Jasmonates play an indispensible role in plant responses to reoxygenation after submergence by regulating the homeostasis of antioxidant defenses.
Abstract
Submergence induces hypoxia in plants; exposure to oxygen following submergence, termed reoxygenation, produces a burst of reactive oxygen species. The mechanisms of hypoxia sensing and signaling in plants have been well studied, but how plants respond to reoxygenation remains unclear. Here, we show that reoxygenation in Arabidopsis (Arabidopsis thaliana) involves rapid accumulation of jasmonates (JAs) and increased transcript levels of JA biosynthesis genes. Application of exogenous methyl jasmonate improved tolerance to reoxygenation in wild-type Arabidopsis; also, mutants deficient in JA biosynthesis and signaling were very sensitive to reoxygenation. Moreover, overexpression of the transcription factor gene MYC2 enhanced tolerance to posthypoxic stress, and myc2 knockout mutants showed increased sensitivity to reoxygenation, indicating that MYC2 functions as a key regulator in the JA-mediated reoxygenation response. MYC2 transcriptionally activates members of the VITAMIN C DEFECTIVE (VTC) and GLUTATHIONE SYNTHETASE (GSH) gene families, which encode rate-limiting enzymes in the ascorbate and glutathione synthesis pathways. Overexpression of VTC1 and GSH1 in the myc2-2 mutant suppressed the posthypoxic hypersensitive phenotype. The JA-inducible accumulation of antioxidants may alleviate oxidative damage caused by reoxygenation, improving plant survival after submergence. Taken together, our findings demonstrate that JA signaling interacts with the antioxidant pathway to regulate reoxygenation responses in Arabidopsis.
Hypoxia, an abiotic stress caused by submergence or waterlogged soils, can have serious detrimental effects on the growth of plants and cause severe agricultural losses. Plants have evolved various strategies to deal with hypoxia, including the development of anatomical and morphological traits, stimulation of ethanolic fermentation, and alterations in transcription and metabolism (Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012a, 2012b; Xie et al., 2015a). Most terrestrial plants are hypersensitive to hypoxic stress, but some plant species (e.g. rice) can survive for weeks in low-oxygen environments (Sasidharan and Voesenek, 2015).
The gaseous phytohormone ethylene functions as a key regulator in plant responses to hypoxic stress (Bailey-Serres et al., 2012a, 2012b; Voesenek and Bailey-Serres, 2013; Voesenek and Sasidharan, 2013; Sasidharan and Voesenek, 2015). Under hypoxic conditions, the activities of several ethylene biosynthetic enzymes are stimulated, which causes an increase in ethylene levels (Sasidharan and Voesenek, 2015). In terrestrial plants, ethylene trapped in the plant due to submergence can trigger cell elongation to facilitate escape from hypoxic conditions. In contrast, plants that have adapted to submergence use ethylene as a signal to sense the hypoxic environment and initiate adaptive responses (Voesenek and Bailey-Serres, 2015). Previous studies revealed that ethylene induces increases in the levels of the transcripts of group VII ethylene response factor (ERF) genes, including HYPOXIA RESPONSIVE ERF1 and RELATED TO AP2.2 (RAP2.2) in Arabidopsis (Arabidopsis thaliana; Hinz et al., 2010; Yang et al., 2011) and SUBMERGENCE1A (SUB1A), SNORKEL1, and SNORKEL2 in rice (Oryza sativa; Xu et al., 2006; Hattori et al., 2009). Five group VII ERF genes have been identified in Arabidopsis, and their regulatory roles in hypoxia sensing have been well characterized (Sasidharan and Mustroph, 2011; Bailey-Serres et al., 2012a, 2012b; van Dongen and Licausi, 2015; Sasidharan and Voesenek, 2015). These five ERF proteins have conserved N-terminal motifs, which are targets for oxygen-dependent proteolytic degradation through the N-end rule pathway (Gibbs et al., 2011; Licausi et al., 2011). Moreover, constitutive overexpression of these ERFs significantly improves plant tolerance to flooding and/or hypoxic stress (Xu et al., 2006; Fukao et al., 2006; Papdi et al., 2008; Hinz et al., 2010; Jung et al., 2010; Licausi et al., 2010; Mustroph et al., 2010), suggesting that this subgroup of ERFs have conserved functions in mediating hypoxia signaling and hypoxia tolerance in monocots and dicots.
Recent findings suggest that in most plant species, re-exposure of the plants to normoxic conditions when the water recedes, the posthypoxic period, termed reoxygenation, causes more severe challenges than the hypoxia itself (Subbaiah and Sachs, 2003). During reoxygenation, many hypoxia-acclimated physiological changes, such as energy shortages, cytoplasmic acidification, and accumulation of toxins produced by anaerobic respiration, are detrimental to plant survival (Biemelt et al., 1998; Pavelic et al., 2000; Branco-Price et al., 2008; Shingaki-Wells et al., 2014; Tamang et al., 2014; Tamang and Fukao, 2015). Moreover, in response to the high-energy demands of the reactivated metabolism in plant cells during reoxygenation, the increased oxygen uptake and accelerated mitochondrial activities are associated with accumulation of reactive oxygen species (ROS; Biemelt et al., 1998; Pavelic et al., 2000; Rawyler et al., 2002; Blokhina et al., 2003). This increased ROS production causes lipid peroxidation and increases membrane leakage, which can lead to excessive water loss (Fukao et al., 2011). A more recent report revealed that in rice, SUB1A1 physically interacts with and is phosphorylated by one member of the MAPK cascade, MPK3 (Singh and Sinha, 2016). In Arabidopsis, MPK3 associates with oxygen deprivation- and reoxygenation-triggered ROS production (Chang et al., 2012).
Reoxygenation activates the antioxidant defense system, which includes enzymatic and nonenzymatic antioxidants, to eliminate ROS-induced damage and maintain cellular redox homeostasis (Monk et al., 1987; Ushimaru et al., 1992; Biemelt et al., 1998; Skutnik and Rychter, 2009). Ascorbate (AsA) and glutathione (GSH) are two potent nonenzymatic antioxidants that can detoxify ROS by the AsA-GSH cycle in plant cells (Foyer and Noctor, 2011). The increased antioxidant capacity to scavenge ROS improves plant tolerance to environmental stresses including drought, salinity, cold, heavy metals, and pathogen infection (Sharma et al., 2012). In rice seedlings, total AsA and GSH levels did not change significantly in submerged seedlings compared to air-grown controls; however, AsA and GSH levels increased markedly upon re-exposure of the submerged seedlings to normoxic conditions (Ushimaru et al., 1992). In contrast, significant increases in the reduced forms of AsA and GSH were observed in the roots of wheat seedlings under hypoxic conditions (Biemelt et al., 1998). Two hours after hypoxia, the contents of both antioxidants had declined rapidly and were restored to high levels after 16 h of reoxygenation (Biemelt et al., 1998). Different antioxidant defense systems, i.e. the AsA-GSH and the alternative oxidase systems, regulate ROS detoxification in the leaves and roots, respectively, of barley subjected to anoxia and postanoxia conditions (Skutnik and Rychter, 2009). Although the essential role of antioxidant defense systems in plants for survival of reoxygenation has been extensively studied, their upstream regulatory signaling pathways remain unknown.
Jasmonates (JAs), a family of lipid-derived phytohormones, function in diverse biological activities including plant growth, development, and defense (Farmer et al., 2003; Browse, 2009; Kazan and Manners, 2012; Wasternack and Hause, 2013; Song et al., 2014). Previous findings indicated that JA may serve as an upstream signal to regulate plant oxidative stress tolerance, possibly by modulating the biosynthesis of antioxidant compounds (Xiang and Oliver, 1998; Sasaki-Sekimoto et al., 2005; Wolucka et al., 2005; Dombrecht et al., 2007; Shan and Liang, 2010; Guo et al., 2012), though how JA regulates the antioxidant defense system remains unknown. Here, we show that in Arabidopsis leaves, the levels of endogenous JA and JA-Ile decreased during submergence but increased rapidly during reoxygenation. We also demonstrate that the application of exogenous methyl jasmonate (MeJA) improved the tolerance of Arabidopsis to reoxygenation stress, while mutants deficient in JA biosynthesis and signaling showed increased sensitivities to reoxygenation. The transcription factor MYC2 is a key regulator of the JA-mediated reoxygenation response. By transcriptomic, biochemical, and genetic analyses, we further confirmed that MYC2 contributes to plant tolerance to reoxygenation by directly activating the expression of VTCs and GSHs, which encode the rate-limiting enzymes of the AsA and GSH biosynthesis pathways. Thus, our findings demonstrate that JA signaling interacts with the antioxidant defense pathway to regulate the response to reoxygenation in Arabidopsis.
RESULTS
Reoxygenation Causes an Increase in Endogenous Levels of JAs and Induces the Expression of JA Biosynthesis Genes
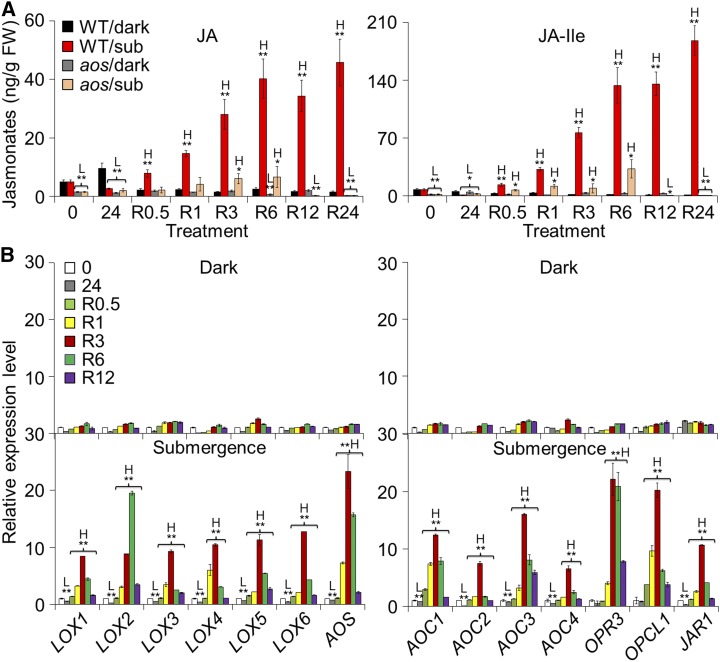
The defense phytohormones ethylene, JA, salicylic acid (SA), and abscisic acid (ABA), play key roles in plant responses to hypoxia and reoxygenation (Voesenek et al., 2003; Tsai et al., 2014; Chen et al., 2015). To investigate the exact mechanisms by which phytohormones regulate plant responses to reoxygenation, we measured the endogenous levels of JA, SA, and ABA at various time points during submergence (Supplemental Fig. S2, A and B) and reoxygenation (Fig. 1A; Supplemental Figs. S1A and S2C) in the wild type and/or the JA-deficient mutant aos, which is defective in the ALLENE OXIDE SYNTHASE (AOS) gene encoding one of the key enzymes in the JA biosynthesis (von Malek et al., 2002). To distinguish the reoxygenation response from that of hypoxia or dark treatment, we also measured these phytohormones in the wild type and aos mutant plants under constant darkness (Fig. 1A; Supplemental Figs. S1A and S2A). During submergence, the levels of JA and JA-Ile increased slightly in the wild-type plants at the early stages (1, 3, and 6 h) but significantly declined at the late stages (12, 24, and 36 h) in comparison to the dark-treated, nonsubmerged wild-type controls (Supplemental Fig. S2A). As expected, the aos mutant showed consistently low levels of JA and JA-Ile at all time points (Supplemental Fig. S2A). We also observed that submergence significantly reduced the levels of SA at 1, 3, 6, 12, and 24 h, and ABA at 3, 6, 12, 24, and 36 h of submergence (Supplemental Fig. S2B). The decreases in SA under dark submergence were consistent with our previous findings (Chen et al., 2015) in which we measured SA levels upon submergence in normal light/dark conditions. By contrast, measurements made during postsubmergence reoxygenation showed increases in JA, JA-Ile, SA, and ABA at differing levels (Fig. 1A; Supplemental Figs. S1A and S2C). In particular, the levels of JA and JA-Ile rapidly increased at 0.5 h and peaked at 6 h of reoxygenation (Fig. 1A; Supplemental Fig. S1A).
Figure 1.
Induction of JA biosynthesis by reoxygenation. A, The endogenous JA levels in the wild type (WT) and aos mutant in response to submersion and reoxygenation. Four-week-old WT and aos mutant plants were treated with dark only or dark submergence for 24 h followed by reoxygenation. Rosette leaf samples were collected for JA extraction at 0 and 24 h of dark or dark submergence as well as 0.5, 1, 3, 6, 12, and 24 h of reoxygenation and then analyzed by liquid chromatography/mass spectrometry. H2-JA was added as an internal quantitative standard. Two biological replicates were conducted with similar results, and the representative data from one replicate is shown. The data are means ± sd (n = 8 technical replicates; 200 mg of leaves harvested from three independent plants were pooled for each technical replicate). B, Quantitative PCR analyses showing the reoxygenation-inducible expression of JA biosynthetic genes (LOX1, LOX2, LOX3, LOX4, LOX5, LOX6, AOS, AOC1, AOC2, AOC3, AOC4, OPR3, OPCL1, and JAR1). Total RNA was isolated from rosettes of 4-week-old soil-grown plants at 0 and 24 h of dark or dark submergence at 0.5, 1, 3, 6, and 12 h of reoxygenation. Transcript levels relative to 0 h (prior to treatment) for each time point were normalized to the levels of TUB3. Three biological replicates were conducted with similar results, and the representative data from one replicate is shown. The data are means ± sd (n = 3 technical replicates). Asterisks with “H” or “L” indicate significant higher or lower than that of WT (*P < 0.05; **P < 0.01 by Student’s t test).
To further understand the reoxygenation-induced accumulation of JA, we examined the transcript levels of genes involved in the JA biosynthesis pathway, including LIPOXYGENASE1 (LOX1), LOX2, LOX3, LOX4, LOX5, LOX6, AOS, ALLENE OXIDE CYCLASE1 (AOC1), AOC2, AOC3, AOC4, 12-OXOPHYTODIENOATE REDUCTASE3 (OPR3), OPC-8:0-CoA LIGASE1 (OPCL1), and JASMONATE RESISTANT1 (JAR1). As expected, all of these genes were upregulated during reoxygenation (Fig. 1B; Supplemental Fig. S1B). Consistent with the JA response to submergence, the transcript levels of most JA biosynthesis genes increased slightly at 1 h of submergence but significantly declined thereafter (Supplemental Fig. S3). These results suggest that reoxygenation triggers the production of endogenous JAs in Arabidopsis.
Exogenous Application of MeJA Enhances Plant Tolerance to Postsubmergence Reoxygenation
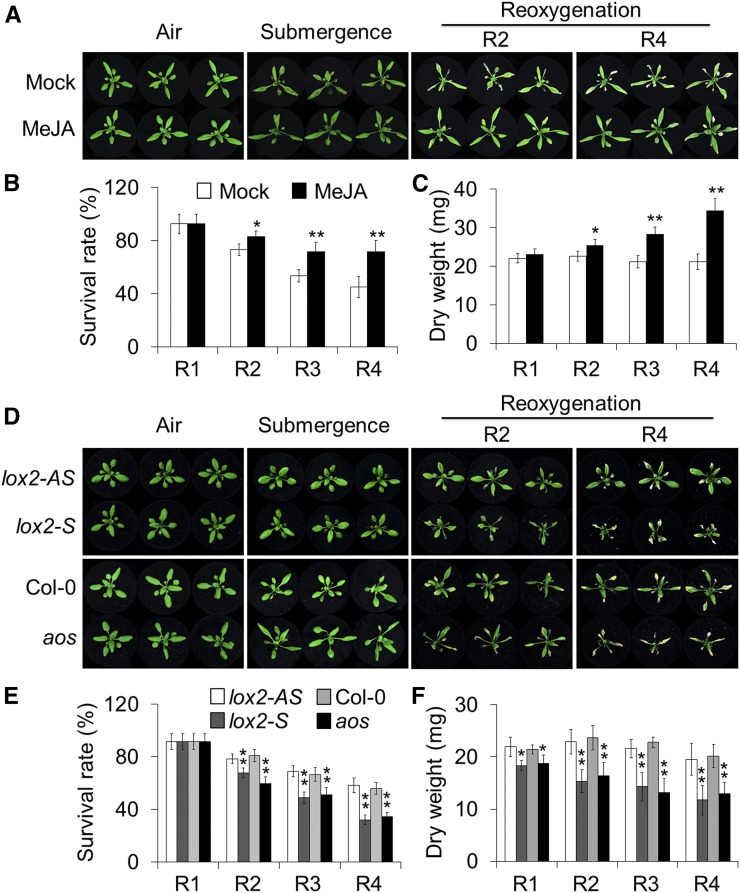
To further explore the role of JAs in the reoxygenation response, we next examined the effect of exogenous application of MeJA on plant tolerance to reoxygenation following submergence. Four-week-old wild-type plants were pretreated with 100 μm MeJA or water control for 24 h and subsequently submerged and maintained in darkness for 48 h followed by reoxygenation for various times. As shown in Figure 2A, the plants displayed few phenotypic differences before or immediately after the submergence treatment. However, following reoxygenation for 2, 3, and 4 d, the MeJA-treated plants showed enhanced tolerance in comparison to the mock-treated controls (Fig. 2A), and had significantly higher survival rates (Fig. 2B) and dry weights (Fig. 2C) than the water-treated controls. As controls, the MeJA-treated and mock-treated plants showed no significant differences upon 48-h darkness followed by 2- and 4-d recovery (Supplemental Fig. S4A). These results suggest that exogenous MeJA improves plant tolerance to postsubmergence reoxygenation.
Figure 2.
Involvement of JA biosynthesis in the plant response to reoxygenation. A to C, phenotypes (A), survival rates (B), and dry weights (C) of wild-type plants treated with exogenous MeJA and then subjected to submergence and reoxygenation. Four-week-old wild-type plants were pretreated with 100 μm MeJA or mock solution (0.1% ethanol in water) for 24 h. Images were photographed before submergence (Air) and after 48 h dark submergence (Submergence) followed by reoxygenation for 1, 2, 3, and 4 d (R1, R2, R3, and R4). D to F, phenotypes (D), survival rates (E), and dry weights (F) of lox-S and aos mutants and their corresponding controls (lox2-AS and Col-0; 4-week-old) before submergence (Air) and after 48 h dark submergence (Submergence) followed by reoxygenation for 1, 2, 3, and 4 d (R1, R2, R3, and R4). The data in B, C, E, and F are means ± sd (n = 3 independent experiments). For each experiment, at least 15 plants were used for each genotype. Asterisks indicate significant differences from the mock treatment or wild-type control (*P < 0.05; **P < 0.01 by Student’s t test).
A Deficiency of JA Biosynthesis and Signaling Confers Hypersensitivity to Postsubmergence Stress
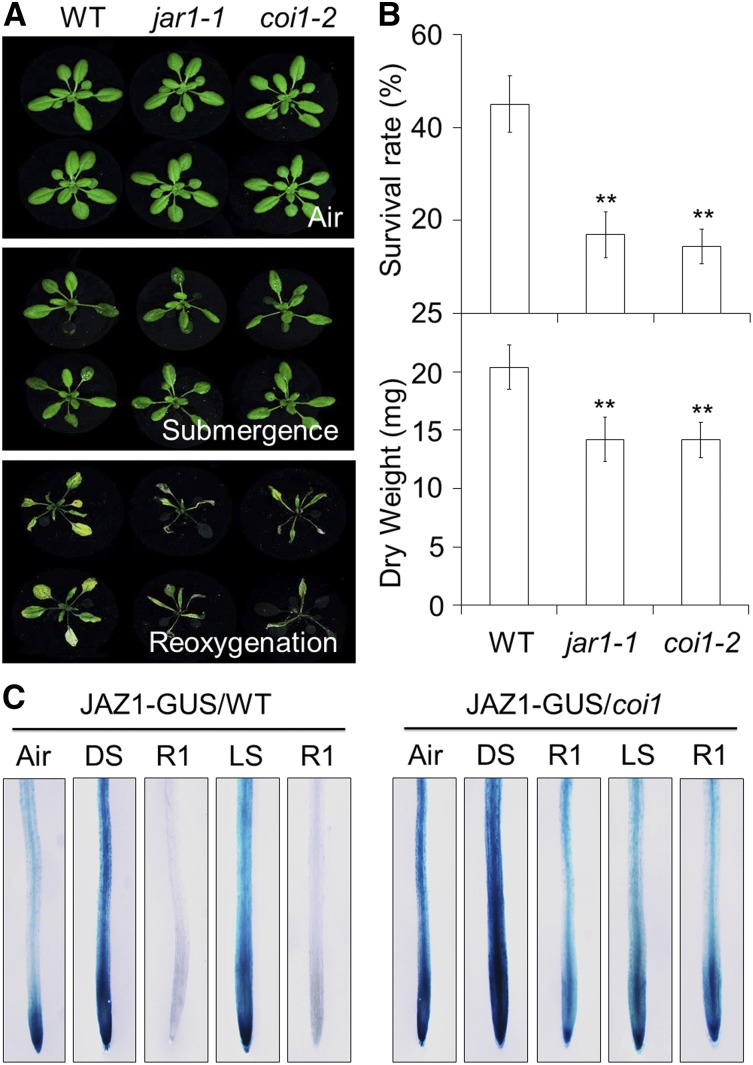
To further investigate the role of JA signaling in plant responses to reoxygenation, mutants deficient in JA biosynthesis (lox2-S and aos) and signaling (jar1; jar1-1 and coronatine insensitive1 [coi1]; coi1-2) were subjected to submergence for 48 h in darkness, and their phenotypes were observed during reoxygenation. The 4-week-old JA-deficient mutants displayed few morphological differences compared to the wild type either prior to and immediately after submergence (Figs. 2D and 3A) or upon dark treatment alone (Supplemental Figs. S4B and S5A). However, following reoxygenation for 2 and 4 d, the JA-deficient mutants exhibited significantly increased sensitivity in comparison to their respective wild-type controls (Figs. 2D and 3A). The JA-deficient mutants had significantly lower survival rates and dry weights than those of the wild type after 2, 3, and 4 d of reoxygenation (Figs. 2, E and F, and 3B). These results indicate that JA deficiency compromises the plant’s tolerance to posthypoxic stress.
Figure 3.
Essential role of JA signaling in the plant response to reoxygenation. A, phenotypes of wild type (WT), jar1-1, and coi1-2 before submergence (Air) and after 48 h dark submergence (Submergence) followed by reoxygenation 4 d (Reoxygenation). B, Survival rates (top) and dry weights (bottom) of WT, jar1-1, and coi1-2 after 2 d dark submergence followed by a 4 d reoxygenation period. The data are means ± sd (n = 3 independent experiments). For each experiment, at least 15 plants were used for each genotype. Asterisks indicate significant differences from the untreated or WT control (**P < 0.01 by Student’s t test). C, Analysis of JAZ1-GUS protein stability before submergence (Air), after dark or light submergence (DS or LS), and during reoxygenation (R1). Ten-day-old transgenic lines expressing JAZ1-GUS in WT and coi1-1 mutant backgrounds were untreated or treated with DS or LS for 24 h followed by a 24 h reoxygenation period. The seedlings were collected and were subsequently stained with X-Gluc.
To test whether the protein stability of jasmonate-ZIM domain (JAZ) proteins was affected by submergence and reoxygenation, histochemical staining was carried out to detect the levels of JAZ1-GUS fusion proteins in the wild type and coi1 backgrounds (Thines et al., 2007). As shown in Figure 3C, the JAZ1-GUS protein levels increased in the roots of the wild-type plants during both dark and light submergence; however, when the plants were exposed to oxygen for 24 h after submergence, the JAZ1-GUS protein levels diminished in the root tissues of the wild-type plants but remained in the coi1 mutant (Fig. 3C), suggesting that reoxygenation promotes COI1-dependent degradation of JAZ1 proteins.
MYC2 Is a Key Regulator in Plant Responses to Reoxygenation
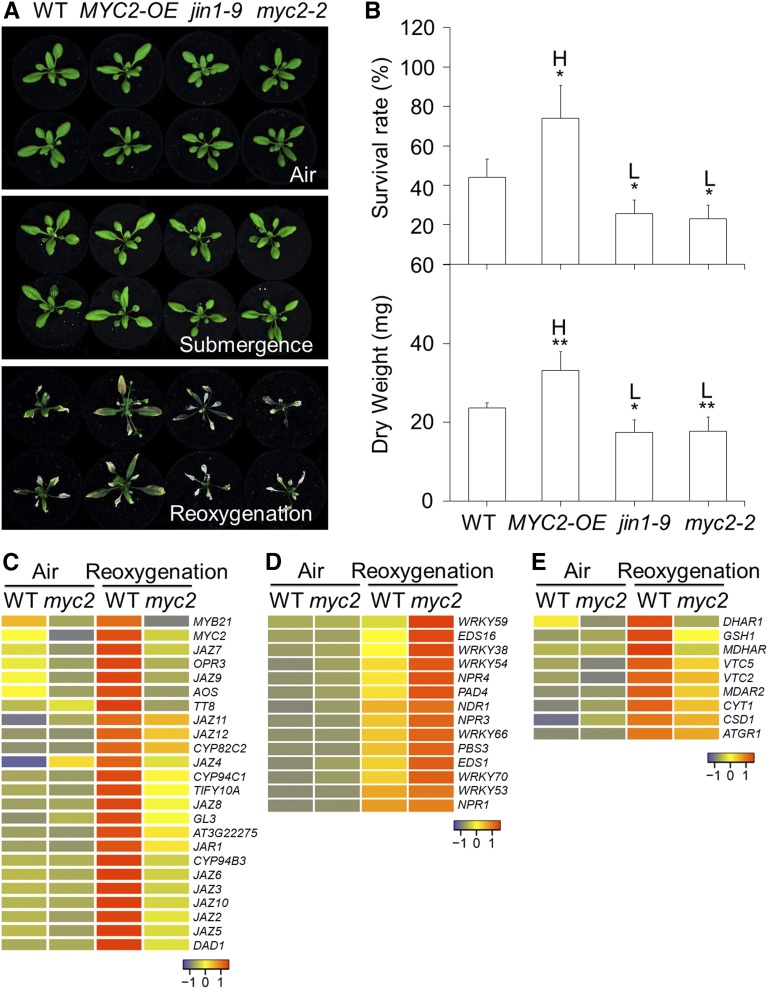
To understand how JA modulates the plant’s response to reoxygenation, we further tested the tolerance to reoxygenation after submergence of overexpression (MYC2-OE) and loss-of-function (jin1-9 and myc2-2) lines of MYC2, which encodes a key transcription factor essential for JA signaling (Boter et al., 2004; Lorenzo et al., 2004). Phenotypic analyses revealed that, similar to the other JA-defective mutants, the MYC2-OE, jin1-9, and myc2-2 lines showed no significant differences compared to the wild type either prior to and immediately after submersion (Fig. 4A) or upon 48-h dark treatment (Supplemental Fig. S5, C and D). However, after 4 d of reoxygenation, the MYC2-OE line displayed less damage while the jin1-9 and myc2-2 mutants displayed more damage compared to the wild type (Fig. 4A). The enhanced tolerance of MYC2-OE and attenuated tolerance of jin1-9/myc2-2 to reoxygenation were further confirmed by the survival rates and dry weights of the plants following reoxygenation (Fig. 4B).
Figure 4.
MYC2 is a key transcription factor in the regulation of plant tolerance to reoxygenation and antioxidant synthesis gene expression. A, Phenotypes of wild type (WT), MYC2-OE, jin1-9, and myc2-2 before submergence (Air) and after 2 d dark submergence (Submergence) followed by 4 d of reoxygenation (Reoxygenation). B, Survival rates (top) and dry weights (bottom) of WT, MYC2-OE, jin1-9, and myc2-2 after 2 d dark submergence followed by a 4 d reoxygenation period. The data are means ± sd (n = 3 independent experiments). For each experiment, 15 plants were used for each genotype. Asterisks indicate significant differences from WT (*P < 0.05 by Student’s t test). “H” and “L” indicate significantly higher or lower values, respectively, than in the WT. C to E, Hierarchical cluster analyses applied to the 24 DEGs (more than 2-fold and FDR < 0.01) in the JA pathway (C), 14 DEGs in the SA pathway (D), and nine DEGs of the antioxidant syntheses (E) in myc2-2 mutant in comparison with WT before treatment (Air) and upon 3 h of reoxygenation (Reoxygenation). The transcriptional profiles of relative gene expression values were analyzed using the heatmap 2.0 command of the R language. Red and blue colors represent upregulated and downregulated genes, respectively. The log2 fold change values from pairwise comparison of myc2-2 mutant with WT under air or upon reoxygenation are shown in Supplemental Tables S1 and S2.
To further investigate the genome-wide regulation of MYC2 during reoxygenation, leaves of 4-week-old wild-type and myc2-2 mutant plants from the nonsubmerged control and the reoxygenation conditions were subjected to transcriptome sequencing. In the plants reoxygenated for 3 h, we identified 1883 genes that were differentially expressed by more than 2-fold, of which 1715 genes were upregulated and 168 genes were downregulated in the myc2-2 mutant in comparison with the wild type (Supplemental Table S1). Further analyses of the differentially expressed genes (DEGs; more than 2-fold and false discovery rate [FDR] < 0.01) revealed that the transcripts of 24 genes involved in either JA biosynthesis or signaling pathways, including OPR3, AOS, DEFECTIVE IN ANTHER DEHISCENCE1, JAR1, and most of the JAZs, were activated by reoxygenation in the wild-type plants, while the levels of these transcripts were significantly lower in the myc2-2 mutant (Fig. 4C). In contrast, the reoxygenation-induced transcripts of 14 SA-associated genes, such as ENHANCED DISEASE SUSCEPTIBILITY1, PHYTOALEXIN DEFICIENT4 (PAD4), NONEXPRESSOR OF PR1, ENHANCED DISEASE SUSCEPTIBILITY16, and WRKY59, were significantly higher in the myc2-2 mutant than in the wild type (Fig. 4D). Moreover, we observed that nine genes involved in antioxidant synthesis were differentially regulated in the reoxygenated wild-type plants (Supplemental Table S2). These genes encoding key enzymes for the synthesis of AsA and GSH, including VITAMIN C DEFECTIVE2 (VTC2), VTC5, and GLUTATHIONE SYNTHETASE1 (GSH1), were markedly upregulated, but the levels of these transcripts were significantly reduced in the reoxygenated myc2-2 mutant plants (Fig. 4E).
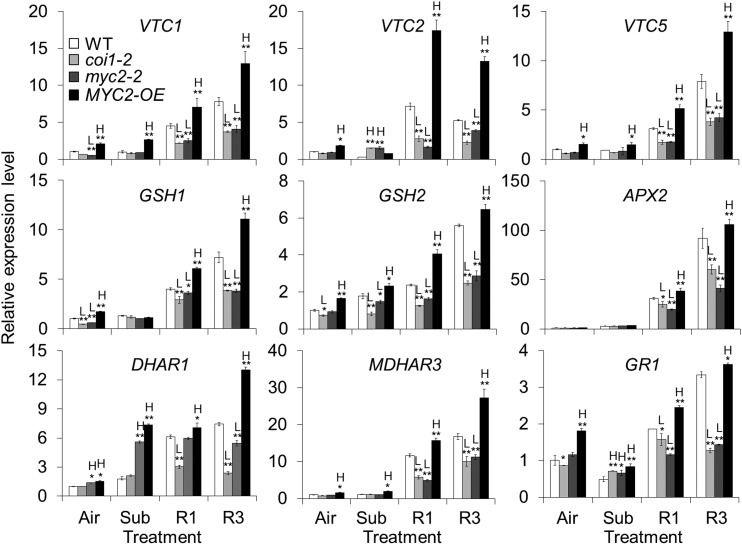
To confirm the transcriptional regulation of antioxidant synthesis by JA signaling, we further analyzed the expression levels of several genes essential for antioxidant homeostasis, including VTC1, VTC2, VTC5, GSH1, GSH2, ASCORBATE PEROXIDASE2 (APX2), DEHYDROASCORBATE REDUCTASE1 (DHAR1), MONODEHYDROASCORBATE REDUCTASE3 (MDHAR3), and GLUTATHIONE-DISULFIDE REDUCTASE1 (GR1), in response to reoxygenation in the wild-type, coi1-2, myc2-2, and MYC2-OE plants. As shown in Figure 5 and Supplemental Figure S6A, the expression levels of these genes were significantly higher in the MYC2-OE line than in the wild type under untreated and submergence conditions. This up-regulation was further enhanced in the MYC2-OE line in response to reoxygenation for 1 and 3 h (Fig. 5). In contrast, this activation was not observed in the dark-treated wild-type and MYC2-OE plants (Supplemental Fig. S6B). During reoxygenation, the expression levels of these antioxidant genes were reduced in the coi1-2 and myc2-2 mutants compared to those in the wild-type plants (Fig. 5). Taken together, these findings indicate that MYC2 is a key regulator in the JA-mediated plant response to reoxygenation, possibly through its transcriptional regulation of genes involved in antioxidant defense responses.
Figure 5.
Relative transcript levels of antioxidant defense-related genes in the wild type (WT), coi1-2, myc2-2, and MYC2-OE lines during reoxygenation. Total RNA was extracted from 4-week-old plants before submergence (Air) and after 24 h dark submergence (Sub) followed by 1 and 3 h reoxygenation (R1 and R3). The relative expression levels of antioxidant synthesis-related genes (VTC1, VTC2, VTC5, GSH1, and GSH2) and genes encoding enzymes involved in ROS detoxification (APX2, DHAR1, MDHAR3, and GR1) were determined by qPCR. Transcript levels relative to the WT at 0 h were normalized to that of TUB3. Three biological replicates were conducted with similar results. The data are means ± sd (n = 3 technical replicates). Asterisks with “H” or “L” indicate significantly higher or lower expression level than that of WT of each time point (*P < 0.05; **P < 0.01 by Student’s t test).
MYC2 Regulates Reoxygenation-Induced Production of Antioxidants
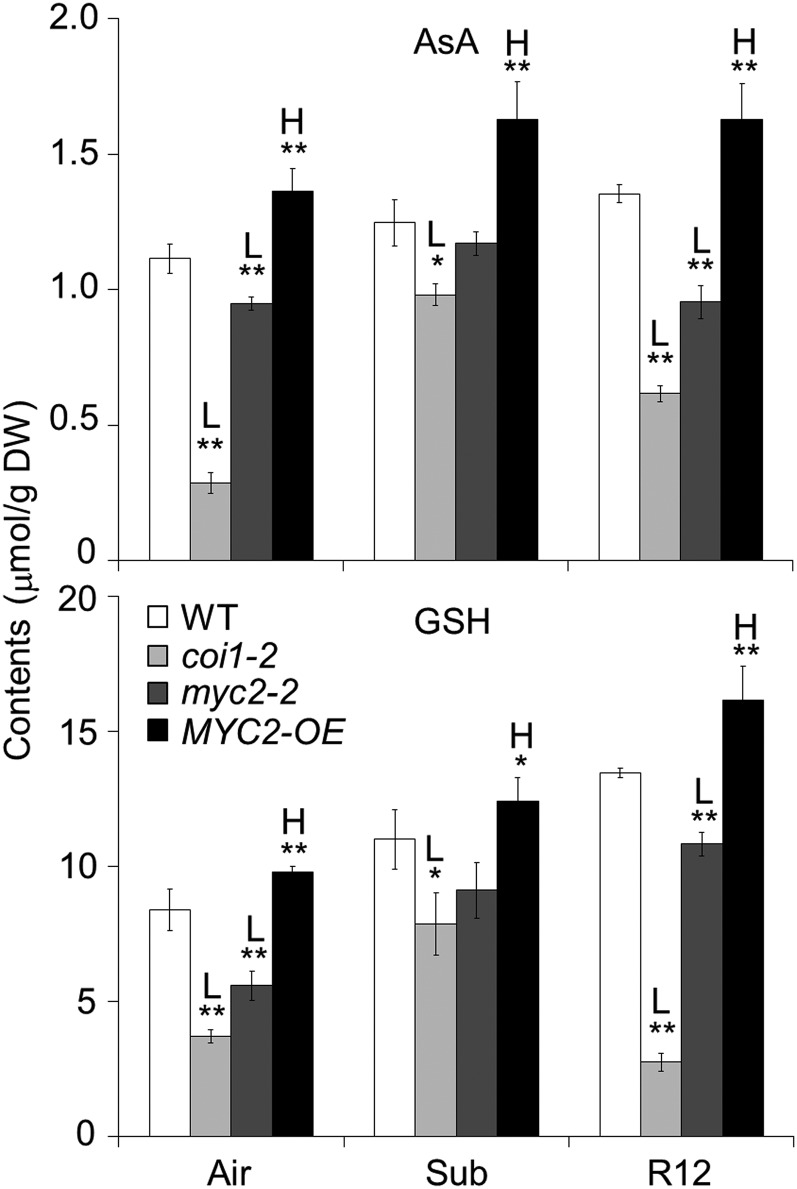
Given that the transcripts of AsA and GSH synthesis-associated genes were down-regulated in response to reoxygenation in the myc2 mutant and upregulated in MYC2-OE plants (Fig. 5), we investigated whether MYC2 contributes to the maintenance of AsA and GSH levels in plant cells by comparing the contents of AsA and GSH in the wild-type, coi1-2, myc2-2, and MYC2-OE plants under untreated, submerged, and reoxygenated conditions. Consistent with the transcriptome data, our analyses showed that reoxygenation, but not dark recovery, elevated the levels of both AsA and GSH in the wild-type plants (Fig. 6; Supplemental Figs. S7, A and B). In the untreated plants, the levels of AsA and GSH were lower in the coi1-2 and myc2-2 mutants and higher in the MYC2-OE plants than in the wild type, and these differences were further enhanced by reoxygenation (Fig. 6).
Figure 6.
MYC2 is required for regulation of reoxygenation-induced accumulation of AsA and GSH. Total AsA and GSH contents were measured in 4-week-old wild-type (WT), coi1-2, myc2-2, and MYC2-OE plants before submergence (Air) and after 24 h dark submergence (Sub) followed by 12 h reoxygenation (R12). Two biological replicates were conducted with similar results. The data are means ± sd (n = 5 technical replicates). Asterisks indicate significant differences from WT (*P < 0.05; **P < 0.01 by Student’s t test). “H” and “L” indicate significantly higher or lower contents, respectively, of AsA and GSH in the mutants or transgenic lines than in the WT.
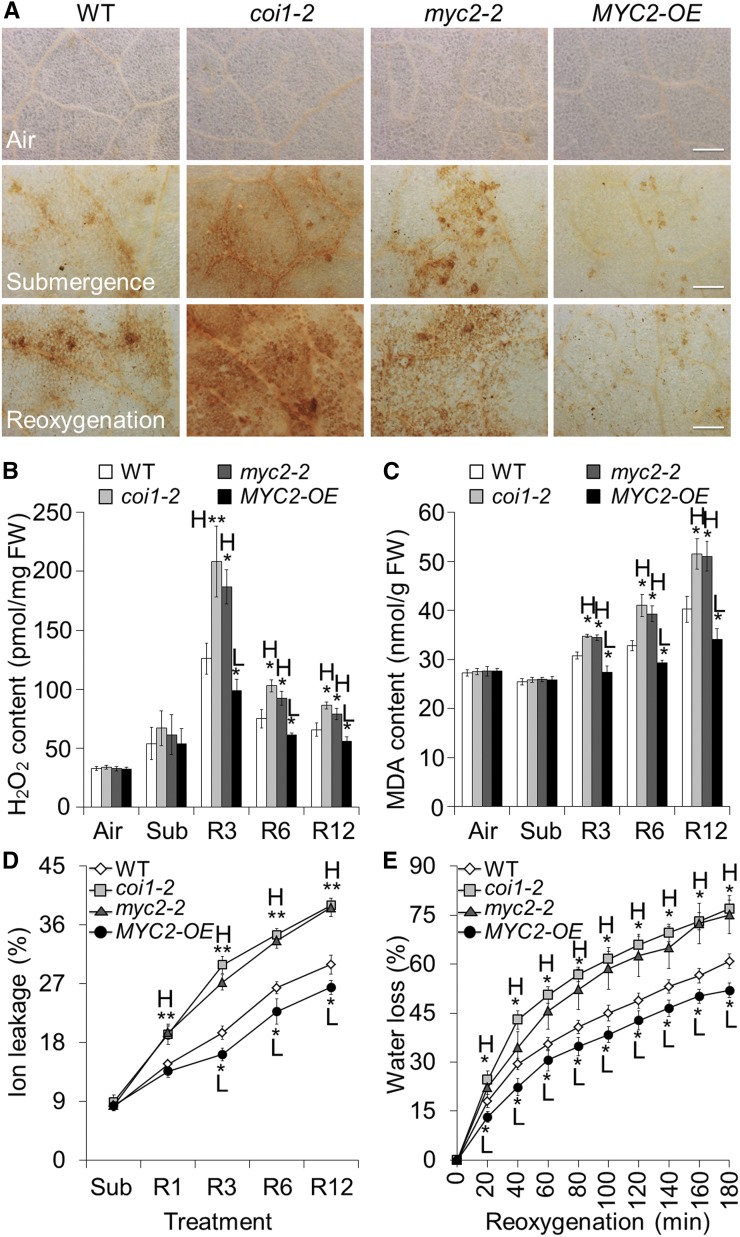
To examine the effect of JA-regulated antioxidants on the reoxygenation-induced accumulation of ROS, we detected in situ hydrogen peroxide (H2O2) levels in the leaves of treated and untreated wild-type, coi1-2, myc2-2, and MYC-OE plants using diaminobenzidine (DAB) staining. In the wild-type leaves, both submergence and reoxygenation triggered the production of H2O2 as indicated by the yellow-brown color (Fig. 7A). However, much higher levels of H2O2 were detected in the leaves of the coi1-2 and myc2-2 mutants in the submergence and reoxygenation treatments in comparison with those of the wild type and untreated or dark-treated controls (Fig. 7A; Supplemental Fig. S9). In contrast, only faint signals were detected in the MYC2-OE leaves from the submergence and reoxygenation treatments (Fig. 7A). The DAB staining results were further confirmed by quantitative measurements of H2O2 and malondialdehyde (MDA) using an Amplex Red-coupled fluorescence assay and a thiobarbituric acid reactive assay, respectively. After 3, 6, and 12 h of reoxygenation, the coi1-2 and myc2-2 mutants contained higher levels of H2O2 and MDA, while the MYC2-OE plants contained lower levels than the wild type (Fig. 7, B and C; Supplemental Fig. S8).
Figure 7.
Analyses of ROS contents in wild-type (WT), coi1-2, myc2-2, and MYC2-OE plants in response to reoxygenation. A, DAB staining showing the accumulation of ROS in the leaves of 4-week-old WT, coi1-2, myc2-2, and MYC2-OE plants before submergence (Air) and after 24 h dark submergence (Submergence) followed by 6 h reoxygenation (Reoxygenation). In contrast to the WT leaves, a weak ROS signal was observed in the MYC2-OE plants while strong ROS accumulation was detected in the coi1-2 and myc2-2 mutants during reoxygenation. Bars = 500 μm. B and C, Contents of H2O2 (B) and MDA (C) in the 4-week-old WT, coi1-2, myc2-2, and MYC2-OE plants before submergence (Air) and after 24 h dark submergence (Sub) followed by 3, 6, and 12 h reoxygenation (R3, R6, and R12). D, Electrolyte leakage in the 4-week-old WT, coi1-2, myc2-2, and MYC2-OE plants after 24 h dark submergence (Sub) and reoxygenation for 1, 3, 6, and 12 h (R1, R3, R6, and R12). E, Water loss in the 4-week-old WT, coi1-2, myc2-2, and MYC2-OE leaves immediately after 24 h dark submergence (which was set to 0 min) and reoxygenation for 20, 40, 60, 80, 100, 120, 140, 160, and 180 min. All of the experiments were performed in triplicate with similar results. The data are means ± sd (n = 6 technical replicates). Asterisks indicate significant differences from WT (*P < 0.05; **P < 0.01 by Student’s t test). “H” and “L” indicate values that are significantly higher or lower, respectively, in the mutants or transgenic lines than in the WT.
Given that the enhanced ROS scavenging capability produced by overexpression of SUB1A improved plant tolerance to rapid dehydration following submergence (Fukao et al., 2011), we further evaluated membrane integrity by measuring electrolyte leakage and water loss in leaves of the wild-type, coi1-2, myc2-2, and MYC2-OE plants before and during reoxygenation. As shown in Figure 7D, reoxygenation significantly induced ionic leakage in the wild-type leaves, which could be the direct result of the lipid peroxidation detected by MDA assay. After 1, 3, 6, and 12 h of reoxygenation, the leaves of the coi1-2 and myc2-2 mutants showed significantly higher ionic leakage than the wild-type leaves (Fig. 7D; Supplemental Fig. S8C). In contrast, ionic leakage in the leaves of the MYC2-OE lines was significantly lower than in the wild type (Fig. 7D; Supplemental Fig. S8C). After 20 to 180 min of reoxygenation, the MYC2-OE lines showed less water loss, and the coi1-2 and myc2-2 mutants showed severe water loss in comparison with the wild-type leaves (Fig. 7E; Supplemental Fig. S8D).
VTC1, VTC2, VTC5, GSH1, and GSH2 Are Direct Targets of MYC2 In Vivo
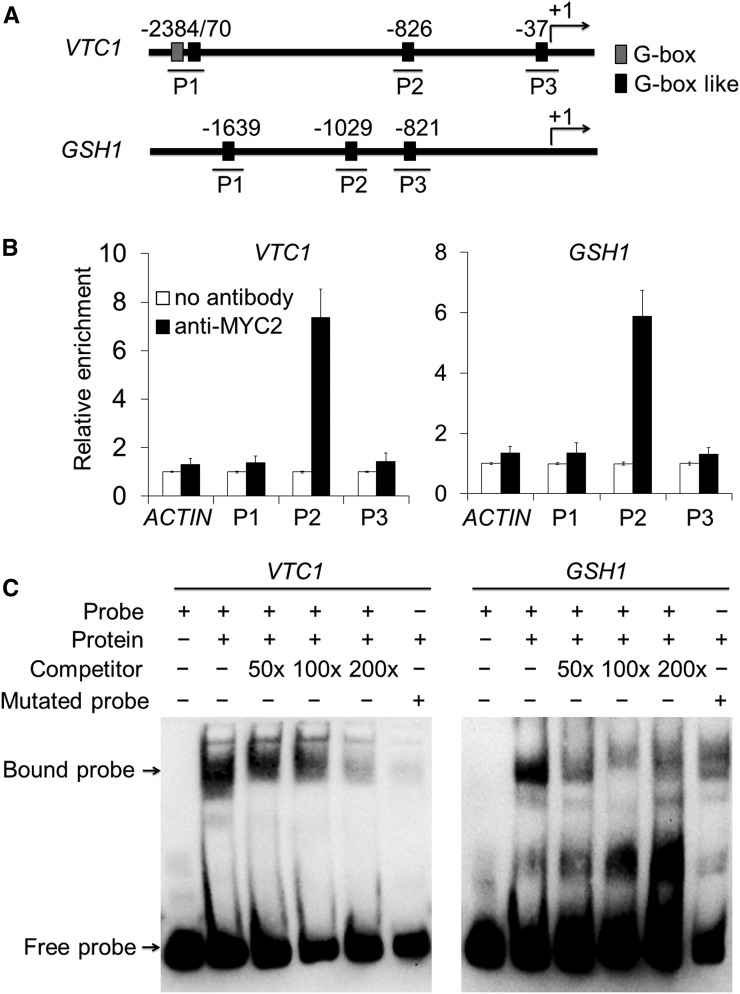
Previous studies have revealed that MYC2 binds to the G-box motif in the promoters of JA-responsive genes to directly regulate their transcription (Dombrecht et al., 2007; Chen et al., 2011; Kazan and Manners, 2013; Qi et al., 2015). We investigated whether MYC2 directly binds to the promoters of the GSH1, GSH2, VTC1, VTC2, and VTC5 genes in vivo and in vitro. First, we analyzed the promoter sequences of these genes and found several G-box or G-box-like motifs in all five promoters (Fig. 8A; Supplemental Fig. S10A). We therefore performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis to determine the in vivo interaction between MYC2 and the potential G-box motifs, using the MYC2-OE line. Our results showed that one DNA fragment of each promoter, particularly P2 in VTC1, VTC2, and GSH1, and P3 in VTC5 and GSH2, was enriched in the DNA immunoprecipitated using the anti-MYC2 antibody (Fig. 8B; Supplemental Fig. S10B).
Figure 8.
MYC2 directly binds to the promoters of VTC1 and GSH1. A, Schematic diagram of the potential G-box and G-box-like motifs in the promoter sequences of the VTC1 and GSH1 genes. Numbers indicate the nucleotide positions relative to their corresponding translational start site (ATG), which is shown as +1. B, ChIP-qPCR analyses showing the in vivo interaction of MYC2 with the DNA fragments in the promoters of VTC1 and GSH1. The protein/DNA complexes isolated from 2-week-old MYC2-OE plants were immunoprecipitated with or without the anti-MYC2 antibodies. For each promoter, three DNA fragments (P1, P2, and P3) were used to determine the enrichment of the DNA fragment containing the G-box and G-box-like motifs. The ACTIN2 promoter fragment was used as a negative control. The data are means ± sd (n = 3 independent experiments). C, EMSA analyses showing the binding of MYC2 to the DNA probes of VTC1 and GSH1 promoters in vitro. Biotin-labeled probes were incubated with nucleoproteins extracted from 2-week-old MYC2-OE seedlings. The free and bound DNAs (arrows) were separated in an acrylamide gel. The excess cold, unlabeled probes were used as competitors (lanes 3, 4, and 5), and the mutated probes (lane 6) were produced by replacing the G-box-like motifs.
To validate the ChIP-qPCR data, we analyzed the interaction between MYC2 and G-box-like motifs in the VTC1 and GSH1 promoters using electrophoretic mobility shift assays (EMSA). When the P2 fragments of the VTC1 and GSH1 promoters were labeled as probes, the mobility of each fragment significantly decreased in the presence of nuclear proteins from MYC2-OE plants (Fig. 8C, lane 2). Moreover, the cold competitors (50-, 100-, and 200-fold of the same fragments, not labeled) were sufficient to reduce the binding of MYC2 to the labeled P2 fragments (Fig. 8C, lanes 3, 4, and 5). When the core G-box sequence (CATGTG or CACATG) of the P2 fragment was mutated to TTTTTT or AAAAAA, the interactions of VTC1 and GSH1 promoters with the MYC2 protein were significantly impaired in the same assay (Fig. 8C, lane 6). Taken together, our results revealed that the antioxidant synthesis genes VTC1, VTC2, VTC5, GSH1, and GSH2 are direct targets of MYC2 in vivo and in vitro.
Overexpression of VTC1 and GSH1 Overcomes the Hypersensitivity to Reoxygenation of the myc2 Mutant
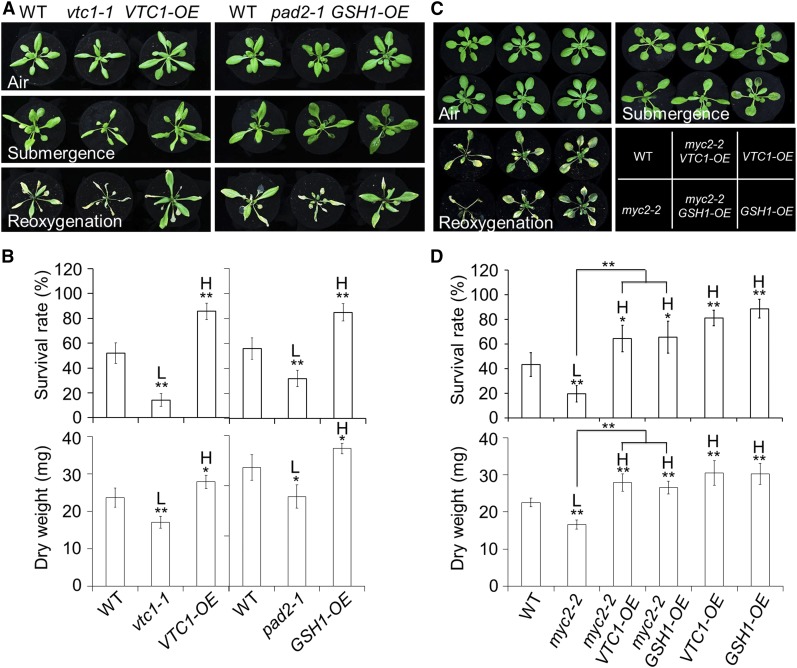
To further investigate the roles of AsA and GSH in plant responses to reoxygenation, we next evaluated the phenotypes of the loss-of-function and overexpression lines of VTC1 and GSH1 to postsubmergence reoxygenation. When the plants were submerged for 48 h under dark conditions or subjected to dark treatment for 48 h followed by recovery, the vtc1-1 and GSH-depleted pad2 (pad2-1) mutants (Glazebrook and Ausubel, 1994; Conklin et al., 1996) as well as the VTC1 and GSH1 overexpression lines (VTC1-OE and GSH1-OE; Hatano-Iwasaki and Ogawa, 2012; Wang et al., 2013) displayed no visible differences compared to the wild type (Fig. 9A; Supplemental Fig. S11). However, after 4 d of reoxygenation, both the vtc1-1 and pad2-1 mutants displayed increased sensitivity to reoxygenation while the VTC1-OE and GSH1-OE lines displayed a level of tolerance comparable to the wild type (Fig. 9, A and B).
Figure 9.
Overexpression of VTC1 or GSH1 rescued the hypersensitivity of myc2-2 mutant to reoxygenation. A, Phenotypes of 4-week-old wild-type (WT), vtc1-1, VTC1-OE, pad2-1, and GSH1-OE plants before submergence (Air) and after 48 h dark submergence (Submergence) followed by a 4 d reoxygenation (Reoxygenation). B, Survival rates (top) and dry weights (bottom) of the WT, vtc1-1, VTC1-OE, pad2-1, and GSH1-OE plants shown in A, following 4 d of reoxygenation. C, Phenotypes of 4-week-old WT, myc2-2, myc2-2 VTC1-OE, VTC1-OE, myc2-2 GSH1-OE, and GSH1-OE plants before submergence (Air) and after 48 h dark submergence (Submergence) followed by a 4 d reoxygenation (Reoxygenation). D, Survival rates (top) and dry weights (bottom) of the WT, myc2-2, myc2-2 VTC1-OE, VTC1-OE, myc2-2 GSH1-OE, and GSH1-OE plants shown in C following 4 d of reoxygenation. The data in B and D are means ± sd (n = 3 independent experiments). For each experiment, 15 plants were used for each genotype. Asterisks indicate significant differences from WT (*P < 0.05; **P < 0.01 by Student’s t test). The differences between myc2-2 and myc2-2 VTC1-OE or myc2-2 GSH1-OE are also significant (**P < 0.01 by Student’s t test). “H” and “L” indicate values that are significantly higher or lower, respectively, in the mutants, double mutants, or transgenic lines than in the WT.
To investigate the genetic interaction between JA signaling and antioxidant defense pathways, the myc2-2 mutant was crossed to the VTC1-OE and GSH1-OE lines and the resulting myc2-2 VTC1-OE and myc2-2 GSH1-OE lines were characterized. As shown in Figure 9, when the plants were treated with dark submergence for 48 h followed by 4 d of reoxygenation, the hypersensitivity of myc2-2 was rescued by overexpression of either VTC1 or GSH1 (Fig. 9C), as indicated by the improved survival rates and dry weights of the myc2-2 VTC1-OE and myc2-2 GSH1-OE lines in comparison with the myc2-2 single mutant (Fig. 9D). These results indicate that JA-mediated antioxidant accumulation in response to reoxygenation occurs through transcriptional regulation of VTC1 and GSH1 by the MYC2 transcription factor.
DISCUSSION
Postsubmergence reoxygenation involves a burst of ROS, which causes oxidative stress in plant cells (Biemelt et al., 1998; Pavelic et al., 2000; Rawyler et al., 2002; Blokhina et al., 2003). To cope with the increased levels of ROS, the cells activate the antioxidant defense systems, including the AsA-GSH cycle and antioxidant enzymes, to maintain cellular redox homeostasis (Monk et al., 1987; Ushimaru et al., 1992; Biemelt et al., 1998; Skutnik and Rychter, 2009). Although the role of antioxidants in the plant response to reoxygenation has been well documented, the regulatory mechanism of the low oxygen-dependent activation of antioxidant defenses is still unclear. Here, we demonstrated that JA is a dominant signaling molecule that is specifically involved in the regulation of antioxidant defense in response to postsubmergence reoxygenation. First, the transcripts of JA biosynthesis genes and the levels of JA and JA-Ile in Arabidopsis leaves were significantly induced by reoxygenation (Fig. 1). Second, the mutants deficient in either JA synthesis or signaling pathways, including aos, lox2-S, jar1, coi1, and myc2, showed hypersensitivity to reoxygenation and the exogenous application of MeJA and overexpression of MYC2 enhanced plant tolerance to reoxygenation (Figs. 2–4). Third, the attenuated and improved tolerance phenotypes of the myc2-2 mutant and MYC2-OE line, respectively, were consistent with the expression levels of antioxidant defense genes and the cellular contents of antioxidant compounds (Figs. 4–6). Fourth, MYC2 was observed to bind to the promoters of VTC and GSH gene family members, to activate their transcription (Fig. 8). Finally, the sensitivity to reoxygenation in the myc2-2 mutant was completely rescued by constitutive overexpression of the VTC1 and GSH1 genes in the myc2 VTC1-OE and myc2 GSH1-OE lines (Fig. 9). Overall, these findings demonstrate that JA is essential for protection from reoxygenation-induced oxidative injuries through direct activation of the antioxidant defense system in Arabidopsis.
Under natural conditions, submergence involves three distinct stages, hypoxia, anoxia, and reoxygenation when the water is removed, as determined by the progressive changes in oxygen levels in plant cells (Blokhina et al., 2003). Increasing evidence suggests that in shoot and root tissues, increased ROS production occurs in plant responses to these stresses, especially to reoxygenation (Blokhina et al., 2003; Fukao et al., 2011; Chang et al., 2012; Voesenek and Sasidharan, 2013; Shingaki-Wells et al., 2014; Campbell et al., 2015). In plants, low levels of ROS serve as signaling molecules to sense low-oxygen environments and to stimulate adaptive processes (Fukao and Bailey-Serres, 2004; Voesenek and Sasidharan, 2013). For example, the most stable ROS, H2O2, is an important signal that controls the activity of ADH1, which subsequently regulates the rates of anaerobic respiration for energy supply under hypoxic conditions (Baxter-Burrell et al., 2002; Yang, 2014). However, excessive ROS may cause irreversible oxidative damage to macromolecules such as lipids, proteins, and nucleic acids, and ultimately lead to programmed cell death and cellular dysfunction (Pérez-Pérez et al., 2012). Therefore, the delicate balance between ROS production and scavenging may determine the damaging or signaling role of ROS and consequently, determine plant survival under posthypoxia reoxygenation (Sharma et al., 2012).
There are several potential mechanisms for plant cells to remove hypoxia-induced ROS or oxidative-damaged organelles. In Arabidopsis, the Rho-like small G protein of plant (ROP) protein is required for the response to oxygen deprivation, and ROP functions to promote ROS generation and ethanolic fermentation (Baxter-Burrell et al., 2002; Fukao and Bailey-Serres, 2004). Fine-tuning of the negative feedback loop of Rop-mediated hypoxia signaling may occur by activating the expression of the GTPase-activating protein RopGAP4 to balance carbohydrate consumption and ROS-induced oxidative damage (Baxter-Burrell et al., 2002; Fukao and Bailey-Serres, 2004). A recent investigation indicated that the submergence-inducible expression of HYPOXIA RESPONSIVE UNIVERSAL STRESS PROTEIN1 (HRU1) is controlled by the ERF-VII transcription factor RAP2.12 and contributes to modulation of ROS production in response to anoxic stress (Gonzali et al., 2015). Further, the HRU1 proteins physically interacts with the GTPase ROP2 and the NADPH oxidase RbohD proteins, suggesting that the HRU1-mediated anoxia-inducible ROS production is controlled by the N-end rule pathway (Gonzali et al., 2015). Our recent findings revealed that Arabidopsis autophagy contributes to hypoxia-induced ROS homeostasis during prolonged submergence, possibly by removing oxidative-damaged cellular components (Chen et al., 2015). The impaired tolerance to submergence and ROS levels of autophagy-defective mutants (atg5 and atg7) were suppressed by the introduction of SA-defective mutants salicylic acid induction deficient2 and nonexpressor of PR1, which suggests that SA is an upstream signal required for the feedback regulation of autophagy-mediated ROS production and sensitivity to hypoxia (Chen et al., 2015).
Several physiological analyses suggested that upon hypoxia/anoxia and reoxygenation, two well-characterized, low-mass antioxidants, AsA and GSH, play major roles in the detoxification of ROS in both leaves and roots of plants (Monk et al., 1987; Ushimaru et al., 1992; Biemelt et al., 1998; Skutnik and Rychter, 2009). In Arabidopsis, VTC1 and GSH1 are representatives of two gene families that encode rate-limiting enzymes for the biosynthesis of AsA and GSH, respectively (Smirnoff and Wheeler, 2000; Noctor et al., 2012). Knockouts of VTC1 and GSH1 in the vtc1-1 and pad2-1 mutants result in significant reductions of AsA or GSH contents (Pavet et al., 2005; Schlaeppi et al., 2008). By phenotypic and genetic analyses, we provided further evidence to show that deletions of VTC1 and GSH1 reduced plant survival after submergence and postsubmergence reoxygenation (Fig. 9). Significant increases of AsA or GSH contents in the VTC1- or GSH1-overexpression lines (Hatano-Iwasaki and Ogawa, 2012; Wang et al., 2013) improved plant tolerance to hypoxic stress and posthypoxia reoxygenation (Fig. 9), suggesting that enhancement of the antioxidant defense system by expressing key genes involved in AsA and GSH metabolism is an efficient way to elevate tolerance to hypoxic stress in plants. The VTC1 and GSH1 overexpression lines have increased tolerance to salt, chilling, and heavy metal stresses in various plant species (Guo et al., 2008; Hatano-Iwasaki and Ogawa, 2012; Wang et al., 2013; Chen et al., 2016). Our observations strengthen previous findings that genetic manipulation of these two gene families in crops has potential applications for improving plant tolerance to stresses, including submergence.
Despite extensive investigations showing the physiological significance of antioxidant defense in plant response to environment cues, little is known about the molecular mechanisms that regulate the AsA and GSH biosynthesis pathways. Increasing evidence indicates that the defense phytohormone JA plays an important role in signaling stress-induced AsA and GSH metabolism (Sasaki-Sekimoto et al., 2005; Wolucka et al., 2005; Shan and Liang, 2010). Moreover, the JA-inducible expression of genes in antioxidant defenses is downregulated in the myc2 mutant, and MYC2 overexpression imparts enhanced tolerance to oxidative stress (Dombrecht et al., 2007), indicating that MYC2 is a key regulator in mediating antioxidant metabolism. In agreement with these findings, our results showed that MYC2 positively regulates the plant response to submergence, the increase in transcripts of genes in antioxidant metabolism, and the levels of AsA and GSH (Figs. 4–6). In comparison to the phenotypes of ropgap4, rop2, and atg mutants, which differed during oxygen deprivation or submergence treatments (Baxter-Burrell et al., 2002; Chen et al., 2015), all of the phenotypic differences in the JA-, AsA-, and GSH-associated lines occurred during reoxygenation, but not during submergence/hypoxia. Based on the data from ChIP-qPCR, EMSA, and genetic analyses, we conclude that as the core transcriptional factor in JA signaling, MYC2 functions in the regulation of reoxygenation-responsive antioxidant metabolism by directly binding to the promoters of VTC1 and GSH1 genes and activating their transcription (Figs. 8 and 9). Thus, our results established a direct link between JA signaling and antioxidant defense pathways, which may coordinately contribute to regulating multiple stress responses. Several independent investigations suggested that both JA and antioxidants are crucial for plant tolerance to diverse ROS-generated stresses (Sharma et al., 2012; Kazan, 2015). These findings support the idea that, at least in Arabidopsis, the stress-responsive antioxidant defense system is likely to be positively regulated by the MYC2 transcription factor in JA signaling. Future investigations on the response of different MYC2 genotypes to the above stresses and their genetic link to the VTC1 and GSH1 will further our understanding of the physiological significance between the JA and antioxidant interactions.
In addition to oxidative stress, dehydration is another challenge that limits plant recovery following submergence (Fukao et al., 2011; Fukao and Xiong, 2013). Overexpression of rice SUB1A up-regulates the expression of genes involved in the acclimation to dehydration and confers enhanced tolerance to both reoxygenation and drought stresses (Fukao et al., 2011). It is still unclear how SUB1A regulates the transcripts of dehydration-acclimated genes. Nonetheless, one member of the AP-family ERFs, RAP2.4, can bind to both the ethylene-responsive GCC-box and the dehydration-responsive element (DRE), which positively affects ethylene signaling and drought tolerance in Arabidopsis (Lin et al., 2008). Thus, it is likely that in response to reoxygenation, ethylene signaling may directly regulate the acclimation of dehydration in plants and that the DREB genes are potential targets of SUB1A. Upon hypoxia and reoxygenation, antagonism between ethylene and ABA has been frequently observed (Bailey-Serres et al., 2012a; 2012b; Tsai et al., 2014). Transcriptomic analyses showed that the ABA-regulated dehydration genes, such as RD20, RD22, KIN1, KIN2, MYB102, MYB121, and P5CS1, are significantly up-regulated in the ethylene-deficient mutants ein2-5 and ein3 eil1 during reoxygenation (Tsai et al., 2014). In contrast, the transcripts of some ABA-independent dehydration genes, including DREB1A, DREB1B, DREB2A, and DREB2B, show pronounced down-regulation in the ein2-5 and ein3 eil1 mutants relative to the wild-type plants (Tsai et al., 2014), suggesting that ethylene is important for the regulation of dehydration response caused by reoxygenation. It is worth mentioning that for the duration of reoxygenation in our study, leaf desiccation was negatively correlated with stress tolerance in the MYC2 genotypes as reflected by measurements of water losses and ion leakage (Fig. 6, D and E), which is indicative of the involvement of JA in regulating dehydration during reoxygenation. Given the various interplays among the ethylene, JA, and ABA signaling pathways, it is of interest to find out how these three plant hormones are coordinately balanced to maintain the cellular homeostasis in response to reoxygenation.
CONCLUSION
In summary, our findings demonstrated that JA acts as a dominant upstream signaling molecule essential for plant responses to postsubmergence reoxygenation by activating the antioxidant defense system, which in turn contributes to alleviation of oxidative damages caused by reoxygenation and subsequently improves plant tolerance to reoxygenation stress.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Treatments
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used in this study unless otherwise indicated. The following mutants and transgenic lines were obtained from the Arabidopsis Information Resource (TAIR; www.arabidopsis.org) and named lox2-S (CS3748; a knockdown line of LOX2; Bell et al., 1995), lox2-AS (CS3749; control of lox2-S; Bell et al., 1995), aos (CS65993; dde2-2; von Malek et al., 2002), jar1-1 (CS8072; Staswick et al., 2002), jin1-9 (SALK_017005; Anderson et al., 2004), myc2-2 (SALK_083483; Boter et al., 2004), vtc1-1 (CS8326; Conklin et al., 1996), and pad2-1 (CS3804; Glazebrook and Ausubel, 1994). Some of the plant materials were described previously: coi1-2 (Xiao et al., 2004), JAZ1-GUS/WT and JAZ1-GUS/coi1 (Thines et al., 2007), MYC2-OE (Chen et al., 2011), VTC1-OE (VTC1-GFP; Wang et al., 2013), and GSH1-OE (Hatano-Iwasaki and Ogawa, 2012). The myc2-2 VTC1-OE and myc2-2 GSH1-OE lines were generated by crosses of myc2-2 to VTC1-OE and GSH1-OE plants, and the resulting lines were characterized by PCR-based genotyping of the F2 population.
Arabidopsis seeds were surface-sterilized with 20% bleach and 0.1% Tween 20 for 15 min and washed with sterile water, then sown on Murashige and Skoog medium (Sigma-Aldrich) with 1% Suc and 0.8% agar (pH 5.8). After chilling at 4°C for 3 d, the plates were transferred to a growth room under a 16-h light/8-h dark (22°C) photoperiod. The 7-d-old seedlings were transplanted into soil in plastic pots for continuous growth under the same conditions.
Submergence treatments were performed under darkness as described previously (Vashisht et al., 2011; Xie et al., 2015a). Briefly, the 4-week-old whole plants with soil were gently submerged under continuous darkness. Control plants were placed in the dark in the same growth room. Plant leaves were 10 cm under the surface of the water. To control the oxygen levels in the submerged leaves and make sure that the environment around the underwater leaves was hypoxic, the levels of dissolved oxygen around the underwater leaves were regularly determined throughout the experiments using a portable dissolved oxygen meter (JPBJ-608, Rex). As previously described (Chen et al., 2015), the oxygen concentration in the water was kept at approximately 8 to 9 mg/L−1 before treatment, but it decreased strongly to approximately ∼2 mg/L−1 within 48 h after dark submergence treatment. After treatment for the indicated times, the water was removed and the plants were re-exposed to normal air for reoxygenation assays. To rule out a possible effect of the circadian clock on the treatments, plants were subjected to submergence or darkness at 9:00 a.m. for all of the experiments and subsequently reoxygenated under normal growth conditions for the indicated times (16-h light [6:00 a.m.–22:00 p.m.]/8-h dark [22:00 p.m.–6:00 a.m.]). For exogenous MeJA application, 4-week-old wild-type Arabidopsis plants were sprayed with 100 μm MeJA (Sigma-Aldrich) or mock solution (0.1% ethanol in water) for 24 h, followed by submergence and reoxygenation for the indicated times.
To calculate the survival rates and dry weights after reoxygenation, 15 plants per genotype were treated with dark submergence for the indicated times followed by exposure to air for the indicated duration, and the average values of three independent experiments were recorded. The survival rates were determined based on the numbers of plants that could produce new leaves and continue to grow after recovery from submergence. The aboveground tissues were collected, heated overnight at 105°C, and weighed to obtain dry weight.
Measurements of Phytohormones
Phytohormones were extracted following the method in Pan et al. (2010). Briefly, fresh leaf tissues were collected and homogenized in liquid nitrogen. The powdered samples (∼200 mg) were sealed in 2-mL tubes containing 0.8 mL extraction buffer (isopropanol:water:concentrated HCl; 2:1:0.002; v/v/v) with internal standards (10 ng D4-SA [OlChemim], H2-JA [Cerilliant], and D6-ABA [OlChemim]). The mixtures were gently agitated for 30 min at 4°C followed by addition of 1 mL CH2Cl2 and agitated for another 30 min. The samples were then centrifuged at 13,000g for 10 min. The 900 μL of solvent from the lower phase was collected and concentrated using a nitrogen evaporator. The samples were dissolved in 100 μL of solution (60 μL methanol and 40 μL distilled water). Ten microliters of each sample was injected into a C18 column and analyzed with the AB Sciex Q-TOF 5600+ system as described previously (Chen et al., 2015).
RNA Extraction, RT-PCR, and qPCR Analysis
Total RNA was extracted with TRIzol reagent (Invitrogen) and reverse transcribed using the PrimeScript RT reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. Quantitative PCR analyses were performed using the StepOne Plus real-time PCR system (Applied Biosystems) with the SYBR Premix ExTaq Mix (Takara) as described previously (Chen et al., 2015). TUB3 was used as a reference gene. Three technical replicates were used for each reaction. The gene-specific primers for the qPCR analysis are listed in Supplemental Table S3.
Histochemical Staining
For GUS staining, whole seedlings (10-d-old) of JAZ1-GUS transgenic lines in Col-0 and coi1 backgrounds (Thines et al., 2007) were harvested after treatment with submergence or reoxygenation at the indicated times. GUS staining was carried out according to Chen et al. (2015). In brief, samples in the GUS staining buffer (100 mm sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 2 mm potassium ferricyanide, and 2 mm potassium ferrocyanide) containing 1 mg mL−1 5-bromo-4-chloro-3-indolyl β-d-glucuronide (Sigma) were vacuum infiltrated for 5 min followed by incubation at 37°C in the dark for 1 h. The samples were washed with 75% ethanol several times to remove the staining buffer and chlorophyll. The GUS activities in roots were observed and photographed.
DAB staining was performed as described previously (Xie et al., 2015b). Leaves were excised and placed in 1 mg mL−1 DAB (Sigma) solution, pH 3.8, for 3 h at room temperature under darkness. The samples were cleared by placing into boiling ethanol (95%) for 10 min.
Measurement of H2O2 and MDA
The H2O2 content was determined using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes) as described previously (Pucciariello et al., 2012) with minor modifications. Briefly, 30 mg of leaves was ground in liquid nitrogen and diluted in a 200 µL reaction buffer. After centrifugation at 10,000g for 10 min at 4°C, 50 µL of supernatant was mixed with 50 µL H2O2 working solution supplied by the kit, followed by incubation for 30 min at room temperature under darkness. Absorbance was detected at 560 nm using a microplate reader (Tecan). The H2O2 concentration was determined using a standard curve generated following the manufacturer’s instructions.
MDA levels were measured by the thiobarbituric acid reaction according to Zhang et al. (2012a). In brief, leaves were homogenized in 1 mL trichloroacetic acid buffer (0.1%, w/v) followed by centrifuging at 10,000g for 10 min at 4°C. The supernatant was further mixed with 4 mL concentrated trichloroacetic acid (20%, w/v) containing 0.5% thiobarbituric acid. After boiling at 95°C for 15 min, the mixtures were quickly cooled on ice. Following centrifugation at 10,000g for 5 min at 4°C, the MDA contents were detected at 532 and 600 nm using a microplate reader (Tecan) and calculated accordingly (Zhang et al., 2012a).
Measurement of Electrolyte Leakage and Water Loss
Electrolyte leakage was measured as previously described (Chen et al., 2015). Water loss was detected following Zhang et al. (2012b) with minor modifications. Briefly, Arabidopsis rosettes were collected after dark submergence treatment for 24 h and placed on plates for various times under normal air conditions. Weights were measured accordingly and the relative loss of fresh weight (%) was used to represent water loss.
RNA Sequencing Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) from Arabidopsis rosettes following the manufacturer’s instructions. Illumina library construction, sequencing, and sequence analysis were conducted as previously described (Yu et al., 2012; van Veen et al., 2016). Four-week-old wild-type and myc2-2 mutant plants were submergence-treated in the dark for 24 h, and rosettes were harvested at 0 and 24 h after reoxygenation. For each genotype, the leaf sample collected from six independent plants was used for RNA sequencing, and the RNA-seq data were further validated by qRT-PCR analysis with three independent technical replicates. The depths of sequencing are as follows: wild type under air (air), 31,591,599 reads; myc2 air, 32,928,175 reads; wild type after 3-h reoxygenation (R3), 30,556,114 reads; and myc2 R3, 25,582,939 reads. Gene expression levels were normalized with the values of gene read counts per kilobase of exon model per million reads (RPKM) data. The FDR was used to correct the P value for multiple hypotheses testing with the Benjamini-Hochberg method. The Z-score was determined based on the RPKM according to the following formula: Z = (X – μ)/σ, where X is the RPKM of a gene for a specific time point, and μ and σ are the mean transcript expression and sd of a gene across all samples, respectively (Yu et al., 2012). The calculations and figures were produced using the R language. Specifically, the RPKM values of the DEGs in the JA, SA, and antioxidant synthesis pathways (Fig. 4, C–E) were compared between the myc2-2 mutant and wild type air or R3 samples, respectively. For the calculation of log2 fold changes (Supplemental Tables 1 and 2), we performed four pairwise comparisons, i.e. wild type R3 to wild type air, myc2 R3 to myc2 air, myc2 air to wild type air, and myc2 R3 to wild type R3. The original data set was deposited in the NCBI GEO database (access no. GSE93393).
ChIP and EMSA Assays
ChIP assays were performed according to published protocols (Yamaguchi et al., 2014) using 2-week-old MYC2-OE seedlings grown on Murashige and Skoog medium. After coating with anti-MYC2 antibodies (Abiocode) or anti-IgG (CST), the MYC2 protein/DNA complexes were immunoprecipitated by Dynabeads Protein G (Invitrogen) for at least 4 h at 4°C. The precipitated DNA was purified using a DNA purification kit (Qiagen) and the enriched DNA fragments were determined by qPCR using the specific primers listed in Supplemental Table S3. The promoter of ACTIN2 was used as a negative control. All ChIP assays were biologically repeated three times.
Nuclear proteins were extracted using a Plant Nuclei Isolation/Extraction Kit (Sigma) from 10 g of 2-week-old MYC2-OE seedlings. Nuclear proteins were isolated following the manufacturer’s instructions. All probes were labeled with biotin using the Biotin 3′ End DNA Labeling Kit (Pierce). The unlabeled oligonucleotides were used as competitors in the binding assay. The oligonucleotide sequences of the probes are listed in Supplemental Table S4.
The EMSA reactions were performed using a LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s instructions. An appropriate amount of enriched MYC2 nuclei protein was incubated in the binding buffer (50 ng μL−1 Poly (dI•dC), 2.5% glycerol, 0.05% NP-40, 10 mm EDTA, 0.5 mm DTT, and probes) in a total volume of 20 µL for 30 min at room temperature. After incubation, the binding reactions were loaded onto the 6% polyacrylamide gel and separated by 0.5× Tris-Borate-EDTA buffer at 4°C. The DNA-protein complex was transferred to a nylon membrane (Pierce). After cross-linking, the biotin activities were detected according to the manufacturer’s instructions (Pierce).
Determination of AsA and GSH
The extraction and determination of AsA and GSH were carried out as previously described with modifications (Wang et al., 2013). Briefly, leaf samples (∼0.1 g) were harvested and immediately homogenized in liquid nitrogen. After adding 1 mL ice-cold 6% (w/v) meta-phosphoric acid, the samples were agitated for 30 min at 4°C and then centrifuged at 12,000g for 15 min at 4°C. The supernatants were collected and diluted 1:3 in mobile phase before injection. AsA and GSH determination was carried out on reverse-phase high performance liquid chromatography by the Waters ACQUITY Arc system. The samples were separated at 40°C over a reverse-phase type C-18 column with an isocratic flow of 1.0 mL min−1 of the mobile phase (25 mm KH2PO4, pH 2.4) and detection occurred between 192 nm and 254 nm via a diode array. Quantification was conducted using the corresponding AsA and GSH standard curves.
Accession Numbers
Sequence data discussed in this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: LOX1 (AT1G55020), LOX2 (AT3G45140), LOX3 (AT1G17420), LOX4 (AT1G72520), LOX5 (AT3G22400), LOX6 (AT1G67560), AOS (AT5G42650), AOC1 (AT3G25760), AOC2 (AT3G25770), AOC3 (AT3G25780), AOC4 (AT1G13280), OPR3 (AT2G06050), OPCL1 (AT1G20510), JAR1 (AT2G46370), JAZ1 (AT1G19180), MYC2 (AT1G32640), VTC1 (AT2G39770), VTC2 (AT4G26850), VTC5 (AT5G55120), GSH1 (AT4G23100), GSH2 (AT5G27380), APX2 (AT3G09640), DHAR1 (AT1G19570), MDHAR3 (AT3G09940), GR1 (AT3G24170), TUB3 (AT5G62700), and ACTIN2 (AT3G18780). The original data set for RNA sequencing was deposited in the NCBI GEO database (access no. GSE93393).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A second biological repeat showing the induction of JA biosynthesis in wild-type leaves during recovery after dark or dark submergence.
Supplemental Figure S2. Measurement of JA, JA-Ile, SA, and ABA during submergence or postsubmergence reoxygenation.
Supplemental Figure S3. The expression of JA biosynthetic genes in wild-type leaves during submergence treatment.
Supplemental Figure S4. The control plants for JA biosynthesis showing no difference between each group before and after 48-h dark treatment followed by recovery.
Supplemental Figure S5. The control plants for JA signaling showing no difference between each group before and after 48-h dark treatment followed by recovery.
Supplemental Figure S6. Expression of antioxidant defense-related genes in response to reoxygenation or dark recovery.
Supplemental Figure S7. Measurements of AsA and GSH in response to reoxygenation or dark recovery.
Supplemental Figure S8. A second biological repeat showing the accumulation of ROS in wild-type, coi1-2, myc2-2, and MYC2-OE plants in response to reoxygenation.
Supplemental Figure S9. ROS contents in wild-type, coi1-2, myc2-2, and MYC2-OE plants in response to dark recovery.
Supplemental Figure S10. MYC2 interacts with the VTC2, VTC5, and GSH2 promoters in vivo.
Supplemental Figure S11. The control plants showing no significant difference among wild type, VTC1- and GSH1-deficient mutants (vtc1-1 and pad2-1), VTC1- and GSH1-overexpressors (VTC1-OE and GSH1-OE), as well as myc2-2 VTC1-OE and myc2-2 GSH1-OE lines in response to dark treatment and dark recovery.
Supplemental Table S1. DEGs in the myc2-2 mutant in response to reoxygenation.
Supplemental Table S2. JA-, SA-, and antioxidant-associated DEGs in the myc2-2 mutant in response to reoxygenation.
Supplemental Table S3. Sequences of primers used in this study.
Supplemental Table S4. Oligonucleotide probes used in EMSA.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing lox2-AS, lox2-S, aos, jar1-1, jin1-9, and pad2-1 mutant seeds, J. Browse (Washington State University, Washington) for the JAZ1-GUS/WT and JAZ1-GUS/coi1 seeds, D.X. Xie (Tsinghua University, Beijing, China) for coi1-2 seeds, C.Y. Li (Chinese Academy of Sciences, Beijing, China) for the MYC2-OE and myc2-2 seeds, R.F. Huang (Chinese Academy of Agricultural Sciences, Beijing, China) for the vtc1-1 and VTC1-OE seeds, and K. Ogawa (RIBS, Okayama, Japan) for the GSH1-OE seeds.
Glossary
- ROS
reactive oxygen species
- JA
jasmonate
- MeJA
methyl jasmonate
- DEG
differentially expressed gene
- FDR
false discovery rate
Footnotes
This work was supported by the National Natural Science Foundation of China (Projects 31670276, 31461143001,and 31370298 to S.X.), Program for New Century Excellent Talents in University (Project NCET-13-0614 to S.X.), Natural Science Foundation of Guangdong Province, China (Project 2015A030313122 to Q.-F.C.), China Postdoctoral Science Foundation (Projects 2015M572400 and 2016T90809 to L.-J.X.), and Sun Yat-sen University (start-up fund to S.X.).
References
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT (2012a) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Lee SC, Brinton E (2012b) Waterproofing crops: Effective flooding survival strategies. Plant Physiol 160: 1698–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ (2008) Flooding stress: Acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemelt S, Keetman U, Albrecht G (1998) Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiol 116: 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann Bot (Lond) 91: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Campbell MT, Proctor CA, Dou Y, Schmitz AJ, Phansak P, Kruger GR, Zhang C, Walia H (2015) Genetic and molecular characterization of submergence response identifies Subtol6 as a major submergence tolerance locus in maize. PLoS One 10: e0120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R, Jang CJH, Branco-Price C, Nghiem P, Bailey-Serres J (2012) Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol 78: 109–122 [DOI] [PubMed] [Google Scholar]
- Chen L, Liao B, Qi H, Xie LJ, Huang L, Tan WJ, Zhai N, Yuan LB, Zhou Y, Yu LJ, et al. (2015) Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11: 2233–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T, Ren Y, Tang X, Xiao F, Liu Y, et al. (2016) Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol 171: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci USA 93: 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2011) Ascorbate and glutathione: The heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J (2004) Plant responses to hypoxia—is survival a balancing act? Trends Plant Sci 9: 449–456 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xiong L (2013) Genetic mechanisms conferring adaptation to submergence and drought in rice: Simple or complex? Curr Opin Plant Biol 16: 196–204 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Cardarelli F, Novi G, Parlanti S, Pucciariello C, Bassolino L, Banti V, Licausi F, Perata P (2015) Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nature Plants 1: 15151. [DOI] [PubMed] [Google Scholar]
- Guo J, Dai X, Xu W, Ma M (2008) Overexpressing GSH1 and AsPCS1 simultaneously increases the tolerance and accumulation of cadmium and arsenic in Arabidopsis thaliana. Chemosphere 72: 1020–1026 [DOI] [PubMed] [Google Scholar]
- Guo J, Pang Q, Wang L, Yu P, Li N, Yan X (2012) Proteomic identification of MYC2-dependent jasmonate-regulated proteins in Arabidopsis thaliana. Proteome Sci 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano-Iwasaki A, Ogawa K (2012) Overexpression of GSH1 gene mimics transcriptional response to low temperature during seed vernalization treatment of Arabidopsis. Plant Cell Physiol 53: 1195–1203 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20: 219–229 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Lin RC, Park HJ, Wang HY (2008) Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant 1: 42–57 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk LS, Fagerstedt KV, Crawford RMM (1987) Superoxide dismutase as an anaerobic polypeptide: A key factor in recovery from oxygen deprivation in Iris pseudacorus? Plant Physiol 85: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH (2012) Glutathione in plants: An integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelic D, Arpagaus S, Rawyler A, Brändle R (2000) Impact of post-anoxia stress on membrane lipids of anoxia-pretreated potato cells. A re-appraisal. Plant Physiol 124: 1285–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139: 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciariello C, Parlanti S, Banti V, Novi G, Perata P (2012) Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol 159: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D (2015) Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27: 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawyler A, Arpagaus S, Braendle R (2002) Impact of oxygen stress and energy availability on membrane stability of plant cells. Ann Bot (Lond) 90: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al. (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44: 653–668 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Mustroph A (2011) Plant oxygen sensing is mediated by the N-end rule pathway: A milestone in plant anaerobiosis. Plant Cell 23: 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Voesenek LACJ (2015) Ethylene-mediated acclimations to flooding stress. Plant Physiol 169: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi K, Bodenhausen N, Buchala A, Mauch F, Reymond P (2008) The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J 55: 774–786 [DOI] [PubMed] [Google Scholar]
- Shan C, Liang Z (2010) Jasmonic acid regulates ascorbate and glutathione metabolism in Agropyron cristatum leaves under water stress. Plant Sci 178: 130–139 [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 2012: 217037. 101155/2012/217037 [Google Scholar]
- Shingaki-Wells R, Millar AH, Whelan J, Narsai R (2014) What happens to plant mitochondria under low oxygen? An omics review of the responses to low oxygen and reoxygenation. Plant Cell Environ 37: 2260–2277 [DOI] [PubMed] [Google Scholar]
- Singh P, Sinha AK (2016) A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE 3 imparts submergence tolerance in rice. Plant Cell 28: 1127–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skutnik M, Rychter AM (2009) Differential response of antioxidant systems in leaves and roots of barley subjected to anoxia and post-anoxia. J Plant Physiol 166: 926–937 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35: 291–314 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Wasternack C, Xie D (2014) Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol 21: 112–119 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM (2003) Molecular and cellular adaptations of maize to flooding stress. Ann Bot (Lond) 91: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang BG, Fukao T (2015) Plant adaptation to multiple stresses during submergence and following desubmergence. Int J Mol Sci 16: 30164–30180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang BG, Magliozzi JO, Maroof MA, Fukao T (2014) Physiological and transcriptomic characterization of submergence and reoxygenation responses in soybean seedlings. Plant Cell Environ 37: 2350–2365 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Chou SJ, Shih MC (2014) Ethylene plays an essential role in the recovery of Arabidopsis during post-anaerobiosis reoxygenation. Plant Cell Environ 37: 2391–2405 [DOI] [PubMed] [Google Scholar]
- Ushimaru T, Shibasaka M, Tsuji H (1992) Development of the O2− detoxification system during adaptation to air of submerged rice seedlings. Plant Cell Physiol 33: 1065–1071 [Google Scholar]
- van Dongen JT, Licausi F (2015) Oxygen sensing and signaling. Annu Rev Plant Biol 66: 345–367 [DOI] [PubMed] [Google Scholar]
- van Veen H, Vashisht D, Akman M, Girke T, Mustroph A, Reinen E, Hartman S, Kooiker M, van Tienderen P, Schranz ME, et al. (2016) Transcriptomes of eight Arabidopsis thaliana accessions reveal core conserved, genotype- and organ-specific responses to flooding stress. Plant Physiol 172: 668–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D, Hesselink A, Pierik R, Ammerlaan JMH, Bailey-Serres J, Visser EJW, Pedersen O, van Zanten M, Vreugdenhil D, Jamar DCL, et al. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Bailey-Serres J (2013) Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol 16: 647–653 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Bailey-Serres J (2015) Flood adaptive traits and processes: An overview. New Phytol 206: 57–73 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Jackson MB, Toebes AHW, Huibers W, Vriezen WH, Colmer TD (2003) De-submergence-induced ethylene production in Rumex palustris: Regulation and ecophysiological significance. Plant J 33: 341–352 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Sasidharan R (2013) Ethylene—and oxygen signalling—drive plant survival during flooding. Plant Biol (Stuttg) 15: 426–435 [DOI] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R (2013) Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot (Lond) 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka BA, Goossens A, Inzé D (2005) Methyl jasmonate stimulates the de novo biosynthesis of vitamin C in plant cell suspensions. J Exp Bot 56: 2527–2538 [DOI] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ (1998) Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10: 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Dai L, Liu F, Wang Z, Peng W, Xie D (2004) COS1: An Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie LJ, Chen QF, Chen MX, Yu LJ, Huang L, Chen L, Wang FZ, Xia FN, Zhu TR, Wu JX, et al. (2015a) Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet 11: e1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie LJ, Yu LJ, Chen QF, Wang FZ, Huang L, Xia FN, Zhu TR, Wu JX, Yin J, Liao B, et al. (2015b) Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J 81: 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D (2014) PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY. (2014) Hydrogen peroxide controls transcriptional responses of ERF73/HRE1 and ADH1 via modulation of ethylene signaling during hypoxic stress. Planta 239: 877–885 [DOI] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC (2011) The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol 156: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LJ, Luo YF, Liao B, Xie LJ, Chen L, Xiao S, Li JT, Hu SN, Shu WS (2012) Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol 195: 97–112 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Zhang R, Huang R (2012a) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan SS (2012b) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69: 667–678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.