DME
degree of methylesterification
- JA
jasmonic acid
- DAMP
damage-associated molecular pattern
- OG
oligogalacturonide
- RT
reverse transcription
- hpi
hours post inoculation
- ET
ethylene
- SA
salicylic acid
- UTR
untranslated region
- OA
oxalic acid
- CoMPP
carbohydrate microarray polymer profiling
- mAb
monoclonal antibody
- PAMPs
pathogen-associated molecular patterns
- qPCR
quantitative PCR
- AIR
alcohol-insoluble residue
- TFA
trifluoracetic acid
- PBS
phosphate-buffered saline
- MPBS
phosphate-buffered saline containing 5% (w/v) low-fat milk powder
Footnotes
This work was supported by the Ministero dell’Istruzione, dell’Università e della Ricerca (PRIN grant no. 2010T7247Z) and by Sapienza University of Rome (grant no. C26A15J34T and Ricerca scientifica 2016).
References
- Abramoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–42 [Google Scholar]
- AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48: 28–44 [DOI] [PubMed] [Google Scholar]
- Alfano JR, Bauer DW, Milos TM, Collmer A (1996) Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol 19: 715–728 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193: 183–187 [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellincampi D, Cervone F, Lionetti V (2014) Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front Plant Sci 5: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Grundman RE, Sreekanta S, Truman W, Katagiri F, Glazebrook J (2014) Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol 164: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Morales-Cruz A, Amrine KCH, Labavitch JM, Powell ALT, Cantu D (2014) Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front Plant Sci 5: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümke A, Sode B, Ellinger D, Voigt CA (2015) Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Mol Plant Pathol 16: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle B, Brisson N (2001) Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF. Plant Cell 13: 2525–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camardella L, Carratore V, Ciardiello MA, Servillo L, Balestrieri C, Giovane A (2000) Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur J Biochem 267: 4561–4565 [DOI] [PubMed] [Google Scholar]
- Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais B, Schumacher J, Moraga J, LE Pêcheur P, Tudzynski B, Collado IG, Viaud M (2011) The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol Plant Pathol 12: 564–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Lacomme C, Morel JB, Roby D (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20: 57–66 [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caroli M, Lenucci MS, Di Sansebastiano GP, Dalessandro G, De Lorenzo G, Piro G (2011) Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J 65: 295–308 [DOI] [PubMed] [Google Scholar]
- De Caroli M, Lenucci MS, Manualdi F, Dalessandro G, De Lorenzo G, Piro G (2015) Molecular dissection of Phaseolus vulgaris polygalacturonase-inhibiting protein 2 reveals the presence of hold/release domains affecting protein trafficking toward the cell wall. Front Plant Sci 6: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38: 473–486 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Komarova TV, Petrunia IV, Frolova OY, Pozdyshev DV, Gleba YY (2012) Airborne signals from a wounded leaf facilitate viral spreading and induce antibacterial resistance in neighboring plants. PLoS Pathog 8: e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie A, Miyazaki S, Bohnert H, John P, Coleman J, Parry M, Haslam R (2004) Expression profiling of the response of Arabidopsis thaliana to methanol stimulation. Phytochemistry 65: 2305–2316 [DOI] [PubMed] [Google Scholar]
- Draeger C, Fabrice TN, Gineau E, Mouille G, Kuhn BM, Moller I, Abdou MT, Frey B, Pauly M, Bacic A, et al. (2015) Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol 15: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J 18: 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nürnberger T (2002) NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32: 375–390 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francocci F, Bastianelli E, Lionetti V, Ferrari S, De Lorenzo G, Bellincampi D, Cervone F (2013) Analysis of pectin mutants and natural accessions of Arabidopsis highlights the impact of de-methyl-esterified homogalacturonan on tissue saccharification. Biotechnol Biofuels 6: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L (1995) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit: purification by affinity chromatography and evidence of a ripening-related precursor. Eur J Biochem 233: 926–929 [DOI] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696: 245–252 [DOI] [PubMed] [Google Scholar]
- Han Y, Joosten HJ, Niu W, Zhao Z, Mariano PS, McCalman M, van Kan J, Schaap PJ, Dunaway-Mariano D (2007) Oxaloacetate hydrolase, the C–C bond lyase of oxalate secreting fungi. J Biol Chem 282: 9581–9590 [DOI] [PubMed] [Google Scholar]
- Hann CT, Bequette CJ, Dombrowski JE, Stratmann JW (2014) Methanol and ethanol modulate responses to danger- and microbe-associated molecular patterns. Front Plant Sci 5: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Vibe Scheller H (2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron SR, Benen JA, Scavetta RD, Visser J, Jurnak F (2000) Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc Natl Acad Sci USA 97: 8762–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MJ, Kim DY, Lee TG, Jeon WB, Seo YW (2010) Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes Genet Syst 85: 97–106 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Van den Ende W, Lammens W, Rybin V, Scheffzek K (2010) Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc Natl Acad Sci USA 107: 17427–17432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16: 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defence. Adv Bot Res 24: 89–166 [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kars I, McCalman M, Wagemakers L, van Kan JAL (2005) Functional analysis of Botrytis cinerea pectin methylesterase genes by PCR-based targeted mutagenesis: Bcpme1 and Bcpme2 are dispensable for virulence of strain B05.10. Mol Plant Pathol 6: 641–652 [DOI] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Müssig C, et al. (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Held MA, Zemelis S, Wilkerson C, Brandizzi F (2015) CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J 82: 208–220 [DOI] [PubMed] [Google Scholar]
- King BC, Waxman KD, Nenni NV, Walker LP, Bergstrom GC, Gibson DM (2011) Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol Biofuels 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klavons JA, Bennett RD (1986) Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J Agric Food Chem 34: 597–599 [Google Scholar]
- Kohorn BD, Kohorn SL, Saba NJ, Martinez VM (2014) Requirement for pectin methyl esterase and preference for fragmented over native pectins for wall-associated kinase-activated, EDS1/PAD4-dependent stress response in Arabidopsis. J Biol Chem 289: 18978–18986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova TV, Sheshukova EV, Dorokhov YL (2014) Cell wall methanol as a signal in plant immunity. Front Plant Sci 5: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Mengiste T (2010) Necrotroph attacks on plants: wanton destruction or covert extortion? The Arabidopsis Book 8: e0136, doi/10.1199/tab.0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA, Kieliszewski MJ, Chen Y, Cannon MC (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Park E, von Arnim AG, Nebenführ A (2009) The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V. (2015) PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front Plant Sci 6: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Cervone F, Bellincampi D (2012) Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J Plant Physiol 169: 1623–1630 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Cervone F, De Lorenzo G (2015a) A lower content of de-methylesterified homogalacturonan improves enzymatic cell separation and isolation of mesophyll protoplasts in Arabidopsis. Phytochemistry 112: 188–194 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Francocci F, Ferrari S, Volpi C, Bellincampi D, Galletti R, D’Ovidio R, De Lorenzo G, Cervone F (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci USA 107: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Giancaspro A, Fabri E, Giove SL, Reem N, Zabotina OA, Blanco A, Gadaleta A, Bellincampi D (2015b) Cell wall traits as potential resources to improve resistance of durum wheat against Fusarium graminearum. BMC Plant Biol 15: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Cervone F, Bellincampi D (2014a) Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol Plant Pathol 15: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Cervone F, Bellincampi D (2014b) How do pectin methylesterases and their inhibitors affect the spreading of tobamovirus? Plant Signal Behav 9: e972863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Mattei B, Bellincampi D (2015c) The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS ONE 10: e0133810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Song F, Goodman RM, Zheng Z (2005) Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol (Stuttg) 7: 459–468 [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Fangel JU, Willats WGT (2014) The role of the cell wall in plant immunity. Front Plant Sci 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JW, Richardson A (1981) The ultrastructure of interactions between Botrytis species and broad bean leaves. Physiol Plant Pathol 19: 41–48 [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Moller I, Marcus SE, Haeger A, Verhertbruggen Y, Verhoef R, Schols H, Ulvskov P, Mikkelsen JD, Knox JP, Willats W (2008) High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj J 25: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller I, Sørensen I, Bernal AJ, Blaukopf C, Lee K, Øbro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, et al. (2007) High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J 50: 1118–1128 [DOI] [PubMed] [Google Scholar]
- Müller K, Levesque-Tremblay G, Bartels S, Weitbrecht K, Wormit A, Usadel B, Haughn G, Kermode AR (2013) Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol 161: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Fimognari L, Sakuragi Y (2015) Interplays between the cell wall and phytohormones in interaction between plants and necrotrophic pathogens. Phytochemistry 112: 63–71 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Akutsu K (2014) Virulence factors of Botrytis cinerea. J Gen Plant Pathol 80: 15–23 [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Osorio S, Bombarely A, Giavalisco P, Usadel B, Stephens C, Aragüez I, Medina-Escobar N, Botella MA, Fernie AR, Valpuesta V (2011) Demethylation of oligogalacturonides by FaPE1 in the fruits of the wild strawberry Fragaria vesca triggers metabolic and transcriptional changes associated with defence and development of the fruit. J Exp Bot 62: 2855–2873 [DOI] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8: 335–342 [DOI] [PubMed] [Google Scholar]
- Patro L, Mohapatra PK, Biswal UC, Biswal B (2014) Dehydration induced loss of photosynthesis in Arabidopsis leaves during senescence is accompanied by the reversible enhancement in the activity of cell wall β-glucosidase. J Photochem Photobiol B 137: 49–54 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G (2008) Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 18: 1943–1948 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Ryden P, Madson M, Smith AC, Carpita NC (2004) The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Balagué C, Bezombes-Marion I, Tronchet M, Deslandes L, Roby D (2001) Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J 26: 495–507 [DOI] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant Microbe Interact 24: 432–440 [DOI] [PubMed] [Google Scholar]
- Rashid A. (2016) Defense responses of plant cell wall non-catalytic proteins against pathogens. Physiol Mol Plant Pathol 94: 38–46 [Google Scholar]
- Reca IB, Lionetti V, Camardella L, D’Avino R, Giardina T, Cervone F, Bellincampi D (2012) A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol Biol 79: 429–442 [DOI] [PubMed] [Google Scholar]
- Reem NT, Pogorelko G, Lionetti V, Chambers L, Held MA, Bellincampi D, Zabotina OA (2016) Decreased polysaccharide feruloylation compromises plant cell wall integrity and increases susceptibility to necrotrophic fungal pathogens. Front Plant Sci 7: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. (2010) The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J 63: 662–669 [DOI] [PubMed] [Google Scholar]
- Röckel N, Wolf S, Kost B, Rausch T, Greiner S (2008) Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J 53: 133–143 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Pradier JM, Simon A, Traeger S, Moraga J, Collado IG, Viaud M, Tudzynski B (2012) Natural variation in the VELVET gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS ONE 7: e47840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TA Jr, Melvin EH (1953) Determination of dextran with anthrone. Anal Chem 25: 1656–1661 [Google Scholar]
- Sénéchal F, Mareck A, Marcelo P, Lerouge P, Pelloux J (2015) Arabidopsis PME17 activity can be controlled by Pectin Methylesterase Inhibitor4. Plant Signal Behav 10: e983351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénéchal F, Wattier C, Rustérucci C, Pelloux J (2014) Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot 65: 5125–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia G, Verdugo I, Yañez M, Ahumada I, Theoduloz C, Cordero C, Poblete F, González E, Ruiz-Lara S (2005) Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol 138: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino M, Abelenda JA, Río-Alvarez I, Navarro C, Vicedo B, Farmaki T, Jiménez P, García-Agustín P, López-Solanilla E, Prat S, et al. (2014) Jasmonate-dependent modifications of the pectin matrix during potato development function as a defense mechanism targeted by Dickeya dadantii virulence factors. Plant J 77: 418–429 [DOI] [PubMed] [Google Scholar]
- ten Have A, Tenberge KB, Benen JAE, Tudzynski P, Visser J, van Kan JAL (2002) The contribution of cell wall degrading enzymes to pathogenesis of fungal plant pathogens. In Kempken F, ed, The Mycota XI, Agricultural Applications. Springer Verlag, Berlin, pp 341–358 [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M (2003) Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol Plant Microbe Interact 16: 360–367 [DOI] [PubMed] [Google Scholar]
- Vandevenne E, Christiaens S, Van Buggenhout S, Jolie RP, González-Vallinas M, Duvetter T, Declerck PJ, Hendrickx ME, Gils A, Van Loey A (2011) Advances in understanding pectin methylesterase inhibitor in kiwi fruit: an immunological approach. Planta 233: 287–298 [DOI] [PubMed] [Google Scholar]
- van Kan JAL. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11: 247–253 [DOI] [PubMed] [Google Scholar]
- Vogel J. (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307 [DOI] [PubMed] [Google Scholar]
- Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant Microbe Interact 24: 1012–1019 [DOI] [PubMed] [Google Scholar]
- Wang M, Yuan D, Gao W, Li Y, Tan J, Zhang X (2013) A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE 8: e72082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B, Tudzynski B, Tudzynski P, van Kan JAL (2007) Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol 8: 561–580 [DOI] [PubMed] [Google Scholar]
- Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins DJ, Penfold CA, Baxter L, Breeze E, et al. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24: 3530–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Rausch T, Greiner S (2009) The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J 58: 361–375 [DOI] [PubMed] [Google Scholar]
- Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schürholz AK, Augustin S, Keinath N, Rausch T, Greiner S, et al. (2014) A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci USA 111: 15261–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydra K, Berl H (2006) Structural changes of homogalacturonan, rhamnogalacturonan I and arabinogalactan protein in xylem cell walls of tomato genotypes in reaction to Ralstonia solanacearum. Physiol Mol Plant Pathol 68: 41–50 [Google Scholar]
- Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandleven J, Sørensen SO, Harholt J, Beldman G, Schols HA, Scheller HV, Voragen AJ (2007) Xylogalacturonan exists in cell walls from various tissues of Arabidopsis thaliana. Phytochemistry 68: 1219–1226 [DOI] [PubMed] [Google Scholar]
- Zega A, D’Ovidio R (2016) Genome-wide characterization of pectin methyl esterase genes reveals members differentially expressed in tolerant and susceptible wheats in response to Fusarium graminearum. Plant Physiol Biochem 108: 1–11 [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GY, Feng J, Wu J, Wang XW (2010) BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 231: 1323–1334 [DOI] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.